Abstract
Ly49 lectin-like receptors and killer cell immunoglobulin-like receptors (KIR) are structurally unrelated cell-surface glycoproteins that evolved independently to function as diverse NK cell receptors for MHC class I molecules. Comparison of primates and various domesticated animals has shown that species have either a diverse Ly49 or KIR gene family, but not both. In four pinniped species of wild marine carnivore, three seals and one sea lion, we find that Ly49 and KIR are each represented by single, orthologous genes that exhibit little polymorphism and are transcribed to express cell-surface protein. Pinnipeds are therefore species in which neither Ly49 nor KIR are polygenic but retain the ancestral single-copy state. Whereas pinniped Ly49 has been subject to purifying selection, we find evidence for positive selection on KIR3DL during pinniped evolution. This selection, which focused on the D0 domain and the stem, points to the functionality of the KIR and likely led to the sea lion’s loss of D0. In contrast to the dynamic and rapid evolution of the KIR and Ly49 genes in other species, the pinniped KIR and Ly49 have been remarkably stable during the > 33 million years since the last common ancestor of seals and sea lions. These results demonstrate that long-term survival of placental mammal species need not require a diverse system of either Ly49 or KIR NK-cell receptors.
Keywords: Comparative Immunology, Comparative Evolution, Natural Killer Cells, Other Animals
Introduction
Natural killer (NK) cells are lymphocytes of the blood that function in the innate immune response to infection, providing a particularly important defense against viruses (1). NK cells also contribute to implantation during human (2) and rodent (3) reproduction and are the most prominent leukocyte in the human uterus (2, 3). Unlike B and T lymphocytes, NK cells do not exploit rearranging genes to create a repertoire of homologous receptors for prospective pathogens, but use a wide variety of different types of receptors, including ones encoded by diverse and rapidly evolving gene families (4).
The killer immunoglobulin-like receptors (KIR) and Ly49 lectin-like receptors are structurally unrelated cell-surface glycoproteins that function as natural killer (NK) cell receptors for major histocompatibility complex (MHC) class I molecules (5). In the mouse a polygenic and polymorphic Ly49 family encodes inhibitory and activating receptors that differ in MHC class I specificity and are present in diverse combinations on NK cell surfaces (6). In humans, where the single Ly49 gene is defective, these functions are borne by an equally diverse family of KIR receptors having specificity for polymorphic determinants of human MHC (HLA) class I (7, 8) The Ly49, KIR and MHC class I genes are each part of a distinct complex of immune system genes that map to different chromosomes and segregate independently (9). In the human population, particular combinations of KIR and HLA class I polymorphisms influence the response of NK cells to viral infection (10), cancer (11), allogeneic hematopoietic cell transplants (12), and trophoblasts during pregnancy (2).
Following the discovery of Ly49 in mice and KIR in humans, the study of other mammals showed rat and horse were like mice, having diverse Ly49 but not KIR, whereas several primates and cattle have diverse KIR genes but not Ly49 (13–19). KIR were also found to comprise two ancient lineages, 3DX and 3DL, that arose by duplication from a common ancestor prior to the radiation of placental mammals 95–113 million years ago (mya) (20). In primates the 3DX lineage remained a single copy gene, while the 3DL lineage expanded and diversified. In cattle the opposite occurred: 3DX expanded and diversified, while 3DL did not. Species comparisons further showed that a majority of components in the diversified Ly49 and KIR families are specific to species, reflecting the rapid evolution of these gene families and the frequent, but transient benefit of novel forms (21). The fundamental differences in the structures of Ly49 and KIR and in the ways they bind to MHC class I argue for their convergent evolution to be NK cell receptors for MHC class I (22–25).
The genetic diversity of these receptors within species and their evolutionary instability can be attributed to constantly changing pressures on the immune and reproductive systems exerted by diverse, rapidly evolving pathogens, such as cytomegalovirus, influenza, hepatitis C and human immunodeficiency virus (26–29). The most well studied KIR/MHC class I system is in humans, where KIR haplotypes can contain between seven and 14 functional genes for which over 200 variants have been characterized (30). There are well over 2000 variants of the highly polymorphic MHC class I loci HLA-A, -B and -C that are main ligands for these KIR receptors (30). These unlinked gene clusters on different chromosomes produce individuality in the KIR/MHC class I mediated NK cell response within populations.
Until now the study of NK cell receptor variability has been confined to terrestrial mammals and, with the exception of the primates, to animals bred and domesticated by humans. To give a new perspective we examined the NK cell receptors of pinnipeds: marine carnivores that diverged from a terrestrial carnivore ancestor 45.4 ± 3.9 million years ago (mya) in North America and adapted to life in the aquatic environment (31). Recent molecular evidence confirms a monophyletic origin of the three pinniped groups, walruses (Odobenidae), sea lions (Otariidae) and true seals (Phocidae). The phocids further divide into the Monachinae (southern true seals and monk seals) and the Phocinae (northern true seals). Although all pinnipeds arose from a common North American ancestor, it is likely that sea lion ancestors subsequently moved into the Pacific, whereas seal ancestors moved to the Atlantic (32).
In our study the otariids are represented by the California sea lion (Zalophus californianus), which shared a common ancestor with the phocids approximately 33 mya (31). The Monachinae are represented by the Weddell seal (Leptonychotes weddellii) and the Phocinae by the grey and harbor seals (Halichoerus grypus and Phoca vitulina respectively). The estimated divergence time between the Monachinae and the Phocinae is between 15 and 22 mya (31, 33). The grey and harbor seals are closely related sympatric species that diverged 5–7 mya (31, 33).
Materials and Methods
Seal sample collection
Seven grey seal blood samples were obtained from captive animals housed temporarily at the Sea Mammal Research Unit (SMRU), University of St Andrews, Scotland. Harbor seal blood was taken from five free-living animals. Both species were captured on the east coast of Scotland in St Andrews Bay (Lat 56° 23 00’:Lon –2° 45 00’) in 2003 and 2004. Weddell seal blood was obtained from three free-living females during the 2003 breeding season at Erebus Bay, Antarctica (77 deg 51 min S, 166 deg 45 min E). California sea lion blood from three rehabilitating animals and archived tissue samples were obtained from the Marine Mammal Center Sausalito, California. All UK Home Office Procedures were carried out under Licence No. 60/3303 following the ethical review procedure. Blood samples and tissue from California sea lions were obtained with permission from the United States National Marine Fisheries Service and collected under the authority of Marine Mammal Protection Act permit number 932–1489–00 by The Marine Mammal Center.
RNA and DNA Extraction
Grey seal, harbour seal and California sea lion peripheral blood was decanted from EDTA Vacutainers (BD Biosciences , Oxford, UK) mixed 1:1 with RPMI 1640, and the mononuclear cells (PBMCs) were separated using histopaque –1077 (Sigma-Aldrich, Dorset, UK). The PBMC cell layer was removed, washed twice in RPMI 1640 and homogenized in TriReagent™ for RNA extraction (Sigma-Aldrich). Weddell seal blood was layered onto 3 % Dextran (Sigma-Aldrich) dissolved in 0.9 % NaCl. The resulting supernatant was centrifuged at 290 × g for 10 min to pellet the white blood cells. Remaining red blood cells were lysed with two subsequent re-suspensions in 0.1 % and 0.2 % NaCl for 30 sec, before restoring the osmolality with an equal volume of 2 % NaCl and centrifugation for 5 min at 290 × g. The resulting pellet was homogenized in TriReagent™. After confirming RNA quality by agarose gel electrophoresis, cDNA was synthesized using Superscript III (Invitrogen, Carlsbad, CA) for use in PCR.
Genomic DNA was isolated from the remaining pellet containing the red blood cells (RBC) and the polymorphonuclear (PMNC) cells. RBC’s were first removed by lysis with two 10 min incubations in a 1 % saponin solution before the remaining PMNCs were digested in 10 ml lysis buffer (0.5 mg/ml proteinase K in 100 mM NaCl, 10 mM Tris, 25mM EDTA, 0.5 % SDS) at 50 °C for at least 6 hours. This lysis solution was also used to digest 5mm2 liver samples from the California sea lion and cattle muscle at 50° C overnight. DNA was extracted from the resulting solutions with two standard phenol/chloroform/isoamyl alcohol steps before precipitation by ethanol. DNA was suspended in TE buffer for use in PCR and Southern blotting.
KIR, Ly49, LILR and FCAR PCR amplification
Degenerate PCR was performed on grey seal cDNA using Advantage™ 2 polymerase mix (BD Biosciences) for 32 cycles. Alignments of all known KIR, Ly49, LILR and FCAR genes were used to design several degenerate primers that corresponded to conserved regions within the extracellular domains of each gene family. In addition, for the related KIR and LILR, primers that were equally likely to amplify both genes were used to increase the chances of detecting divergent receptors. Primers specific for the 3DX lineage KIR were also included. For each gene, every possible primer combination was used and all amplicons of approximately the correct size were cloned using TOPO® TA (Invitrogen). At least eight clones of each were sequenced using a Beckman Coulter (Fullerton, CA) CEQ 2000 instrument and assembled using the STADEN PACKAGE (34). This sequence information enabled gene specific RACE using the SMART RACE system (BD Biosciences) to amplify the full 5’ and 3’ ends according to the manufacturers’ guidelines. These grey seal KIR and Ly49 sequences were then included in the multi-species alignments to facilitate the design of additional degenerate primer sets that were used to amplify these genes in other pinniped species. Full-length gene-specific PCR was ultimately performed using cDNA from all the pinniped species with the Advantage™ 2 polymerase mix for 32 cycles. Genomic DNA PCR was performed with BD Advantage Genomic Polymerase (BD Biosciences) for 28 cycles using 150 ng DNA. Full details of the primers and PCR conditions used are available upon request. All amplicons were cloned, sequenced and assembled as above. Sequences are deposited in GenBank with the accession numbers FJ190084-FJ190095 (KIR), FJ190096-FJ190101 (Ly49), FJ190102 (FCAR) and FJ190103 & FJ190104 (LILR).
Generation of pinniped KIR and Ly49 NKL transfectants
NKL, a human leukemia-derived cell line with NK cell-like properties, was maintained as previously described (35). Full-length coding regions of grey seal HgKIR3DL*002 and HgLy49*001 and California sea lion ZcKIR2DL and ZcLy49*001 were amplified by PCR and used to generate FLAG epitope-tagged constructs by recombinant PCR. For KIR, constructs containing an N-terminal CD8 leader peptide and FLAG epitope before the first Ig domain were amplified. Substituting the KIR leader peptide with that of CD8 increases the stability of FLAG tag expression (personal communication, Lewis Lanier, University of California San Francisco, CA). A human KIR2DL1 construct was also manufactured in the same way to confirm that the addition of the CD8 leader and FLAG did not effect cell surface expression. For Ly49, a C-terminal FLAG motif was added after the extracellular lectin domain. FLAG constructs were cloned into the PBMN-IRES-eGFP retroviral vector (a gift from Gary Nolan, Stanford University, Stanford, CA) and transfected into Phi-NX cells using standard protocols. The supernatant containing recombinant amphotrophic retrovirus was used to transfect growing NKL cells which were sorted for GFP expression after 48 hours using a FACSVantage cell sorter (BD Biosciences). To determine if KIR and Ly49 were expressed on the cell surface, GFP positive NKL were incubated with the mouse anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) followed by staining with a PE labeled anti-mouse IgG1 antibody (BD Biosciences) and analysed for FLAG expression by flow cytometry using a FACScan (BD Biosciences). A mouse IgG1 isotype control (BD Biosciences) was used to assess the specificity of the reactions with the M2 antibody.
Southern blot analysis
Four µg of genomic DNA were digested with EcoRV and SphI restriction enzymes (New England Biolabs, Beverley, MA), separated by 0.8 % agarose gel electrophoresis at 40 V overnight and blotted onto Hybond-N+ nylon membrane (Amersham, Piscataway, NJ) using an alkaline transfer buffer (1.5M NaCl, 2 % NaOH). The membrane was then prehybridized with hybridization solution (5 × SSC, 5 × Denhardt’s solution, 0.5 % SDS) for at least one hour at 68 °C before a single exon probe radiolabeled with P32 using the Prime-It® II Random Primer Labeling Kit (Stratagene, La Jolla, CA) and sheared salmon sperm DNA were added and incubated overnight. To probe for Ly49 a 143 bp exon 4 probe was amplified from grey seal cDNA and corresponded to the N-terminal end of the lectin domain. For the 3DL lineage KIR a 288 bp D0 or 300 bp D2 domain probe amplified from grey seal cDNA was used. To probe for the 3DX lineage KIR in pinnipeds, a mixture of 3DX lineage D0 domain probes amplified from cattle gDNA were used. After hybridization, four low stringency washes were performed (2 × SSC, 0.1 % SDS) before exposure to Biomax MR film (Kodak, Rochester, NY) for 24 hours and 120 hours.
Phylogenetic analysis
Sequences were aligned using CLUSTAL X (36) and manually edited if necessary using Bioedit version 7.0.1 (37). Neighbor-joining phylogenetic analysis was performed with MEGA version 3.1 (38) using the Tamura-Nei method with 1000 replicates. Tree topologies were compared using the Templeton test as implemented in PAUP (39).
Selection analysis
Estimation of dN/dS (ω)ratios was performed by maximum likelihood using PAML v3.14 (40). Both site and branch analyses were carried out using the F3×4 model of codon frequencies. In the site analysis, the likelihood of tree topology was estimated for four site-specific models in which the selective pressure varied among the different sites but the site-specific patterns were identical across all lineages. A likelihood ratio test (LRT) was then conducted to compare a null model that does not allow ω>1 with a model that does. LRTs were performed for M1a (nearly neutral) vs. M2a (selection) and M7 (beta) vs. M8 (beta and ω). The Bayes Empirical Bayes (BEB) approach was then used to identify codons with ω>1 (41). For the branch analysis a LRT was performed to compare the likelihood of a tree topology given a null model that does not allow ω>1 for the branch of interest to the likelihood of the same tree topology given an alternative model that does. A BEB was also used to identify codons with ω>1. The CODEML program was modified for this branch analysis to provide a better approximation of the distribution when the ω2 for the branch of interest was estimated to be >10.5.
Results
Single expressed KIR and Ly49 genes in four pinniped species
KIR and Ly49 cDNA were characterized from one species of otariid pinniped, the California sea lion, and three species of phocid pinniped: the grey, harbor and Weddell seals. Using PCR amplification primers based upon conserved, KIR- or Ly49-specific sequences of other mammalian species, we isolated partial cDNA clones from the grey seal, which were then extended by 3’ and 5’ RACE to give full-length clones. With knowledge of the grey seal sequences, additional PCR were designed that permitted isolation of KIR and Ly49 cDNA from the other species.
Pinniped KIR
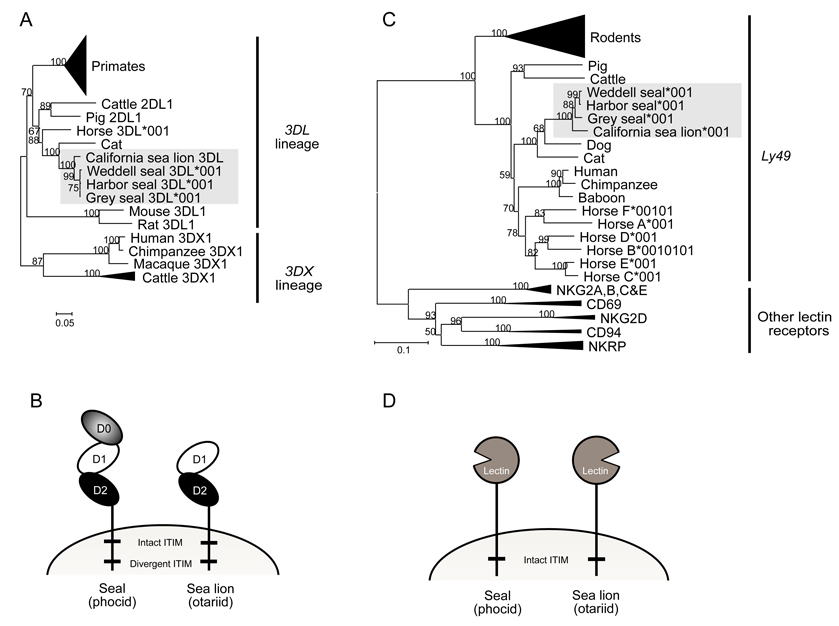
For each pinniped species cDNA clones corresponding to a single KIR gene were obtained. Phylogenetic analysis and sequence similarity (95.8–99.8%) showed that these pinniped KIR were orthologous (Fig.1A). For the three seal species the cDNA encodes a KIR3DL having three extracellular Ig-like domains (D0, D1 and D2), whereas the sea lion cDNA encodes a KIR2DL having only D1 and D2 (Fig. 1B). The lack of a D0 domain in sea lions was further investigated by sequencing the genomic region from the leader peptide exon 2 to the D1 domain. This revealed a ~1.6kb deletion in the sea lion gene, which included exon 3 encoding the D0 domain. Since divergence of the phocid and otariid pinnipeds, ~ 33 MYA (32), exon 3 encoding the D0 domain has been deleted from the KIR3DL gene in the otariid lineage leading to the California sea lion. The transmembrane domains of the pinniped KIR2DL and KIR3DL contain one intact and one non-canonical ITIM motif and are likely inhibitory receptors. A single membrane proximal ITIM can generate inhibitory signals (42) and the non-canonical membranedistal ITIM might also be functional, because its substitutions from the norm are conservative (42).
Figure 1. Pinniped KIR and Ly49 group with the carnivore KIR and Ly49.
(A): Pinniped KIR group with other non-primate KIR of the 3DL lineage. Neighbor-joining bootstrap analysis comparing the full length KIR cDNA sequences isolated from the pinnipeds with primate and non-primate KIR. The tree was rooted at the midpoint and the support for each node (expressed as a percentage) is shown when >50%. The dataset used to generate this phylogenetic tree is available upon request. The primate KIR branch has been collapsed and includes all the primate species KIR apart from the 3DX1 lineage. The cattle KIR3DX lineage branch has been collapsed and includes all the cDNA sequences publicly available with the exception of cattle KIR2DL1 which is of the KIR3DL lineage (15, 18–20). The shaded box contains all the pinniped KIR.
(B): Comparison of seal and sea lion KIR domain structure.
(C): Neighbor-joining bootstrap analysis comparing the full length Ly49 cDNA sequences isolated from the pinnipeds with related families of lectin-like receptors. The tree was rooted at the midpoint and the support for each node (expressed as a percentage) is shown when >50%. The dataset used to generate this phylogenetic tree is available upon request. The rodent Ly49 branch has been collapsed and includes all the rat and mouse Ly49 genes. The other collapsed branches contain representative genes from several primate and non-primate species. The pinniped Ly49 sequences are contained in the shaded box.
(D): Comparison of seal and sea lion Ly49 domain structure.
Mammalian KIR genes form two ancient lineages, 3DL and 3DX, that are situated at different locations in the LRC (43, 44) and were differentially expanded in primates and cattle (20). Phylogenetic comparison shows pinniped KIRs are part of the 3DL lineage (Fig. 1A), which includes the highly variable primate KIR gene family and are situated between the LILR and FCAR genes. Within this lineage, the pinniped KIR cluster with the cattle, pig, horse and cat KIR, being closest to the cat KIR, consistent with pinnipeds and felids both being carnivores. Our analysis reveals that the genome of the dog, another carnivore, does not contain a KIR gene of either lineage. Further, the region between the LILR and FCAR where a 3DL lineage KIR is likely to be located shows evidence of genomic deletion as the 5’ end of the FCAR gene is truncated.
Pinniped Ly49
Complementary DNAs corresponding to a single Ly49 gene were obtained from all four pinniped species. Phylogenetic analysis shows that pinniped Ly49 cluster with other species Ly49 and form a pinniped specific-group (Fig. 1C). Among the pinniped Ly49 the sequence similarities are 94.2–99.9%, and in comparison to other carnivores, dog and cat, the sequence similarities are 76.4–78.4 % and 73.2–74.6 %, respectively. The few differences between the Ly49 genes in seals causes the tree topology to incorrectly represent these species’ relationships. It is unlikely that this is caused by selection as manual modification to correct for this did not create a tree that was significantly better. The pinniped Ly49 all appear to be functional inhibitory receptors, having the cysteines needed for disulphide bond formation and proper protein folding (24) and an intact, canonical ITIM in the cytoplasmic tail (Fig. 1D).
Pinniped KIR and Ly49 are cell surface receptors
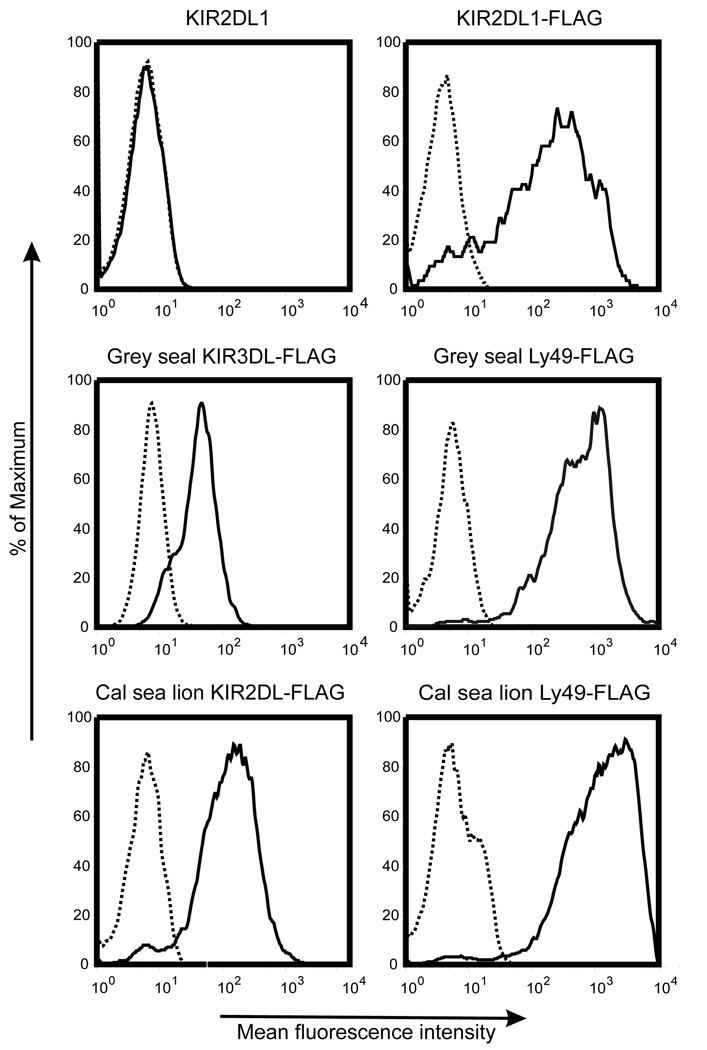
The predicted protein products of pinniped KIR and Ly49 cDNAs appear to be functional cell surface receptors. We tested cell surface expression by transfecting the NKL cell line with constructs of grey seal and California sea lion KIR and Ly49 coupled to the FLAG-epitope. Antibodies detected FLAG on the surface of transfected NKL cells containing the grey seal 3 Ig domain KIR, the California sea lion 2 Ig KIR and Ly49 from both species (Fig. 2). This confirmed that the intracellular machinery traffics these proteins to the cell membrane as predicted for functional NK cell receptors. This cell surface expression as well as the high sequence identity between both pinniped KIR (95.8–99.8%) and Ly49 (94.2–99.9%) supports their functionality. Greater sequence divergence has likely been prevented by functional constraints for the 33 million years since the otariid and phocid species diverged.
Figure 2. Pinniped KIR and Ly49 can be expressed on the cell surface.
NKL cells transfected with grey seal and California sea lion KIR and Ly49 constructs containing the FLAG-epitope were incubated with a mouse IgG1 isotype control (broken line) or M2 anti-FLAG antibody (unbroken line) before washing and incubation with a PE-conjugated anti-mouse IgG1 antibody. NKL cells transfected with human KIR2DL1 constructs with and without the FLAG-epitope were used as positive and negative controls respectively. The expression of the FLAG-epitope was determined by gating on the GFP-positive NKL cell population.
Pinniped genomes contain single KIR and Ly49 genes
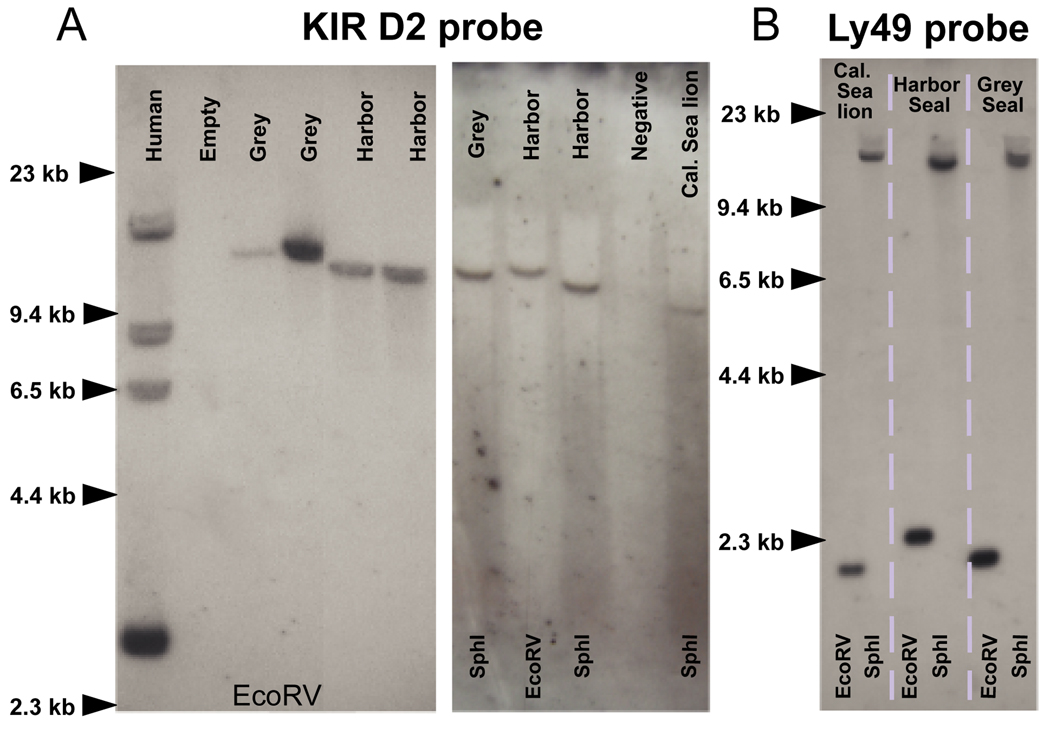
As a further assessment of the number of KIR and Ly49 genes in pinniped genomes, low-stringency Southern blots were performed on genomic DNA cut either with the EcoRV, SpeI or SphI restriction endonucleases (Fig. 3). With a probe corresponding to the sequence encoding D2 of grey seal KIR3D (HgKIR3DL), one hybridizing band was observed for each phocid species (Fig. 3A). Similar data were obtained with a probe encoding the D0 domain (data not shown). These results are consistent with the cDNA analysis and show that the seal genomes contain a single 3DL-lineage KIR gene.
Figure 3. Southern blotting confirms a single KIR and Ly49 locus.
(A). Membranes were probed with a single exon KIR D2 probe based on the grey seal KIR sequence. One hybridizing band in each species was observed. The same membranes were stripped and probed with a single exon grey seal KIR D0 probe which produced an identical pattern of hybridization (data not shown). The only exception was the California sea lion which showed no hybridization with the D0 probe, consistent with the absence of a D0-encoding exon in the single KIR3DL gene in this species.
(B). One hybridization band was also detected using a single exon Ly49 probe based on the grey seal sequence.
To search for genes of the 3DX lineage in grey and harbor seals, membranes previously hybridized with KIR3DL probes were hybridized with a mixed DNA probe corresponding to the D0 domains of the cattle KIR3DX genes (3DS1, 3DL1, 3DL1-like and 2DS1). A mixed cattle D0 probe presented the best chance of hybridizing to a KIR3DX in seals as D0 domain is the most divergent domain between the KIR lineages. Single faint bands of hybridization were observed, but they corresponded precisely to the band containing the 3DL-lineage gene detected with the seal KIR probes (data not shown). This cross-reactivity shows that the cattle 3DX-lineage D0 probe can hybridize to 3DL-lineage genes with 63–67% sequence similarity and should therefore have hybridized to any pinniped 3DX-lineage KIR. In addition, degenerate KIR3DL, KIR3DX and KIR/LILR primers amplified only a single KIR gene of the 3DL-lineage in each species. In conclusion, the results indicate that pinniped genomes lack 3DX-lineage KIR and that the single expressed 3DL-lineage KIR gene is their only KIR gene.
When EcoRV or Sph1 digested pinniped DNA was probed with the sequence encoding the lectin-like domain of the expressed grey seal Ly49 gene, single hybridizing bands were observed (Fig. 3B). As in the case of KIR, pinniped genomes have a single Ly49 gene that corresponds to the cDNA characterized.
The grey seal has a minimal KIR locus flanked by FCAR and LILR
In all mammalian species except mouse, the 3DL - lineage KIR genes are flanked on one side by FCAR, the gene encoding the IgA receptor of myeloid cells (FcαRI), and on the other by the leukocyte immunoglobulin-like receptor (LILR) gene family. To determine if this organization is conserved in the grey seal, we first characterized FCAR and LILR cDNA in this species, using the strategy that worked for KIR and Ly49.
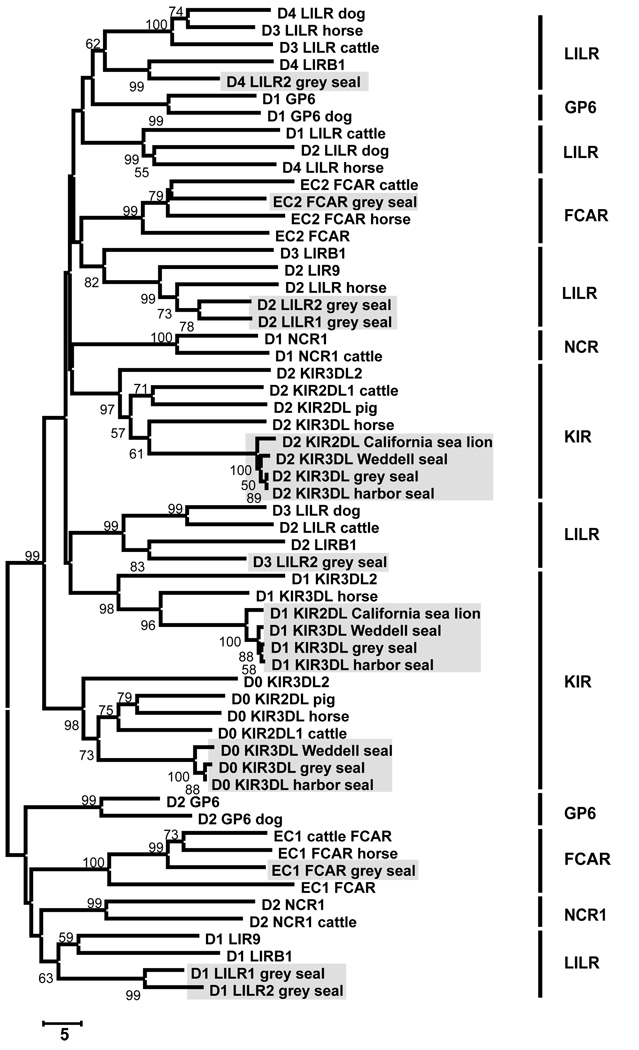
The 1181bp cDNA sequence encoding grey seal FcαRI (HgFCAR) is orthologous to FCAR from other species (Fig 4), the highest sequence similarities being 77.7% with cattle and 74.9% with horse. The domain organization is the same as in other species, as is the arginine residue of the transmembrane region that recruits the signal transducing FcRγ subunit. Two full-length LILR cDNA sequences were characterized using PCR primers corresponding to conserved regions in the LILR Ig domains of other species. HgLILR1 encodes a receptor with two Ig domains and HgLILR2 encodes a receptor with four Ig domains. The cytoplasmic tails of HgLILR1 and HgLILR2 both contain two ITIM motifs. The Ig domains of these receptors align with the LILR Ig domains of other species (Fig 4). Several incomplete Hg-LILR cDNA were also identified.
Figure 4. Pinniped NK cell receptor Ig domains group with the equivalent domains from other species.
Neighbor-joining analysis comparing the amino acid sequence of individual Ig domains from a range of LRC receptors from different species. The tree was rooted at the midpoint and the support for each node (expressed as a percentage) is shown when >50%.The shaded boxes contain all the pinniped Ig domains. Where no species is given this is a human gene.
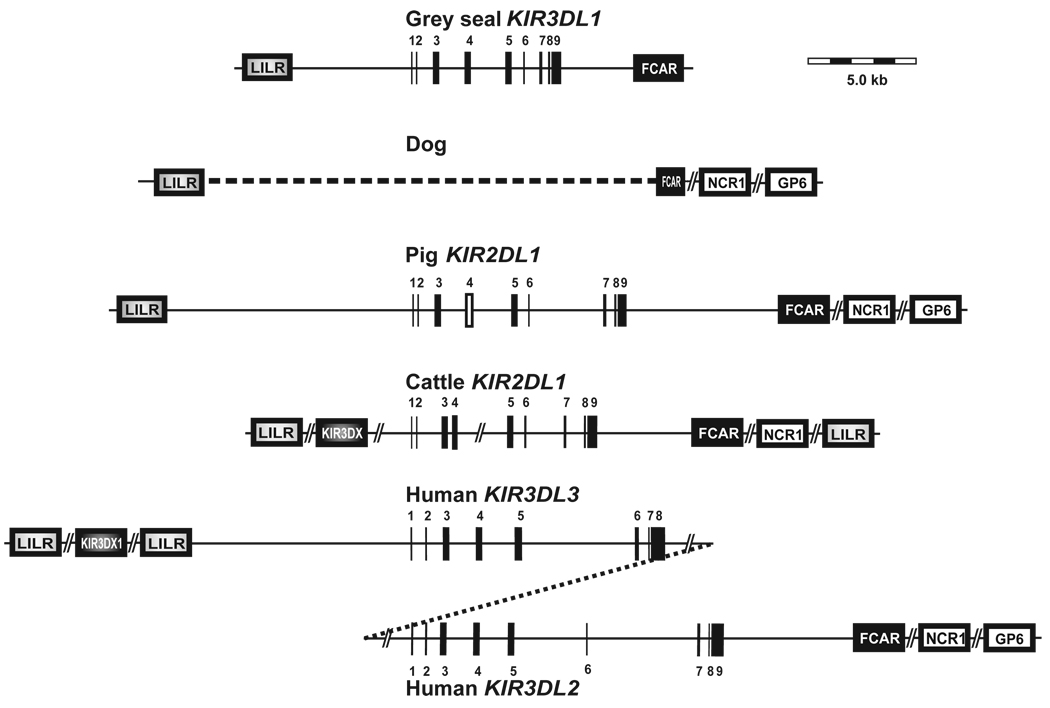
Knowledge of the HgFCAR and HgLILR sequences facilitated the design of specific primers that, in combination with HgKIR3DL-specific primers, allowed the entire ~16.2kb region between HgFCAR and Hg-LILR to be isolated in two overlapping PCR amplifications of ~10.5kb each. The only gene in this region is HgKIR3DL, which is separated from the nearest HgLILR gene by 5.8kb and from HgFCAR by 3.4kb. This analysis demonstrates that the single grey seal KIR gene maps to the LRC like other mammalian 3DL lineage KIRs and as such constitutes a minimal KIR locus. It is >12.kb shorter than the pig KIR locus, which also contains a single gene, and is about the same size as a single human KIR gene (Fig. 5). The size of the bands observed on Southern blots and their similarity in the different pinnipeds indicates that the size and organization of their KIR locus is similar to that of the grey seal.
Figure 5. The grey seal has the smallest KIR locus so far defined.
The content, organization and size of the KIR locus in the grey seal is compared to the KIR locus in the pig, cattle and human. The KIR gene is drawn to scale and exons are indicated by vertical bars. Gaps in the sequence or between genes are represented by line breaks. The pseudo-exons 4 (D1) for the cattle KIR2DL1 and pig are represented by white bars. The dog and cattle KIR gene size and LRC organization were determined from the latest version of the genome assemblies (dog build 2.1 and cattle build 3.1) as well as the BAC sequences and information from (20). Our provisional cattle LRC organization is very similar to that reported by Dobromylskyj and Ellis in 2007 (19). However, other KIR2DL fragments have also been identified in the genome and many LRC genes are not yet able to be mapped.
Consequently these maps may be subject to changes in the future as The FCAR locus in the dog in truncated and does not include the sequence encoding the upstream Ig domain (EC1 domain). The gene organization in the pig is taken from (50). For the human KIR locus, only the framework genes at each end of the haplotype are shown.
Polymorphism and diversity in pinniped Ly49 and KIR
Whereas the hominoid KIR and rodent Ly49 families have evolved a striking diversity in haplotype gene content (6, 8), the pinniped KIR and Ly49 loci have preserved a minimal gene content over at least 33 million years of evolution. As allelic polymorphism, species specificity and splice variants are other features of primate KIR and rodent Ly49 diversity (4, 6), we studied allelic polymorphism in the KIR and Ly49 genes of seven grey, five harbor, three Weddell seals and three California sea lions.
Pinniped KIR
Five grey seal HgKIR3DL alleles were defined by six nucleotide substitutions, and four Weddell seal LwKIR3DL alleles, also by six nucleotide substitutions. Least polymorphic were the harbor seal, for which a single non-synonymous substitution defines two PvKIR3DL alleles, and the California sea lion with a single nucleotide polymorphism in intron four. Harbor seal PvKIR3DL*001 and grey seal HgKIR3DL*002 encode identical proteins, but differ by a synonymous difference.
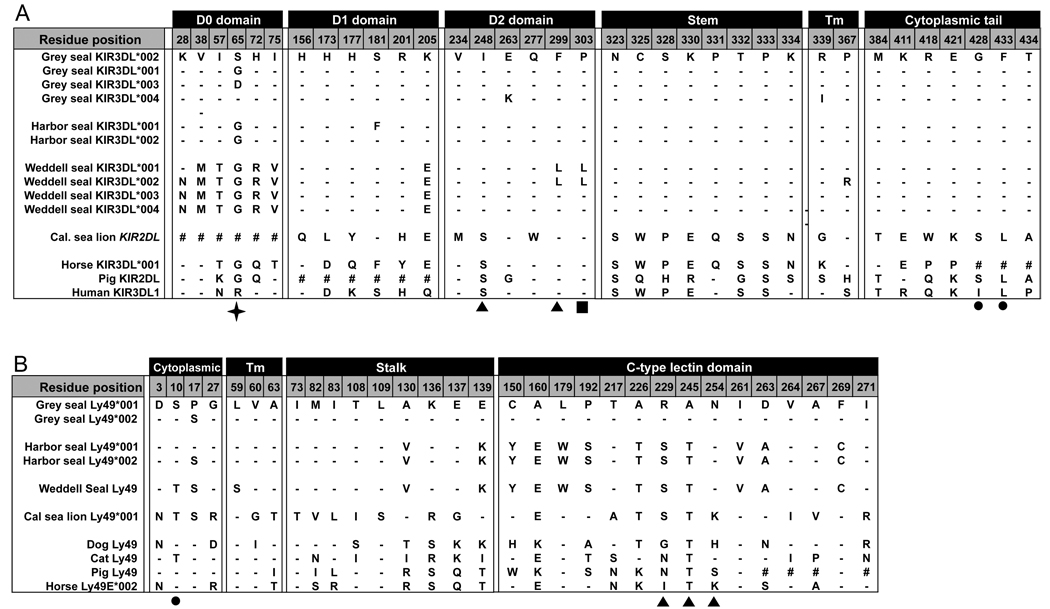
A total of 34 amino acid substitutions diversify the pinniped KIR, which does not include the absence of D0 from sea lion KIR2DL (Fig. 6A). Of these only positions 248, 299 and 303 in D2 are predicted to contact MHC class I, based upon the three dimensional structures of human KIR2D bound to HLA-C (22, 45). Position 248 is under positive selection in hominoid KIR3DL (46), and positions 299 and 303 are both in a loop predicted to contact MHC, which has alternative motifs in the Weddell seal: L299 + L303 or F299 + P303 (Fig. 6A).
Figure 6. Variable residues between pinniped KIR and Ly49.
(A) KIR protein residues that are variable between all the pinniped species alleles are shown with the horse KIR3DL*001, pig KIR2DL and human KIR3DL1*001 for comparison. The symbol ‘#’ indicates there is no equivalent residue in that species and ‘-’ indicates the same residue as the reference sequence HgKIR3DL*002. Amino acids are numbered according to the full length predicted protein from the cDNA with position 1 being the start codon. Filled arrows indicate predicted MHC contacting residues and the filled square indicates residues within the MHC contacting loop according to the known KIR2D structures in complex with their ligand (22, 45). The filled circle indicates a residue in the ITIM motif. The star indicates the residue found to be under positive selection in the grey seal.
(B) Ly49 protein residues that are variable between all the pinniped species alleles are shown with the sequences from the dog, cat, pig and horse for comparison. The symbol ‘#’ indicates there is no equivalent residue in that species and ‘-’ indicates the same residue as the reference sequence HgLy49*001. Amino acids are numbered according to the full length predicted protein with position 1 being the start codon. Filled arrows underneath indicate predicted MHC contacting residues according to the known mouse structures with their ligands (23, 25) and the filled circle indicates a residue within the ITIM motif but one of the conserved residues of the consensus motif.
Likelihood ratio tests that consider codon variation in dN/dS (ω) found strong evidence of positive selection for pinniped KIR cDNA sequences (α=0.001). Branch analysis also detected evidence for positive selection on the branch leading to the pinnipeds (α=0.01) (Fig. 1A). One focus of selection has been the D0 domain. Site analysis detected evidence for positive selection on this domain (α=0.05) and analysis of either the complete sequence or just the D0 domain identified position 65, for which the grey seal has three alternative residues, as a target for selection. Deletion of the D0 domain in the sea lion further points to the D0 domain having been subject to selective pressure in pinniped evolution. The D0 domain functions as a binding enhancer of the KIR3D to MHC class I (47).
The second focus for diversity is the stem region. Within its 17 residues there are eight substitutions between seals and the sea lion (Fig. 6A). The seals have the more divergent stem, whereas sea lions have a stem more like other mammals, it being identical to the horse and differing by only six non-synonymous substitutions from human KIR3DL1 (Supplementary fig. 1). The short length precludes using dN/dS methods to assess selection on the stem. Although these selective pressures have not produced a multigene family they show the pinniped KIR have served a valuable function.
Complementary DNA corresponding to alternatively spliced forms of seal KIR mRNA were identified with some frequency. Most common was a cDNA lacking exon 6 that encodes the stem, which accounted for >50% of the grey seal cDNA clones, ~30% of harbor seal clones and was also obtained from the Weddell seal. Another variant observed in the grey and Weddell seals lacks 66bp from the 5’ end of exon 9. This deletion eliminates the conserved ITIM motif from the cytoplasmic domain therefore signaling function. Such alternative splicing is a mechanism of functional regulation induced by other genes, as is seen with FCAR splice variant expression during the inflammatory response (48).
Pinniped Ly49
Analysis of pinniped Ly49 revealed less variation than in KIR. (Fig. 6B). No Ly49 polymorphism was detected in the Weddell seal and the two Ly49 alleles in the California sea lion differ by a synonymous substitution in the ITIM motif. Two Ly49 alleles that differ by one non-synonymous change (S17P) in the cytoplasmic tail, but not in the ITIM motif, were detected in both grey and harbor seals. The only common Ly49 splice variant was obtained from the California sea lion: it lacks exon 2 encoding the transmembrane domain and accounts for ~50% of the Ly49 cDNA clones analyzed.
Between the four pinniped species there are 31 variable positions in the Ly49 amino-acid sequence (Fig. 6B). Extrapolating from the three-dimensional structures of mouse Ly49 bound to MHC class I (23, 25), only three of these positions. 229, 245 and 254 are predicted to contact MHC class I (Fig. 6B). Analysis of dN/dS by maximum likelihood methods gave, however, no evidence for positive selection in the evolution of differences in Ly49 between the pinniped species.
Discussion
This study examined genetic diversity in the KIR and Ly49 genes of four pinniped species: grey, harbor and Weddell seals, and the California sea lion. All four species have single, orthologous KIR and Ly49 genes, which are transcribed and likely encode functional receptors. The major difference between the species is deletion of exon 3 encoding the D0 domain from the KIR gene of the California sea lion, resulting in the expression of a KIR2DL in that species compared to a KIR3DL in the three seal species. The slow morphological evolution of these species in the marine environment make it likely that the otariids and phocids differentiated before they fully entered the oceans (32). Consequently the sea lion’s loss of the D0 domain may have preceded their almost complete marine reliance. Overall, the constancy in the number and nature of pinniped KIR and Ly49 during the ~33 million years since seals and sea lions shared a common ancestor, contrasts dramatically with the extensive changes wrought in the KIR gene families of the higher primates over much shorter time periods, and also with the diversity of the rodent Ly49 gene families (6, 13). This study clearly demonstrates that pinnipeds are species in which neither KIR nor Ly49 form a polygenic system of receptors, and that the long-term survival of mammalian species need not be dependent upon having either highly variable KIR or Ly49 NK cell receptors.
In the pinnipeds studied, we found no evidence for positive selection on Ly49, either for the differences between species or the few polymorphisms within species. Although KIR polymorphism within species is low, there is statistically significant evidence for positive selection along the pinniped branch of evolution, since they separated from the terrestrial carnivores. This selection has not focused on the KIR3DL’s predicted binding site for MHC class I, but on the D0 domain - the binding enhancer (47) - and to lesser extent on the stem region between the extracellular domains and the transmembrane region. The complete loss of D0 from sea lion KIR is a likely consequence of a past selection upon this domain. Discarding an Ig domain from KIR3D is a common event in KIR evolution, for example in humans all eight genes of lineage III KIR suppress the expression of exon 3 encoding the D0 domain (49).
Among the carnivore genome projects, assembly of the domestic dog genome is complete and for the domestic cat it is in progress. Both genomes contain a single Ly49 gene; cat Ly49 appears functional, whereas the dog Ly49 lacks a conserved cysteine and may be defective. The dog LRC appears to be on chromosome 1 where several LILR genes juxtapose a truncated FCAR gene. Consistent with the absence of KIR genes between LILR and FCAR, no KIR-like genes were detected in the genome build 2.1 assembly (Fig. 5). In conclusion the dog genome seems to lack KIR genes. In contrast the cat genome has an arrangement like the grey seal, in which a single KIR3DL gene is flanked by LILR and FCAR. The KIR3DL gene is non-functional, due to a two nucleotide deletion in exon 1 that causes frameshift and premature termination. The absence of any evidence for 3DX-lineage KIR in pinnipeds, dog and cat points to the Carnivora having deleted these genes entirely. Of note the cats (Felidae) and dogs (Canidae) are both groups that do not delay implantation and appear to lack any functional KIR.
Apart from the pinnipeds, the domestic pig is the only other species to have single Ly49 and KIR genes, but here too they do not appear functional (16, 50). Although the porcine KIR was predicted to have only D0 and D2, the gene contains a D1-like exon with intact splice sites but a divergent, truncated 3’ end that is predicted cause a premature termination in translation. No cDNA evidence exists to reveal if this is an expressed 2D KIR or a 3D KIR pseudogene. Pig Ly49 lacks a conserved cysteine in the CTLD so is considered likely non functional (16). An emerging question is whether the frequency of defective KIR and Ly49 genes in domesticated species is a result of natural processes or of their inbreeding and selection by man. Although cattle have an expanded KIR3DX receptor family, domestication may have also increased the frequency of non-functional receptors in these animals as gene expansion likely preceded selective breeding. To answer this question will require additional studies to examine these genes in wild populations of terrestrial carnivores and other species.
In conclusion the pinnipeds are the first species shown to have single, functional Ly49 and KIR3DL genes, and as such likely to represent the ancestral state of these gene families in placental mammals. The pinnipeds lack of Ly49 and KIR diversity does not represent an adaptation to the marine environment as the evidence available suggests that diverse KIR or Ly49 receptors are not a characteristic of terrestrial carnivores. MHC class I molecules are the ligands for a majority of primate KIR and rodent Ly49 but the content and nature of these genes in pinnipeds has yet to be determined. The Hawaiian monk seal (Monachus schauinslandi) MHC class I is the best studied and contains at least two loci with limited polymorphism, however this is an endangered and likely inbred population which may not be reflective of pinniped species with larger populations and ranges (51). It has yet to be determined if the state of Ly49 and KIR in pinnipeds reflects the number and nature of their MHC class I gene content. Given the selective expansion of KIR and Ly49 in mammalian species, it is possible that one of the many other types NK cell receptors has evolved to be diverse and polygenic in the pinnipeds.
Supplementary Material
Acknowledgements
We thank The Marine Mammal Centre and the United States National Marine Fisheries Service for their generous help.
Abbreviations
- CTLD
C-type lectin domain
- FCAR
Fc alpha receptor
- Ig-SF
immunoglobulin superfamily
- ITIM
inhibitory tyrosine-containing immunomotif
- KIR
killer immunoglobulin-like receptor
- LILR
leukocyte immunoglobulin-like receptor
- LRC
leukocyte receptor complex
- MHC
major histocompatibility complex
- MYA
million years ago
- NK
natural killer
- NKC
natural killer cell receptor gene complex.
Footnotes
This work was supported by NIH grant AI024258 to P.P. Weddell seal samples were obtained as part of a United Kingdom NERC funded collaborative grant between the Sea Mammal Research Unit, Antarctica New Zealand and Macquarie University, Sydney, Australia; NER/B/S/2002/00271
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Andoniou CE, Andrews DM, Degli-Esposti MA. Natural killer cells in viral infection: more than just killers. Immunol. Rev. 2006;214:239–250. doi: 10.1111/j.1600-065X.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 2.Sargent IL, Borzychowski AM, Redman CWG. NK cells and human pregnancy - an inflammatory view. Trends Immunol. 2006;27:399–404. doi: 10.1016/j.it.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Croy BA, Chantakru S, Esadeg S, Ashkar AA, Wei Q. Decidual natural killer cells: key regulators of placental development (a review) Journal of Reproductive Immunology. 2002;57:151–168. doi: 10.1016/s0165-0378(02)00005-0. [DOI] [PubMed] [Google Scholar]
- 4.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 5.Barten R, Torkar M, Haude A, Trowsdale J, Wilson MJ. Divergent and convergent evolution of NK-cell receptors. Trends Immunol. 2001;22:52–57. doi: 10.1016/s1471-4906(00)01802-0. [DOI] [PubMed] [Google Scholar]
- 6.Anderson SK, Ortaldo JR, McVicar DW. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- 7.Westgaard IH, Berg SF, Orstavik S, Fossum S, Dissen E. Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28:1839–1846. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Norman PJ, Parham P. Complex interactions: the immunogenetics of human leukocyte antigen and killer cell immunoglobulin-like receptors. Semin Hematol. 2005;42:65–75. doi: 10.1053/j.seminhematol.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 10.French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol. 2003;15:45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- 11.Diefenbach A, Raulet DH. The innate immune response to tumors and its role in the induction of T-cell immunity. Immunol Rev. 2002;188:9–21. doi: 10.1034/j.1600-065x.2002.18802.x. [DOI] [PubMed] [Google Scholar]
- 12.Dupont B, Hsu KC. Inhibitory killer Ig-like receptor genes and human leukocyte antigen class I ligands in haematopoietic stem cell transplantation. Curr Opin Immunol. 2004;16:634–643. doi: 10.1016/j.coi.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Nylenna O, Naper C, Vaage JT, Woon PY, Gauguier D, Dissen E, Ryan JC, Fossum S. The genes and gene organization of the Ly49 region of the rat natural killer cell gene complex. Eur J Immunol. 2005;35:261–272. doi: 10.1002/eji.200425429. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi T, Yawata M, Raudsepp T, Lear TL, Chowdhary BP, Antczak DF, Kasahara M. Natural killer cell receptors in the horse: evidence for the existence of multiple transcribed LY49 genes. Eur J Immunol. 2004;34:773–784. doi: 10.1002/eji.200324695. [DOI] [PubMed] [Google Scholar]
- 15.McQueen KL, Wilhelm BT, Harden KD, Mager DL. Evolution of NK receptors: a single Ly49 and multiple KIR genes in the cow. Eur J Immunol. 2002;32:810–817. doi: 10.1002/1521-4141(200203)32:3<810::AID-IMMU810>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Gagnier L, Wilhelm BT, Mager DL. Ly49 genes in non-rodent mammals. Immunogenetics. 2003;55:109–115. doi: 10.1007/s00251-003-0558-9. [DOI] [PubMed] [Google Scholar]
- 17.Mager DL, McQueen KL, Wee V, Freeman JD. Evolution of natural killer cell receptors: coexistence of functional Ly49 and KIR genes in baboons. Curr Biol. 2001;11:626–630. doi: 10.1016/s0960-9822(01)00148-8. [DOI] [PubMed] [Google Scholar]
- 18.Storset AK, Slettedal IO, Williams JL, Law A, Dissen E. Natural killer cell receptors in cattle: a bovine killer cell immunoglobulin-like receptor multigene family contains members with divergent signaling motifs. Eur J Immunol. 2003;33:980–990. doi: 10.1002/eji.200323710. [DOI] [PubMed] [Google Scholar]
- 19.Dobromylskyj M, Ellis S. Complexity in cattle KIR genes: transcription and genome analysis. Immunogenetics. 2007;59:463–472. doi: 10.1007/s00251-007-0215-9. [DOI] [PubMed] [Google Scholar]
- 20.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 21.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 23.Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, Schuck P, Margulies DH, Mariuzza RA. Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b) Nat Immunol. 2003;4:1213–1222. doi: 10.1038/ni1006. [DOI] [PubMed] [Google Scholar]
- 24.Sawicki MW, Dimasi N, Natarajan K, Wang J, Margulies DH, Mariuzza RA. Structural basis of MHC class I recognition by natural killer cell receptors. Immunol Rev. 2001;181:52–65. doi: 10.1034/j.1600-065x.2001.1810104.x. [DOI] [PubMed] [Google Scholar]
- 25.Tormo J, Natarajan K, Margulies DH, Mariuzza RA. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature. 1999;402:623–631. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- 26.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 28.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Fujiwara M, Nikaido M, Okada N, Hasegawa M. Interordinal relationships and timescale of eutherian evolution as inferred from mitochondrial genome data. Gene. 2000;259:149–158. doi: 10.1016/s0378-1119(00)00427-3. [DOI] [PubMed] [Google Scholar]
- 32.Arnason U, Gullberg A, Janke A, Kullberg M, Lehman N, Petrov EA, Vainola R. Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol Phylogenet Evol. 2006 doi: 10.1016/j.ympev.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Fyler CA, Reeder TW, Berta A, Antonelis G, Aguilar A, Androukaki E. Historical biogeography and phylogeny of monachine seals (Pinnipedia: Phocidae) based on mitochondrial and nuclear DNA data. Journal of Biogeography. 2005;32:1267–1279. [Google Scholar]
- 34.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 35.Robertson MJ, Cochran KJ, Cameron C, Le JM, Tantravahi R, Ritz J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 36.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and alalysis program for windows 95/98/NT. Nucleic acids symposium series. 1999;41:95–98. [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 39.Swofford DL. PAUP*:Phylogenetic analysis using parsimony (*and other methods) version 4.0. Sinauer. Sunderland: Massachusetts; 2001. [Google Scholar]
- 40.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 42.Burshtyn DN, Lam AS, Weston M, Gupta N, P. Warmerdam AM, Long EO. Conserved Residues Amino-Terminal of Cytoplasmic Tyrosines Contribute to the SHP-1-Mediated Inhibitory Function of Killer Cell Ig-Like Receptors. J Immunol. 1999;162:897–902. [PubMed] [Google Scholar]
- 43.Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, Lifson JD, Carrington M, Beck S. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 45.Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. PNAS. 1999;96:3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 47.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 Domain of KIR3D Acts as a Major Histocompatibility Complex Class I Binding Enhancer. J. Exp. Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Togo S, Shimokawa T, Fukuchi Y, Ra C. Alternative splicing of myeloid IgA Fc receptor (Fc[alpha]R, CD89) transcripts in inflammatory responses. FEBS Lett. 2003;535:205–209. doi: 10.1016/s0014-5793(02)03891-7. [DOI] [PubMed] [Google Scholar]
- 49.Vilches C, Pando MJ, Parham P. Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000;V51:639–646. doi: 10.1007/s002510000184. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook JG, Sehra H, Coggill P, Humphray S, Palmer S, Sims S, Takamatsu HH, Wileman T, Archibald AL, Beck S. Identification of a single killer immunoglobulin-like receptor (KIR) gene in the porcine leukocyte receptor complex on chromosome 6q. Immunogenetics. 2006;58:481–486. doi: 10.1007/s00251-006-0110-9. [DOI] [PubMed] [Google Scholar]
- 51.Aldridge BM, Bowen L, Smith BR, Antonelis GA, Gulland F, Stott JL. Paucity of class I MHC gene heterogeneity between individuals in the endangered Hawaiian monk seal population. Immunogenetics. 2006;58:203–215. doi: 10.1007/s00251-005-0069-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.