Abstract
The gap junction protein connexin36 (CX36) has been well studied in the mature central nervous system, but there has been little information regarding its possible roles in embryonic development. We report here the isolation of the full-length chick CX36 coding sequence (predicted Mr 35.1 kDa) and its strikingly restricted pattern of gene expression in the mesoderm of the chick embryo. In situ hybridization experiments demonstrated CX36 expression in somites by embryonic day 2. The transcripts first appeared dorsomedially within the somite and expanded ventrolaterally to form stripes in the middle of each somite. The CX36 stripes fell within somitic territories enriched in MYOD and FGF8 expression and impoverished in PAX3 transcripts, establishing that CX36 mRNA is expressed in the myotome. We compared the somitic expression pattern of CX36 with those of three other connexins, CX42, CX43, and CX45. At embryonic day 4, CX42 transcripts were localized to the myotome in a pattern resembling that of CX36. In contrast, CX43 was enriched in the dermomyotome, and CX45 was detected in both the myotome and the dermomyotome. Immunoblotting using Cx36 antibodies demonstrated bands of identical electrophoretic mobilities in trunk and retinal homogenates, and Cx36 immunostaining detected punctate immunoreactivity in the myotome. These results demonstrate that some connexins in the developing mesoderm are broadly expressed whereas others are highly localized, and suggest that CX36, CX42, and CX45 are involved in intercellular communication among developing muscle cells.
Keywords: Gap junction, FGF8, MYOD, PAX3, Myotome
Introduction
Intercellular communication through gap junction channels may be important for defining cell compartments during embryonic development. Gap junctions are membrane specializations containing clusters of intercellular channels that allow transfer of ions and molecules up to 1 kDa between adjacent cells. Gap junction channels are made of oligomeric assemblies of homologous proteins named connexins (CX). Most cells in the adult animal are coupled through gap junctions. One exception is skeletal muscle, in which cells are initially coupled but become uncoupled as development and differentiation proceed (Rash and Staehelin, 1974; Keeter et al., 1975; Kalderon et al., 1977; Schmalbruch, 1982).
During development, trunk paraxial mesoderm becomes segmented into somites. Somites give rise to distinct cell populations, including the sclerotome, the dermomyotome, and the myotome, from which skeletal muscle cells originate. Experiments using injection of a gap junction permeant dye established the presence of several stage-dependent communication compartments within somites. Myotome cells are coupled as an ensemble, as are dermomyotome cells; sclerotome cells are subdivided into rostral and caudal compartments separated by a 3rd communication compartment made of cells from the intrasclerotomal fissure (Bagnall et al., 1992). The molecular basis of these compartments has not been extensively studied. In the mouse, Cx43 transcripts have been detected in the dermatome and sclerotome (Ruangvoravat and Lo, 1992) and Cx40 transcripts in myoblasts and myotubes (Dahl et al., 1995). In the rat, Cx43 protein has been detected in the dermatome (Yancey et al., 1992).
The present experiments explored the expression of connexins during somite development in the chicken embryo with special emphasis on CX36, the ortholog of mammalian Cx36 (Condorelli et al., 1998; Söhl et al.,1998) and fish Cx35 (O’Brien et al., 1996; 1998). Here, we report the cloning of chicken CX36, its expression pattern in somites during development, and the relation of its expression pattern to those of signaling molecules, transcription factors, and other connexins.
Materials and methods
Chick embryos
Fertilized White Leghorn chicken eggs obtained from Charles River SPAFAS (Connecticut, USA) and local suppliers were incubated at 99.5°F in a humidified incubator. Embryos were staged according to trunk morphology (Hamburger and Hamilton, 1951) and somite maturation (Ordahl, 1993).
Connexin36 cloning
A DNA fragment of chicken CX36 was obtained by PCR using genomic DNA and a set of primers based on the mouse, human, skate and perch connexin 36 sequences (accession numbers: AF016190, AF153047, U43290, and AF059183, respectively) using the CODEHOP program (Rose et al., 1998). Total cellular RNA was isolated from brain and retina by the method of Chomczynski and Sacchi (1987). Complementary DNAs spanning the full CX36 coding sequence were obtained using the SMART RACE cDNA Amplification Kit (Clontech, BD Biosciences, Palo Alto, CA). The CX36 sequence was deposited into the GeneBank database (accession number: AF458098).
Wholemount in situ hybridization
Wholemount in situ hybridization was performed as previously described (Agarwala and Ragsdale, 2002) using embryos from HH stage 10 until day 7 with probes for CX36, CX42 (Beyer, 1990), CX43 (Musil et al., 1990), CX45 (Beyer, 1990), FGF8 (kindly provided by Dr. Gail Martin), MYOD and PAX3 (kindly provided by Dr. Martyn Goulding). Some stained embryos were embedded in gelatin and sectioned at 32–40 µm on an SM 2000R sliding microtome (Leica, Houston, TX). Sections were dried onto glass slides, dehydrated, and mounted with coverslips and Eukitt Mounting Medium (Electron Microscopy Sciences, Fort Washington, PA). Sections were studied with an Axioplan 2 microscope (Carl Zeiss, Thornwood, NY), and images were captured with an AxioCam digital camera (Carl Zeiss, Thornwood, NY). Composite figures were assembled using Adobe Photoshop (Adobe Systems, San Jose, CA)
Protein determination
Protein concentrations were determined using the BioRad Protein Assay (BioRad, Hercules, CA) based on the Bradford dye-binding procedure (Bradford, 1976).
Immunoblotting
Homogenates from embryonic day 4 (E4) trunk, E12 retina, and E12 liver were prepared in 4 mM EDTA, 2 mM phenylmethylsul-fonylfluoride in phosphate buffered saline (PBS), pH 7.4. One hundred µg of protein were resolved on an 11% SDS-containing poly-acrylamide gel and transferred to Immobilon-P (Millipore, Billerica, MA). Membranes were blocked in 5% nonfat milk in Tris-buffered saline (TBS), pH 7.4, and incubated overnight in rabbit polyclonal anti-Cx36 antibodies (Zymed Laboratories, South San Francisco, CA). Membranes were rinsed in TBS and incubated in peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) for 1 h and rinsed in TBS. Binding of secondary antibody was detected using enhanced chemiluminescence (ECL; Amersham, Arlington Heights, IL).
Immunofluorescence
Chicken embryos at day 6 were fixed in 4% paraformaldehyde in PBS for 4 h at room temperature. They were transferred to 30% sucrose in PBS and left at 4°C until they sank. Twelve-µm cryostat sections were incubated in 0.2% Triton X-100 for 30 min at room temperature, followed by incubation in blocking solution (5 mM EDTA, 1% fish gelatin, 0.05% NP40, 1% essentially immunoglobulin- free bovine serum albumin, 1% normal goat serum in PBS). Sections were incubated overnight at 4°C with anti-Cx36 antibodies (Zymed Laboratories, South San Francisco, CA) and the mouse monoclonal anti-chicken pectoralis myosin antibody MF20 (NICHD/University of Iowa Developmental Studies Hybridoma Bank). Sections were rinsed four times with PBS and then incubated in Cy2-conjugated goat anti-rabbit IgG and Cy3-conjugated goat anti-mouse IgG antibodies (Jackson ImmunoResearch, West Grove, PA) for 1.5 h at room temperature. Sections were rinsed four times with PBS, and coverslips were mounted with 2% n-propyl-gallate in PBS:glycerol (1:1).
Results
DNA sequence for a fragment of chicken CX36 encoding the 1st extracellular loop through the 4th transmembrane domain was obtained by PCR from genomic template DNA. Full coding sequence was obtained by RACE using total cellular RNA from brain and retina with primers based on the DNA fragment obtained by PCR. Analysis of the chicken CX36 sequence predicted a 304 amino-acid protein of a molecular mass of 35.1 kDa showing a high degree of sequence identity to CX36 sequence from mammals and fishes (Fig. 1).
Fig. 1.
Alignment of the DNA sequences of connexin36 from chicken (cCX36), skate (sCx35), rat (rCx36), and human (hCX36). Amino acids that are identical in all sequences are indicated in dark green, while those that differ among the sequences are indicated in red. The transmembrane domains (TM) predicted by the HMM TOP server (http://www.enzim.hu/hmmtop/) are indicated with lines. There are 259, 271, and 276 identical amino acids between chicken CX36 and skate Cx35, rat Cx36, and human CX36, respectively
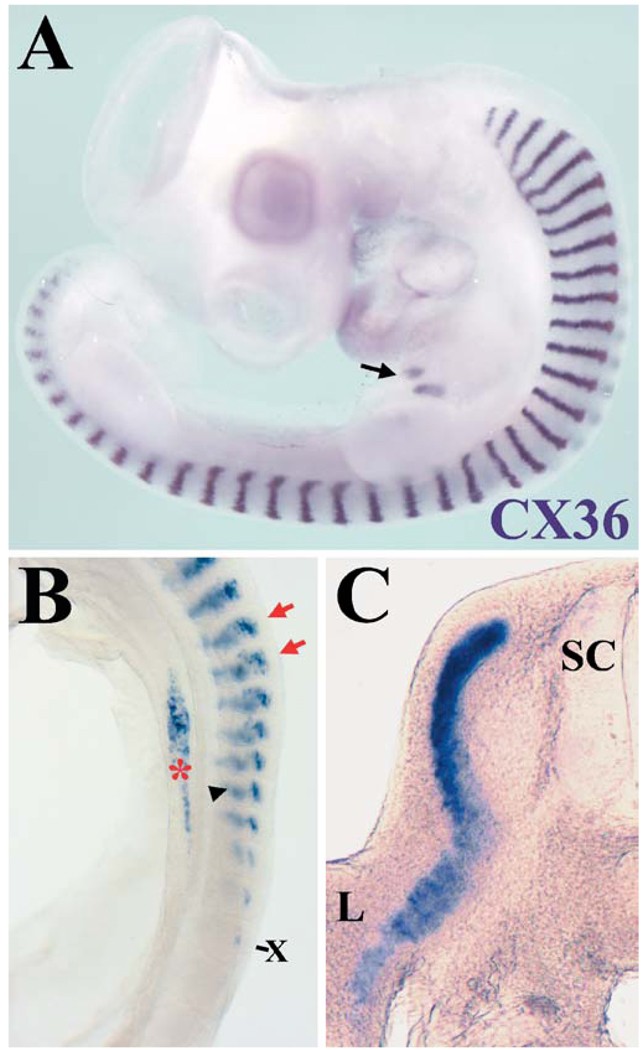
The expression pattern of CX36 was studied by wholemount in situ hybridization. A strong and striking pattern of hybridization was detected in the somites of the chick embryo. The CX36 probe hybridized to a narrow stripe in the middle of the somite (illustrated in Fig. 2A for E4). At this stage, CX36 hybridization was also detected in pancreas and heart (Fig. 2A). The hybridizing region in somites initially formed a triangular area in a dorsomedial position, within which labeled cells formed a “mosaic” pattern (E2; from HH stage 13 and illustrated for HH stage 15 in Fig. 2B). As somite differentiation proceeded, the hybridizing region extended ventrolaterally and became more restricted to the middle of the somite. The striped pattern of expression of CX36 was observed by wholemount in situ hybridization through embryonic day 7, the latest stage studied; at this stage, hybridization in the tail region was much stronger than in the trunk region. Sectioned embryos showed labeling in the myotomal region of the somite and in cells that had populated the limb bud (Fig. 2C).
Fig. 2A–C.
Expression pattern of CX36 in somites. A Photomicrograph of an embryonic day 4 (E4) chick (HH stage 24) demonstrates CX36 expression in stripes in the middle of the somites, in the pancreas (arrow), and in the heart. B Lateral view of the trunk region of an E2 chicken (HH stage 15). Hybridization of CX36 in the somites is first detected in the 10th-most distal somite (X). The mosaic pattern of CX36 hybridization is indicated by the red arrows. Somitic hybridization in the contralateral side is seen out of focus (black arrowhead). Endoderm expression (red asterisk) can be detected by HH stage 14 and will later (HH stage 16–18) coalesce to mark the ventral and dorsal rudiments of the pancreas. C Transverse section through the trunk region of an E5 chicken (HH stage 27) demonstrates hybridization of the CX36 probe to the myotome and to cells that have populated the limb. (SC spinal cord, L limb)
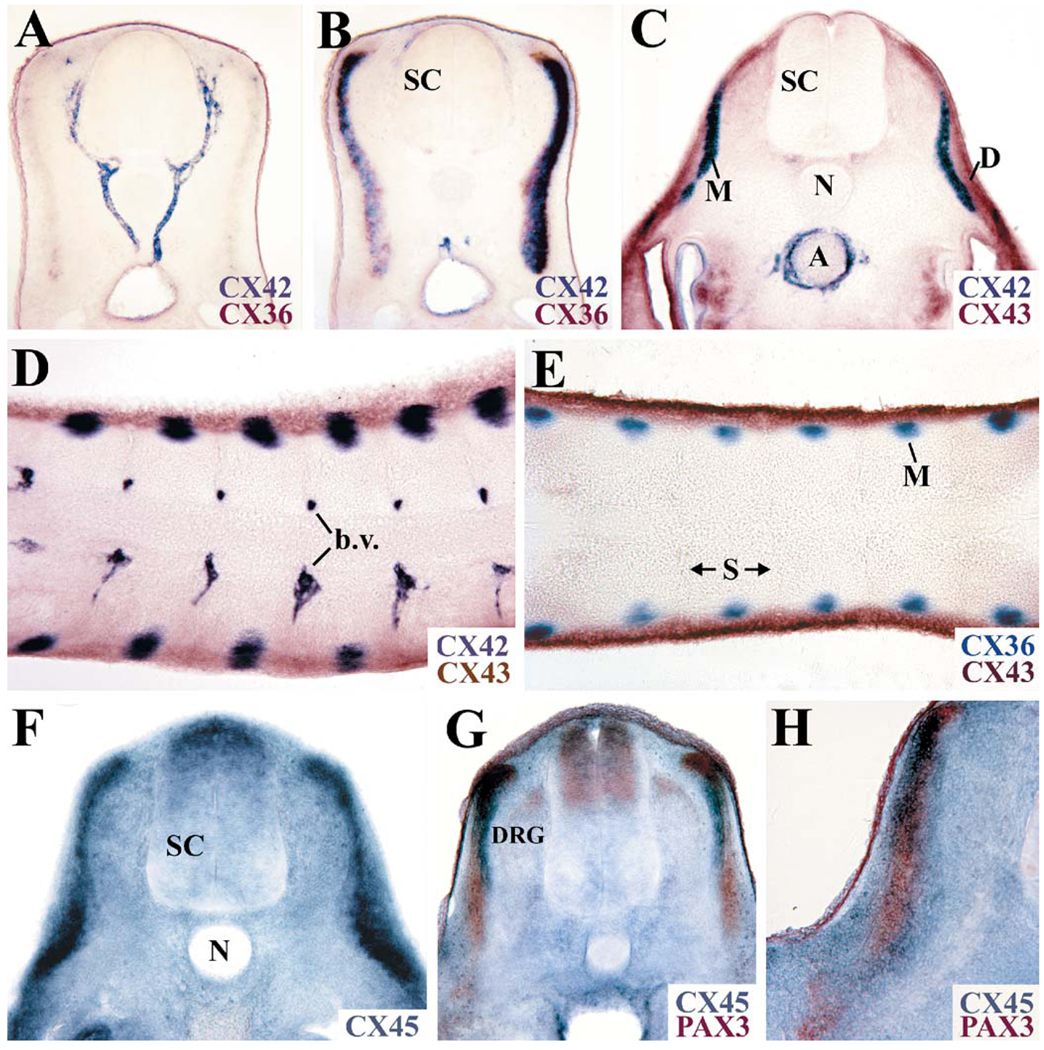
We compared the expression pattern of CX36 to that of signaling molecules and transcription factors known to be expressed in somites. Two-color in situ hybridization was performed using probes for CX36 and PAX3, FGF8, or MYOD on wholemount embryos. The gene expression patterns for CX36 and the transcription factor PAX3 were strikingly different. PAX3 was detected in the mesoderm before the onset of CX36 expression. At embryonic day 4, PAX3 was strongly expressed in the dermomyotome (Goulding et al., 1994; Williams and Ordahl, 1994), whereas CX36 appeared to identify only the myotome (Fig. 3A, B, E). Very little overlap at the dorsomedial and ventrolateral regions was observed between the PAX3- and CX36-hybridizing domains (Fig. 3B). In contrast, substantial overlap of the gene expression patterns was observed in experiments using probes for CX36 and the muscle-specific transcription factor MYOD, which is expressed in the myotome (Pownall and Emerson, 1992), although the area of MYOD expression was broader than that of CX36 (Fig. 3C, F). At this developmental age, FGF8 transcripts were found in the myotome in agreement with the findings of Stolte et al. (2002), and the expression domains of CX36 and FGF8 showed extensive overlap (Fig. 3D).
Fig. 3A–F.
Connexin36 is expressed in the myotome. Transverse (A–D) and horizontal (E, F) sections through somites from stage 24 embryos prepared for two-color in situ hybridization with probes to PAX3 and CX36 (A, B, E), MYOD and CX36 (C, F), and FGF8 and CX36 (D). Hybridization of the CX36 probe is marked by a blue-purple reaction product and that of the PAX3, MYOD, and FGF8 probes appears in brown. (D dermomyotome, DRG dorsal root ganglion, M myotome, N notochord, SC spinal cord)
The expression pattern of CX36 was also compared to that of three connexins, CX42, CX43, and CX45, previously reported to be expressed in somites based on mRNA, protein, and lacZ reporter expression experiments (Ruangvoravat and Lo, 1992; Yancey et al., 1992; Dahl et al., 1995; Krüger et al., 2000). Connexin42 was concentrated in the heart and blood vessels from early stages of development, but by embryonic day 4, the wholemount in situ hybridization pattern of CX42 in somites showed a striking similarity to that of CX36. Serial sectioning demonstrated extensive overlap between the CX42 and CX36 signals in the myotome (Fig. 4B) and strong CX42 labeling of intersomitic vessels (Fig. 4A). Connexin43, in contrast, was widely expressed in the chicken embryo. At embryonic day 4, CX43 transcripts in the somites were found within the dermomyotome and sclerotome. No overlap between the CX36- and CX43-hybridizing regions was observed (Fig. 4E). Similarly, no significant overlap between the CX42- and the CX43-rich territories was observed (Fig. 4C, D). Connexin45 was also widely expressed during development (Fig. 4F). Two-color in situ hybridization with PAX3 and CX45 established that CX45 transcripts were expressed in both the dermomyotome and the myotome (Fig. 4G, H).
Fig. 4A–H.
Multiple connexins are expressed in developing somites. A, B Adjoining serial transverse sections from an embryo probed for CX36 (brown) and CX42 (blue-purple) expression. Connexin42 and CX36 are both expressed in the myotome (B) and CX42 is enriched in the vasculature (A). C, D Transverse (C) and horizontal (D) sections from an embryo after two-color in situ hybridization with CX42 (blue-purple) and CX43 (brown) probes. E Horizontal section from an embryo hybridized with CX36 (blue-purple) and CX43 (brown) probes. F Transverse section from an embryo hybridized using a CX45 probe. G, H Transverse sections from an embryo after in situ hybridization with CX45 (blue) and PAX3 (brown) probes. All tissues were obtained from HH stage 24 embryos. (A aorta, b.v. blood vessel, D dermomyotome, DRG dorsal root ganglion, M myotome, N notochord, S somite, SC spinal cord)
By embryonic day 5, hybridization of the CX36 probe was also observed in the limb buds (Fig. 5A), eye muscles, and the central nervous system (not illustrated). The CX36-hybridizing domain in the limb bud showed substantial overlap with that of the PAX3 (Fig. 5B) and MYOD (Fig 5C). This result suggests that CX36 is also expressed in limb muscles; overlap of PAX3 and MYOD expression has been previously reported in the limb bud (Goulding et al., 1994).
Fig. 5A–C.
Connexin36 is expressed in the limb buds. A Whole-mount in situ hybridization of an embryonic day 6 (HH stage 29) hindlimb probed for CX36 expression shows hybridization in the dorsal and ventral regions of the proximal and distal parts of the limb bud. B, C Transverse sections of an embryonic day 5 forelimb after two-color in situ hybridization with probes to CX36 and PAX3 (B) and CX36 and MYOD (C). Hybridization of the CX36 probe is seen in blue-purple and that of PAX3 and MYOD is shown in brown. Dorsal is oriented towards the top
Expression of Cx36-like protein in the trunk region was confirmed by immunoblotting. A band with an electrophoretic mobility indistinguishable from that detected in homogenates of 12th embryonic day retina was detected in homogenates from the embryonic day 4 trunk region (Fig. 6). The localization of this protein was studied by immunofluorescence using anti-Cx36 antibodies. Immunoreactive spots with a distribution mimicking the shape of the myotome were observed in transverse sections (not illustrated). In horizontal sections, double immunofluorescence experiments using a monoclonal antibody that recognizes sarcomeric myosin showed punctate Cx36-like immunoreactivity at the plasma membrane of myotomal cells that was more evident at the caudal and rostral limits of each myotome (Fig. 7).
Fig. 6.
Immunoblot pattern of CX36. Homogenates of E12 liver, E12 retina, and E4 trunk were resolved by SDS-PAGE and subjected to immunoblotting using anti-Cx36 antibodies. The migration position of molecular mass standards is indicated
Fig. 7A, B.
Localization of Cx36-like immunoreactivity. A, B Photomicrographs of double immunofluorescence staining of a 12-µm horizontal section from an E6 chicken. Rabbit polyclonal anti-Cx36 antibodies and Cy2-conjugated goat anti-rabbit IgG antibodies (green) were used to detect Cx36-like protein, and a mouse monoclonal anti-chicken sarcomeric myosin (MF20) antibody and Cy3-conjugated goat anti-mouse IgG antibodies (red) were employed to label myotome cells. Connexin36-like staining (arrows) is shown in A (green channel alone), and its relationship to sarcomeric myosin is demonstrated in B (superposition of green and red channels). Punctate Cx36-like immunoreactivity (arrows) was observed between cells of the myotome (M). The lack of an extensive superposition between the red and green signals is due to the different cellular localizations of the proteins: Cx36-like immunoreactivity localizes to the plasma membrane, whereas MF20 immunoreactivity is present in the cytoplasm
Discussion
Expression of CX36 has previously been detected in neural cells (Condorelli et al., 1998) and β-cells of the pancreas (Serre-Beinier et al., 2000). In this paper, we report that during development, CX36 is expressed within the myotome, limb and eye muscles, the heart, and the pancreas. By embryonic day 4, CX36 mRNA expression was restricted to the middle of the somite, while staining for CX36 protein was evident in horizontal sections at the rostral and caudal ends of the myotome.
The myotome is formed from myoblasts derived from the dermomyotome. The first terminally differentiated myotome cells begin to appear by somite stage X (Ordahl, 1993). The onset of CX36 expression (as determined by wholemount in situ hybridization) would thus correlate with the appearance of terminally differentiated myotomal cells. At this early stage, the hybridization pattern of CX36 had a triangular shape. This transient triangular domain is reminiscent of the shape of the developing myotome defined by other muscle markers (i.e., desmin and acetylcholinesterase) (Kaehn et al., 1988), and it would result from migration of mesenchymal myoblasts to the rostral margin of the somite to form the primary myotome (Kahane et al., 2002).
The nuclei of the earliest postmitotic myotomal progenitors become reorganized by E4 into a narrow stripe restricted to the middle part of the myotome length (Kahane et al., 1998). Similarly, CX36 transcripts became restricted between E3 and E4 to the central region of the somite corresponding to the position of myotomal fiber nuclei. Thus, it is possible that CX36 transcripts are always restricted to the nuclear domain. A restricted localization of transcripts has been previously shown for the initial stages of slow myosin heavy chain expression (Sacks et al., 2003).
The finding of a mosaic pattern of hybridization for any gene expressed in somites is unexpected. However, nuclear localization of transcripts during migration of mesenchymal myoblasts and redistribution of nuclei during elongation of myotomal cells may explain the mosaicism observed in the hybridization pattern of CX36 at early stages of development. Because a role for gap junctions containing Cx43 has been previously proposed for neural crest cell migration (Huang et al., 1998), it is also plausible that CX36 may be involved in the migration events associated with myotome formation.
In contrast to the restricted localization of CX36 transcripts within the nuclear domain, immunoreactive Cx36-like protein was found between myotome cells and appeared concentrated at the sites of attachment of myotome cells to the rostral and caudal ends of somites. The fine punctae of Cx36-like protein observed in cross-sections and the linear-like pattern of staining observed in horizontal sections are reminiscent of the distribution of gap junctions during amphibian development as seen by transmission electron microscopy (Keeter et al., 1975; Blackshaw and Warner, 1976). In these embryos, gap junctions have been observed between muscle cells within a given somite and between muscle cells in adjacent somites (Keeter et al., 1975; Blackshaw and Warner, 1976). The linear-like appearance of staining at the rostral and caudal ends of the myotome as observed in horizontal sections might result from the convergence of myotomal cells to their sites of attachment which produces the fusiform shape of the myotome.
Skeletal muscle cells are initially coupled through gap junctions, but become uncoupled and lose these structures after formation of muscle fibers and innervation (Rash and Staehelin, 1974; Keeter et al., 1975; Kalderon et al., 1977; Schmalbruch, 1982). We have found that CX36, CX42, and CX45 are expressed in the myotome. Because rodent Cx40 and Cx43 have been previously reported to be expressed in somites (Ruangvoravat and Lo, 1992; Yancey et al., 1992; Dahl et al., 1995), it is possible that their expression in somites is conserved across species. Thus, our data confirm and extend previously published results and localize CX42 (ortholog of mouse Cx40) to the myotome, CX43 to the dermomyotome and CX45 to both the dermomyotome and the myotome. The extensive degree of overlap between the CX36- and CX42-expressing regions of the myotome suggests that both connexins could participate in the dye coupling observed by Bagnall et al. (1992) between myotome cells.
During myotome formation, expression of PAX3 precedes the appearance of MYOD transcripts (Goulding et al., 1994; Williams and Ordahl, 1994). Similarly, we found that PAX3 expression preceded expression of CX36. Myotomal expression of FGF8, which is detected in somite stages IX–XII at stage 12 and somite stages VII–XII at stage 13 (Stolte et al., 2002), also preceded expression of CX36. The transient expression of FGF8 in the myotome defining a subpopulation of muscle precursor cells (Stolte et al., 2002) suggests that FGF8 may be involved in regulation of CX36 expression in the myotome.
Acknowledgements
The authors are indebted to Anna Mae Greenle for technical assistance. These studies were funded by NIH grants (HD09402 to ECB; NS RO1 NS35680 to CWR) and the Bernice Meltzer Pediatric Research Fund. PCR products were sequenced at the University of Chicago Cancer Research Center DNA Sequencing Facility. The mouse monoclonal anti-chicken pectoralis myosin antibody (MF20) developed by D.A. Fischman was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA, USA.
Contributor Information
Viviana M. Berthoud, Department of Pediatrics, Section of Hematology/Oncology, University of Chicago, 5841 S. Maryland Ave., MC 4060, Chicago, IL 60637, USA, vberthou@peds.bsd.uchicago.edu, Tel.: +1-773-7021205, Fax: +1-773-7029881
Rashmi Singh, Department of Pediatrics, Section of Hematology/Oncology, University of Chicago, 5841 S. Maryland Ave., MC 4060, Chicago, IL 60637, USA, Tel.: +1-773-7021205, Fax: +1-773-7029881.
Peter J. Minogue, Department of Pediatrics, Section of Hematology/Oncology, University of Chicago, 5841 S. Maryland Ave., MC 4060, Chicago, IL 60637, USA, Tel.: +1-773-7021205, Fax: +1-773-7029881
Clifton W. Ragsdale, Department of Neurobiology, Pharmacology and Physiology, University of Chicago, Chicago, IL, USA
Eric C. Beyer, Department of Pediatrics, Section of Hematology/Oncology, University of Chicago, 5841 S. Maryland Ave., MC 4060, Chicago, IL 60637, USA, Tel.: +1-773-7021205, Fax: +1-773-7029881
References
- Agarwala S, Ragsdale CW. A role for midbrain arcs in nucleogenesis. Development. 2002;129:5779–5788. doi: 10.1242/dev.00179. [DOI] [PubMed] [Google Scholar]
- Bagnall KM, Sanders EJ, Berdan RC. Communication compartments in the axial mesoderm of the chick embryo. Anat Embryol. 1992;186:195–204. doi: 10.1007/BF00174957. [DOI] [PubMed] [Google Scholar]
- Beyer EC. Molecular cloning and developmental expression of two chick embryo gap junction proteins. J Biol Chem. 1990;265:14439–14443. [PubMed] [Google Scholar]
- Blackshaw SE, Warner AE. Low resistance junctions between mesoderm cells during development of trunk muscles. J Physiol. 1976;255:209–230. doi: 10.1113/jphysiol.1976.sp011276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Condorelli DF, Parenti R, Spinella F, Salinaro AT, Belluardo N, Cardile V, Cicirata F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian neurons. Eur J Neurosci. 1998;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Dahl E, Winterhager E, Traub O, Willecke K. Expression of gap junction genes, connexin40 and connexin43, during fetal mouse development. Anat Embryol. 1995;191:267–278. doi: 10.1007/BF00187825. [DOI] [PubMed] [Google Scholar]
- Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Huang GY, Cooper ES, Waldo K, Kirby ML, Gilula NB, Lo CW. Gap junction-mediated cell-cell communication modulates mouse neural crest migration. J Cell Biol. 1998;143:1725–1734. doi: 10.1083/jcb.143.6.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehn K, Jacob HJ, Christ B, Hinrichsen K, Poelmann RE. The onset of myotome formation in the chick. Anat Embryol. 1988;177:191–201. doi: 10.1007/BF00321131. [DOI] [PubMed] [Google Scholar]
- Kahane N, Cinnamon Y, Kalcheim C. The origin and fate of pioneer myotomal cells in the avian embryo. Mech Dev. 1998;74:59–73. doi: 10.1016/s0925-4773(98)00066-5. [DOI] [PubMed] [Google Scholar]
- Kahane N, Cinnamon Y, Kalcheim C. The roles of cell migration and myofiber intercalation in patterning formation of the postmitotic myotome. Development. 2002;129:2675–2687. doi: 10.1242/dev.129.11.2675. [DOI] [PubMed] [Google Scholar]
- Kalderon N, Epstein ML, Gilula NB. Cell-to-cell communication and myogenesis. J Cell Biol. 1977;75:788–806. doi: 10.1083/jcb.75.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeter JS, Pappas GD, Model PG. Inter- and intramyotomal gap junctions in the Axolotl embryo. Dev Biol. 1975;45:21–33. doi: 10.1016/0012-1606(75)90237-7. [DOI] [PubMed] [Google Scholar]
- Krüger O, Plum A, Kim J-S, Winterhager E, Maxeiner S, Hallas G, Kirchoff S, Traub O, Lamers W, Willecke K. Defective vascular development in connexin 45-deficient mice. Development. 2000;127:4179–4193. doi: 10.1242/dev.127.19.4179. [DOI] [PubMed] [Google Scholar]
- Musil LS, Beyer EC, Goodenough DA. Expression of the gap junction protein connexin43 in embryonic chick lens: molecular cloning, ultrastructural localization, and post-translational phosphorylation. J Membr Biol. 1990;116:163–175. doi: 10.1007/BF01868674. [DOI] [PubMed] [Google Scholar]
- O’Brien J, Al-Ubaidi MR, Ripps H. Connexin 35: a gap-junctional protein expressed preferentially in the skate retina. Mol Biol Cell. 1996;7:233–243. doi: 10.1091/mbc.7.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, Bruzzone R, White TW, Al-Ubaidi MR, Ripps H. Cloning and expression of two related connexins from the perch retina define a distinct subgroup of the connexin family. J Neurosci. 1998;18:7625–7637. doi: 10.1523/JNEUROSCI.18-19-07625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordahl CP. Myogenic lineages within the developing somite. In: Bernfield M, editor. Molecular basis of morphogenesis. New York: Wiley-Liss; 1993. pp. 165–176. [Google Scholar]
- Pownall ME, Emerson CP., Jr Sequential activation of three myogenic regulatory genes during somite morphogenesis in quail embryos. Dev Biol. 1992;151:67–79. doi: 10.1016/0012-1606(92)90214-2. [DOI] [PubMed] [Google Scholar]
- Rash JE, Staehelin LA. Freeze-cleave demonstration of gap junctions between skeletal myogenic cells in vivo. Dev Biol. 1974;36:455–461. doi: 10.1016/0012-1606(74)90066-9. [DOI] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangvoravat CP, Lo CW. Connexin 43 expression in the mouse embryo: localization of transcripts within developmentally significant domains. Dev Dyn. 1992;194:261–281. doi: 10.1002/aja.1001940403. [DOI] [PubMed] [Google Scholar]
- Sacks LD, Cann GM, Nikovits W, Jr, Conlon S, Espinoza NR, Stockdale FE. Regulation of myosin expression during myotome formation. Development. 2003;130:3391–3402. doi: 10.1242/dev.00541. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H. Skeletal muscle fibers of newborn rats are coupled by gap junctions. Dev Biol. 1982;91:485–490. doi: 10.1016/0012-1606(82)90056-2. [DOI] [PubMed] [Google Scholar]
- Serre-Beinier V, Le Gurun S, Belluardo N, Trovato-Salinaro A, Charollais A, Haefliger J-A, Condorelli DF, Meda P. Cx36 preferentially connects β-cells within pancreatic islets. Diabetes. 2000;49:727–734. doi: 10.2337/diabetes.49.5.727. [DOI] [PubMed] [Google Scholar]
- Söhl G, Degen J, Teubner B, Willecke K. The murine gap junction gene connexin36 is highly expressed in mouse retina and regulated during brain development. FEBS Lett. 1998;428:27–31. doi: 10.1016/s0014-5793(98)00479-7. [DOI] [PubMed] [Google Scholar]
- Stolte D, Huang R, Christ B. Spatial and temporal pattern of Fgf-8 expression during chick development. Anat Embryol. 2002;205:1–6. doi: 10.1007/s00429-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Williams BA, Ordahl CP. Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development. 1994;120:785–796. doi: 10.1242/dev.120.4.785. [DOI] [PubMed] [Google Scholar]
- Yancey B, Biswal S, Revel J-P. Spatial and temporal patterns of distribution of the gap junction protein connexin43 during mouse gastrulation and organogenesis. Development. 1992;114:203–212. doi: 10.1242/dev.114.1.203. [DOI] [PubMed] [Google Scholar]