Abstract
The development of in vivo brain imaging has lead to the collection of large quantities of digital information. In any individual research article, several tens of gigabytes-worth of data may be represented – collected across normal and patient samples. With the ease of collecting such data, there is increased desire for brain imaging datasets to be openly shared through sophisticated databases. However, very often the raw and pre-processed versions of these data are not available to researchers outside of the team that collected them. A range of neuroimaging databasing approaches has streamlined the transmission, storage, and dissemination of data from such brain imaging studies. Though early sociological and technical concerns have been addressed, they have not been ameliorated altogether for many in the field. In this article, we review the progress made in neuroimaging databases, their role in data sharing, data management, potential for the construction of brain atlases, recording data provenance, and value for re-analysis, new publication, and training. We feature the LONI IDA as an example of an archive being used as a source for brain atlas workflow construction, list several instances of other successful uses of image databases, and comment on archive sustainability. Finally, we suggest that, given these developments, now is the time for the neuroimaging community to re-prioritize large-scale databases as a valuable component of brain imaging science.
Introduction
The increasing ability to obtain digital information in medical and biological neuroimaging research has lead to a vast increase of scientific data from across a variety of spatial and temporal scales (Van Essen 2002). With each new technological advance neuroscientific data may be collected with finer resolution per unit time and render more detailed forms of biologically relevant information (Bandettini 2007). Occurring simultaneously with advances in imaging technology has been the advancement of the World Wide Web - whose original purpose was to permit ease of data exchange between collaborating scientists but now links people, computers, and information on an unprecedented global scale. From this co-evolution of neuroscientific and computer network technology is an increased expectation that primary scientific data be openly shared via readily accessible databases (Koslow 2000). One particularly notable example is from the domain of human brain imaging where large, three and four dimensional, volumes of structural and functional brain data are obtained using high-resolution magnetic resonance scanners. In any individual research publication, several tens of gigabytes-worth of data may be represented – collected across normal and patient populations. However, very often the raw and pre-processed versions of these data are not available to researchers outside of the team that collected them. Concerns over the sharing of the primary data may exist that prohibits their availability (Koslow 2002). What are available may only be lists of local “hot spots” of activity referenced with respect to a triplet of brain atlas spatial coordinates, perhaps tables of region volumetric results, other summary statistics, and some very selective graphical renderings. Study meta-data (the data that describes how the data were obtained, the parameters, experimental design, etc) may be incomplete and limit the scope of future use. The raw and preprocessed versions of those data may end up being lost should the post-doc who did the work leave the lab, if the data are archived onto media that soon becomes outdated, or are unrecoverable following a computer mishap.
If, on the other hand, the data from published as well as ongoing studies can be archived using a reliable and well maintained framework, then the utility of the data can extend beyond the intent of their original collection (Van Horn and Gazzaniga 2005). Datasets from diverse subjects or between patient groups can be mined to examine patterns among the data that would otherwise go unseen in any individual investigation wherein combining datasets can increase statistical power to observe more subtle effects. Using centralized (Van Horn, Woodward et al. 2002) or distributed databasing approaches (Grethe, Baru et al. 2005), research consortia can better manage work being performed across distant research centers. Importantly, through the use of databases, federally funded collections of neuroimaging data can reach the widest numbers of researchers who can turn that data into new knowledge, thereby maximizing their utility and justifying the cost of their collection.
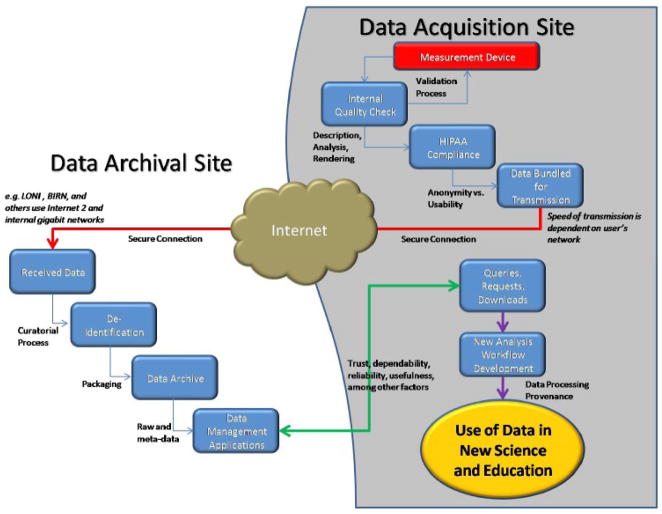
With the rapid advances being made in neuroimaging technology, data acquisition, and computer networks the successful organization and management of neuroimaging data has become more important than ever before (Poliakov, Hertzenberg et al. 2007; Hasson, Jeremy et al. 2008). Technological advances in computer network throughput, disk storage, and archival capabilities can be brought to bear so that databases can truly be a resource for exchange and future use in computational anatomy and modeling (Figure 1). However, databases still suffer from some reluctance on the part of the community who harbor doubts about their trustworthiness, the difficulties associated with sharing, and how their data will be used by others.
Figure 1. Neuroimage Data Flow via the LONI Imaging Data Archive (IDA).
The Laboratory of Neuro Imaging (LONI) has established a highly reliable framework for the efficient archiving, querying, and dissemination of brain imaging data. This model for data deposition has proven successful across a number of NIH-funded projects and is adaptable to a wide range of new applications.
During the early years of this decade, considerable attention was given to neuroimaging databases from the Organization of Human Brain Mapping (OHBM) (Governing Council of the Organization for Human Brain Mapping 2001), who expressed concern about the quality of brain imaging data being deposited into such archives, how such data might be re-used, and the potential for their being represented in new publication. The question of data ownership, in particular, was a primary concern in initial attempts to archive data (Editorial 2000). A recent data ownership controversy (Abbott 2008) has highlighted anew the still tenuous nature of data ownership, re-use, research ethical standards, and the pivotal role that peer-reviewed journals play in this process (Fox, Bullmore et al. 2008). The implications of disagreements concerning appropriate data re-use and new publication also impacts the users of neuroscience data archives and how researchers might independently draw from archives and publish results independently. While some might view the threat of similar disputes as an argument against data sharing or large-scale archiving, we believe that this need not be the case and that open access to primary neuroscience data through curated archives can enhance collaborations, not hinder them. Leading scientific organizations, working closely with government organizations and journal publishers, are poised to enact policies that promote the use of databases while being sensitive to intellectual priority and research ethics. There are many positives to databasing neuroimaging data and it is helpful to review these aspects and how they contribute to the health of the field and encourage new thinking.
In this commentary we examine the various roles that neuroimaging databases play in scientific data sharing, data re-use, and consider some of the characteristics of trusted data archives. A spectrum of database models has been proposed that range from simple FTP sites to fully curated efforts containing data from published studies. We note the importance of thorough data management and organization. We discuss population-level brain atlases as one natural outcome of databases - essential for understanding normal and abnormal brain form and function. We detail our own experiences developing databases and give examples of successful utility for several large scale neuroimaging initiatives. The processing and examination of datasets from multiple subjects necessitates clever workflow design, optimization, and provenance with a view toward promoting independent re-analysis and study replication. We observe that, among other metrics, databases are only as good as how they are being used and their effectiveness in generating new science and contributions to education are important benchmarks of their success. The data present in neuroimaging archives also forms a basis for content-driven comparison representing new and interesting computational challenges. As many these resources are of immeasurable value to neuroscience, their long-term sustainability is imperative. Finally, we discuss the lessons that we, and the community, have learned in the creation and maintenance of these essential neuroscientific resources. We believe that such aspects strongly favor scientific organizations, such as OHBM, re-examining the role of neuroimaging databases and their use in promoting a healthy research enterprise.
The Continuum of Neuroimaging Data Resources
The various levels of data archiving can be seen as forming a continuum (Van Horn, Grethe et al. 2001). At one extreme, sample or “test” data sets might be located on an anonymous FTP site that other scientists may use for instruction or for the early assessment of new image processing algorithms. This may include datasets and their accompanying meta-data from a particular empirical study which may be publicly advertized with a view toward encouraging competition between new analysis techniques (as has been done in the OHBM-sponsored FIAC competition (2006), for instance). At the next level there is the archiving of data from within one’s own laboratory. Where greater expertise seems necessary to interpret results, a researcher may wish to share data with a colleague from different laboratory. This may require the establishment of formal database privileges and a more advanced level of meta-data description. Finally, following the publication of primary results and their inclusion in the collective scientific body of work, researchers might submit their data set to a formal archive specifically designed to accommodate published neuroimaging data. These may not only include collections of raw data but also summary results (e.g. lists of activation local maxima) or derived representations (e.g. extracted surface-based representations). These may be publically accessible databases where other researchers are able to access the data that generated reported findings and on which they may perform their own independent analyses. These new analyses may confirm the reported results or offer a new interpretation not discussed in the original article and can form part of a new published article themselves.
Anonymous FTP sites need little more than an accessible disk space needed to store the representation of the raw image data. Such databases are low-cost and need little human supervision. On the other hand, repositories of raw data from published research articles necessitate detailed demographic and experimental meta-data, considerable computational resources, and curatorial effort to maintain them. Those that provide minimal curatorial activities, sparse database normalization, etc. and simply provide a data warehousing service may also necessitate little supervision. However, these resources are all costly and necessitate infrastructure considerations as well as support from funding agencies. But it is at this more complex and costly end of the spectrum that the greatest potential for advancing neuroscience exists. It is here that bridges may be constructed to other neuroscientific databases (e.g. molecular, electroencephalographic, genomic, biobehavioral, etc.) as well as other types of databases, thereby enabling researchers to gather and cross-reference data descriptions and identify convergence of findings. The repositories at this end of the spectrum must build a trust with the communities they seek to serve and provide dependable services to researchers that are not provided by anonymous FTP sites.
Several factors contribute to a database’s utility, including whether it actually contains viable data and these are accompanied by a detailed description of their acquisition (e.g. meta-data); whether the database is well-organized and the user interface is easy to navigate; whether the data are derived versions of raw data or the raw data itself; the manner in which the database addresses the sociological and bureaucratic issues that can be associated with data sharing; whether it has a policy in place to ensure that requesting authors give proper attribution to the original collectors of the data; and the efficiency of secure data transactions. Several authors have proposed considerations for how formalized neuroimaging databases might be best constructed (Van Essen 2002; Keator 2006) and have developed useful implementations of these systems (Evans 2006; Olabarriaga, Nederveen et al. 2006; Marcus, Olsen et al. 2007). Clearly, large-scale relational databases offer a highly flexible means for describing data and the relatedness of their various meta-data characteristics (Hasson, Jeremy et al. 2008). Moreover, those that have been specifically designed to serve a large and diverse audience with a variety of needs and that possess the qualities described above, represent the types of databases that can have the greatest benefit to neuroscientists looking to assess new methods, examine previously published data, or with interests in exploring novel ideas in cognitive or patient data (Van Horn, Grethe et al. 2001).
Characteristics of Trusted Data Repositories
Given this range of database formats, the motivation to deposit or obtain data from a digital resource often comes down to a matter of trust in the resource itself. Arzberger and colleagues (2004) have noted several characteristics of successful data archiving and exchange efforts that can form the basis of operating principles for any such archive of scientific data. These include: 1) the openness of the data archive – that access to information contained in a database is generally unrestricted with respect to its user-base; 2) the database is transparent and there is evidence of active data dissemination where it is clear what the database contains and that its contents experience ongoing access over a period of time; 3) that there is an assignment and assumption of formal responsibilities among the stake holders; 4) that technical and semantic interoperability exists between the database in question and other online resources; 5) curation systems governing quality control, data validation, authentication, and authorization are in place; 6) there is demonstrated operational efficiency and flexibility; 7) the database insists upon respect for intellectual property and other ethical and legal requirements; 8) there exists management accountability which includes approaches to funding; 9) the archive is built upon a solid technological architecture; and 10) users of the archive receive reliable support in data deposition and access. Beaulieu (2001) has elaborated on many of these characteristics and how they relate to what constitutes a trusted neuroscience digital resource. Additional issues involve HIPAA compliance (Kulynych 2002), concern over incidental findings (Illes, Kirschen et al. 2006), anonymization of facial features (Bischoff-Grethe, Fischl et al. 2004; Neu and Toga 2008), and skull stripping (Zhuang, Valentino et al. 2006). Given the degree of effort required to curate active data deposition as well as for comprehensively addressing these issues, the most trusted archives tend to be those whose infrastructure and archival processes are sufficiently mature and specifically dedicated to the goals of long term community-oriented databasing.
Sharing Neuroimaging Data
The National Institute of Health policy on data sharing has recognized that “data sharing is essential for expedited translation of research results into knowledge, products, and procedures to improve human health.” (http://grants.nih.gov/grants/policy/data_sharing/data_sharing_guidance.htm). With this mandate to share data there has been considerable interest concerning the databasing the results of and the raw data from studies of human neuroimaging (Fox and Lancaster 2002; Van Horn, Grafton et al. 2004). However, there are as many barriers to sharing primary neuroimaging data with established data repositories as there are investigators performing these studies. The reasons for this have been discussed by us elsewhere (Van Horn and Ball 2008) but it suffices to say that these reasons all involve the issue of trust: trust in the repository, its utility, in who is using the data, and in how that data is being used.
But when data have been shared, there are instances of highly positive outcomes. In one example, data from a large database of functional MRI studies were re-examined to explore specific hypotheses concerning the differences between young and older subjects as compared to those with pre-Alzheimer’s dementia related to resting state (“default mode”) processing (Greicius, Srivastava et al. 2004). Prominent co-activation of the hippocampus, detected in all groups, implied that the so-called default-mode network (Raichle and Gusnard 2002) may be closely involved with episodic memory processing. However, the older subjects with dementia showed decreased resting-state activity in the posterior cingulate and hippocampus, suggesting that disrupted connectivity between these two regions accounts for the posterior cingulate hypometabolism commonly detected in positron emission tomography studies of early dementia. Such a re-analysis of data from a public repository provided a clinically significant finding beyond the intent of the original investigation and whose outcomes speak directly to the purpose behind the NIH mandates on data availability.
Neuroimaging Databases and Their Role in Creating Brain Atlases
A detailed listing of a number of leading relevant examples of neuroscientific databases and their attributes is provided in Table 1. We draw these, admittedly selective examples, from more comprehensive catalogs of neuroscience database resources which may be found at the Society for Neuroscience Database Gateway (http://ndg.sfn.org/) and at the Neuroscience Information Framework (NIF; http://neurogateway.org) websites. Detailed statistics on the usage, uploads, downloads, etc. of some on this list are maintained by the Neuroimaging Informatics Tools and Resources Clearinghouse (http://www.nitrc.org/), an important source for a vast array of neuroimaging tools as well as data. Many of these neuroimaging data resources cover a variety of funded projects or research studies, provide raw as well as derived versions of electrophysiological and neuroimaging data, with a few also containing data from non-human samples. Additionally, as these archives have developed, some have preferred to remain focused on one human population, for example, where as other have sought to represent data from multiple diagnostic groups as well as multiple data types (e.g. MRI, EEG). Some require user registration to obtain data files whereas others serve data directly from within a web browser interface. This table illustrates a cross-section of the types of neuroimaging databases, representing intrepid attempts to serve their communities, and which can be utilized to obtain data from across a range of samples, methods, and applications. Further details for many of the resources listed here can be located on the NITRC website.
Table 1.
Selected Neuroimaging Databases
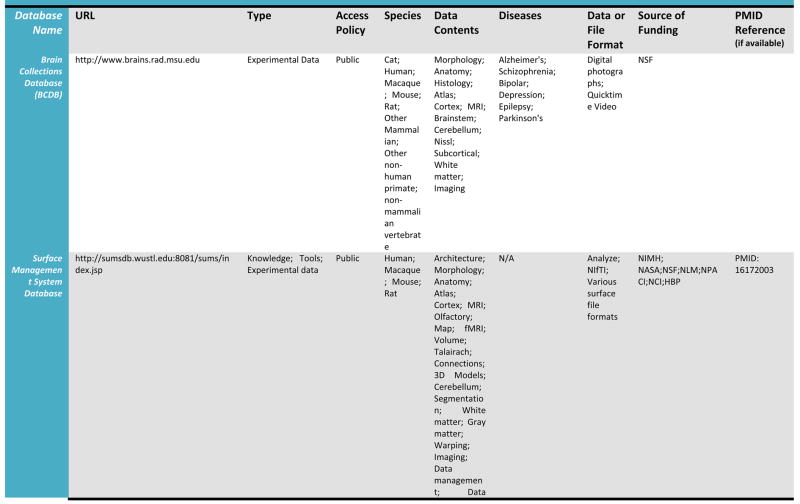 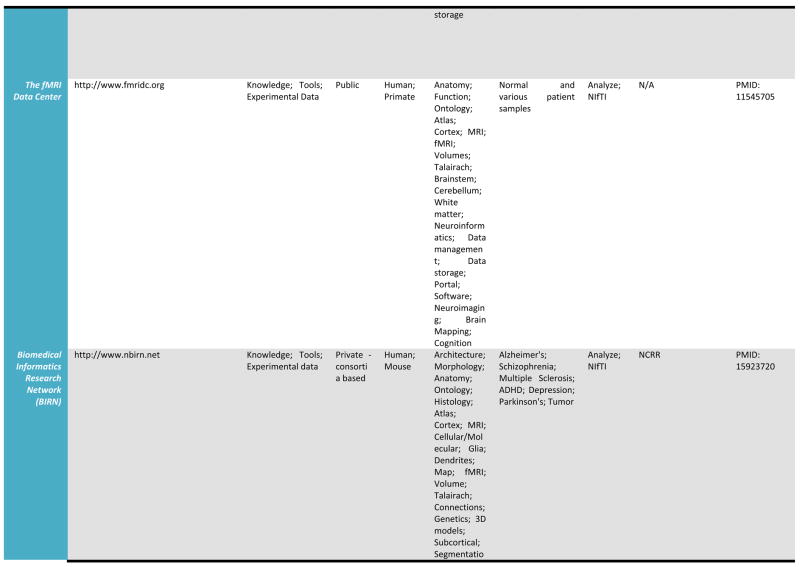 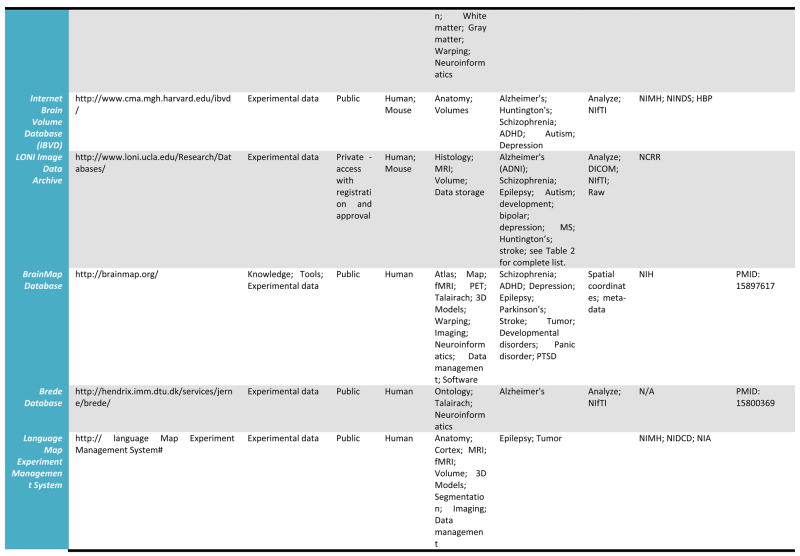 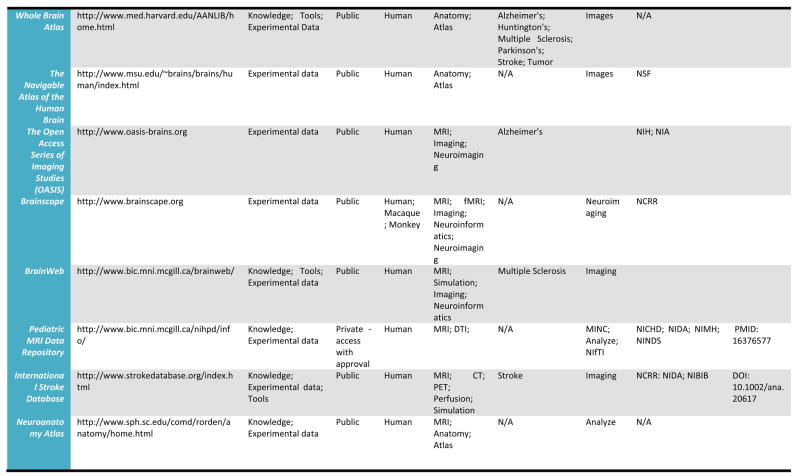
|
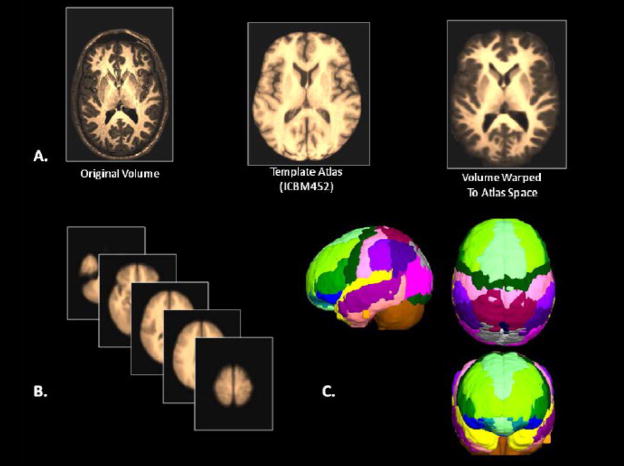
Databases such as these provide a wealth of structural and functional data obtained from across a range of subjects or specific patient groups. The advantage of having such large collections of data in one place is that they can be used to construct detailed population-level atlases of brain morphometry or function. By population-level, we mean any form of brain atlas assembled by drawing from neuroimaging data voxel intensity, geometry, or other attributes from across large, representative samples of human subjects that is warped to fit a known spatial reference frame. These include probabilistic anatomical atlases (Mazziotta, Toga et al. 1995; Toga, Thompson et al. 2001; Toga, Thompson et al. 2006), white matter fiber atlases (Wakana, Jiang et al. 2004; Mori, Oishi et al. 2008), and cortical surface atlases (Van Essen 2005). These can also refer to functional maps (such as group-level results of function analysis) or to the relation between functional results and anatomical features. Brain atlases can be constructed to incorporate data describing multiple aspects of brain structure or function at different scales from different subjects, at different times, yielding a comprehensive description of the organ in normal or disease populations (Roland and Zilles 1994; Toga and Thompson 2001). However, the complexity and variability of brain structure, especially in the gyral patterns of the human cortex, can present challenges in creating standardized brain atlases that reflect the anatomy of a population (Toga and Thompson 2002). Based on well characterized subject groups, age-specific atlases can potentially contain thousands of structure models, composite maps, average templates, and visualizations of structural variability, asymmetry and group-specific differences. They correlate the structural, metabolic, molecular and histologic hallmarks of the disease (Narr, Thompson et al. 2000; Thompson 2003). Figure 2 shows an example of a typical registration against the ICBM452 average brain atlas. Rather than simply arithmetically averaging information from multiple subjects and sources, however, new mathematical workflows can be introduced to resolve group-specific features not apparent in individual scans (Davatzikos 1996; Thompson and Apostolova 2007). Figure 3, for instance, demonstrates averaged regional geometric shape parcellation obtained using a boost-probabilistic voxel assignment approach. High-dimensional elastic mappings, based on covariant partial differential equations, are developed to encode patterns of cortical variation (Davatzikos 1997; Weaver, Healy et al. 1998; Thompson, Woods et al. 2000). In the resulting brain atlas, age-stratified features and regional asymmetries emerge that are not apparent in individual anatomies. Recently developed pediatric structural and white matter brain atlases form notable examples (Huang, Zhang et al. 2006; Jelacic, de Regt et al. 2006; Shan, Parra et al. 2006). Figure 4 illustrates how processing workflows can be specifically designed to draw from large archives of individual subjects in order to produce customized age-stratified average brain spaces. The resulting probabilistic atlas can be used to identify patterns of altered structure and function, and can guide algorithms for knowledge-based image analysis, automated image labeling, tissue classification, data mining and functional image analysis. These integrative approaches and their dependence on rich databases of primary data have provided significant motivation for the human brain mapping initiatives, and have important applications in health and disease.
Figure 2.
A) The basic warping of data obtained during imaging experiments has become the de facto first step for many data processing workflows involving both normal and patient samples. Affine and non-linear approaches for warping anatomical image volumes (A) to standardized spaces (B) have been developed, each having certain advantages and disadvantages in terms of accuracy or computational load. However, the warping of patient data to “normal” template atlases may lead to improper conclusions concerning localization of brain activity, the degree of alteration under voxel-based morphometry in patients when contrasted against normative samples, and other confounds. B) Averaged normal subjects (a–e) drawn from the LONI IDA registered against an atlas space permits automated extraction of C) “average” brain regional shapes using sophisticated voxel position and intensity sorting algorithm (Tu, Narr et al. 2008).
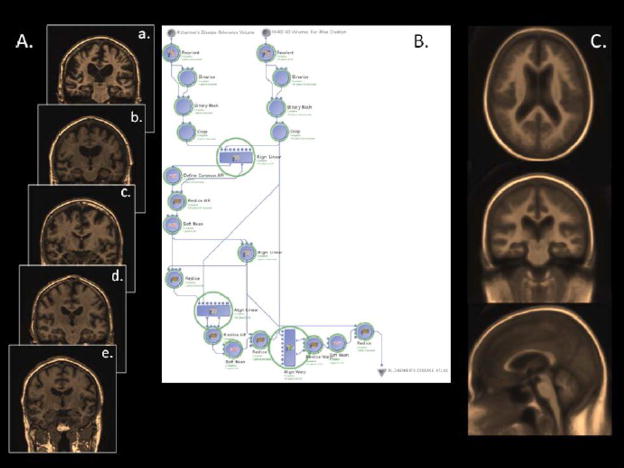
Figure 3.
N=400 Alzheimer’s disease subjects were drawn from the ADNI database housed at LONI/UCLA, of which examples of N=5 are shown here (A). These data were systematically submitted to the LONI grid computer cluster via the use of a purpose-built LONI Pipeline workflow comprised of a heterogeneous collection of processing modules from commonly available software packages, e.g. AIR, FSL, and related toolsets. The data were spatially reoriented, warped against an Alzheimer’s disease subject reference volume, a common realignment solution identified, and so on, to produce a disease-specific Alzheimer’s atlas volume (C) against which individual variability may be assessed or used for specific studies underway at other laboratories. The workflow does not contain any operations against a “normal” brain atlas. The processing of this number of subjects required approximately 50 minutes using the LONI Pipeline and LINUX computational grid.
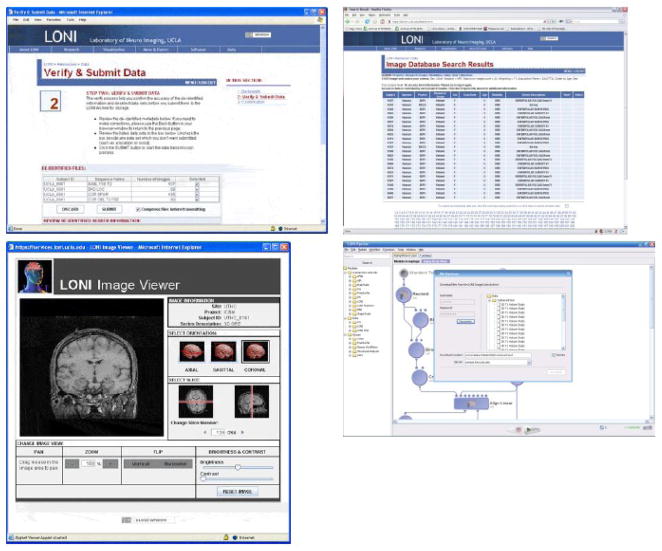
Figure 4. An example of end-to-end data sharing, databasing, and data processing: A. Archiving Data.
Contribution to the LONI IDA involves two basic steps: De-identification - a HIPAA compliant manner for removing patient-identifying information involving the LONI Debabeler (Neu, Valentino, et al., 2005), capable of de-identifying common medical imaging file formats; and Data Transmission - a storage server that enables an efficient transfer of many files at once, bundling files together so that file I/O is minimized when accessing multiple associated files. All data are securely transferred using the HTTPS protocol. B. Meta-Data Search. Meta-data searches include the species, research project, the diagnostic group, gender, scan dates, age, scanner modality and accompanying scan parameters, among other criteria. C. The LONI Image Viewer: From the database as a whole or from searches thereof, users can examine data in multiple planes using a convenient and intuitive web-based viewer. Finally, D. Access via The LONI Pipeline: The LONI Pipeline interface permits users direct access to the LONI IDA to create shareable processing workflows on data residing there. These workflows may themselves be shared with others and serve to record data and processing provenance.
Practically speaking, these examples represent standard atlas spaces that can be obtained without access to any particular database or that are provided without any database schema, per se. However, with the availability of data repositories containing large numbers of subjects, it is possible for researchers to create new atlases for specialized purposes or that are representative of specific disease populations. The creation of stratified brain atlases of the normal aging process, for example, would be a highly desirable resource for the community doing research on aging and the study of functional and structural brain changes associated with aging (for instance, as the leading edge of the “Baby Boomer” generation enters retirement). Such atlases could be created on an “as needed” basis by drawing from the data comprising large-scale archives of shared neuroimaging data and be added to the growing collection of available brain templates against which to spatially warp subject data. They could be updated periodically as new methods for image warping and synthesis become available. Additionally, atlases from similar patient samples could be compared across scanners, centers, countries, or other variables to determine the effects of these variables on atlas construction. Thus, atlases need not be static entities but, with the aid of available data archives, form the basis for continually refining knowledge about brain structure and function and play important roles in understanding those variables that influence the characteristics of “standard” atlas spaces.
Data Management and Organization
Digital archiving of scientific information is also an important element for research secondary to initial publication or description involving large-scale image analysis (mega-analysis), interactive visualization, and data exploration (Amari, Beltrame et al. 2002). Each of these areas is large and worthy of its own article describing their dependence on databases. Briefly, however, the informatics of medical image processing and analysis is subdivided into several research areas of intense activity (Kanaan, Kim et al. 2005; Maxim, Sendur et al. 2005; Kriegeskorte, Goebel et al. 2006; Moorhead, Harris et al. 2006). These include; acquisition, processing and recording of acquisition meta-data; the management of complete study information, including image summarization and subject anonymization; and the integration of clinical and other biologically-relevant information. Collectively, each area of informatics seeks to contribute to a strong web-driven database infrastructure that the community may seamlessly take advantage of (Grethe, Baru et al. 2005). Yet, the most immediate and important challenge for many neuroimaging laboratories is the end-to-end scientific data management from data acquisition and data integration, to data treatment, provenance and persistence.
Local data management architectures have been developed over the past few years that assist research teams with the management of their acquired primary data. For neuroimaging, notable examples include BIRN XCEDE (Keator, Gadde et al. 2006), BrainMap (Laird, Lancaster et al. 2005), and XNAT (Marcus, Olsen et al. 2007). XCEDE is the basis of the Biomedical Informatics Resource Network’s (BIRN) neuroimage data repository that organizes data from across the contributing Function BIRN sites. XNAT, on the other hand, was developed to be deployed at individual sites for local data management. Each of these provides a schema for containing subject demographic information, analysis annotations, activation threshold parameters, as well as cluster- and voxel-level statistics. BrainMap, is itself a database of published activation coordinates and is built around an extensive means for characterizing the specifics of the underlying cognitive paradigm under study (Fox, Laird et al. 2005). Additionally, BrainMap tools provide user-friendly means for interacting with prominent neuroimaging statistical packages such as Statistical Parametric Mapping software (http://www.fil.ion.ucl.ac.uk/spm/), or for anatomical labeling is via the Talairach Daemon (http://ric.uthscsa.edu/projects/talairachdaemon.html). Where there exists a well arranged data and meta-data organization, including linkages to other external resources, this significantly help to maximize the utility of data present in digital repositories, provide it appropriate context, and helps to preserve the specific details typically only known to the original collectors of the data. We note now the approach we have taken toward addressing these goals for the management of primary neuroimaging research data which we believe is a particularly unique and successful deployment for data management, organization, and subsequent data re-use.
The LONI Image Data Archive (IDA)
The Laboratory of Neuro Imaging (LONI) has long had a strategy of combining collaborative research with robust computational resources to foster and environment in which the exchange of ideas, data, and techniques may flow freely (Toga 2002). An initial component of this strategy was to develop an infrastructure for storing collaborator data such that the lab’s computational resources could be leveraged in performing image analysis. The advent of large, multi-site neuroimaging initiatives in more recent years exposed the need for a reliable and robust large-scale data repository. As a result, this led to the development of the LONI Image Data Archive (IDA).
The LONI IDA serves as a central relational-database repository for dozens of single and multi-site neuroimaging research studies. The IDA was designed as a long-term archive with considerable hardware and software resources devoted to ensuring: 1) protection of patient privacy through integrated data de-identification components. This provides for HIPAA compliant stripping of sensitive patient meta-data; 2) strict access controls to ensure data are only accessible to authorized individuals; 3) tracking of all data accesses to provide an audit trail so that project managers may understand who and in what way their data are being accessed; 4) ease of use through a platform-independent, user friendly interface; 5) automated of semiautomated capture of image acquisition and image viewer for evaluating image quality and a file format translation engine which supplies on-demand image file format conversions. These qualities help to address the issue of trust in the archive by satisfying depositors that the data are being securely maintained, dealing expressly with issues of subject identification, and providing users with easy to use tools for viewing and manipulating the image data.
The contents of the LONI IDA represent several national and international initiatives where the need for pooling and protecting data were deemed paramount. For instance, the National Institute on Aging (NIA) funded National Alzheimer’s Coordinating Center (NACC; http://www.alz.washington.edu/) maintains its own database of demographic, clinical and pathological data collected by the 29 Alzheimer’s disease Research Centers (ADRC). Many of the ADRCs also participate in the Alzheimer’s disease Neuroimaging Initiative (ADNI) in which detailed brain imaging data is gathered analyzed and then shared with the scientific community. The ADNI program was established to increase knowledge of the mechanisms of AD through the use of neuroimaging, thereby informing the development of treatment strategies aimed at slowing down or preventing neuronal death. With the help of the LONI IDA, ADNI has been instrumental in helping to identify clinical, neuroimaging, and biomarker outcome measures and longitudinal changes and the prediction of disease transitions. Users from around the world have obtained data from this collection for use in new research into the subtleties of AD (Fletcher, Powell et al. 2007; Boyes, Gunter et al. 2008; Yanovsky, Thompson et al. 2008). Additionally, the International Consortium for Brain Mapping (ICBM) is a project involving investigators from the US, Canada, and Europe seeking to combine multi-modal neuroimaging data to form population-based probabilistic brain atlases as standard references (Mazziotta, Toga et al. 2001). Table 2 provides a complete listing of the various research projects and their primary institutions, classified by research domain, that currently take advantage of the LONI IDA for image data archiving and availability between project members. Overall, the contents of the LONI IDA encompass nearly 30 large-scale research projects, containing upwards of 75,000 images (>39,000 raw scans; >29,000 pre-processed images; ~2,200 post-processed images). There are over 220 users actively uploading images into the archive on a regular basis and in excess of 350 registered users downloading images to their local sites (as of 03/09/2009).
Table 2.
Research Projects Utilizing the LONI Image Data Archive (IDA)
| Domain | Project Utilizing LONI IDA Resources | Primary Institution | Reference |
|---|---|---|---|
| Agingand Dementia | |||
| * Age Moderates HIV - Related CNS Dysfunction | University of Pittsburgh School of Medicine | (Lepore, Brunet al. 2006) | |
| * Alzheimer’s Disease Neuroimaging Initiative (ADNI) | University of California San Francisco | (Mueller, Weiner et al. 2005) | |
| Modifiers of Brain Aging and Alzheimer’s Disease | University of Kansas | (Burns, Church et al. 2005) | |
| PET Study of 5 - HT1AReceptors in AD | University of Toronto, Ontario, Canada | (Lanctot, Hussey et al. 2007) | |
| UCLA Alzheimer’s Disease Research Center (ADRC) | University of California Los Angeles | (Cummings, Miller et al. 2008) | |
| Anatomy | |||
| Anatomy of the Corpus Callosum | Université de Montréal, Québec, Canada | N/A | |
| * Brain Info | University of Washington | (Bowden and Dubach2003) | |
| BrainAtlasing | |||
| Chinese Digitized Standard Brain Atlas | Shandong University, China | N/A | |
| International Consortium for Brain Mapping | University of California Los Angeles | (Mazziotta, Toga et al. 2001) | |
| Development | |||
| Development | University of California San Diego | (Sowell, Trauner et al. 2002) | |
| Caring for a child with a serious illness | University of California Los Angeles | N/A | |
| MentalHealth | |||
| * Bipolar Endophenotypes in Population Isolates | University of California Los Angeles | N/A | |
| * Childhood Onset Schizophrenia | National Institutes of Mental Health | (Gogtay, Giedd et al. 2004) | |
| * First Episode Schizophrenia | University of California Los Angeles | (Narr, Bilder et al. 2007) | |
| Neurogenomics | |||
| ApoE& Alzheimer’s Disease | University of California Los Angeles | (Chou, Lepore et al.2008) | |
| Genetic influences on the brain: A twin study | University of Queensland, Australia | ||
| Neurogenomics Investigation of Depression | Oxford University, UK | N/A | |
| Neurology | |||
| Hippocampal Volume Loss in Multiple Sclerosis | University of California Los Angeles | (Sicotte, Kern et al. 2008) | |
| * Human Epileptic Brain: 3DMRI | University of California Los Angeles | (Lin, Salamon et al. 2007) | |
| Huntington’s Disease Neuroimaging Project | University College London, UK | (Holmes, Tsang et al.2006) | |
| * Neural Bases of Lexical Processes in H. Stroke | Johns Hopkins University | (Hillis, Newhart et al. 2005) | |
| * Neuropsychological Progession - New Onset Epilepsy | University of Wisconsin Madison | (Hermann, Jones et al. 2007) | |
| * Volumetrics in Brain Trauma | University of California Los Angeles | (Wu, Huang et al. 2004) | |
| Non-Human | |||
| Cryosection | University of California Los Angeles | N/A | |
| Mouse BIRN | University of California Los Angeles | (MacKenzie-Graham, Boline et al. 2007) | |
| Other | |||
| Minority Populations TS-Marina | University of California Los Angeles | N/A | |
| * Multi-center Estriol Study | University of California Los Angeles | N/A | |
| Santa Fe Institute Consortium | University of California Los Angeles | N/A |
indicates project having current NIH funding
The issue of developing trust in the archive has been of paramount importance. This has been accomplished in several ways. There is no charge to obtain data from the LONI IDA and anyone may request IDA access via the LONI website. The decision to share data, however, belongs to the PI or Liaison for the project whose data is present in the archive. LONI curators then only grant access authorization to study data once the PI and/or Liaison have given expressed permission for that person. This provides depositors of data with the knowledge that they have control over who can see and work with the data. In general, there is also no formal charge to deposit data into the LONI IDA. However, as part of the sustainability model for the IDA, funded investigators approved as LONI collaborators contribute funds to offset the curatorial costs associated with data deposition.
As we noted above, we recognize that many other database models exist that cover the spectrum of potential needs and use cases. The LONI IDA is just one of these models and other approaches may provide different or alternative services. But the LONI IDA example serves as a particularly compelling example of how neuroimaging databases can be developed, can grow, and can form trusted elements in major scientific initiatives.
Neuroimaging Workflow Description and Optimization
Drawing from databases for the purposes of mining their contents, re-analyzing reported effects, and combining data from across separate studies requires a careful consideration of the workflow of processing steps needed to generate an informative final result. Data frequently need to be registered within and between modalities, to correct inhomogeneities, and for the fitting of statistical models (Van Horn and Gazzaniga 2002). Ordering these steps in a logical sequence wherein the output from one step of processing becomes the input for another step represents a data workflow and is the underlying basis of all the major data processing packages currently available. Increased interest in the development of automated workflow environments, APIs, and graphical methods for the design and execution of data processing streams has led to the emergence of a range of workflow approaches. For instance, the Swift system offers a simple scripting language, SwiftScript, seeking to provide a concise high-level specification of workflows that invoke various application programs drawn from large quantities of data (Stef-Praun, Clifford et al. 2007). Workflows of linked scripts can be submitted to distributed computer grids to provide parallel processing performance and increase analysis throughput (see Hasson et al., 2008, for overview). Likewise, XNAT also provides a set of routines for the detailed specification of processing steps to be performed on data contained in an XNAT-based data archive. A final example is that of the FBIRN Image Processing Scripts (FIPS), a package for the comprehensive management of large-scale multi-site fMRI projects, and including data analysis using SPM, FSL, and FreeSurfer packages (Keator, Gadde et al. 2006). It also provides a modular set of scripts so that the user can flexibly set up their own standardized analysis. Each of these approaches offers users a way to string together those processing operations that are best suited to their data analysis needs and that can then be run in an unsupervised fashion over many archived data sets.
The LONI Pipeline (http://pipeline.loni.ucla.edu), for instance, is a simple, efficient, and distributed computing environment, enabling software inclusion from different laboratories in different environments (Toga, Rex et al. 2001; Rex, Ma et al. 2003). The primary goals of the LONI Pipeline are: 1) to create a robust environment for scientific software tool interoperability, Grid integration and low-cost interactive user interface. For maximum portability, scalability and efficiency, this environment is built in Java and utilizes XML for storing and communication of meta-data, and descriptors for tools and services; 2) To enable expert researchers to quickly design, test and validate novel experimental designs and to rapidly examine new data analysis protocols. This is achieved via dynamic, responsive and extensible graphical user interface; and 3) to provide the necessary means for integration of LONI Pipeline XML workflow descriptions with other established graphical environments for scientific Grid computing.
The LONI Pipeline provides a visual programming interface for the design, execution, and dissemination of neuroimaging analyses. Individual executables are represented as “modules” that can be included, deleted, and substituted for other modules within a user-friendly graphical user interface. Connections between the modules that establish an analysis methodology are represented as “workflows”. The environment handles the bookkeeping, controls the details of the computation, and information transfer between modules and within the workflow. It permits files, intermediate results, and other information to be accurately passed between individually connected modules. The DRMAA API (http://www.drmaa.net), backed by the Sun Grid Engine (http://gridengine.sunsource.net), acts as an interface to grid environments. Modules and workflows can be saved to disk at any stage of development and recalled at a later time for modification, use, or distribution. This functionality facilitates the translation of existent analysis paradigms from other environments to the LONI Pipeline and vice-versa. An XML description protocol allows any command-line driven process, web-service or data-server to be encapsulated into the environment by reference. This is a deliberate design we have imposed to reduce the integration/utilization costs of including new resources within the LONI Pipeline environment. This approach provides the benefit of quick and easy management of large and disparately located resources and data. In addition, this choice significantly minimizes the hardware requirements for user-client machine (e.g., memory, storage, CPU). Database server connectivity is a specific design feature that enables a user to construct workflows that directly act upon data archived in the LONI IDA as well as potentially other relational-database architectures. Tools such as this will be critical for a future in which data management and mining are based in web-driven access in addition to infrastructures aimed at allowing researchers access via GRID/PetaScale computing. Finally, though tools like the LONI Pipeline are primarily used in the context of neuroimaging, the underlying data-models can be made agnostic to any particular scientific domain or data type, and so is suitable for use with many types of scientific data archive, most notably for storing how the data sets were processed, e.g. the “provenance” of the data.
Neuroimaging Provenance Information and Data Archives
In the biological sciences, a description of how data was obtained is often crucial for assessing its quality and usefulness, as well as enabling analysis in an appropriate context. Additionally, the analysis of raw data in neuroimaging has become a computationally rich process with many individual operations run on increasingly larger datasets (Liu, Meier et al. 2005). Many commonly available software packages exist that provide either complete analyses or enable specific steps in neuroimaging data analysis. The recording of the data generation and processing provenance (Bidgood, Horii et al. 1997; Mackenzie-Graham, Van Horn et al. 2008) is, however, not often practiced. Many software packages, for instance, possess diverse input and output requirements, utilize different file formats, run only under particular computer environments, or are appropriate for only certain types of data. The accurate preservation of data integrity during study data transactions or to document any database normalization operations also falls under the domain of provenance. Recording the provenance of data, its processing, curation, alterations or addendums to it and including this information in databases can aide in the fidelity of the independent reproduction of results or, if viewed as meta-data itself, can be used as predictor variables in multi-center trials to examine how acquisition or processing parameters influence experimental results.
Indeed, the provenance of neuroimaging data has recently begun to receive attention in the fields of neuroimaging (Hasson, Jeremy et al. 2008) and computer science (Moreau, Ludäscher et al. 2007). Several databases and analysis platforms such as XNAT (Marcus, Olsen et al. 2007), Swift (see above), and Fiswidgets (Fissell, Tseytlin et al. 2003; Fissell 2007) provide such capabilities. We have addressed this issue ourselves through the use of the LONI Pipeline (http://pipline.loni.ucla.edu) which has been developed to generate a detailed description of the data and processing executables used by the LONI Pipeline into the workflow description files (MacKenzie-Graham, Payan et al. 2008). The efficient but detailed documentation of neuroimaging provenance description is a presently rich area for neuroimaging databases and a topic of mutual interest to brain, computer, and information sciences and one that can help to better capture those details concerning how data are processed that are often not provided in published research articles.
Use of Databases in New Scientific Publication
The use of neuroimaging archives such as these to produce new scientific publications has been noted as being one hallmark of the success of a databasing effort. One example of where this has been true is that of the BrainMap database of brain activation local maxima. Using mean activity locations as a basis and considering a rigorous Bayesian probabilistic framework called activation likelihood estimation (ALE), several novel meta-analyses have appeared that explore the effects of experimental predictor variables on motor activity (Witt, Laird et al. 2008), differences in patterns of language-related activity in stutters (Brown, Ingham et al. 2005), as well as in the cerebellum’s contributions in auditory function (Petacchi, Laird et al. 2005). A very large number of studies have emerged from LONI that draw from the LONI IDA, especially from the contributions from the ADNI project (Leow, Klunder et al. 2006; Boyes, Gunter et al. 2008; Jack, Bernstein et al. 2008; Yanovsky, Thompson et al. 2008). Other examples also exist from other archives in which previously published studies contained in databases have been reused or re-purposed to produce new published results (Liou, Su et al. 2006; Chen, Samuraki et al. 2008). The use of databases to broaden the extent of the original research findings represents their most important advantages to the scientific community. These successful outcomes are further enhanced with the involvement of scientific publishers.
The relationship between the neuroscience databases and peer-reviewed journals is an important one. Databases and journals have partnered to examine how such processes might work in practice. For instance, the partnership between the fMRI Data Center (fMRIDC) and the Journal of Cognitive Neuroscience (JOCN) was one that other journals publishing neuroimaging data could adopt for their for their own contributions to the fMRIDC or other formal data archive (Van Horn, Grethe et al. 2001). As well, the basic model of this partnership could be adapted easily to accommodate studies and study data from other neuroscientific domains and modalities. Having software tools that facilitate and make easy the data contribution process are essential. The process and its eventual outcome represents value-added for the journal in terms of enhancing what is being made available with each published article. It also represents the opportunity for researchers in the field to obtain and examine primary data from the published literature itself, confirming results, testing new hypotheses, or exploring emerging analytic approaches.
Several articles have appeared in the literature whose secondary analyses of data drawn from neuroimaging data archives has extended the scope of the original research (Carlson, Schrater et al. 2003; Mechelli, Price et al. 2003; Aizenstein, Clark et al. 2004) findings (Ishai, Ungerleider et al. 2000). Recently, Van Horn and Ishai (Van Horn and Ishai 2007) examined how the data from an earlier article by Ishai and coworkers (Ishai, Ungerleider et al. 2000) had been re-analyzed by others after that data was made available through an open data archive. It was observed that the data, originally collected in an experiment of categorical object visual processing, had been used in the further understanding of underlying cognitive processes but also new methods development and statistical analyses. The authors argued that the dataset from the original Ishai article, as evident from these new applications, took on greater value because its data were openly available. The re-use and re-interpretation of data from published studies helps to inform and energize subsequent published literature and that helps to enhance the value of the original research. Importantly, access to data used in a journal article can expose analysis errors when secondary parties attempt to replicate the published computational and statistical procedures. We note that such re-analyses are not new or independent studies, per se, but are complementary treatments of the same data that can provide additional detail on underlying effects and alternative points of view. Over time, corrected or revised conclusions about the effects present in the data might be drawn as more people examine them from these different perspectives.
Role for Databases in Education
Digital repositories of primary data are particularly well suited to play a role in the education of the next generation of neuroscientists. To present students with actual data from published articles or major research initiatives can broaden journal club article reviews in which, not only do students have the author’s interpretation of the results, they can perform analyses of their own to confirm the results of the published article or apply alternative methods to look at data in novel ways. Students may perform meta-analyses that utilize databases as a means for identifying interesting avenues for subsequent research. Distillation of data into clusters of similar studies may reveal patterns that would have otherwise gone un-recognized, leading a student to consider new, testable hypotheses. Additionally, drawing from digital repositories may aid in computing the statistical power required to find particular effects and can be used to justify a student’s intentions to obtain new data as part of a Ph.D. dissertation. Already, genomic and proteomic studies have demonstrated that meta-analytic and informatics-based research is an important new element for discovery science (Hood 2003) and it is not unreasonable to consider such approaches as a prelude to formal experimentation and new data collection. Though, very few papers have been written discussing this role for neuroscience databases, use in medical education (Gutmark, Halsted et al. 2007) and research training (http://www.sfn.org/index.cfm?pagename=PublicEducationOutreach_NeurosciEduResources) is likely to be one of their most important attributes.
Databases as a Basis for Comparison and Validation
Beyond simply accumulating more and more data, databases also need to look toward providing a useful basis for comparison with newly collected data. In applying an informatics based approach to examining new information against large quantities of genomic data, BLAST was at the forefront of the emerging bioinformatics field which continues to develop into new domains (Phoebe Chen and Chen 2008). However, database technologies do not yet permit this type of content-based search. As has been noted by others, databases permit meta-data searches based on statements to the effect of “show me all the scans from right-handed, male, schizophrenics, age 30 and over”. These are very useful searches and lists of such use cases is of particular importance for database developers and ontologists as they decide what meta-data to gather that describes the main data of interests. However, few, if any, tools are available that utilize the data itself as the basis of comparison among database records or for permitting a comparison of an unknown example with the database to identify most similar cases. For instance, a user might upload a recently collected MPRAGE anatomical volume to a server where it is digitally dissected, standardized measurements are made upon its elements, and these are systematically compared against the entire contents of the archive. Results returned to the user might indicate the degree of geometric similarity between their uploaded data and its closest counterparts in the archive. Examination of the meta-data for these similar records from the archive may help to better understand the newly obtained image volume. For instance, if the uploaded data were most morphologically similar to Alzheimer’s disease patients from the archive, it may be concluded, along with other evidence, that the uploaded data are also from a patient with AD. One example of this type of work is presently underway in LONI to develop online image registration validation tools in which a user can validate the results of volume registration against the contents of the LONI IDA (Yanovsky, Thompson et al. 2008). A related approach was recently applied toward the development of a new whole brain human atlas (Shattuck, Mirza et al. 2008). In these ways, neuroimaging databases could provide a similar role to that played by conducting BLAST searches for genomic data (Altschul, Gish et al. 1990). Thus, development of comparable tools that can evaluate newly obtained brain imaging data, not just meta-data, against large digital brain archives will do much to energize the discipline of neuroinformatics.
Neuroimaging Database Sustainability
Governmental scientific agencies are encouraging the development and use of these resources but tend not to be interested in long term support, per se. Curiously, the opposite appears to be true for brain tissue banks (Haroutunian and Pickett 2007; Graeber 2008). While these are different types of brain archive, to be sure, both forms of information, physical brain specimens and their digital representation, have significant value to the neuroscience community. Once a database has been developed, questions arise as to who is benefiting from the resource, how is it being maintained, what is the data model underlying its organization, and is it interoperable with other resources. Failure in any of these areas may mean that the database cannot be sufficiently continued as a resource.
Many variables come into play when considering database sustainability not the least of which is ongoing governmental support for database curation and tools development. Others include an engaged process of curation, systems support, and ongoing scholarly activity that draws from the resource. Moreover, people are increasingly using these resources to conduct novel scientific discovery and have come to rely on them. This means that, should they fail to maintain their sustainability, a certain segment of the neuroscience research enterprise (not just the database in question) will be affected. Their use in the training of the next generation of neuroscientists also cannot be overstated. Funding agencies must examine carefully the impact of initiating such programs, what it will take to continue their momentum following their initial construction, and who will be inconvenienced should they falter. Very often it is the community that suffers not just the proprietors of the database in question.
The fMRIDC is an example of what can happen when resources cannot be sustained. Begun with ample funding, the effort quickly became one of the success stories for the community in gaining open access to the data from published fMRI studies. Raw, processed, and results image data and the accompanying meta-data were deposited with the fMRIDC by the authors upon acceptance of their papers (Van Horn, Grafton et al. 2004). Curatorial experts examined, catalogued, and packaged the data for dissemination. Complete study data were provided to users in countries around the world and, as we noted above, re-analyzed to answer new questions about the cognitive domain during which they were collected or re-purposed to address new and promising lines of thought. But when funding lapsed, critical curation and computer systems personnel were lost, and this valuable archive has since struggled.
The issue of database sustainability is of such concern that the International Neuroinformatics Coordinating Facility (INCF) (Bjaalie and Grillner 2007) recently organized an INCF Workshop on Neuroscience Database Sustainability (Van Horn and van Pelt 2008). The goal of the workshop was to deliberate issues related with sustainability of neuroscience databases, to identify problems, to discuss solutions or approaches to these problems, and to formulate recommendations to the INCF. The recommendations of meeting participants included greater transparency into the contents of databases, enhancement of the tools needed to explore the data collections, development of ways to leverage databases for meta- and mega-analyses, and the interoperability of databases with each other.
Lessons Learned in Database Development
The intentions and processes behind the creation of databases have been varied and the lessons learned in their development are important to consider for how new databases might be designed de novo or how existing resources might be extended. While there are clear sociological concerns about the impact of databases (Barinaga 2003), we believe that with careful consideration of several elements some of these concerns can be mitigated. Firstly, despite interpretive value applied to summary results from neuroimaging studies, the raw data provides the opportunity for reprocessing as new methods are developed. One of the first in the pioneering group of databases emerging during the Decade of the Brain, BrainMap (Fox, Mikiten et al. 1994) was designed with a focus for meta-analysis but not data mining from the image data themselves (Fox, Laird et al. 2005). While invaluable for obtaining a meta-analytic assessment of local maxima results from the published literature and the study factors that influence them (see discussion above), users are not able to re-apply uniform data processing workflows or apply alternative approaches to the raw data. Secondly, one must keep in mind the constituency of the data archive and who is expected to benefit as a result. Under XNAT, for example, the constituent is the individual laboratory, in which there is interest in better organization of local data collection activities. In contrast, the fMRI Data Center effort sought to serve the community more generally by providing users with complete, organized, and packaged data sets from published studies for them to conduct new analyses or provide alternative interpretations. Such a model can be successful for data dissemination, but the individual contributing their data may not see a return on their investment until their data has been re-used successfully several times in new peer-reviewed publications. Thirdly, focus on a specific set of achievable deliverables with maximal utility. The BIRN project has been enormously successful in coordinating effort across collaborating centers with ambitious goals for databasing and tool delivery. Trying to solve too many problems, however, may result in overly ambitious expectations at the expense of consortium productivity. Finally, look for general purpose solutions. The IDA is simply one application but one that has many instances applied to a number of different large-scale disease-oriented projects. While each sub-project may remain contained in size, with its own specific needs, the collective IDA database gets richer with the increasing addition of more projects. Such lessons were learned conjointly with the growth of the internet as a major means for collaboration and information availability. But we now know better the potential, as well as the limitations, of the internet and what these may mean for databases in terms of what data should be made available, how tools for databases use the web to their advantage without becoming enslaved by it, and what user expectations are for interacting with data. On this basis, we suggest that it is time for the field to seriously revisit the notion of neuroscience databases and their potential for the community.
Is it Time to Re-Prioritize Neuroimaging Databases?
In this article, we have reviewed many of the ways in which neuroimaging databases have been designed and how these resources have been used by others. Despite considerable attention during the 1990’s through the first several years of the 21st century, neuroimaging databases have not been fully adopted by the community in the way many proponents had anticipated. In part, this may be due to lingering sociological concerns or to having no clear message about the role of these resources from leading neuroimaging organizations. At one time the OHBM had formed a committee on neuroinformatics whose responsibilities included supporting database development, content quality and meta-data description, accessibility, standards, and community interactions (Governing Council of the Organization for Human Brain Mapping 2001). However, in the time since these interests were articulated and the committee formed, the organization has been mute on its position concerning databases and their advantages and caveats for the community. Yet, societies like the OHBM are in an important position to raise awareness of useful data resources, validate leading neuroimaging databases, promote deposition of data into them, and encourage their use for methods development and in education. Given the developments of the past several years in the realm of neuroscience databases, as reviewed here, now may be a good time for the leading organization for brain mapping to re-visit neuroimaging databases. In addition, recent activities in US biomedical research funding under the American Recovery and Reinvestment Act (http://www.nih.gov/recovery) seek to support not only new research and but also major infrastructural projects, including construction of new imaging centers. One can expect that, in the next two years, neuroimaging research in the US alone will be significantly increased and with it the production of large amounts of new data. A revived OHBM informatics committee could provide guidance standards for new imaging studies and minimal information requirements necessary for published reports (see, for example, Poldrack, Fletcher et al. 2008), as well as serve to evaluate and endorse neuroimaging databases that meet strict archival and curatorial standards. The committee could also carry-on what began under the FIAC competition and encourage contestants to use data from openly available databases to validate, optimize, and justify their proposed computational approaches. With a concerted effort, the OHBM can be instrumental in promoting data archives as the indispensible resources for science and education that they are.
Conclusions
Large-scale neuroscientific archival efforts have now begun to produce significant scientific rewards for cellular and cognitive neuroscience, and most notably, brain mapping. Other databases, too, hold great promise for linking images of brain structure and activity with other useful biological information (e.g. GenBank, GENSAT). The involvement of brain researchers as well as multiple scientific communities in examining published brain imaging data must be welcomed and encouraged as this will strengthen and improve the inferences and conclusions that can be made from these data. As a result of these infrastructural and data resources, novel research, hypotheses and education using existing data can reach across scientific disciplines—engaging workers from other fields to apply sophisticated new tools for data analysis and integration. The human scale of these projects is not insignificant, however, often requiring a dedicated curatorial staff to manage study deposition and to keep computer systems operational. The example of the LONI IDA is but one successful effort showing how databases can benefit the field. The examples provided in Table 1 clearly illustrate that many different models exist that can satisfy the unique needs of specialized neuroimaging domains. No one-size-fits-all solution exists, despite what some might contend, and nor should it -- having a range of databasing approaches represents the health of the field and the interest in exploring alternative solutions. However, those archives which have a demonstrated long-term commitment to detailed neuroimaging curation, that have gained the confidence of the community, and generated are likely to be those that are the most successful examples. But, despite this healthy intellectual effort to construct useful and trustworthy neuroimaging data archives, only through a sustained national and international effort will the vision of using, mining, analyzing, and synthesizing the vast amounts of data being obtained by these rapidly advancing technologies be realized. Now is the time for the neuroimaging community and its representative organization to re-prioritize databasing efforts and take stock in their value for neuroimaging science.
Acknowledgments
The authors express their gratitude to our colleagues in the Laboratory of Neuro Imaging (LONI) and those collaborators at our partner laboratories who utilize LONI database and computational resources. This article was supported by an NIMH P41 grant (5 P41 RR013642) to AWT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott A. Neuroscientist: my data published without authorization are ‘misleading’: Max Planck researchers charged with misusing data. Nature. 2008;454:6–7. doi: 10.1038/454006a. [DOI] [PubMed] [Google Scholar]
- Aizenstein HJ, Clark KA, et al. The BOLD Hemodynamic Response in Healthy Aging. J Cogn Neurosci. 2004;16(5):786–793. doi: 10.1162/089892904970681. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amari S, Beltrame F, et al. Neuroinformatics: the integration of shared databases and tools towards integrative neuroscience. J Integr Neurosci. 2002;1(2):117–28. doi: 10.1142/s0219635202000128. [DOI] [PubMed] [Google Scholar]
- Arzberger P, Schroeder P, et al. Science and government. An international framework to promote access to data. Science. 2004;303(5665):1777–8. doi: 10.1126/science.1095958. [DOI] [PubMed] [Google Scholar]
- Bandettini P. Functional MRI today. International Journal of Psychophysiology. 2007;63:138–145. doi: 10.1016/j.ijpsycho.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Barinaga M. Neuroimaging. Still debated, brain image archives are catching on. Science. 2003;300(5616):43–5. doi: 10.1126/science.300.5616.43. [DOI] [PubMed] [Google Scholar]
- Beaulieu A. Voxels in the brain: neuroscience, informatics and changing notions of objectivity. Soc Stud Sci. 2001;31(5):635–80. doi: 10.1177/030631201031005001. [DOI] [PubMed] [Google Scholar]
- Bidgood WD, Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc. 1997;4(3):199–212. doi: 10.1136/jamia.1997.0040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff-Grethe A, Fischl B, et al. A Technique for the Deidentification of Structural Brain MR Images. Human Brain Mapping. 2004 doi: 10.1002/hbm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjaalie JG, Grillner S. Global neuroinformatics: the International Neuroinformatics Coordinating Facility. J Neurosci. 2007;27(14):3613–5. doi: 10.1523/JNEUROSCI.0558-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden DM, Dubach MF. NeuroNames 2002. Neuroinformatics. 2003;1(1):43–59. doi: 10.1385/NI:1:1:043. [DOI] [PubMed] [Google Scholar]
- Boyes RG, Gunter JL, et al. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage. 2008;39(4):1752–62. doi: 10.1016/j.neuroimage.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Ingham RJ, Ingham JC, Laird AR, Fox PT. Stuttered and fluent speech production: an ALE meta-analysis of functional neuroimaging studies. Hum Brain Mapp. 2005;25(1):105–17. doi: 10.1002/hbm.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JM, Church JA, et al. White matter lesions are prevalent but differentially related with cognition in aging and early Alzheimer disease. Arch Neurol. 2005;62(12):1870–6. doi: 10.1001/archneur.62.12.1870. [DOI] [PubMed] [Google Scholar]
- Carlson TA, Schrater P, He S. Patterns of Activity in the Categorical Representations of Objects. J Cogn Neurosci. 2003;15(5):704–717. doi: 10.1162/089892903322307429. [DOI] [PubMed] [Google Scholar]
- Chen WP, Samuraki M, et al. Effect of sample size for normal database on diagnostic performance of brain FDG PET for the detection of Alzheimer’s disease using automated image analysis. Nucl Med Commun. 2008;29(3):270–6. doi: 10.1097/MNM.0b013e3282f3fa76. [DOI] [PubMed] [Google Scholar]
- Chou YY, Lepore N, et al. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. Neuroimage. 2008;40(2):615–30. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JL, Miller BL, Christensen DD, Cherry D. Creativity and dementia: emerging diagnostic and treatment methods for Alzheimer’s disease. CNS Spectr. 2008;13(2 Suppl 2):1–20. quiz 22. [PubMed] [Google Scholar]
- Davatzikos C. Spatial normalization of 3D brain images using deformable models. J Comput Assist Tomogr. 1996;20(4):656–65. doi: 10.1097/00004728-199607000-00031. [DOI] [PubMed] [Google Scholar]
- Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput Vis Image Underst. 1997;66(2):207–22. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- Editorial. Whose scans are they anyway? Nature. 2000;406:443. doi: 10.1038/35020214. [DOI] [PubMed] [Google Scholar]
- Editorial. Proceedings of the Functional Imaging Analysis Contest (FIAC) Workshop held at the 11th Annual Meeting of the Organization for Human Brain Mapping. 2005. Toronto, Canada. Hum Brain Mapp. 2006;27(5):351–470. [PubMed] [Google Scholar]
- Evans AC. The NIH MRI study of normal brain development. Neuroimage. 2006;30(1):184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Fissell K. Workflow-based approaches to neuroimaging analysis. Methods Mol Biol. 2007;401:235–66. doi: 10.1007/978-1-59745-520-6_14. [DOI] [PubMed] [Google Scholar]
- Fissell K, Tseytlin E, et al. Fiswidgets: a graphical computing environment for neuroimaging analysis. Neuroinformatics. 2003;1(1):111–25. doi: 10.1385/ni:1:1:111. [DOI] [PubMed] [Google Scholar]
- Fletcher PT, Powell S, Foster NL, Joshi SC. Quantifying metabolic asymmetry modulo structure in Alzheimer’s disease. Inf Process Med Imaging. 2007;20:446–57. doi: 10.1007/978-3-540-73273-0_37. [DOI] [PubMed] [Google Scholar]
- Fox P, Lancaster J. Mapping context and content: the BrainMap model. Nature Reviews Neuroscience. 2002;3(April):319–321. doi: 10.1038/nrn789. [DOI] [PubMed] [Google Scholar]
- Fox PT, Bullmore E, Bandettini PA, Lancaster JL. Protecting peer review: Correspondence chronology and ethical analysis regarding Logothetis vs. Shmuel and Leopold. Hum Brain Mapp. 2008 doi: 10.1002/hbm.20682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Laird AR, et al. BrainMap taxonomy of experimental design: description and evaluation. Hum Brain Mapp. 2005;25(1):185–98. doi: 10.1002/hbm.20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PT, Mikiten S, Davis G, Lancaster J. BrainMap: A database of human function brain mapping. In: Thatcher RW, Hallett M, Zeffiro T, John ER, Heurta M, editors. Functional Neuroimaging Technical Foundations. San Diego: Academic Press; 1994. pp. 95–105. [Google Scholar]
- Gogtay N, Giedd JN, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101(21):8174–9. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Governing Council of the Organization for Human Brain Mapping. Neuroimaging Databases. Science. 2001;292:1673–1676. doi: 10.1126/science.1061041. [DOI] [PubMed] [Google Scholar]
- Graeber MB. Twenty-first century brain banking: at the crossroads. Acta Neuropathol. 2008;115(5):493–6. doi: 10.1007/s00401-008-0363-5. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grethe JS, Baru C, et al. Biomedical informatics research network: building a national collaboratory to hasten the derivation of new understanding and treatment of disease. Stud Health Technol Inform. 2005;112:100–9. [PubMed] [Google Scholar]
- Gutmark R, Halsted MJ, Perry L, Gold G. Use of computer databases to reduce radiograph reading errors. J Am Coll Radiol. 2007;4(1):65–8. doi: 10.1016/j.jacr.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Haroutunian V, Pickett J. Autism brain tissue banking. Brain Pathol. 2007;17(4):412–21. doi: 10.1111/j.1750-3639.2007.00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson U, Jeremy J, Wilde M, Nusbaum H, Small S. Improving the analysis, storage and sharing of neuroimaging data using relational databases and distributed computing. NeuroImage. 2008;39(693–706) doi: 10.1016/j.neuroimage.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Jones J, Sheth R, Seidenberg M. Cognitive and magnetic resonance volumetric abnormalities in new-onset pediatric epilepsy. Semin Pediatr Neurol. 2007;14(4):173–80. doi: 10.1016/j.spen.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis AE, Newhart M, et al. Anatomy of spatial attention: insights from perfusion imaging and hemispatial neglect in acute stroke. J Neurosci. 2005;25(12):3161–7. doi: 10.1523/JNEUROSCI.4468-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Tsang TM, Tabrizi SJ. The application of NMR-based metabonomics in neurological disorders. NeuroRx. 2006;3(3):358–72. doi: 10.1016/j.nurx.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L. Systems biology: integrating technology, biology, and computation. Mech Ageing Dev. 2003;124(1):9–16. doi: 10.1016/s0047-6374(02)00164-1. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang J, et al. White and gray matter development in human fetal, newborn and pediatric brains. NeuroImage. 2006;33(1):27–38. doi: 10.1016/j.neuroimage.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Illes J, Kirschen MP, et al. Ethics. Incidental findings in brain imaging research. Science. 2006;311(5762):783–4. doi: 10.1126/science.1124665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishai A, Ungerleider LG, Martin A, Haxby JV. The representation of objects in the human occipital and temporal cortex. J Cogn Neurosci. 2000;12(Suppl 2):35–51. doi: 10.1162/089892900564055. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelacic S, de Regt D, Weinberger E. Interactive digital MR atlas of the pediatric brain. Radiographics. 2006;26(2):497–501. doi: 10.1148/rg.262055009. [DOI] [PubMed] [Google Scholar]
- Kanaan RA, Kim JS, et al. Diffusion Tensor Imaging in Schizophrenia. Biol Psychiatry. 2005 doi: 10.1016/j.biopsych.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Keator DB, Gadde S, Grethe JS, Taylor DV, Potkin SG. A general XML schema and SPM toolbox for storage of neuro-imaging results and anatomical labels. Neuroinformatics. 2006;4(2):199–212. doi: 10.1385/ni:4:2:199. [DOI] [PubMed] [Google Scholar]
- Keator DB, Ozyurt B, Wei D, Gadde S, Potkin SG, Brown G, Grethe J Morphometry BIRN, Function BIRN. A General and Extensible Multi-Site Database and XML based Informatics System for the Storage, Retrieval, Transport, and Maintenance of Human Brain Imaging and Clinical Data. Human Brain Mapping; Florence, Italy. 2006. [Google Scholar]
- Koslow SH. Should the neuroscience community make a paradigm shift to sharing primary data? Nature Neuroscience. 2000;3(4):863–865. doi: 10.1038/78760. [DOI] [PubMed] [Google Scholar]
- Koslow SH. Opinion: Sharing primary data: a threat or asset to discovery? Nat Rev Neurosci. 2002;3(4):311–3. doi: 10.1038/nrn787. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Goebel R, Bandettini P. Information-based functional brain mapping. Proc Natl Acad Sci U S A. 2006;103(10):3863–8. doi: 10.1073/pnas.0600244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulynych J. Legal and ethical issues in neuroimaging research: human subjects protection, medical privacy, and the public communication of research results. Brain Cogn. 2002;50(3):345–57. doi: 10.1016/s0278-2626(02)00518-3. [DOI] [PubMed] [Google Scholar]
- Laird AR, Lancaster JL, Fox PT. BrainMap: the social evolution of a human brain mapping database. Neuroinformatics. 2005;3(1):65–78. doi: 10.1385/ni:3:1:065. [DOI] [PubMed] [Google Scholar]
- Lanctot KL, Hussey DF, et al. A positron emission tomography study of 5-hydroxytryptamine-1A receptors in Alzheimer disease. Am J Geriatr Psychiatry. 2007;15(10):888–98. doi: 10.1097/JGP.0b013e3180488325. [DOI] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31(2):627–40. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore N, Brun CA, et al. Multivariate statistics of the Jacobian matrices in tensor based morphometry and their application to HIV/AIDS. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2006;9(Pt 1):191–8. doi: 10.1007/11866565_24. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Salamon N, et al. Reduced neocortical thickness and complexity mapped in mesial temporal lobe epilepsy with hippocampal sclerosis. Cereb Cortex. 2007;17(9):2007–18. doi: 10.1093/cercor/bhl109. [DOI] [PubMed] [Google Scholar]
- Liou M, Su HR, et al. A method for generating reproducible evidence in fMRI studies. Neuroimage. 2006;29(2):383–95. doi: 10.1016/j.neuroimage.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Liu L, Meier D, et al. Multiple sclerosis medical image analysis and information management. Neuroimaging. 2005;15(4 Suppl):103S–117S. doi: 10.1177/1051228405282864. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Boline J, Toga AW. Brain atlases and neuroanatomic imaging. Methods Mol Biol. 2007;401:183–94. doi: 10.1007/978-1-59745-520-6_11. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Payan A, Dinov I, Van Horn JD, Toga AW. Neuroimaging Data Provenance Using the LONI Pipeline Workflow Environment. Provenance and Annotation of Data International Provenance and Annotation Workshop, IPAW; 2008; Salt Lake City, UT: University of Utah; 2008. [Google Scholar]
- Mackenzie-Graham AJ, Van Horn JD, Woods RP, Crawford KL, Toga AW. Provenance in neuroimaging. Neuroimage. 2008 doi: 10.1016/j.neuroimage.2008.04.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Maxim V, Sendur L, et al. Fractional Gaussian noise, functional MRI and Alzheimer’s disease. Neuroimage. 2005;25(1):141–58. doi: 10.1016/j.neuroimage.2004.10.044. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356(1412):1293–322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga AW, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) Neuroimage. 1995;2(2):89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Price C, Noppeney U, Friston K. A dynamic causal modelling study on category effects: Bottom-up or top-down mediation? Journal of Cognitive Neuroscience. 2003;15(7):925–934. doi: 10.1162/089892903770007317. [DOI] [PubMed] [Google Scholar]
- Moorhead TW, Harris JM, et al. Automated computation of the Gyrification Index in prefrontal lobes: methods and comparison with manual implementation. Neuroimage. 2006;31(4):1560–6. doi: 10.1016/j.neuroimage.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Moreau L, Ludäscher B, et al. Special Issue: The First Provenance Challenge. Concurrency and Computation: Practice & Experience 00 2007 [Google Scholar]
- Mori S, Oishi K, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15(4):869–77. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr K, Bilder R, et al. Asymmetries of cortical shape: Effects of handedness, sex and schizophrenia. Neuroimage. 2007;34(3):939–948. doi: 10.1016/j.neuroimage.2006.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Thompson PM, et al. Mapping morphology of the corpus callosum in schizophrenia. Cereb Cortex. 2000;10(1):40–9. doi: 10.1093/cercor/10.1.40. [DOI] [PubMed] [Google Scholar]
- Neu SC, Toga AW. Automatic Localization of Anatomical Point Landmarks for Brain Image Processing Algorithms. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olabarriaga SD, Nederveen AJ, Snel JG, Belleman RG. Towards a virtual laboratory for FMRI data management and analysis. Stud Health Technol Inform. 2006;120:43–54. [PubMed] [Google Scholar]
- Petacchi A, Laird AR, Fox PT, Bower JM. Cerebellum and auditory function: an ALE meta-analysis of functional neuroimaging studies. Hum Brain Mapp. 2005;25(1):118–28. doi: 10.1002/hbm.20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoebe Chen YP, Chen F. Identifying targets for drug discovery using bioinformatics. Expert Opin Ther Targets. 2008;12(4):383–9. doi: 10.1517/14728222.12.4.383. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Fletcher PC, et al. Guidelines for reporting an fMRI study. Neuroimage. 2008;40(2):409–14. doi: 10.1016/j.neuroimage.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov AV, Hertzenberg X, et al. Unobtrusive integration of data management with FMRI analysis. Neuroinformatics. 2007;5(1):3–10. doi: 10.1385/ni:5:1:3. [DOI] [PubMed] [Google Scholar]
- Raichle ME, Gusnard DA. Appraising the brain’s energy budget. Proc Natl Acad Sci U S A. 2002;99(16):10237–9. doi: 10.1073/pnas.172399499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex D, Ma J, Toga AW. The LONI Pipeline Processing Environment. Neuroimage. 2003;19(3):1033–48. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Roland PE, Zilles K. Brain atlases--a new research tool. Trends Neurosci. 1994;17(11):458–67. doi: 10.1016/0166-2236(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Shan ZY, Parra C, et al. A digital pediatric brain structure atlas from T1-weighted MR images. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2006;9(Pt 2):332–9. doi: 10.1007/11866763_41. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, et al. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39(3):1064–80. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicotte NL, Kern KC, et al. Regional hippocampal atrophy in multiple sclerosis. Brain. 2008;131(Pt 4):1134–41. doi: 10.1093/brain/awn030. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Trauner DA, Gamst A, Jernigan TL. Development of cortical and subcortical brain structures in childhood and adolescence: a structural MRI study. Dev Med Child Neurol. 2002;44(1):4–16. doi: 10.1017/s0012162201001591. [DOI] [PubMed] [Google Scholar]
- Stef-Praun T, Clifford B, et al. Accelerating Medical Research using the Swift Workflow System. Stud Health Technol Inform. 2007;126:207–16. [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci. 2003;23(3):994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Apostolova LG. Computational anatomical methods as applied to ageing and dementia. Br J Radiol. 2007;80(Spec No 2):S78–91. doi: 10.1259/BJR/20005470. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Hum Brain Mapp. 2000;9(2):81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga A, Rex DE, Ma J. A graphical interoperable processing pipeline. Organization for Human Brain Mapping Annual Meeting; Brighton, England, Academic Press. 2001. [Google Scholar]
- Toga A, Thompson P. Maps of the brain. Anat Rec. 2001;265(2):37–53. doi: 10.1002/ar.1057. [DOI] [PubMed] [Google Scholar]
- Toga AW. The Laboratory of Neuro Imaging: what it is, why it is, and how it came to be. IEEE Trans Med Imaging. 2002;21(11):1333–43. doi: 10.1109/TMI.2002.806432. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. New approaches in brain morphometry. Am J Geriatr Psychiatry. 2002;10(1):13–23. [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Mega MS, Narr KL, Blanton RE. Probabilistic approaches for atlasing normal and disease-specific brain variability. Anat Embryol (Berl) 2001;204(4):267–82. doi: 10.1007/s004290100198. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Mori S, Amunts K, Zilles K. Towards multimodal atlases of the human brain. Nat Rev Neurosci. 2006;7(12):952–66. doi: 10.1038/nrn2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Narr KL, et al. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans Med Imaging. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. Windows on the brain: the emerging role of atlases and databases in neuroscience. Curr Opin Neurobiol. 2002;12(5):574–9. doi: 10.1016/s0959-4388(02)00361-6. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005 doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Ball CA. Domain-Specific Data Sharing in Neuroscience: What Do We Have to Learn from Each Other? Neuroinformatics. 2008 doi: 10.1007/s12021-008-9019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Gazzaniga MS. Databasing fMRI Studies - Toward a ‘Discovery Science’ of Brain Function. Nature Reviews Neuroscience. 2002;3(4):314–318. doi: 10.1038/nrn788. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Gazzaniga MS. Maximizing Information Content in shared Neuroimaging Studies of Cognitive Function. In: Koslow SH, Subramanian A, editors. Databasing the Brain: From Data to Knowledge. New York: John Wiley and Sons; 2005. [Google Scholar]
- Van Horn JD, Grafton ST, Rockmore D, Gazzaniga MS. Sharing neuroimaging studies of human cognition. Nat Neurosci. 2004;7(5):473–481. doi: 10.1038/nn1231. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Grethe JS, et al. The Functional Magnetic Resonance Imaging Data Center (fMRIDC): The Challenges and Rewards of Large-Scale Databasing of Neuroimaging Studies. Philos Trans R Soc Lond B Biol Sci. 2001;356:1323–1339. doi: 10.1098/rstb.2001.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Ishai A. Mapping the human brain: new insights from FMRI data sharing. Neuroinformatics. 2007;5(3):146–53. doi: 10.1007/s12021-007-0011-6. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, van Pelt J. 1st INCF Workshop on Sustainability of Neuroscience Databases. Nature Precedings. 2008 < http://dx.doi.org/10.1038/npre.2008.1983.1>.
- Van Horn JD, Woodward JB, et al. The fMRI Data Center: Software Tools for Neuroimaging Data Management, Inspection, and Sharing. In: Kotter R, editor. A Practical Guide to Neuroscience Databases and Associated Tools. Amsterdam: Kluwer; 2002. pp. 221–235. [Google Scholar]
- Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PC, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230(1):77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- Weaver JB, Healy DM, Jr, Periaswamy S, Kostelec PJ. Elastic image registration using correlations. J Digit Imaging. 1998;11(3 Suppl 1):59–65. doi: 10.1007/BF03168261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt ST, Laird AR, Meyerand ME. Functional neuroimaging correlates of finger-tapping task variations: an ALE meta-analysis. Neuroimage. 2008;42(1):343–56. doi: 10.1016/j.neuroimage.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HM, Huang SC, et al. Subcortical white matter metabolic changes remote from focal hemorrhagic lesions suggest diffuse injury after human traumatic brain injury. Neurosurgery. 2004;55(6):1306–15. doi: 10.1227/01.neu.0000143028.08719.42. discussio 1316–7. [DOI] [PubMed] [Google Scholar]
- Yanovsky I, Thompson PM, et al. Validating Unbiased Registration on Longitudinal MRI Scans from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; Paris, France, IEEE. 2008. [Google Scholar]
- Zhuang AH, Valentino DJ, Toga AW. Skull-stripping magnetic resonance brain images using a model-based level set. Neuroimage. 2006;32(1):79–92. doi: 10.1016/j.neuroimage.2006.03.019. [DOI] [PubMed] [Google Scholar]