Abstract
Cell lineage commitment and differentiation are governed by a complex gene regulatory network. Disruption of these processes by inappropriate regulatory signals and by mutational rewiring of the network can lead to tumorigenesis. Cancer cells often exhibit immature or embryonic traits and dysregulated developmental genes can act as oncogenes. However, the prevailing paradigm of somatic evolution and multi-step tumorigenesis, while useful in many instances, offers no logically coherent reason for why oncogenesis recapitulates ontogenesis. The formal concept of “cancer attractors”, derived from an integrative, complex systems approach to gene regulatory network may provide a natural explanation. Here we present the theory of attractors in gene network dynamics and review the concept of cell types as attractors. We argue that cancer cells are trapped in abnormal attractors and discuss this concept in the light of recent ideas in cancer biology, including cancer genomics and cancer stem cells, as well as the implications for differentiation therapy.
1. INTRODUCTION
Pathologists have long suggested, based on cell morphology, that malignant tumors represent an aberrant form of cellular development [1]. The degree of immaturity of cancer cell phenotype indeed roughly scales with malignancy. That tumor cells express genetic programs of immature or of embryonic cells has now consistently been confirmed at the molecular level by genome-wide analysis of gene expression profiles (‘transcriptomes”) of tumor tissues and cancer cells [2–4]. Cellular programs that play essential roles during development overlap with those traits of tumor cells that have been defined as “hallmarks of cancer” [5], including rapid proliferation and clonal expansion, migration and invasion, stimulation of angiogenesis, etc. The prevailing paradigm that these hallmarks are acquired in a multistep fashion [6], driven by somatic evolution [7, 8], however, fails to account for the consistency of the embryonic cell phenotype. Moreover, the old observation that a population of cancer cells is heterogeneous, continuously producing a variety of distinct cell phenotypes, also defies the concept of clonal expansion of a mutated cell [7, 8]. Instead, it has stimulated renewed attention to the idea of cancer as a developmental disease, with a strong non-genetic component, as best epitomized in the revived interest in epithelial-mesenchymal transition [9, 10] or the “cancer stem cell” hypothesis [11, 12]. Furthermore, the list of developmental genes whose mutational (in)activation contributes to cancer continues to grow [13]. Thus, after decades of characterizing mutated signaling pathways that act as proximate cause in tumorigenesis, the larger picture that tumors are a consequence of a disrupted regulation of normal cell and tissue development is re-emerging [14–16].
The recent technological advances in genomics has led to a deluge of gene expression profiles and whole-genome sequences of tumors, fostered for instance by the Cancer Genome Anatomy Project (http://www.ncbi.nlm.nih.gov/ncicgap) [17]. But if in making sense of the data, we aspire to go beyond just embellishing the systematic enumeration of oncogenic pathways with functional annotations pointing to “development”, then we will need to see such high-throughput molecular characterization through the combined lens of systems and developmental biology. It is also not sufficient to extend the notion of oncogenic mutations to covalent chromatin marks and DNA methylation [16] or to introduce ad hoc, qualitative models [18, 19]. One concrete way to begin to embrace such a integrative view that considers development is to ask: Why does oncogenesis mimic ontogenesis? How can a handful of random genetic mutations and subsequent selection so rapidly and consistently generate the molecular organization within tumors that activates the unfathomably sophisticated developmental phenotype? In what precise sense is cancer really a “derailment of development”?
With the integrative spirit of systems biology and recent advances in understanding how gene regulatory networks (GRN) govern cell fate switching and differentiation, the time is ripe for discussing a unifying formal framework that connects tumorigenesis with development, and thereby, explains the seemingly spontaneous order and patterns in tumor formation as an inevitable consequence of errors in the machinery that so robustly produces the diversity of cell types during metazoan development. Therefore, we present here the idea of “cancer attractors”, which was first suggested by Stuart Kauffman 40 years ago [20] as a corollary of an encompassing theory from complex systems studies that has only recently begun to find experimental support thanks to genomic technologies. It explains how a genome-spanning GRN affords in one single genome the capacity to produce a diversity of stable, discretely distinct cell phenotypes in the metazoan body that we recognize as “cell types” [21]. In this review we will explain the formal concept of attractors in a permissively simplifying language and without taking refuge in abstract mathematical equations. We then discuss its wide-reaching implications for cellular development and tumorigenesis and demonstrate the link between these two. The integrative approach will also embrace the ideas of tumor as a tissue disease [15] with distributed genetic and non-genetic abnormalies [16, 19, 22] and involving “cancer stem cells” [11, 12] and the non-neoplastic tumor stroma [23–25].
2. TOWARDS AN INTEGRATIVE VIEW: CANCER CELLS AS ABNORMAL CELL TYPES
2.1. A central paradox: Explaining phenotype differences by mutations
The common epistemological habit of modern molecular biology is to reduce an observed phenotype or function to a molecular entity, such as a gene, protein or pathway, which have become the embodiment of causation in biology. The quest for a molecular basis of tumorigenesis in this tradition of “proximate explanation” has led to the model of tumorigenesis as a linear, multi-step, evolutionary process [5, 6, 26].
This reductionist–mechanistic and Darwinist view, supported by countless examples of “plausible mutations”, obviates the need for a more encompassing and integrative view for examining tumorigenesis in the broader context of development. Instead, any novel phenotypic feature (“hall mark”) that a malignant cell acquires is conveniently explained by a specific lasting molecular alteration: a genetic mutation or more recently, an “epigenetic” change, such as a histone modification or DNA methylation.
But here is a rarely articulated paradox: While one will automatically seek to determine the gene that is mutated to explain an incremental malignant trait nobody will doubt that normal cells as distinct as a stem cell, a mature neuron, a blood cell or an epithelial cell, all share the very same genome. No mutations are invoked to explain the vastly different phenotypes and their inheritance within a lineage. This opens the first fundamental question: how can the same set of genetic instructions produce a variety of discrete, persistent (non-genetically inherited) cell phenotypes? (As discussed below, the covalent “epigenetic” chromatin marks invoked in cell-type specific gene expression are actually another layer of proximate causation and strictly speaking, not truly explanatory for the establishment and stability of specific gene expression patterns since they are reversible and lack locus specificity [27–29]).
We suggest here to view tumors from an opposite vantage point: consider each type of tumor cell as a cell type in its own right whose phenotype would result from a “functional” error in the complex cell developmental process that has evolved to allow the very same genome to produce a broad diversity of discretely distinct cell phenotypes. This mental picture will mitigate our dependence on genetic mutations in explaining the tumor phenotype.
2.2. Genome-wide studies of cancer cells
Indeed, the recently available DNA sequences of tumor genomes shed light on the hidden cracks in the paradigm of genetic mutations as prima causa of cancer. Cancer genome sequencing projects [30–35] provide a general estimate of the extent of genetic alterations. For instance, tumor cell genomes contain approximately ~10–200 mutations not found in the wild type tissue – with a large variability between tumor types. (The majority of the genetic alterations found in tumor cells are “passenger” rather than “driver” mutations which contribute to tumorigenesis and progression). Tumors of the same type typically show no congruently overlapping but quite differing sets of driver mutations. Only a few mutations are found in a majority of tumors whereas the majority of mutations are observed in less than 5% of tumors [36]. This lack of overlap has been explained away by arguing that driver mutations affect genes which belong to the same functional (gene ontology) class and hence, need not be identical although the functional classes (gene ontology annotation) are rather broadly defined [37].
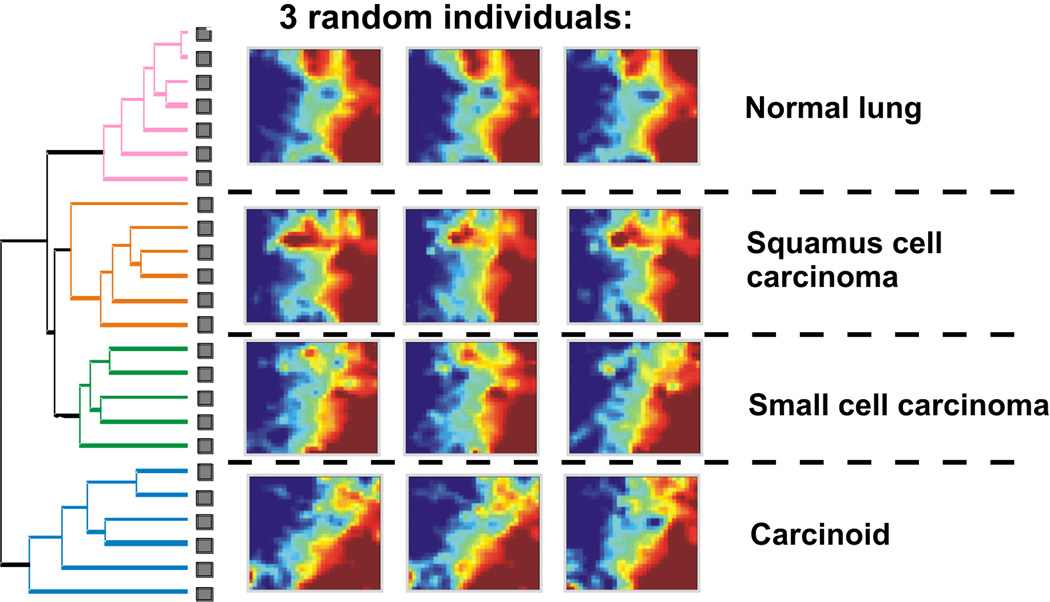
In moving beyond genomic alterations, the abundance of transcriptome (gene expression profiles) of tumor tissues [38, 39] now offers a view on emergent patterns that will help to establish the link to development. If instead of using expression profiling just to identify individual “tumor specific” genes or prognostic subtypes of cancer [40–43] one takes a step back to catch a “holistic view” of the entire transcriptome as a whole [44], a stunning degree of organization and order of global expression patterns will spring to eye (see Fig. 1). Similarly, cluster analysis of tumor transcriptomes readily and robustly classifies tumors into a small number of discretely distinct groups. This molecular clustering correlates well with the traditional classification of tumor types derived from morphology and biochemical markers. The discrete clusters of pathological transcriptomes are reminiscent of the organization of transcriptomes of normal cell types into groups of related tissues [45, 46]. For instance, despite individual variability, lung cancers can be divided into four clearly and discretely distinct main groups that encompass > 95 % of all pulmonal neoplasia and whose members share highly similar expression profiles [44].
FIGURE 1.
Example of clustering of tumor transcriptomes into discrete, well separated clusters. Left: Dendrogram showing hierarchical clustering of the lung (tumor) tissue sample of 24 individuals. Right: GEDI-maps for three randomly chooses tissue samples, showing the transcriptome patterns. Note the characteristic global pattern for each tumor type. Each map represents a transcriptome, with pixels representing the same genes in each map and their color the expression level. Adapted from ref. [44].
2.3. Cancer cells as a discrete phenotype
Why isn’t there a “continuum” of tumor transcriptomes, given that they do not represent physiological states but result from hundreds of random mutations scattered throughout the genome? Whence the organization into discrete subtypes in a chaotic world of random mutations? Is all order due to converging somatic selection? Why is there order in the realm of the abnormal, chaotic? Is this all due to convergent evolution of the most fit cancer cell? Perhaps the over-crediting of the creative power of natural selection seen in organismal evolution [47, 48] is also creeping into tumor biology. In fact, strikingly, even at an early stage, tumor cells not yet exposed to the formative forces of adaptive evolution exhibit abnormal yet characteristically recurring types of gene expression profiles that can predict metastasis potential [49, 50].
The idea that the tumor cell is an aberrant cell type is most lucidly embodied by small cell-lung cancer which despite the diversity of its etiology and patterns of mutation presents to the pathologist with a distinctively discrete, easily recognizable, characteristic phenotype not found in normal tissue.
The notion of “cancer stem cells” (discussed in Section 7) and their preserved but ineffective ability to produce a variety of more differentiated cells [12] also points to a pathological cell development that affects the machinery of cell type generation. Hence, cancer is likely not simply the product of (somatic) evolution that sculpts the malignant phenotype, starting from a normal cell as a blank slate following the dictate of “survival of the fittest”.
In order to appreciate that cancer cells represent abnormal cell types we need to first understand what is the essence of a cell type and how its development is regulated. How does the genome so reliably produce the exact patterns of expression of ten thousands of genes that define each (normal) cell type?
3. CELL TYPES AS ATTRACTORS
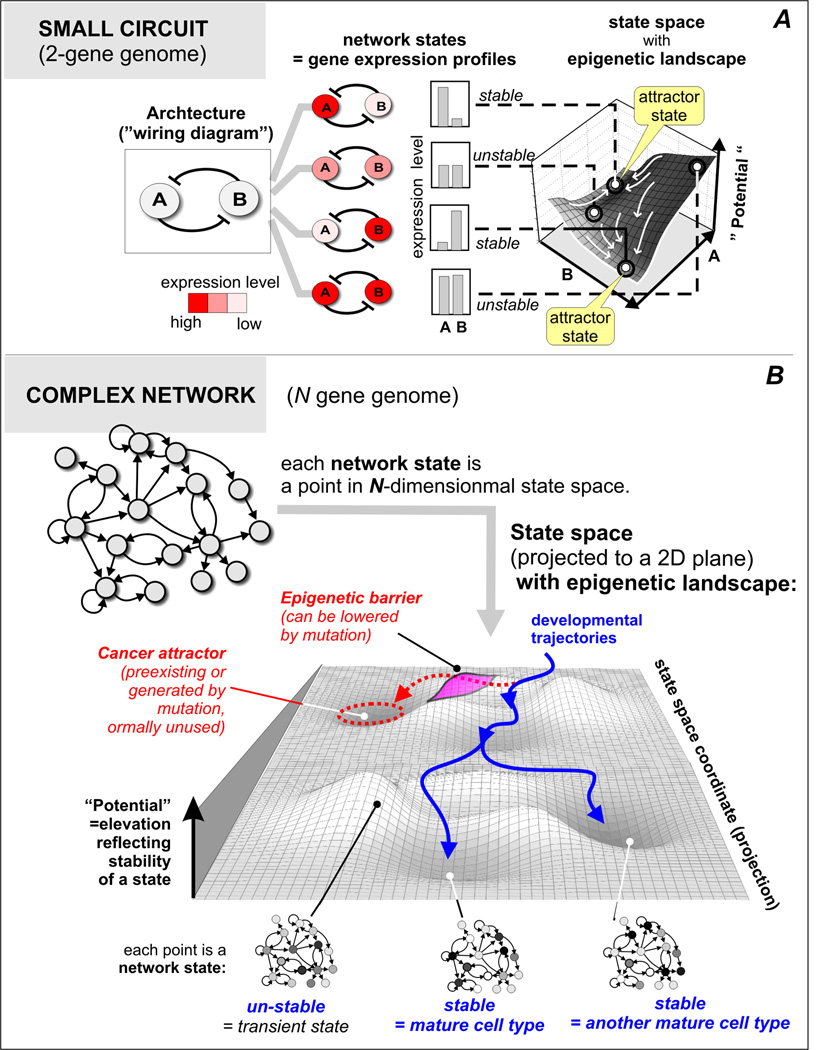
A gene expression pattern reflects the state of a gene regulatory network (= GRN), and as a whole, is dynamic: it develops in time due to the mutual regulation between the genes of each others’ expression and settles down into an “equilibrium state” that complies with the regulatory interactions (reviewed in [21]). The ability of small gene regulatory circuits to produce more than one stable equilibrium state (=stable pattern of expression of all genes in the circuit) has first been proposed by Max Delbruck in 1948 [51], and later, by Jacob and Monod [52] and others to explain differentiation into a multitude of stable phenotypic states (see Primer in Supplementary Material, A, for detailed explanation). In the 1960s Kauffman showed that a complex network of up to hundred thousands of mutually regulating genes can under certain conditions produce hundreds of stable equilibrium states, termed attractors [53, 54]. Kauffman proposed that attractor states correspond to the gene expression profiles associated with each cell type [54, 55]. (Suppl. material, Fig. 2A.)
To understand the essence of an attractor the concept the state space is necessary. The state space of a GRN, as explained in the Suppl. material (A) is the space that contains all theoretically possible gene expression patterns = network states of that GRN. Each point in the (high-dimensional) state space represents one gene expression pattern of the GRN and moves around as the expression patterns change (Fig. 2A, B). The attractor state is a particular point in the state space and has particular properties: as a stable equilibrium state it resembles that of a “lowest energy state” at the bottom of a “potential well” which represents the “basin of attraction”. Thus the attractor state is surrounded by unstable states and is “self-stabilizing”.
FIGURE 2.
From network dynamics to epigenetic landscape and attractors.
A. Pedagogical example of a 2-gene network (mutual inhibition of genes A and B) and associated epigenetic landscape in the two-dimensional state space. Note that each network state maps to one point in the state space (for details see Suppl. Material). B. Epigenetic landscape for a complex network with N genes. Overall slope from back to front represent developmental progression. The landscape is a schematic projection from N to a two-dimensional state space. Normal developmental trajectories (blue solid line) lead to attractors that represent distinct cell types and are shielded from entering the unused “abnormal attractors” (red dashed circle) by epigenetic barriers (pink hill). Mutations can lower this barrier, thus opening access to the unused, attractors that encode an abnormal, immature phenotype = cancer attractor (red dashed arrow). Differentiation therapy aims at perturbing the network state to move back to the normal trajectories of development (blue).
This notion of a potential is captured in Waddington’s “epigenetic landscape” [56] which was introduced as a metaphor in the 1950s to explain the discrete nature of cell types (Fig. A, Suppl. material). According to Waddington, each valley represents a cell type, which in the modern state space formalism correspond to the potential wells with their basins of attraction. At their bottom are the attractor states. Since each point in state space represents a state of the GRN and hence a gene expression pattern, the attractor state defines a stable gene expression pattern. Hills that separate the attractors represent the unstable network states. Thus, attractors encode specific “genetic programs” of the cell “pre-programmed” in the GRN, including those which produce a stable cell type-specific gene expression pattern. Accordingly, a switch between two cell phenotypes is represented by the transition of a cell state between valleys. Such transitions are triggered by regulatory signals, such as cytokines, that change the expression status of a set of genes in a concerted manner or by gene expression noise which produce random fluctuations in the expression of the genes [21, 57].
As it turns out, Waddington’s landscape is more than a metaphor, for it can be formally explained as a generalized potential landscape in state space that represents the global dynamics of a GRN. The basis for the “elevation” at each state space position that gives rise to the picture of a landscape topography is explained in more detail in Suppl. material, A.
The existence of high-dimensional attractor states (Fig. 2B) defined by N=thousands of genes across the genome and their correspondence to particular cell types has recently been experimentally verified. Using microarrays for dynamic gene expression profiling the “attraction” of trajectories from different directions in state space towards a common final state of a differentiated cell, as well as the relaxation back to the bottom of the potential well after local perturbations are both indicative of attractors [58, 59]. Such self-propelled convergence of high-dimensional trajectories (gene expression profile change) is a necessary signature of an attractor.
4. BIOLOGY ON THE EPIGENETIC LANDSCAPE
We shall note that the term “epigenetic” as in Waddington’s “epigenetic landscape” (Fig. A, Suppl. material) used here is distinct from that used by molecular biologists to describe covalent chromatin and DNA modifications (Section 2). It is closer to the physicists notion of an “epigenetic state” [60], a system-level stable state that arises from genetic interactions, which in turn directly reflects Waddington’s original meaning.
4.1. Properties of cell fate regulation
The attractor nature of distinct cell phenotypes, most obviously, the cell types, explains a series of cell behaviors that are not easily accounted for by linear molecular pathways. It explains why cell-type specific genome-wide expression profiles, defined by the values of thousands of variables, are so reliably established during differentiation, as if orchestrated by an invisible hand: The self-organizing and self-stabilizing property of biologically significant gene expression profiles is a natural feature conferred by attractors. Hence, cell-type specific gene expression patterns are robust to noise, re-establishing themselves after small perturbations (=imposed changes of expression levels of individual genes) and can be reached in principle via an almost infinite number of paths [21]. Conversely, they are capable of undergoing drastic quasi-discontinuous transitions to other specific stable expression profiles via genome-wide changes of gene expression. Such transitions occur when cells encounter the proper cell fate regulatory signals that, via branching signal transduction pathways change the expression of a specific set of genes of the network, or due to sufficiently high random fluctuations of gene expression levels. In a simplified picture, attractor transitions underlie the cell phenotype switching during development [21, 57, 61].
The presence of a landscape with preexisting neighboring valleys is well illustrated in the classical experiment of treatment with 5'Azacytidine (5’Aza). This chemical demethylates DNA and thereby affects the expression at gene loci across the genome to impose a genome-wide perturbation. Treatment of a fibroblast population with 5'Aza induces their differentiation into adipocytes, muscle cells and chondrocytes [62] within the very same cultural environment. This observation, repeated in many other cell lineages since, is recapitulated in the model: cells placed on top of a mountain top or at a ‘watershed’ in the epigenetic landscape will roll down into just the few distinct valleys accessible to them, driving the spontaneous separation into discrete fates.
4.2. From pluripotent embryonic cell to the mature cell type
The epigenetic landscape offers an integrative view of development. Pluripotent embryonic stem cells are in a metastable state at the highest point (“summit”) in a landscape. They roll down the landscape as cells multiply and swarm out to fill the state space of the genomic network, driven by gene expression noise and regulatory interactions, and come to rest as they occupy the various valleys and subvalleys in the “lower regions” of the landscape, representing terminal differentiation [21, 56]. The associated inevitable loss of “potential energy” is consistent with the progressive “lineage-restriction” in the course of development. The discernible intermediate states of maturation that correspond to metastable multipotent progenitor cells are represented by small groves located at watersheds, where a cell has to make a binary fate decision to roll further down into either one of the two adjacent valleys.
Experimental and theoretical analysis of bipotent progenitor or stem cells are consistent with this picture [63, 64]. In other words, development is the distribution of cells into a set of (“low energy”) attractors and their balanced occupation by cell populations – much as water draining a landscape fills up a distinct number of lakes in the valleys. Another layer of regulatory network not discussed here, namely, that of cell-cell communication implemented by paracrine, iuxtacrine and humoral interaction may serve to regulate the relative occupancy of neighboring attractors to be commensurate for the formation of tissues by a balanced set of distinct cells. (Mathematically, such cell-cell regulation can be formalized as a modification of the landscape topography that deepens an attractor).
Taken together, the epigenetic landscape is the stage on which the play of cell fate decisions and cell type maturation is choreographed. Its detailed topography must be such as to guide the coordinated production of the appropriate proportions of distinct cell types at the right place and time. Driving this graphical representation further, each of the expanding number of cells, rolling down the landscape during development, has to be channeled to the appropriate valleys and subvalleys without “spilling” over into unphysiological side valleys. It is important to keep in mind, before we move on to discuss cancer attractors, that the epigenetic landscape is more than a metaphor – it is mathematically derived from the dynamics of the gene regulatory network (Suppl. material, A): The GRN of a particular genome maps into one landscape and each geographic position in it represents a unique gene expression profile, i.e., a cell state, in the high-dimensional state space of the genome-wide network. Then, one may speculate, evolution of development fine-tunes the wiring diagram of the genomic network to shape the topography of the epigenetic landscape such as to guarantee a smooth flow of the multiplying cells down into the destined valleys of mature cell types without leaving them behind at inappropriate positions.
5. CANCER AS ATTRACTORS
5.1. Logical and system level justification
If cell types are attractors (Section 3) and cancer cells warrant being viewed as abnormal cell types (Section 2), then cancer cells should also be represented by attractors. But in addition to this logical clause, a global view of the network dynamics offers a formal and more compelling reason to postulate the existence of “cancer attractors”: If a given GRN (say, the human genome) produces a particular epigenetic landscape with valleys and hills then the landscape may be so complex (“rugged”) that, in the implementation of the network, not all attractors are occupied by those cells that represent physiological cell types in the body. The “unused” attractors would be inevitable byproducts of the complex dynamics of the GRN, and the majority of them most likely represent abnormal, non-viable gene expression patterns, perhaps due to “conflicting signals” (see Fig. 2B). The genome may have not evolved to integrate them in the roadmap of development. However, if an unused attractor is associated with a viable proliferative phenotype it could represent a cancer attractor. A first outline of this is simple yet profound theoretical idea was first proposed by Kauffman in 1971 [20], inspired by the statistical properties of large ensembles of boolean networks which for the first time established a relationship between network size and the typical number of attractors.
One first question that the cancer attractor hypothesis raises is: If the cancer phenotype is to be explained by attractors that pre-exist in the epigenetic landscape because of the way the GRN is wired, what then is the role of genomic mutations in the etiology of tumors? The formalism of GRN and the associated epigenetic landscape places mutations in a new perspective not available in the traditional paradigm of linear causative pathways.
5.2. The role of genetic mutations
In the framework of gene network architecture a genetic mutation is an architectural change that removes a node or a connection in the network (loss-of-function mutation), strengthens or adds a network connection (gain of function), or changes the interaction properties of a protein, including its target gene or its interaction modality (e.g., generation of a dominant negative protein by deletion of the activation domain), etc. In short - mutations introduce changes of the wiring diagram of the GRN. Since each network architecture uniquely maps into one particular landscape, mutations will translate into a change in the landscape topography.
But to what extent do mutations reshape the epigenetic landscape? Computer simulations of large boolean networks have shown that individual mutations (affecting one network element = gene at a time) have a relatively mild effect on the overall landscape topography [65]. This may not be surprising given the complexity of a vast web of interactions that can well buffer considerable structural changes and maintain integrated architectural properties, such as global connectivity [66]. With respect to dynamics, while the overall topography of the landscape is maintained following mutation of a single gene, attractor basins can be distorted. Only occasionally, may attractors disappear, merge, or new ones created following mutation [65]. Moreover, routine theoretical analysis of small gene networks modeled as continuous dynamical systems readily reveals that changes in the interaction parameters (as could be introduced by genetic mutations), also have mostly distorting (gradual) effects because of what is called “structural stability” in parameter space of these network [63]: Changes in network interaction parameters typically (=when not encountering bifurcation events) affect the size of the basins of attraction, the height of hills or depths of valleys [67] or they change the relative position of attractor states. These landscape changes would affect the transition probabilities between attractors or shift the nature of the expressed cellular program of an attractor state, respectively. Only occasionally do such parameter changes lead to birth or death of stable attractor states (= bifurcation events) [63].
Given these consequences of mutations on the landscape topography we now propose the following role for mutations in cancer: Cancer attractors define stable gene expression profiles that implement a malignant cell phenotype and pre-exist in the healthy genome; however, they are normally not accessible and hence, not occupied. Developmental trajectories pass by them much as the course of well planned roads avoid abysses. However, tumorigenic mutations may create a side-path on the landscape, e.g., by tilting a slope or lowering a separating hill, which allows cells to accidentally enter normally unoccupied attractors whose associated gene expression profiles may encode a malignancy program (Fig. 2B). Alternatively, mutations may create de novo pathological attractors in state space regions corresponding to expression patterns that encode malignant phenotypes that are normally unstable states on hill and mountains. Thus, mutation-induced reshaping of the landscape topography interferes with Waddington’s smooth “buffering” and “canalization” [56, 68] and increases the probability that developing cells deviate from the normal ontogenetic trajectories and enter side valleys that facilitate oncogenesis.
6. OTHER CANCER CELL PROPERTIES EXPLAINED BY THE CANCER ATTRACTOR HYPOYHESIS
The idea of cancer attractors in an epigenetic landscape can account for many non-genetic aspects of tumorigenesis which appear to defy the view of genetic mutations as the primary and proximate cause of cancer [22]. In other words, if a cancerous state is an attractor state – much as a normal cell type is an attractor state – then tumor progression may be more aptly regarded as an abnormal developmental process and to a lesser extent as an evolutionary process.
Cancer attractors provide a simple formal framework for integrating non-genetic (regulatory) and genetic contributions to tumorigenesis, and hence, can help to more coherently formalize this rarely articulated dichotomy. In the perspective of the epigenetic landscape with (cancer) attractors, non-genetic factors perturb the network state S(t) by transiently altering the expression of genes and thereby trigger a jump of the network state from one site in the landscape to another. Such perturbations of gene expression can occur as a consequence of stochastic fluctuations in the cellular abundance of regulatory molecules (“gene expression noise”) [57] or of aberrant regulatory signals. In principle, non-genetic perturbations could place cells into the basins of cancer attractors without mutations (Fig. 2B), [16, 22, 60].
Importantly, attractor transitions embody the elementary principle that a cell can stay in the new attractor even after the stimulus triggering the transition has vanished. Such “memory effect” in a landscape with multiple attractors (multistability) explains why often the transient experimental expression of a tumor-promoting gene suffices to trigger a lasting malignant phenotype [69–71]. The irreversibility effectively implies the loss of dependence on the tumorigenic protein and thus, hampers target-selective anti-cancer drugs therapy. [Its opposite is the acquisition of new dependencies, or “oncogene addition” (see below)].
In contrast, genetic alterations in the genome permanently change the landscape topography (see 5.2.), and thereby, can lower the “energy barrier” separating the cancer attractor from normal attractor basins or enlarge the basin of the cancer attractor. The relative “structural stability” of the epigenetic landscape to random rewiring (= mutations, see 5.2) readily explains the “constructive” capacity of random mutations in generating distinct phenotypes without much selection: they only have to facilitate access to the normally unused but pre-existing attractors which spontaneously establish specific stable gene expression patterns and need not create gene expression programs de novo.
The attractor concept affords a consistent model, free of ad hoc argumentation, that naturally accounts for a large array of observations that appear paradoxical or to defy mechanistic (proximate) explanation based on causative molecular pathways. A few key examples are further detailed in the Supplementary Material; see also [22]):
Non-genetic (environmental) and genetic factors synergize in tumorigenesis
Multiple distinct molecular pathways lead to similar types of tumors
Tumor viruses reprogram the stem cell state in differentiated cells and induce tumors that mimic non-viral tumors
Transformed cells can exhibit a reversed response to regulatory signals and novel dependency on regulatory molecules for survival (“oncogenes addition”)
Cancer cells relatively readily undergo EMT and the latter is linked to the stem cell program
7. REFINING THE CANCER ATTRACTOR HYOPTHESIS: THE EMBRYONIC PHENOTYPE
The second fundamental challenge, beside integrating the role of mutations, that the cancer attractor hypothesis faces is to explain how cells so rapidly and consistently produce the specific phenotype of an immature cell state which somatic evolution clearly cannot explain. In many areas of biology one is often tempted to attribute the unfathomably creative force in the biosphere to natural selection. As discussed so far, the self-organizing property of attractors can account for the rapid generation of robust cancerous states that exhibit a degree of coordination of gene expression so improbable as to challenge the explanation by natural selection alone. However, cancer attractors do not provide an answer to the question: Why do the stable gene expression profiles of cancer attractors often encode an immature or embryonic program?
To explain the preference for attractors that encode an immature state the context of the entire epigenetic landscape needs to be considered (Fig. 2B). Aberrant attractors that are accessed by chance (due to mutational reshaping of the landscape and unphysiological perturbations) are more likely to be located close to the developmentally immature states in the regions of “higher mountains” of the landscape because these regions are, by necessity, traversed by all trajectories during normal cell development. Specifically, the most simple way by which a newly accessible pathological attractor can trap cells is to disrupt the normal developmental trajectory by diverting developing cells into a dead-end side valley, thereby preventing them from completing their predestined journey down to the physiological cell type attractors (see Fig. 2B). This sideward deviation prevents developing cells from efficient downward movement. By necessity then, the associated phenotype not only is abnormal but also more immature.
Much as normal stem cells, the hence trapped immature cells reside in regions of higher potential energy of the epigenetic landscape and not only exhibit “self-renewal” but also some degree of multi-potency. In fact, these cancer stem cells [11] can give rise to an entire hierarchy of differentiated cells that mirrors that of physiological development, as best characterized in the case of acute leukemia [12]. Consistently, increased plasticity of tumor cells is often observed [72, 73]. Such intra-tumoral aberrant (trans)differentiation, driven by cells exploring the state space neighborhood of the cancer attractor, leads to the production of a variety of more differentiated, often abnormal cells that are not capable of propagating the tumor but contribute to the non-genetic heterogeneity of tumors [60].
In summary, the “hallmarks of cancer” almost “come for free”: They are directly encoded by latent cancer attractors and need not evolve via natural selection. Cancer is immanent to the genomic GRN; it is the price the organism pays for evolving the machinery for multi-cellular development. Mutations and selection do not create tumor cells from scratch but rather, either enhance the probability for a cell to find the cancer attractors or they fine-tune the neoplastic phenotype that they encode.
8. CONCLUSION
At the center of the integrative view presented here is the epigenetic landscape whose topography is determined by the GRN. Although biologists not used to the concepts of network dynamics, state space and generalized potentials may mistake the intuitive landscape picture for an overstretched cartoon, it should be stressed that the landscape has a formal basis in the theory of dynamical, non-equilibrium systems [55, 74] even if the specific details are not known yet. The landscape may epitomize the substrate of evolution which sculpts its topography: the architecture of the GRN may have evolved to produce a landscape that assures a smooth journey of the developing cells so that they descend through the complex high-dimensional state space to reach the gene expression pattern conferring the mature phenotype without spilling into inappropriate dead-end valleys. While most phenotypes associated with side-valleys are not viable, some may confer proliferative potential of developing cells. Changes in the landscape’s fine-topography by mutations can disrupt the developmental trajectory in such a way that cells are trapped in these abnormal (=cancer) attractors before maturity. Somatic Darwinian evolution can further deepen the cancer attractor and their occupancy by cancer cells. This framework allows us to naturally place tumorigenesis in the context of normal development and to explain the intuitively unlikely, yet inevitable and highly specific nature of cancer following accidental events.
While the network view and landscape concept are consistent with many elementary, counter-intuitive properties of cancer, they are far from a complete theory of cancer. Two aspects of cancer for which we refer to the Supplementary material can benefit from the concepts introduced here: (i) non cell-autonomous dynamics of tumors due the influence of non-neoplastic cells in the tumor stroma (see Supp. material, C); and (ii) the implications of cancer attractors for cancer therapy (Suppl. material, D). Viewing cancer stem cells as trapped in attractors raises the following question: how can one perturb the malignant cells back to the trajectory that leads to the non-malignant, more differentiated cell? This will place the old, underexplored idea of differentiation therapy in a new light.
To predict the actual, specific epigenetic landscape of the human genome, the detailed knowledge of the actual wiring diagram of the genomic GRN is required. The increasing pace of advance in genomic technologies for analyzing global regulatory interactions offers hope that this state of knowledge can be achieved in the foreseeable future. Meanwhile, we shall learn to appreciate the importance of an integrative, systems view and refine the cancer attractor hypothesis to incorporate new observations of tumor behavior. As we continue to gather the pieces of specific information needed to construct the GRN’s wiring diagram, we shall hence be guided by the broad vision of systems dynamics and the epigenetic landscape rather than succumb to the narrow urge for finding in a new molecular pathway an ad hoc explanation for a particular tumor behavior.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants to S.H. from the U.S. AFSOR and the NIH and to S.K. from iCore (Alberta, Canada). I.E. is supported by XXX
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Virchow RLK. Cellular pathology. 1859 special ed. London, UK: John Churchill; 1978. [Google Scholar]
- 2.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naxerova K, Bult CJ, Peaston A, Fancher K, Knowles BB, Kasif S, et al. Analysis of gene expression in a developmental context emphasizes distinct biological leitmotifs in human cancers. Genome Biol. 2008;9:R108. doi: 10.1186/gb-2008-9-7-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 7.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 8.Curtis HJ. Formal discussion of: somatic mutations and carcinogenesis. Cancer Res. 1965;25:1305–1308. [PubMed] [Google Scholar]
- 9.Baum B, Settleman J, Quinlan MP. Transitions between epithelial and mesenchymal states in development and disease. Seminars in cell & developmental biology. 2008;19:294–308. doi: 10.1016/j.semcdb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 11.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 95-6. [DOI] [PubMed] [Google Scholar]
- 12.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 13.Xie K, Abbruzzese JL. Developmental biology informs cancer: the emerging role of the hedgehog signaling pathway in upper gastrointestinal cancers. Cancer Cell. 2003;4:245–247. doi: 10.1016/s1535-6108(03)00246-0. [DOI] [PubMed] [Google Scholar]
- 14.Rubin H. On the nature of enduring modifications induced in cells and organisms. Am J Physiol. 1990;258:L19–L24. doi: 10.1152/ajplung.1990.258.2.L19. [DOI] [PubMed] [Google Scholar]
- 15.Clark WH., Jr The nature of cancer: morphogenesis and progressive (self)-disorganization in neoplastic development and progression. Acta Oncol. 1995;34:3–21. doi: 10.3109/02841869509093632. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 17.Strausberg RL, Buetow KH, Greenhut SF, Grouse LH, Schaefer CF. The cancer genome anatomy project: online resources to reveal the molecular signatures of cancer. Cancer Invest. 2002;20:1038–1050. doi: 10.1081/cnv-120005922. [DOI] [PubMed] [Google Scholar]
- 18.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 19.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauffman S. Differentiation of malignant to benign cells. J Theor Biol. 1971;31:429–451. doi: 10.1016/0022-5193(71)90020-8. [DOI] [PubMed] [Google Scholar]
- 21.Huang S, Kauffman S. Complex gene regulatory networks - from structure to biological observables: cell fate determination. In: Meyers RA, editor. Encyclopedia of Complexity and Systems Science. Springer; 2009. Vol. in press. [Google Scholar]
- 22.Huang S, Ingber DE. A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast Dis. 2006;26:27–54. doi: 10.3233/bd-2007-26104. [DOI] [PubMed] [Google Scholar]
- 23.Liotta LA, Kohn KC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 24.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 25.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boland CR, Goel A. Somatic evolution of cancer cells. Semin Cancer Biol. 2005;15:436–450. doi: 10.1016/j.semcancer.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Ptashne M. On the use of the word 'epigenetic'. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 28.Huang S. Reprogramming cell fates: reconciling rarity with robustness. Bioessays. 2009;31:546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 29.Bonifer C, Hoogenkamp M, Krysinska H, Tagoh H. How transcription factors program chromatin--lessons from studies of the regulation of myeloid-specific genes. Semin Immunol. 2008;20:257–263. doi: 10.1016/j.smim.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 31.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1816. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 37.Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nat Rev Genet. 2008;9:509–515. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- 38.Elfilali A, Lair S, Verbeke C, La Rosa P, Radvanyi F, Barillot E. ITTACA: a new database for integrated tumor transcriptome array and clinical data analysis. Nucleic acids research. 2006;34:D613–D616. doi: 10.1093/nar/gkj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 41.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 42.Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol. 2004;31:4–19. doi: 10.1053/j.seminoncol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 43.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 44.Guo Y, Eichler GS, Feng Y, Ingber DE, Huang S. Towards a holistic, yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. J Biomed Biotechnol. 2006;2006:69141. doi: 10.1155/JBB/2006/69141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI) Proc Natl Acad Sci U S A. 2005;102:2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodwin BC, Webster GC. Rethinking the origin of species by natural selection. Riv Biol. 1999;92:464–467. [PubMed] [Google Scholar]
- 48.Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 49.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 50.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 51.Delbrück M. Unités biologiques douées de continuité génétique Colloques Internationaux du Centre National de la Recherche Scientifique: CNRS. Paris: CNRS; 1949. Discussion. [Google Scholar]
- 52.Monod J, Jacob F. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 53.Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 54.Kauffman S. Homeostasis and differentiation in random genetic control networks. Nature. 1969;224:177–178. doi: 10.1038/224177a0. [DOI] [PubMed] [Google Scholar]
- 55.Kauffman SA. The Origins of Order. New York: Oxford Univ. Press; 1993. [Google Scholar]
- 56.Waddington CH. The strategy of the genes. London: Allen and Unwin; 1957. [Google Scholar]
- 57.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett. 2005;94:128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- 59.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 61.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 63.Huang S, Guo YP, May G, Enver T. Bifurcation dynamics of cell fate decision in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 64.Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 65.Aldana M, Balleza E, Kauffman S, Resendiz O. Robustness and evolvability in genetic regulatory networks. J Theor Biol. 2007;245:433–448. doi: 10.1016/j.jtbi.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 66.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Xu L, Wang EK, Huang S. The “potential” landscape of genetic circuits imposes the arrow of time in stem cell differentiation in review. doi: 10.1016/j.bpj.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 69.Lochter A, Bissell MJ. An odyssey from breast to bone: multi-step control of mammary metastases and osteolysis by matrix metalloproteinases. Apmis. 1999;107:128–136. doi: 10.1111/j.1699-0463.1999.tb01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plattner R, Anderson MJ, Sato KY, Fasching CL, Der CJ, Stanbridge EJ. Loss of oncogenic ras expression does not correlate with loss of tumorigenicity in human cells. Proc Natl Acad Sci U S A. 1996;93:6665–6670. doi: 10.1073/pnas.93.13.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–571. doi: 10.1016/j.ccr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Z, Yuan XM, Li LH, Xie FP. Transdifferentiation in neoplastic development and its pathological implication. Histol Histopathol. 2001;16:1249–1262. doi: 10.14670/HH-16.1249. [DOI] [PubMed] [Google Scholar]
- 73.Rapp UR, Ceteci F, Schreck R. Oncogene-induced plasticity and cancer stem cells. Cell Cycle. 2008;7:45–51. doi: 10.4161/cc.7.1.5203. [DOI] [PubMed] [Google Scholar]
- 74.Nicolis G, Prigogine I. Exploring Complexity: An Introduction W.H. Freeman & Company. 1989 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.