Abstract
The presence of multiple clusters of runs of asymmetric adenine or thymine is a feature commonly found in eukaryotic replication origins. Here we report that the helicase and ATPase activities of the mammalian Mcm4/6/7 complex are activated specifically by thymine stretches. The Mcm helicase is specifically activated by a synthetic bubble structure which mimics an activated replication origin, as well as by a Y-fork structure, provided that a single-stranded DNA region of sufficient length is present in the unwound segment or 3′ tail, respectively, and that it carries clusters of thymines. Sequences derived from the human lamin B2 origin can serve as a potent activator for the Mcm helicase, and substitution of its thymine clusters with guanine leads to loss of this activation. At the fork, Mcm displays marked processivity, expected for a replicative helicase. These findings lead us to propose that selective activation by stretches of thymine sequences of a fraction of Mcm helicases loaded onto chromatin may be the determinant for selection of initiation sites on mammalian genomes.
Keywords: initiation of DNA replication/Mcm helicase/replication bubble or fork/replication origin/thymine stretch
Introduction
Initiation of DNA replication at the onset of S phase requires prior assembly of pre-replicative complexes (preRCs) during the preceding late M phase (Stillman, 1996). The regulation of preRC assembly plays a key role in restricting DNA replication to once per cell cycle. The protein components of the preRC complex include origin recognition complex (ORC), Cdc6, Cdt1 and minichromosome maintenance proteins (Mcms). At the G1–S boundary, the cyclin-dependent kinase (CDK) and the Cdc7–Dbf4 kinase trigger initiation of DNA replication by phosphorylating the preRC components (Bell and Dutta, 2002; Masai and Arai, 2002). Following the origin activation, the duplex DNA is continuously unwound by a DNA helicase to permit processive DNA synthesis.
DNA replication is generally initiated from a specific site on the template DNA. In bacteria, a replicon-specific initiator binds to a minimum sequence required for initiation, normally called the ‘replication origin’, and actual DNA chain elongation is initiated within or near the replication origin (Kornberg and Baker, 1992). In the budding yeast, Saccharomyces cerevisiae, the ORC specifically recognizes an essential 11 bp sequence (5′-A/TTTTATG/ATTTA/T-3′) within ARS (autonomously replicating sequences), and this specific recognition plays a major role in selection of replication initiation sites in this organism (Marahrens and Stillman, 1992). In contrast, the mechanisms of selection of initiation sites in higher eukaryotes are still elusive. Distinct sites of leading-to-lagging strand transition were detected in the lamin B2 origin of human cells, and mapping with a competitive PCR method also led to identification of fairly narrow segments of the chromosome as initiation sites (Abdurashidova et al., 2000). In contrast, two-dimensional gel electrophoresis mapping indicated the presence of a ‘broad initiation zone’ on which replication appears to be initiated at many sites (Wang et al., 1998). An additional feature of DNA replication in higher eukaryotes is its flexibility. The most notable example is sequence-independent initiation in amphibian eggs (Hyrien and Mechali, 1993). Although replication does appear to initiate at specific loci in mammalian somatic cells, no ‘essential and conserved’ sequence motifs, such as the 11 bp sequence of budding yeast origin described above, for origin functions have been identified.
Structural and functional analyses of replication origins from various species have indicated that the AT-richness is a general feature of the sequences surrounding the initiation sites for DNA replication in eukaryotes (Boulikas, 1996; DePamphilis, 1996; Kelman, 2000). A start site of the human lamin B2 origin was located in an AT-rich segment (Abdurashidova et al., 2000). A fragment of several kilobases from the β-globin origin, containing sequences highly enriched in AT, can promote initiation of replication (Aladjem et al., 1998). The peak of nascent DNAs in the DHFR origin of CHO cells is located adjacent to an AT-rich sequence (Kobayashi et al., 1998). The replication origins from which the chorion genes in Drosophila melanogaster are amplified are also AT-rich and contain sequences very similar to the 11 bp ARS core (Spradling, 1999). A broad replication origin of Drosophila, oriDα, consists of multiple discrete initiation sites with AT-rich sequences (Ina et al., 2001). Another point to be made is the asymmetric distribution of A and T on each strand. This has been shown most clearly with origins of fission yeast, Schizosaccharomyces pombe. Clusters of T stretches were shown to be essential for origin function (Okuno et al., 1999). Although the ORC, which is structurally and functionally conserved from yeasts to human, is the prime candidate that may recognize the replication origins for initiation, mammalian ORC binds to DNA without any apparent sequence specificity (Gilbert, 2001; Vashee et al., 2003). Therefore, it is still an open question how the sequences with asymmetric T-rich and A-rich strands are selected for initiation sites of DNA replication in mammalian cells.
The six Mcm proteins, Mcm2, Mcm3, Mcm4, Mcm5, Mcm6 and Mcm7, contain highly conserved DNA-dependent ATPase motifs in their central regions (Tye, 1999). The Mcm proteins form several stable subassemblies including Mcm2/3/4/5/6/7, Mcm2/4/6/7, Mcm4/6/7 and Mcm3/5 complexes (Thömmes et al., 1997; You et al., 1999; Lee and Hurwitz, 2000). Among them, DNA helicase activity was identified in the Mcm4/6/7 complexes of human, mouse and fission yeast (Ishimi, 1997; You et al., 1999; Lee and Hurwitz, 2000, 2001). Mcm2 and Mcm3/5 were shown to inhibit its helicase activity by converting its double trimer structure into a heterotetramer or heteropentamer (You et al., 1999; Sato et al., 2000). Mcm4, Mcm6 and Mcm7 proteins make a distinct contribution to its helicase activity (You et al., 1999, 2002). Chromatin immunoprecipitation assays in S.cerevisiae suggested that Mcm proteins are associated with moving replication forks (Aparicio et al., 1997). Analyses using degradable mutants of Mcm in S.cerevisiae showed that all the six Mcm subunits are required for elongation of DNA chains, suggesting that Mcm could be involved not only in initiation but also in the DNA chain elongation stage as a replicative helicase (Labib et al., 2000). Consistent with this notion, the archae homologue of Mcm has a helicase activity which can unwind duplex DNA up to 500 bp (Kelman et al., 1999; Chong et al., 2000). In contrast, the processivity of eukaryotic Mcm4/6/7 complexes is rather low, and it can displace only 30 nucleotides on a conventional partial heteroduplex substrate in vitro. However, Lee and Hurwitz (2001) reported that the processivity of the S.pombe Mcm4/6/7 complex is significantly stimulated on forked DNA structures and it can unwind duplex DNA of ∼600 bp by forming a double hexameric structure on SSB-coated substrates containing a 5′ tail (Lee and Hurwitz, 2001).
In this report, we have shown that Mcm4/6/7 can bind to a bubble structure resembling the unwound replication origin and displace the duplex region flanking the single-stranded bubble. We discovered that the T-rich strand of the ARS core or poly(dT) is a potent activator of the Mcm helicase on both bubble and fork structures. Consistent with this, oligo(dT) but not other oligonucleotides stimulated the ATPase activity of Mcm4/6/7 to a significant extent. Based on the specific ability of T-rich sequences to activate the Mcm helicase, we propose a novel model for selection of initiation sites of mammalian DNA replication.
Results
The presence of a 5′ tail stimulates the helicase activity of the Mcm4/6/7 complex on a partially heteroduplex single-stranded circular DNA
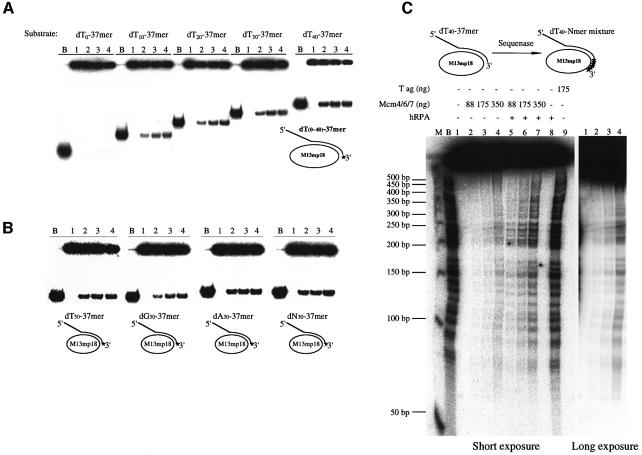
Previous studies have shown that the mouse Mcm4/6/7 complex, translocating along single-stranded DNA (ssDNA) in the 3′ to 5′ direction (Ishimi, 1997), can displace duplex DNA only up to 30 nucleotides. Recently, it was reported that the fission yeast Mcm4/6/7 complex displays high processivity on forked DNA substrates (Lee and Hurwitz, 2001). We therefore examined the ability of mouse Mcm4/6/7 to unwind partial heteroduplex M13 ssDNA containing a 37 bp duplex region in the presence or absence of the 5′ dT tail of various lengths. Although it barely displaced the annealing oligonucleotide without a 5′ tail (dT0-37mer; Figure 1A), the presence of a 10 nucleotide dT tail significantly stimulated the displacement of the 37mer fragment, and the efficacy of displacement at a low concentration of Mcm increased as the length of the tail increased. We next examined whether the nucleotide composition of the tail would affect the helicase action. The helicase substrates containing the 37 bp duplex regions with a 30 nucleotide 5′ tail of dT, dA, dG or a random sequence (dN) were constructed and were used for helicase assays. The Mcm4/6/7 complex displaced all the substrates with similar efficiency, although dG tail was a somewhat less efficient substrate than the others (Figure 1B). This result suggests that the Mcm helicase does not display significant sequence preference for the 5′ tail.
Fig. 1. Stimulation of helicase activity of the Mcm4/6/7 complex by the presence of a 5′ tail on the annealed oligonucleotide. (A) DNA helicase assays were performed with 6 fmol of helicase substrates containing a 5′ dT tail of various lengths as indicated. (B) Helicase assays were conducted on 4 fmol of helicase substrates containing a 30 nucleotide 5′ tail of various nucleotide sequences as shown. Oligo(dC) tail substrate was unstable and could not be constructed. In (A) and (B), the stars represent the radioactive G residues used for labelling of the annealed oligonucleotides. B, boiled substrate; lanes 1–4 contain 0, 50, 100 and 150 ng of the Mcm4/6/7 complex, respectively. (C) Processivity of the Mcm4/6/7 helicase. dT40-Nmer mixture/M13mp18 represent the labelled duplex regions of various lengths (indicated by stars). M, 50 bp ladder DNA (denatured); B, boiled substrate. + and – indicate the presence and absence, respectively, of each factor indicated. T ag, SV40 T-antigen. The right panel is a long exposure of lanes 1–4 to visualize the bands of weak intensities.
Helicase processivity of the Mcm4/6/7 complex
In the experiments described above, we have shown that a 37 nucleotide duplex region on a tailed substrate is efficiently displaced by the Mcm4/6/7 complex. In order to determine the extent of the duplex region which can be displaced by the Mcm4/6/7 complex, we prepared a 5′-tailed partial heteroduplex DNA containing duplex regions of variable lengths. The result shows that it can displace duplex DNA up to 350 nucleotides long on its own (Figure 1C, lanes 2–4). The presence of a ssDNA-binding protein, human RPA protein (hRPA), increased the extent of unwinding up to 450 nucleotides and also the efficiency of unwinding (Figure 1C, lanes 5–7). Presumably, once the duplex region is melted, hRPA can bind to the exposed ssDNA regions to prevent its reannealing, contributing to the increased processivity of the Mcm helicase. Alternatively, RPA may facilitate the helicase activity of Mcm by removing secondary structures on the substrate or by protein–protein interactions. It could also be possible that RPA increases the turnover rate by stabilizing partially unwound substrates. These results are consistent with the notion that Mcm plays an important role as a helicase at eukaryotic replication forks. However, it should be noted that the processivity of SV40 T-antigen protein, as measured on the same template DNA, is much higher than that of the Mcm helicase (Figure 1C, lane 9).
The Mcm4/6/7 complex can bind and unwind synthetic DNA replication bubbles
The above helicase assays utilized partially duplex ssDNA with 5′ tail sequences. However, under physiological conditions, free ends of DNA would not be available for loading of Mcm helicase. Therefore, we have constructed synthetic bubble-like substrates mimicking an activated replication origin, in which unpaired segments of various lengths are flanked by 21 bp duplex regions. We first examined the binding and helicase activities of Mcm4/6/7 on a set of synthetic bubble-like substrates [Bub-0 (37mer duplex DNA), Bub-20 and Bub-60] that differed in the length of the central unpaired segment. Mcm4/6/7 was incubated with each radiolabelled synthetic bubble substrate in the presence of 0.25 mM AMP-PNP to allow complex formation. Half of these reaction mixtures were analysed for DNA binding in gel shift assays (Figure 2A), and the remainder were incubated further in the presence of 10 mM ATP to measure DNA helicase activity (Figure 2B). Bub-0 (37mer) generated very little complex with Mcm4/6/7, and Bub-20 generated primarily a single major mobility-shifted form, which may contain a single hexamer of the Mcm4/6/7 complex. SV40 T-antigen generated a complex on Bub-0 containing a single hexamer, and more slowly migrating forms on Bub-20 and Bub-60, which are likely to represent a double hexamer (Figure 2A, lanes 6; Smelkova and Borowiec, 1998). With Mcm4/6/7, Bub-60 gave rise to shifted complexes, which migrated more slowly than those generated on Bub-20 at high concentrations of Mcm4/6/7, indicating that more than two molecules of Mcm4/6/7 trimers can bind to Bub-60 but not to Bub-20 (Figure 2A, lanes 5). At a low concentration, Mcm4/6/7 generated a single hexamer complex on Bub-60 (Figure 2A, lane 2 in Bub-60).
Fig. 2. DNA binding and helicase assays on synthetic bubble DNAs with the Mcm4/6/7 helicase: stimulation of helicase activity by T-rich bubble sequences. (A) DNA binding activity of the Mcm4/6/7 helicase was measured in gel shift assays using synthetic bubble DNAs (10 fmol), Bub-0, Bub-20 and Bub-60, as substrates. Complexes were separated on a 5% native polyacrylamide gel. The drawings below the panels show schematic representations of the substrates used in this assay. Lanes 1–5 contain 0, 100, 200, 400 and 800 ng of the Mcm4/6/7 protein, respectively; lane 6, 400 ng of SV40 T-antigen. (B) DNA helicase assays were conducted on the same set of synthetic bubble DNAs as in (A). At higher concentrations of Mcm4/6/7 with Bub-20 and Bub-60, some of the substrate DNAs are mobility shifted, resulting in reduction of band intensities of the intact substrates (lanes 3 and 4). Lanes 1–4 contain 0, 200, 400 and 800 ng of the Mcm4/6/7 protein, respectively; lane 5, 400 ng of SV40 T-antigen. DNA helicase (C) and gel shift (D) assays were performed using synthetic bubble DNAs, Bub-60, Bub-66/T-rich and Bub-66/G-rich, as substrates (Table I). In (C) and (D), lanes 1–5 contain 0, 100, 200, 400 and 600 ng of the Mcm4/6/7 protein, respectively; lane 6, 400 ng of SV40 T-antigen. B, boiled substrate.
In DNA helicase assays, SV40 T-antigen unwound Bub-0 only very inefficiently (Figure 2B, lane 5 in Bub-0), but efficiently unwound Bub-20 and Bub-60 (Figure 2B, lanes 5 in Bub-20 and Bub-60). In contrast, Mcm4/6/7 did not show helicase activity on either Bub-0 or Bub-20 substrate, while weak but significant helicase activity was observed on Bub-60 (Figure 2B, lanes 2–4 in Bub-60). These results are consistent with the notion that the Mcm4/6/7 complex, loaded onto DNA through the bubble on Bub-60, unwinds the duplex region of the substrate DNA.
T-rich sequences in the single-stranded bubble region greatly stimulate the DNA-unwinding activity of Mcm
Most replication origins are rich in A and T residues and contain A/T-rich stretches essential for origin function (Boulikas, 1996; DePamphilis, 1996; Kelman, 2000). The S.cerevisiae origins are best characterized, which include essential 11 bp ARS consensus sequences, A/TTTTATG/ATTTA/T (Marahrens and Stillman, 1992). The initiation sites, defined as transition points from lagging to leading strand, were mapped right next to the core sequence (Bielinsky and Gerbi, 1998). Therefore, a synthetic bubble containing six repeats of the T-rich strand of the ARS core (Bub-66/T-rich), and a G-rich bubble in which all the T residues of Bub-66/T-rich were substituted by G (Bub-66/G-rich) were constructed (Table I), and the helicase and DNA-binding activities of Mcm4/6/7 were examined on these substrates as well as on Bub-60 containing random sequences used in previous assays. As shown in Figure 2C, compared with Bub-60, the Bub-66/T-rich substrate was unwound by the Mcm complex to a much higher level, whereas no unwinding was detected on the Bub-66/G-rich substrate. These results are consistent with the idea that the T-rich bubble facilitates the helicase activity of Mcm. In gel shift assays, Bub-66/T-rich formed complexes with much higher affinity than Bub-60 (Figure 2D). Unexpectedly, Bub-60/G-rich also generated complexes with affinity higher than Bub-60, in spite of its lower unwindability. These results indicate that higher affinity for a T-rich bubble can at least partially explain the readiness with which Bub-66/T-rich can be unwound by Mcm. However, the behaviour of Bub-66/G-rich suggests that binding is not sufficient and that some other features of the T-richness specifically activate the Mcm helicase.
Table I. Oligonucleotides used for construction of DNA substrates.
| No. | Code | Sequence (5′–3′) |
|---|---|---|
| 1 | Bub-0 top | CTTCTGTGACTACCTGGACGACCGGGTGACTAGTTGC |
| 2 | Bub-0 bottom | GCAACTAGTCACCCGGTCGTCCAGGTAGTCACAGAAG |
| 3 | Bub-20 top | GTTTTCCCAGTCACGACGTTGATTTTGCTGCCGGTCACGGTAGCTTGCATGCCTGCAGGTCG |
| 4 | Bub-20 bottom | CGACCTGCAGGCATGCAAGCTTGGCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAAC |
| 5 | Bub-60 top | GTTTTCCCAGTCACGACGTTGATTTTGCTGCCGGTCACGGTTCGAACGTACGGACGTCCAGCTGAGATCTCCTAGGGGCCCTACCGAGCTCGAATTCGTAAT |
| 6 | Bub-60 bottom | ATTACGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAAC |
| 7 | Bub-66/T-rich top | GTTTTCCCAGTCACGACGTTGTTTTATGTTTATTTTATGTTTATTTTATGTTTATTTTATGTTTATTTTATGTTTATTTTATGTTTATACCGAGCTCGAATTCGTAAT |
| 8 | Bub-66/T-rich bottom | ATTACGAATTCGAGCTCGGTAATTTGTATTTTATTTGTATTTTATTTGTATTTTATTTGTATTTTATTTGTATTTTATTTGTATTTTCAACGTCGTGACTGGGAAAAC |
| 9 | Bub-66/G-rich top | GTTTTCCCAGTCACGACGTTGGGGGCGTGGGCGGGGCGTGGGCGGGGCGTGGGCGGGGCGTGGGCGGGGCGTGGGCGGGGCGTGGGCTACCGAGCTCGAATTCGTAAT |
| 10 | Bub-66/G-rich bottom | ATTACGAATTCGAGCTCGGTACGGGTGCGGGGCGGGTGCGGGGCGGGTGCGGGGCGGGTGCGGGGCGGGTGCGGGGCGGGTGCGGGGCAACGTCGTGACTGGGAAAAC |
| 11 | Bub-50/T-rich top | GTTTTCCCAGTCACGACGTTGTAAAACGACTTTTATGTTTATTTTATGTTTATTTTATGTTTATTTTATGTTTATTTTATTACCGAGCTCGAATTCGTAATCATGGTCAT |
| 12 | Bub-50/T-rich bottom | ATGACCATGATTACGAATTCGAGCTCGGTATATTTTATTTGTATTTTATTTGTATTTTATTTGTATTTTATTTGTATTTTGTCGTTTTACAACGTCGTGACTGGGAAAAC |
| 13 | Lamin B2 top | AGAAGATGCATGCCTAGCGTGTTCTTTTTTTTTTCCAATGATTTGTAATATACATTTTATGACTGGAAACTTTTTTGTACAACACTCCAATAAACATTTTGATTTTAGGTTCTGCCTCTGAGTTTATTCCTG |
| 14 | Lamin B2 bottom | CAGGAATAAACTCAGAGGCAGAACCATTTTAGTTTTACAAATAACCTCACAACATGTTTTTTCAAAGGTCAGTATTTTACATATAATGTTTAGTAACCTTTTTTTTTAGAACACGCTAGGCATGCATCTTCT |
| 15 | Lamin B2-G top | AGAAGATGCATGCCTAGCGTGTTCTGGGGGGGGGCCAATGAGGGGTAATATACAGGGGATGACTGGAAACGGGGGGGTACAACACTCCAATAAACAGGGGGAGGGGAGGTTCTGCCTCTGAGTTTATTCCTG |
| 16 | Lamin B2-G bottom | CAGGAATAAACTCAGAGGCAGAACCAGGGGAGGGGGACAAATAACCTCACAACATGGGGGGGCAAAGGTCAGTAGGGGACATATAATGGGGAGTAACCGGGGGGGGGAGAACACGCTAGGCATGCATCTTCT |
| 17 | dT(0–40)-37mer | dT(0–40)-GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT |
| 18 | dA30-37mer | dA30-GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT |
| 19 | dG30-37mer | dG30-GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT |
| 20 | dN30-37mer | ACGTTCCGCTAATTCAACCCATTGCGGTCCGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT |
| 21 | 37mer-dT(10–50) | GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT-dT(10–50) |
| 22 | 37mer-dA50 | GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT-dA50 |
| 23 | 37mer-dC50 | GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT-dC50 |
| 24 | 37mer-dG50 | GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGT-dG50 |
| 25 | dT30–50mer | dT30-GGTTGGCCGATCAAGTGCCCAGTCACGACGTTGTAAAACGAGCCCGAGTG |
| 26 | dA30-50mer | dA30-GGTTGGCCGATCAAGTGCCCAGTCACGACGTTGTAAAACGAGCCCGAGTG |
| 27 | dC30-50mer | dC30-GGTTGGCCGATCAAGTGCCCAGTCACGACGTTGTAAAACGAGCCCGAGTG |
| 28 | 50mer-dT60 | CACTCGGGCTCGTTTTACAACGTCGTGACTGGGCACTTGATCGGCCAACC-dT60 |
| 29 | 50mer-dA60 | CACTCGGGCTCGTTTTACAACGTCGTGACTGGGCACTTGATCGGCCAACC-dA60 |
| 30 | 50mer-dC60 | CACTCGGGCTCGTTTTACAACGTCGTGACTGGGCACTTGATCGGCCAACC-dC60 |
| 31 | A-rich50 | AAAATATAAAAAAAATATAAAAAAAATATAAAAAAAATATAAAAAAAATA |
The underlines indicate the unpaired segments of synthetic bubble substrates. dT(0–40)-37mer indicates the dT0-37mer, dT10-37mer, dT20-37mer, dT30-37mer and dT40-37mer. 37mer-dT(10–50) indicates the 37mer-dT10, 37mer-dT20, 37mer-dT30, 37mer-dT40 and 37mer-dT50.
T stretches on the 3′ tail preferentially activate Mcm helicase
The affinity of Mcm4/6/7 for T-rich sequences in the bubble appears to be inconsistent with the apparent absence of the sequence preference for the 5′ tails on partial heteroduplex substrates (Figure 1A). This could be due to the 3′ to 5′ polarity of the Mcm complex and, therefore, we have constructed helicase substrates containing the 37 bp duplex regions with a 50 nucleotide 3′ tail of various sequences and tested them in helicase assays. On these substrates, dT-tailed substrate was significantly displaced by Mcm4/6/7, but only very weak activity was detected on other substrates (Figure 3A). Quantification of the results indicated that Mcm4/6/7 showed helicase activity on the dT-tailed substrate ∼10-fold higher than that on dC-, dG- or dA-tailed substrates (Figure 3B). We also examined the effects of the length of the 3′ tail on helicase activity (Figure 3C). In contrast to the 5′ tail, very little activity was observed on a substrate with a 10 nucleotide 3′ tail. The efficiency of displacement only slightly increased as the length of the 3′ tail increased up to 40 nucleotide. With a 50 nucleotide 3′ tail, significant displacement was observed. This indicates that a single-stranded segment of at least 50 nucleotides is required for efficient loading of the Mcm helicase on a 3′-tailed substrate.
Fig. 3. Helicase assays of the Mcm4/6/7 complex on 3′-tailed substrates: preference for T residues. (A) DNA helicase assays were performed with 4 fmol of helicase substrates containing a 50 nucleotide 3′ tail of various nucleotide sequences as shown. Lanes 1–4 contain 0, 50, 100 and 200 ng of the Mcm4/6/7 complex, respectively; B, boiled substrate. The stars represent the radioactive 5′ end of the annealed oligonucleotide. (B) Quantification of the fractions of oligonucleotide displaced in (A). Shown are the mean values for three independent experiments ± SEM. (C) DNA helicase assays were performed with 4 fmol of helicase substrates containing a 3′ dT tail of various lengths as indicated. Lanes 1–4 contain 0, 25, 50 and 100 ng of the Mcm4/6/7 complex, respectively; B, boiled substrate.
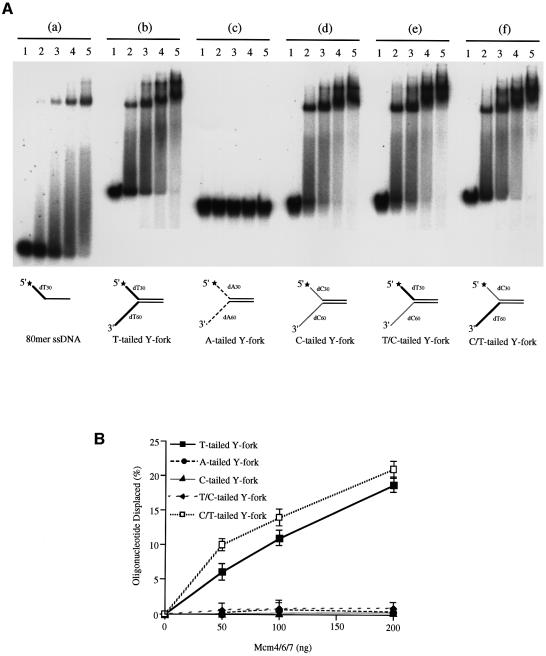
We then examined DNA binding and helicase activities of Mcm4/6/7 with a Y-fork-like structure. We first compared binding of Mcm4/6/7 to various Y-fork structures and ssDNA. The 80mer ssDNA mainly gave rise to a single mobility-shifted band (Figure 4A, a, lanes 2–5), known to contain a single hexamer of Mcm4/6/7 (You et al., 1999). At a high concentration of Mcm4/6/7, complexes with higher molecular weight, albeit at a low level, could be seen (Figure 4A, a, lane 5). This may represent the complexes containing more than two Mcm4/6/7 trimers. On a T-tailed fork DNA, a single mobility-shifted band, co-migrating with that observed on ssDNA, appeared at a low concentration of Mcm4/6/7 (Figure 4A, b, lane 2), and a more slowly migrating form was generated as more protein was added (Figure 4A, b, lanes 3–5). Comparison with the position of a double hexameric T-antigen–DNA complex suggested that the higher band might contain a double hexamer of Mcm4/6/7 (data not shown; Smelkova and Borowiec, 1998; Lee and Hurwitz, 2001). Similar complexes were generated on Y-forks with different combinations of dT and dC tail at the 5′ or 3′ end of DNA (Figure 4A, d, e and f), but very few complexes were formed on the A-tailed fork (Figure 4A, c), indicating that Mcm4/6/7 can bind dT or dC tail, but not dA tail. This also shows that a fork structure per se may not be sufficient for binding of Mcm.
Fig. 4. Binding of the Mcm4/6/7 complex to Y-fork substrates. (A) Gel shift assays were conducted in the presence of 0.5 mM ATP-γ-S on 80mer ssDNA (a) or on Y-fork substrates (b–f, 15 fmol each) composed of a 50 nucleotide duplex region as well as a 30 nucleotide 5′ tail and a 60 nucleotide 3′ tail with various nucleotide compositions [(b) T/T; (c) A/A; (d) C/C; (e) T/C; (f) C/T]. The amounts of Mcm4/6/7 used were 0 (lanes 1), 25 (lanes 2), 50 (lanes 3), 100 (lanes 4) and 200 ng (lanes 5). (B) DNA helicase assays were performed with 5 fmol of Y-fork substrates used in (A). Shown are the mean values for three independent experiments ± SEM.
These Y-fork substrates were next examined in DNA helicase assays. The Mcm4/6/7 efficiently unwound the T-tailed Y-fork and C/T-tailed Y-fork, but not other Y-forks (Figure 4B). The failure to unwind the A-tailed fork is expected from the inability of Mcm4/6/7 to bind to this substrate. Although C-tailed Y-fork and T/C-tailed Y-fork can be bound by Mcm4/6/7, they were not displaced. This indicates that a thymine stretch on a 3′ tail is essential to activate the DNA helicase activity of Mcm, consistent with the results of helicase assays on 5′- or 3′-tailed partial heteroduplexes (Figures 1B and 3A).
T-rich single-stranded DNA preferentially activates the ATPase activity of Mcm4/6/7
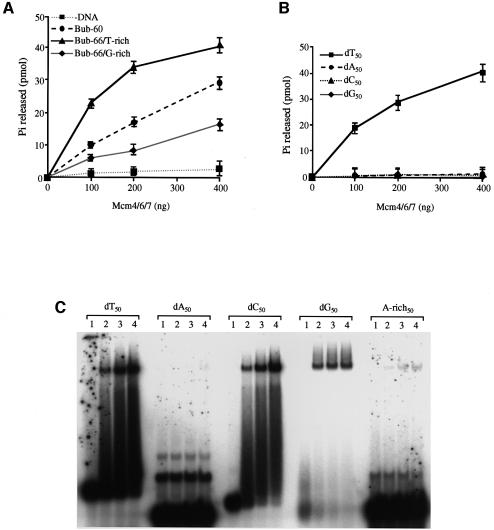
Hydrolysis of a nucleotide triphosphate is necessary for helicase activity. We next examined whether a T-rich bubble could activate the ATPase activity of Mcm4/6/7 more efficiently than others (Figure 5A). We used Bub-60, Bub-66/T-rich and Bub-66/G-rich substrates as effector DNA in ATPase assays of Mcm4/6/7. The level of ATP hydrolysis was the highest with Bub-66/T-rich. ATP was hydrolysed to a significant level with Bub-60 DNA but only to a limited extent with Bub-66/G-rich. These results indicate that the ATP hydrolysis activity of Mcm can be also stimulated by the T-rich bubble substrate.
Fig. 5. Oligo(dT) DNA stimulates the ATPase activity of Mcm4/6/7 protein. The effects of various synthetic bubble DNAs (A), or oligonucleotide DNAs (B) on the ATPase activity of Mcm4/6/7 were examined. The released phosphate (Pi in pmol) was quantified, and the values, corrected by subtracting the background level in the absence of added protein, are presented. In (A) and (B), the mean values for three independent experiments ± SEM are presented. (C) The gel shift assays with Mcm4/6/7 were conducted on various oligonucleotides (10 fmol each) as substrate. Lane 1, no protein added; lanes 2–4 contain 50, 100 and 200 ng of the Mcm4/6/7 protein, respectively. Band intensities are weaker with dG50 than the others due to its lower specific radioactivity. This is because polynucleotide kinase has difficulties in end-labelling this oligonucleotide.
In order to clarify sequence specificity for ATPase activation, we examined various homopolymers for their ability to stimulate the ATPase activity of the Mcm4/6/7 complex. Consistent with the results of helicase assays, dT50 stimulated the ATPase activity of Mcm4/6/7 protein to a high level, whereas very little stimulation was seen with dA50 and dC50, and no stimulation with dG50 (Figure 5B). These results are consistent with those of the helicase assays and indicate that T-rich ssDNA serves as a preferred effector DNA for ATP hydrolysis activity of the Mcm4/6/7 complex.
We also examined binding of the Mcm4/6/7 protein with these homo-oligonucleotides. We have found that the Mcm4/6/7 can bind not only to dT50 but also to dC50 and dG50 with similar efficiency (Figure 5C). However, it showed very little binding to dA50, as was the case for A-tailed Y-fork. Binding was slightly increased with A-rich50 containing nine T residues. These results indicate that Mcm4/6/7 can bind to ssDNA, except for polyadenine, but that its ATPase and helicase activities can be activated efficiently only by thymine stretches.
A DNA fragment spanning the replication initiation region of the lamin B2 origin also supports efficient helicase activity of Mcm4/6/7 when present in a bubble
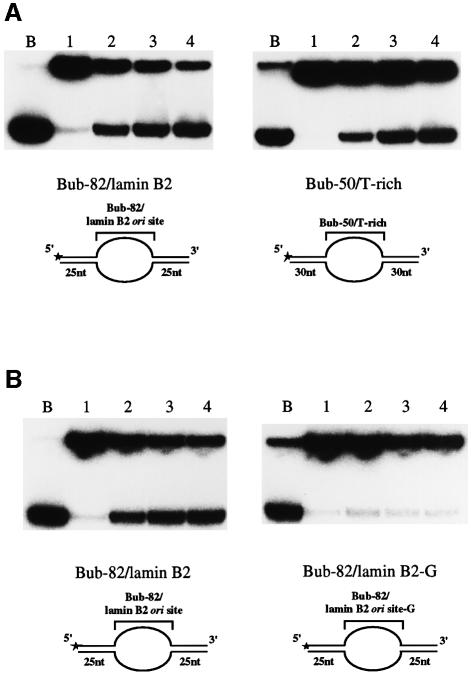
In order to examine the possibility that the T-rich sequences in natural replication origins also function to facilitate loading and activation of the Mcm helicase, we constructed a new bubble substrate (Bub-82/lamin B2) containing sequences derived from the human lamin B2 origin, in which lagging to leading strand junctions were mapped within periodic (dT)n tracts (Abdurashidova et al., 2000). The unpaired 82 nucleotide region in the Bub-82/lamin B2 spans the OBI (origin of bidirectional initiation) and is believed to coincide with the initially melted region of the lamin B2 origin. As a control, Bub-50/T-rich carrying 4.5 repeats of the ARS core (50 nucleotide) in the bubble and flanking 30mer duplex regions was also constructed. As shown in Figure 6A, Mcm4/6/7 unwound Bub-50/T-rich, indicating that the loaded Mcm can displace 30 nucleotides in both directions. The Bub-82/lamin B2 substrate was also unwound with similar efficiency under the same condition, indicating that the sequences from a naturally occurring origin can serve as a loading site for the Mcm helicase in vitro.
Fig. 6. Activation of Mcm4/6/7 DNA helicase activity by sequences derived from the human lamin B2 origin. (A) DNA helicase assays of Mcm4/6/7 were conducted using Bub-82/lamin B2 and Bub-50/T-rich substrates (4 fmol). T contents in the unpaired region of Bub-82/lamin B2 and Bub-50/T-rich are 48 and 74%, respectively. Lanes 1–4 are reactions with 0, 100, 200 and 400 ng of the Mcm4/6/7 complex, respectively. (B) The clusters of T in Bub-82/lamin B2 are replaced by G in Bub-82/lamin B2-G (see Table I). Lanes 1–4 are reactions with 0, 50, 100 and 200 ng of the Mcm4/6/7 complex, respectively. Schematic drawings of the substrates used in this assay are shown at the bottom of (A) and (B). B, boiled substrate.
Since the size of the bubble may affect the efficiency of unwinding, we constructed a derivative of Bub-82/lamin B2 in which six clusters of runs of thymines (33 in total) were replaced with guanines (Bub-82/lamin B2-G), and compared the unwinding efficiency. As shown in Figure 6B, very little unwinding was observed on Bub-82/lamin B2-G, indicating the critical role of thymine stretches in the lamin B2 origin for activation of the Mcm helicase.
Discussion
In order to clarify the roles of Mcm proteins during the course of DNA replication, we have been biochemically characterizing the mouse Mcm4/6/7 complex. The association of DNA helicase activity with the Mcm4/6/7 complex now appears to be conserved from mammals to fission yeast. In this report, we have characterized DNA binding and helicase activities of the Mcm4/6/7 complex on various substrate DNAs. The most remarkable finding in this study is that the Mcm4/6/7 complex displays a marked preference for T-rich sequences for helicase activation.
Mcm4/6/7 helicase can unwind bubble-like DNAs through loading onto a single-stranded DNA region
Initiation of DNA replication is associated with localized melting of duplex DNA near replication origins. Helicases are loaded onto replication forks through the melted region, induced by initiator binding, in bacteria (Bramhill and Kornberg, 1988). Therefore, we have examined whether Mcm4/6/7 can be loaded onto a bubble-like structure and can serve as a DNA helicase at the forks. The ability of Mcm4/6/7 to unwind the bubble substrate (Figure 2) indicates that Mcm can be loaded through the melted duplex DNA. Both Bub-60 and Bub-20 synthetic bubble substrates were bound by Mcm4/6/7 in gel shift assays, indicating that a 20 nucleotide single-stranded segment may be sufficient for Mcm4/6/7 to bind. However, Bub-20 generated mainly a complex containing a single hexamer and could not be unwound by Mcm4/6/7, whereas Bub-60, generating larger complexes, was unwound, albeit to a small extent (Figure 2A and B). The larger complex may include a double hexamer of Mcm4/6/7, as judged by comparison of its mobility with that of double hexameric T-antigen–DNA complex (Smelkova and Borowiec, 1998). Therefore, the helicase action of Mcm4/6/7 on synthetic bubbles may depend on the presence of an unpaired region of sufficient length, which may permit assembly of a double hexameric complex on the substrate DNA. The minimum bubble size that serves as an entry site for active helicase action appears to be 40–50 nucleotides long (data not shown).
T-rich single-stranded DNA activates Mcm helicase
The occurrence of AT-rich sequences, with asymmetric distribution of A and T, near the replication origins of many organisms prompted us to examine the effect of sequences of the bubble on the helicase activity of Mcm. A striking preference for T-rich sequences was observed in helicase assays on bubble substrates (Figure 2C) or on 3′-tailed substrates including Y-fork substrates (Figures 3 and 4). Our data indicate that the helicase and ATPase activities of Mcm4/6/7 are specifically activated by thymine stretches of certain lengths. It is known that the poly(dT) sequences may be associated with the least level of secondary structure compared with other polynucleotides (Bhattacharya et al., 2002). The absence of secondary structure may partly contribute to the efficient loading of the MCM helicase onto thymine stretches.
It was reported previously that single-stranded dT homopolymers stimulated the ATPase activity of the SV40 T-antigen (Giacherio and Hager, 1979). The SV40 core origin of replication consists of three functional domains, one of which is a 17 bp AT-rich segment associated with a bent DNA structure (Deb et al., 1986). Borowiec and Hurwitz (1988) have shown that thymine residues at the SV40 core origin were strongly modified by KMnO4, a chemical specifically reacting with unpaired thymine residues, upon T-antigen binding, suggesting untwisting or a conformational change of the T-rich region at the SV40 origin. Thus, helicase activation by thymine clusters may be common to the virus and cellular helicases.
Possible roles of Mcm in selection of replication initiation sites in mammalian genomes
In prokaryotic replicons, the sites of initial unwinding were localized at AT-rich sequences adjacent to the initiator binding sites (Bramhill and Kornberg, 1998). In eukaryotes, the sites of unwinding have not been determined for any specific origins, but clusters of AT-rich sequences are almost always found near the replication origins or initiation regions, leading to speculation that they may play crucial roles in initial unwinding (DePamphilis, 1996; Kelman, 2000). However, one of the biggest unsolved problems in mammalian DNA replication is how replication origins are selected and how this selection process is differentially regulated during developmental stages as well as in various cell types. Although ORC-binding sites dictate where replication can be initiated in budding yeast due to its strict sequence specificity (Marahrens and Stillman, 1992), mammalian ORC proteins do not show sequence-specific DNA binding (Vashee et al., 2003). Therefore, it does not appear that ORC per se can determine the sites of replication initiation. It is also difficult to explain the flexibility of the origin selection process during developmental stages (Hyrien and Mechali, 1993). Our results indicate that activation of the Mcm helicase requires its specific interaction with T-rich sequences. This led us to propose an exciting possibility that Mcm may play crucial roles in selection of replication initiation sites, as explained below (Figure 7).
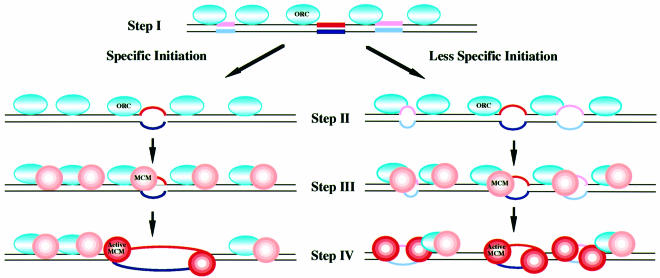
Fig. 7. A model on selection of replication initiation sites in mammalian cells. Blue ovals, pink and red circles represent ORC, pre-activated Mcm and activated Mcm helicases, respectively. Red and pink bars indicate highly and moderately T-rich segments, respectively, and blue and pale blue bars represent their complementary strands. For two divergent active Mcm helicases to be generated, T-rich segments may need to be present on both strands near the replication initiation sites. This is not shown in the figure. For details, see text.
The ORC may be bound to the genomic DNA due to its DNA binding activity, although the binding sites may not be specific along the genome (Figure 7, step I). At late telophase, Mcm is loaded onto chromatin with the aid of loading factors such as Cdt1 and Cdc6 and through direct or indirect protein–protein interactions with ORC (Figure 7, step III; Donovan et al., 1997; Nishitani et al., 2000). The loading step is not dictated by the genome sequences and presumably the number of Mcm complexes loaded would far exceed the number of the initiation sites (Edwards et al., 2002). The loaded Mcm protein is not active as helicase until it is activated by interacting with T-rich sequences which may happen to be present in the vicinity of ORC-binding sites (Figure 7, step IV, left pathway). Phosphorylation of Mcm by Cdc7 kinase may also play crucial roles in this activation step (Masai and Arai, 2002). Since double-stranded DNA is not capable of activating the Mcm helicase, the T-rich sequences need to adopt a single-stranded conformation to act as an Mcm activator. This could be possible by spontaneous melting or breathing of the AT-rich sequences or may be induced by ORC binding or by topological changes associated with nearby transcriptional activity (step II; Ohba et al., 1996). We propose that only a subset of Mcm proteins loaded onto DNA are activated, and the probability of activation of loaded Mcm complexes would be dependent on how frequently the conformational change of DNA occurs that exposes the single-stranded T stretches. Helicase activation of Mcm4/6/7 requires only 40–50 nucleotides of ssDNA in vitro, and the extent of activation would depend on the sequence composition; the more T stretches, the more activation (Figures 3 and 6). There is a fair chance of encountering the T stretch sequences along the genome, and, therefore, if the localized conformational change occurs at more locations on the genome, more Mcm may be activated (Figure 7, right pathway) and serves as initiation sites for DNA replication, as found in amphibian eggs. We speculate that the readiness with which this initial conformational change occurs may be determined by local chromatin structures, methylation or other modification of template DNA. The ‘initiation zone’ has been identified in some of the origin regions of mammalian genomes (Aladjem et al., 1998; Ina et al., 2001; Kobayashi et al., 1998). This may coincide with the clustered AT-rich stretches which are frequently found in the intergenic regions.
Consistent with this model, the nucleotide sequences spanning the initiation sites of the human lamin B2 origin served as an efficient activator for the Mcm helicase in bubble helicase assays, and substitutions of thymine stretches with guanine led to almost complete loss of unwinding (Figure 6B). The ability of a given sequence to activate Mcm helicase or ATPase activity could be a very convenient assay system to determine whether it can serve as an initiation site on the genome.
Interaction of Mcm4/6/7 with DNA and mode of helicase activation
Nuclease protection analyses of the interaction of Mcm4/6/7 with bubble structures revealed that Mcm4/6/7 is in contact with the entire ssDNA region but not with the duplex region (data not shown). The protection pattern supports the idea that Mcm4/6/7 recognizes the single-stranded region and not the specific structures such as a branched DNA. This is consistent with the prediction from the results of gel shift assays that Mcm recognizes the sequence of the ssDNA region rather than the forked structure per se (Figure 4A). In that sense, Mcm may not be a structure-specific binding protein, as is the case for other helicases such as PriA or RecG of bacteria (McGlynn et al., 1997). The higher affinity of Mcm4/6/7 for forked substrates than for ssDNA (Figure 4A) may be due to stabilization of the Mcm–DNA complex by a fork structure.
Although the precessivity of mammalian Mcm helicase is significantly increased by a fork structure and the presence of RPA (Figure 1C), the full level of activation of its helicase at the forks may require modification by phosphorylation and/or interaction with other replication factors, including Cdc45, Mcm10 and others (Zou et al., 1997; Homesley et al., 2000). Although Mcm2, Mcm3 and Mcm5 inhibit Mcm4/6/7 helicase in vitro, they are also essential for DNA replication (Labib et al., 2000). Their roles in promotion of replication forks also need to be investigated.
The Mcm helicase is likely to be loaded onto DNA through a ‘ring opening’ mechanism, as advocated by Patel and Picha (2000). Electron microscopic studies of the Mcm4/6/7 complex indicate a doughnut shape with a central hole and that it forms a beads-on-a-string structure on ssDNA, consistent with the above possibility (Sato et al., 2000; Fletcher et al., 2003). Oligo(dT) DNA may penetrate through the hole during the helicase action. The failure of Mcm to bind to oligo(dA) (Figure 5C) indicates that it interacts with only one strand at unwound T stretch sequences. More detailed biochemical and structural studies of the Mcm complex as well as genetic evaluation of the potential Mcm-activating sequences present in the known mammalian replication origins for their role in origin activation would shed new light onto how initiation of mammalian DNA replication is regulated.
Materials and methods
Reagents
Labelled and unlabelled dNTPs/rNTPs were purchased from Amersham Pharmacia. M13mp18 ssDNA, T4 polynucleotide kinase and the Klenow fragment were purchased from Takara. Anti-FLAG M2 Ab (antibody)–agarose and FLAG peptide were from Sigma.
Expression and purification of mouse Mcm4/6/7 complex
The recombinant baculovirus expressing His6-Mcm4/Mcm6 proteins was described previously (You et al., 1999). Sf9 and High 5 insect cells were cultured at 27°C in Sf-900 II SFM (Life Technologies, Inc.) and EX-CELL 405 media (JRH Biosciences), respectively. For expression of the Mcm4/6/7 proteins, recombinant baculoviruses expressing the His6-Mcm4/Mcm6 proteins and those expressing the Mcm7-FLAG were co-infected and then cells were collected at 48 h post-infection. The recombinant Mcm4/6/7 complexes in infected High 5 cell lysates were purified as previously described (You et al., 2002).
DNA substrates
The sequences for all the oligonucleotides used for constructions of DNA substrates are listed in Table I. 5′-tailed partial heteroduplex substrates were constructed by annealing an oligonucleotide, dT(0–40)-37mer carrying oligo(dT) tails of various lengths at the 5′ end of the invariable 37mer sequence, to M13mp18 ssDNA and labelling its 3′ end with [α-32P]dGTP and the Klenow fragment. Similar substrates, the 5′ tails of which were replaced by oligo(dA30) oligo(dG30) or randomized oligonucleotides (dA30-37mer, dG30-37mer, dN30-37mer), were also constructed. To determine the maximal length of duplex DNA displayed by the Mcm4/6/7 helicase, the substrates containing longer duplex regions were prepared by extending DNA chains with Sequenase (Amersham Pharmacia Biotech). For 3′-tailed partial heteroduplex substrates, 37mer-dT(10–50) was labelled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase and the labelled oligonucleotide was then annealed with M13mp18 ssDNA. The 3′-tailed substrates in which oligo(dT) was replaced by oligo(dA50), oligo(dC50) or oligo(dG50) were also constructed similarly. The labelled substrates were purified by Sepharose CL4B column chromatography (Amersham Pharmacia Biotech.).
The DNA bubble substrates were assembled from two partially complementary oligonucleotides with top and bottom strand sequences (see Table I). For each substrate, the top strand oligonucleotide was labelled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase and was annealed with the bottom strand oligonucleotide in a reaction mixture (50 µl) containing 20 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 25 mM NaCl. A 3 pmol aliquot of top strand plus 6 pmol of unlabelled bottom strand was used for annealing reaction mixtures (100 µl), which were heated to 95°C, kept at 67°C for 1 h, and then allowed to slowly cool down to 37°C. The bubble substrates were purified from the polyacrylamide gel by elution into TE buffer (Sambrook et al., 1989).
Y-fork substrates were prepared by annealing a 5′-end-labelled oligonucleotide, dT30-50mer, dA30-50mer or dC30-50mer (3 pmol), and an unlabelled oligonucleotide, 50mer-dT60, 50mer-dA60 or 50mer-dC60 (6 pmol), in various combinations and purification of the annealed products (T-tailed Y-fork, A-tailed Y-fork, C-tailed Y-fork, T/C-tailed Y-fork and C/T-tailed Y-fork) from the polyacrylamide gel, as above (Sambrook et al., 1989). Oligo(dT50), oligo(dA50), oligo(dC50), oligo(dG50) and A-rich50 oligonucleotides were synthesized and purified from the polyacrylamide gel.
Bub-82/lamin B2 was constructed by annealing the T-rich strand of the lamin B2 origin region (lamin B2 top; nucleotides 3877–4008 in GenBank accession No. M94363) with the opposite strand (lamin B2 bottom) in which the central 82 nucleotides (A-rich strand) were changed to those of the T-rich strand. The Bub-82/lamin B2-G was constructed from lamin B2-G top and lamin B2-G bottom, in which runs of thymine within the 82 nucleotide unpaired region were replaced by guanine.
Preparation of SV40 T-antigen and RPA, helicase processivity assay
SV40 T-antigen was purified from baculovirus-infected insect cells as reported (Ishimi and Matsumoto, 1993). Recombinant three-subunit human RPA was purified from Escherichia coli (Henricksen et al., 1994). To determine the maximal length of duplex DNA displaced by the Mcm4/6/7 helicase, the substrates containing longer duplex regions were prepared by extending DNA chains with Sequenase (Amersham Pharmacia Biotech) from the 3′ end of the dT40-37mer annealed to M13mp18. The 5′-tailed substrate (dT40-37mer/M13mp18) was incubated with Sequenase, first to a limited extent in the presence of [α-32P]dGTP and the other three dNTPs at 2 µM, followed by further extension after addition of 8 µM ddGTP and all four dNTPs at 80 µM, resulting in substrates containing labelled duplex regions of various lengths (dT40-Nmer mixture/M13mp18). The substrate (5 fmol) was first pre-incubated with the indicated amount of the Mcm4/6/7 complex at 30°C for 10 min, and then human RPA (0.4 µg) was added where indicated. In the reaction with T-antigen, human RPA was not added. After incubation at 37°C for 1 h, reactions were stopped, and aliquots were electrophoresed on a 7.5% polyacrylamide gel in 1× TBE at 300 V for 4 h.
DNA helicase, gel shift and ATPase assays
DNA helicase assays on partially heteroduplex circular DNAs were conducted as described previously (You et al., 1999). In the gel shift assay, Mcm proteins were incubated at 30°C for 30 min in reaction mixtures (15 µl) containing 25 mM HEPES-NaOH pH 7.5, 50 mM sodium acetate, 10 mM Mg(CH3COO)2, 1 mM dithiothreitol (DTT), 0.25 mg/ml bovine serum albumin (BSA), 0.25 mM AMP-PNP or 0.5 mM ATP-γ-S, and labelled forked substrates or bubble substrates in the amounts indicated. After addition of 2 µl of 50% glycerol, the reaction mixtures were directly applied to a polyacrylamide gel (acrylamide: bisacylamide 79:1) containing 6 mM magnesium acetate and 5% glycerol in 0.5× TBE, and electrophoresis was conducted at room temperature. For DNA helicase assays on Y-fork and synthetic bubble DNAs, the reaction mixtures for the gel shift assay, as described above, were incubated at 30°C for 30 min, and then the ATP was added at 10 mM, followed by incubation at 37°C for an additional 30 min to 1 h. The reactions were terminated by addition of EDTA (20 mM), SDS (0.1%) and 2 µg of proteinase K, and were incubated for an additional 30 min. The samples were separated by electrophoresis on a 15% (acrylamide: bisacylamide 19:1) non-denaturing polyacrylamide gel in 1× TBE. ATPase activity was measured as previously described (You et al., 2002), except that the concentration of ATP was 1 mM.
Acknowledgments
Acknowledgements
We thank Taku Tanaka and other members of our laboratory for helpful discussion. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Rodewaldm,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S.cerevisiae: redistribution of Mcm proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Bhattacharya P.K., Cha,J. and Barton,J.K. (2002) 1H NMR determination of base-pair lifetimes in oligonucleotides containing single base mismatches. Nucleic Acids Res., 30, 4740–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinsky A.K. and Gerbi,S.A. (1998) Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science, 279, 95–98. [DOI] [PubMed] [Google Scholar]
- Borowiec J.A. and Hurwitz,J. (1988) Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J., 7, 3149–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. (1996) Common structural features of replication origins in all life forms. J. Cell. Biochem., 60, 297–316. [DOI] [PubMed] [Google Scholar]
- Bramhill D. and Kornberg,A. (1988) A model for initiation at origins of DNA replication. Cell, 54, 915–918. [DOI] [PubMed] [Google Scholar]
- Chong J.P., Hayashi,M.K., Simon,M.N., Xu,R.M. and Stillman,B. (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 97, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., DeLucia,A.L., Koff,A., Tsui,S. and Tegtmeyer,P. (1986) The adenine–thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol. Cell. Biol., 6, 4578–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L. (1996) Origin of DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 45–86. [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.C., Tutter,A.V., Cvetic,C., Gilbert,C.H., Prokhorova,T.A. and Walter,J.C. (2002) MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem., 277, 33049–33057. [DOI] [PubMed] [Google Scholar]
- Fletcher R.J., Bishop,B.E., Leon,R.P., Sclafani,R.A., Ogata,C.M., Chen,X.S. (2003) The structure and function of MCM from archaeal M.thermoautotrophicum.Nature Struct. Biol., 10, 160–167. [DOI] [PubMed] [Google Scholar]
- Giacherio D. and Hager,L.P. (1979) A poly(dT)-stimulated ATPase activity associated with simian virus 40 large T antigen. J. Biol. Chem., 254, 8113–8116. [PubMed] [Google Scholar]
- Gilbert D.M. (2001) Making sense of eukaryotic DNA replication origins. Science, 294, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- Homesley L., Lei,M., Kawasaki,Y., Sawyer,S., Christensen,T. and Tye,B.K. (2000) Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev., 14, 913–926. [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. and Mechali,M. (1993) Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J., 12, 4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ina S., Sasaki,T., Yokota,Y. and Shinomiya,T. (2001) A broad replication origin of Drosophila melanogaster, oriDα, consists of AT-rich multiple discrete initiation sites. Chromosoma, 109, 551–564. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. and Matsumoto,K. (1993) Model system for DNA replication of a plasmid DNA containing the autonomously replicating sequence from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 5399–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z. (2000) The replication origin of archaea is finally revealed. Trends Biochem. Sci., 25, 521–523. [DOI] [PubMed] [Google Scholar]
- Kelman Z., Lee,J.K. and Hurwitz,J. (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum ΔH contains DNA helicase activity. Proc. Natl Acad. Sci. USA, 96, 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Rein,T. and DePamphilis,M.L. (1998) Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol., 18, 3266–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd edn. W.H.Freeman and Co., New York. [Google Scholar]
- Labib K., Tercero,J.A. and Diffley,J.F. (2000) Uninterrupted MCM2–7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Lee J.K. and Hurwitz,J. (2000) Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem., 275, 18871–18878. [DOI] [PubMed] [Google Scholar]
- Lee J.K. and Hurwitz,J. (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6 and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA, 98, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y. and Stillman,B. (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science, 255, 817–823. [DOI] [PubMed] [Google Scholar]
- Masai H. and Arai,K. (2002) Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell. Physiol., 190, 287–296. [DOI] [PubMed] [Google Scholar]
- McGlynn P., Al-Deib,A.A., Liu,J., Marians,K.J. and Lloyd,R.G. (1997) The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol., 270, 212–221. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Ohba R., Matsumoto,K. and Ishimi,Y. (1996) Induction of DNA replication by transcription in the region upstream of the human c-myc gene in a model replication system. Mol. Cell. Biol., 16, 5754–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Satoh,H., Sekiguchi,M. and Masukata,H. (1999) Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol. Cell. Biol., 19, 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Labortary Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sato M., Gotow,T., You,Z., Komamura-Kohno,Y., Uchiyama,Y., Yabuta,N., Nojima,H. and Ishimi,Y. (2000) Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol., 300, 421–431. [DOI] [PubMed] [Google Scholar]
- Smelkova N.V. and Borowiec,J.A. (1998) Synthetic DNA replication bubbles bound and unwound with twofold symmetry by a simian virus 40 T-antigen double hexamer. J. Virol., 72, 8676–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C. (1999) ORC binding, gene amplification and the nature of metazoan replication origins. Genes Dev., 13, 2619–2623. [DOI] [PubMed] [Google Scholar]
- Stillman B. (1996) Cell cycle control of DNA replication. Science, 274, 1659–1664. [DOI] [PubMed] [Google Scholar]
- Thömmes P., Kubota,Y., Takisawa,H. and Blow,J.J. (1997) The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J., 16, 3312–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- Vashee S., Cvetic,C., Lu,W., Simancek,P., Kelly,T.J. and Walter,J.C. (2003) Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev., 17, 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Dijkwel,P.A. and Hamlin,J.L. (1998) Lagging-strand, early-labelling and two-dimensional gel assays suggest multiple potential initiation sites in the Chinese hamster dihydrofolate reductase origin. Mol. Cell. Biol., 18, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Komamura,Y. and Ishimi,Y. (1999) Biochemical analysis of the intrinsic Mcm4–Mcm6–Mcm7 DNA helicase activity. Mol. Cell. Biol., 19, 8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Ishimi,Y., Masai,H. and Hanaoka,F. (2002) Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J. Biol. Chem., 277, 42471–42479. [DOI] [PubMed] [Google Scholar]
- Zou L., Mitchell,J. and Stillman,B. (1997) CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol. Cell. Biol., 17, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]