Abstract
Respiratory oxidative phosphorylation is a cornerstone of cellular metabolism in aerobic multicellular organisms. The efficiency of this process is generally assumed to be maximized, but the presence of dynamically regulated nonphosphorylating bypasses implies that plants can alter phosphorylation efficiency and can benefit from lowered energy generation during respiration under certain conditions. We characterized an Arabidopsis (Arabidopsis thaliana) mutant, ndufs4 (for NADH dehydrogenase [ubiquinone] fragment S subunit 4), lacking complex I of the respiratory chain, which has constitutively lowered phosphorylation efficiency. Through analysis of the changes to mitochondrial function as well as whole cell transcripts and metabolites, we provide insights into how cellular metabolism flexibly adapts to reduced phosphorylation efficiency and why this state may benefit the plant by providing moderate stress tolerance. We show that removal of the single protein subunit NDUFS4 prevents assembly of complex I and removes its function from mitochondria without pleiotropic effects on other respiratory components. However, the lack of complex I promotes broad changes in the nuclear transcriptome governing growth and photosynthetic function. We observed increases in organic acid and amino acid pools in the mutant, especially at night, concomitant with alteration of the adenylate content. While germination is delayed, this can be rescued by application of gibberellic acid, and root growth assays of seedlings show enhanced tolerance to cold, mild salt, and osmotic stress. We discuss these observations in the light of recent data on the knockout of nonphosphorylating respiratory bypass enzymes that show opposite changes in metabolites and stress sensitivity. Our data suggest that the absence of complex I alters the adenylate control of cellular metabolism.
In eukaryotes, cellular energy is provided in the form of ATP and is mainly produced in mitochondria through oxidative phosphorylation (OXPHOS). Reducing equivalents resulting from the degradation of sugars or fats are oxidized by the respiratory chain, protons are pumped to form a membrane potential, and ATP synthase uses this membrane potential to synthesize ATP. The respiratory chain is composed of four complexes localized in the inner mitochondrial membrane. Complex I and complex II are, respectively, a NADH-ubiquinone oxidoreductase and a FADH2-ubiquinone oxidoreductase that transfer electrons from matrix-localized reducing equivalents to ubiquinone. One-third of the proton translocation of OXPHOS is linked to electron transfer through complex I. Complex III (ubiquinone-cytochrome c oxidoreductase) and complex IV (cytochrome c oxidase) represent the cytochrome pathway and transfer electrons from ubiquinone to molecular oxygen. The transfer of electrons through complex III and IV is coupled to the remaining two-thirds of the proton translocation.
Plant respiratory chains also contain additional, non-proton-translocating, entry points for electrons known as nonphosphorylating bypasses. These NAD(P)H dehydrogenases have been identified and localized either on the cytosolic or the matrix side of the inner membrane and transfer electrons from cytosolic or matrix NADH pools directly to ubiquinone (Rasmusson and Moller, 1991; Rasmusson et al. 2008). In addition, the alternative oxidase (AOX) can bypass the cytochrome pathway by oxidizing ubiquinol and reducing molecular oxygen, and it does this in a nonphosphorylating manner (Lambers, 1982).
The potential for plants to dynamically alter their efficiency of phosphorylation is remarkable when one considers that even small changes in OXPHOS efficiency in animals are associated with disease, aging, and cellular dysfunction (Gellerich et al., 2004). For example, deficiency in complex I function has been associated with several human diseases such as Leber's heredity optic neuropathy (Wallace et al., 1988), Leigh syndrome (Hoefs et al., 2008), and leukodystrophy (Holt et al., 1988), and complete absence of complex I is considered lethal. A number of plant mutants lacking respiratory chain components have been studied, including mutants in complex I (Marienfeld and Newton, 1994; Pla et al., 1995; Lee et al., 2002; Perales et al., 2005) and mutants removing specific nonphosphorylating bypasses such as AOX (Giraud et al., 2008). Mutants lacking complex II are embryo lethal in plants (Leon et al., 2007) and mutants lacking complex III or IV have not been reported, presumably as these are lethal in plants, as are mutants in the assembly of c-type cytochromes (Meyer et al., 2005).
Complex I is especially interesting for respiratory remodeling studies, as its absence will lower the maximal efficiency of OXPHOS by one-third, thus forcing a recalibration of a primary driver in cellular energy homeostasis. Mutants in genes encoding complex I subunits in Arabidopsis (Arabidopsis thaliana) include the 18-kD subunit (Lee et al., 2002) and the plant-specific CA2 subunit (Perales et al., 2005). Natural mitochondrial mutants lacking some of the mitochondrial nad genes as a result of genome rearrangements have been described in maize (Marienfeld and Newton, 1994) and Nicotiana sylvestris (Pla et al., 1995). Finally, several mutants have been identified in genes encoding splicing factors involved in the expression of nad4 in N. sylvestris (Brangeon et al., 2000) and Arabidopsis (Nakagawa and Sakurai, 2006) and of nad1 in Arabidopsis (de Longevialle et al., 2007). These mutants display a whole range of phenotypes, but the only one that has been studied to characterize the effects of the absence of complex I is the cytoplasmic male sterile II (CMSII) mutant in N. sylvestris.
The CMSII mutant is a natural mutant that has lost the nad7 gene after a rearrangement of the mitochondrial genome (Pla et al., 1995). CMSII mutant growth is retarded compared with wild-type plants (Gutierres et al., 1997). In this mutant, complex I is absent, the respiration is rotenone insensitive (rotenone is an inhibitor of complex I), the oxidation of Gly is reduced, and the capacity of the alternative pathway is increased (Sabar et al., 2000). Photosynthesis is reduced by 20% to 30% under atmospheric conditions. This effect can be canceled under high CO2 or low O2 (Dutilleul et al., 2003a). The photorespiratory pathway is increased in CMSII via an increased internal resistance to CO2 diffusion (Priault et al., 2006). A metabolomic study showed that nitrogen metabolism was affected in the mutant with accumulation of nitrogen-rich amino acids (Dutilleul et al., 2005) and that the shoot-root ratio is higher in the CMSII mutant at low nitrogen (Pellny et al., 2008). Finally, the expression of many stress-related genes was modified, resulting in a higher tolerance to ozone and Tobacco mosaic virus (Dutilleul et al., 2003b).
Another approach used to understand the role of complex I is to use inhibitors such as rotenone. Rotenone is an isoflavonoid produced by plants in the legume family and is classically used as an inhibitor of complex I (Ayala et al., 2007). In mammals, rotenone treatment inhibits mitochondrial complex I by blocking electron transfer to ubiquinone, which leads to superoxide production from the dissipation of electrons from complex I and oxidative damage to cell components (Kussmaul and Hirst, 2006). Chronic exposure of rodents to rotenone causes Parkinson-like degenerative syndrome, suggesting a link between this disease and complex I function (Betarbet et al., 2000). We used rotenone treatment on Arabidopsis cell culture to understand the short-term effects of complex I inhibition in plants (Garmier et al., 2008). A transitory decrease in respiration was observed immediately following the treatment, but a return to pretreatment steady-state levels was observed within a few hours. The glycolytic activity associated with mitochondria (Giegé et al., 2003) was lost and tricarboxylic acid (TCA) cycle activity was altered, as shown by the accumulation of many intermediates (Garmier et al., 2008). These data demonstrated that short-term inhibition of complex I caused a remodeling of metabolism.
In order to study the long-term metabolic adaptations resulting from lowering the phosphorylating efficiency of respiration, we have studied an Arabidopsis mutant lacking NDUFS4 (the 18-kD subunit of complex I). This mutant shows Suc-sensitive germination, delayed growth, a modified respiration pathway, and altered stress responses. The lack of complex I has no major influence on the mitochondrial proteome or transcriptome but leads to a lowering of growth-related nuclear transcripts and clearly influences central metabolism, with strong differences between night and day. These results indicate that complex I plays an important role in energy production and the control of metabolite levels, especially at night, and the responses to its absence are consistent with altered adenylate control of cellular metabolism.
RESULTS
ndufs4 Lacks Complex I and Has a Small Plant Phenotype That Is Recovered by Complementation
Screening of the T-DNA Express database (http://signal.salk.edu/cgi-bin/tdnaexpress) for insertions within exons of nuclear genes encoding complex I subunits in Arabidopsis revealed a mutant from the Syngenta collection (Sessions et al., 2002), SAIL_596_E11, containing a T-DNA inserted in the first exon of the NDUFS4 gene (At5g67590). We obtained plants homozygous for the insertion as well as wild-type plants (ecotype Columbia [Col-0]) by screening plants obtained after self-pollination of heterozygous plants. We confirmed the mutation by reverse transcription (RT)-PCR, showing that transcripts covering the first exon cannot be detected (Supplemental Fig. S1). Grown under normal conditions, these plants are smaller than wild-type plants (Fig. 1A). To confirm that the observed phenotype was a result of the insertion in NDUFS4, we complemented the homozygous mutant with a full-length cDNA of NDUFS4 expressed under the control of the 35S promoter. The complemented plant does not show the delayed-growth phenotype, indicating that it is caused by the inactivation of the NDUFS4 gene (Fig. 1A). We performed western-blot analysis using a polyclonal antibody against the NDUFS4 protein to confirm the absence of NDUFS4 from a mitochondrial fraction of the mutant (Fig. 1B). Blue-Native gel electrophoresis (Schägger and von Jagow, 1991) was used to investigate the effect of the absence of NDUFS4 on complex I assembly and activity. No complex I activity was detected in the ndufs4 mutant (Fig. 1C, lane 2). In contrast, in the complemented line, complex I activity was restored to wild-type levels (Fig. 1C, lane 3).
Figure 1.
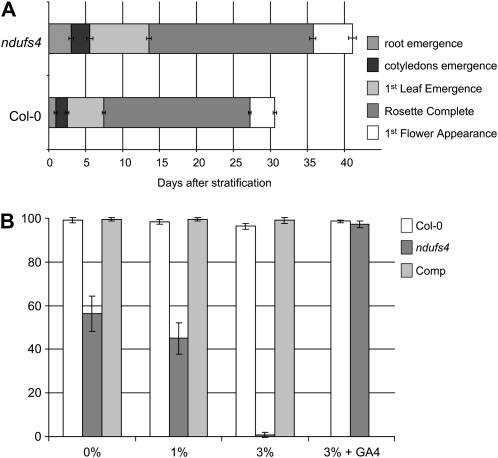
Characterization of the ndufs4 mutant. A, Image of 4-week-old plants grown under a long-day photoperiod. Comp, ndufs4 mutant complemented with a cDNA corresponding to the NDUFS4 gene. B, Western blots of mitochondrial fractions purified from Col-0 and ndufs4 plants. The mitochondrial matrix enzyme PDC-E1α is used as a loading control. C, Mitochondrial membrane complexes were separated on a Blue Native gel. Following migration, the gel was stained for complex I activity. The stars indicate the position of complex I. The activity stain on the lower part of the gel corresponds to the activity of the dehydrolipoamide dehydrogenase (E.H. Meyer, unpublished data) and can act as a loading control.
Seed Dormancy, Slow Germination, and Delayed Development Coincide with Suppressed Transcript Pools for Growth Processes in ndufs4 Plants
As ndufs4 plants are smaller than wild-type plants of the same age (Fig. 1A), we analyzed the growth of the mutant in more detail in order to identify if a specific developmental stage is impaired. Seeds were sown on agar plates, stratified for 2 d at 4°C, and transferred to a long-day photoperiod. The time when the different organs emerged was monitored. The ndufs4 mutant was delayed in all developmental stages of its growth (Fig. 2A). We analyzed the nuclear mRNA pools using ATH1 GeneChip microarrays in triplicate. Only a small portion of the Arabidopsis genome is differently expressed in the ndufs4 mutant compared with the wild type (1,479 genes; P ≤ 0.05). We used MapMan (Thimm et al., 2004) with the Ath_AFFY_TAIR7 mapping file to study functional groups of genes (Supplemental Table S2). Processes involving protein metabolism (BIN 29), the cell cycle (BIN 31.3), as well as RNA metabolism (BIN 27.1, BIN 27.3, and BIN 27.4) and development (BIN 33) were significantly down-regulated. In parallel microarray experiments, similar results were obtained for a ndufa1 mutant, an insertion mutant in At3g08610, a gene encoding another complex I subunit (Supplemental Fig. S4), indicating that these transcript changes are likely due to the lack of complex I rather than the NDUFS4 subunit per se (Supplemental Table S2). These observations are in agreement with the slower growth of both ndufs4 and ndufa1 mutants.
Figure 2.
Growth analysis of the ndufs4 mutant. A, Comparison of the length of different growth stages in the ndufs4 mutant and wild-type plants (Col-0). Values are means ± sd for n = 50. B, Effect of Suc concentration on germination after 2 d. The data are expressed as percentages of germinated seeds. Means ± sd of three independent experiments are shown. Each experiment involved between 150 and 400 seeds. Comp, Complemented mutant.
The germination of the ndufs4 mutant is the growth stage that was the most affected (three times slower than in the wild type; Fig. 2A). During the germination of oil seeds like Arabidopsis seeds, fatty acids are degraded by the β-oxidation that produces carbon skeletons for Suc synthesis. This Suc is then degraded to power germination (Hayashi et al., 1998). We tried to rescue the germination phenotype by sowing seeds on increasing concentrations of Suc. Interestingly, the germination of the ndufs4 mutant is inhibited by high concentration of Suc (Fig. 2B), indicating that the ndufs4 mutant could be producing hyperdormant seeds. Indeed, the Suc-sensitivity phenotype of ndufs4 can be complemented by addition of gibberellic acid (GA4) in the medium (Fig. 2B). As the complemented line behaves like the wild-type control (Fig. 2B), this indicates that the absence of complex I modulates hormone balance and thus influences the germination of the ndufs4 mutant.
ndufs4 Has an Electron Transport Chain Impairment But Wild-Type Respiratory Rate
The ndufs4 mutant is lacking a fully assembled and active complex I based on measurements of NADH-dependent reduction of nitroblue tetrazolium (NBT) in native gels (Fig. 1C). We further investigated the consequences of this mutation on the respiratory activity of the mutant. The total leaf respiration rate was not significantly modified in ndufs4, as shown by measurement of oxygen consumption of leaf discs in the dark (Fig. 3A). As oxygen reduction is proportional to electron transfer rates, this indicates that a similar electron transfer rate is occurring through the respiratory chains of the ndufs4 mutant and wild-type plants.
Figure 3.
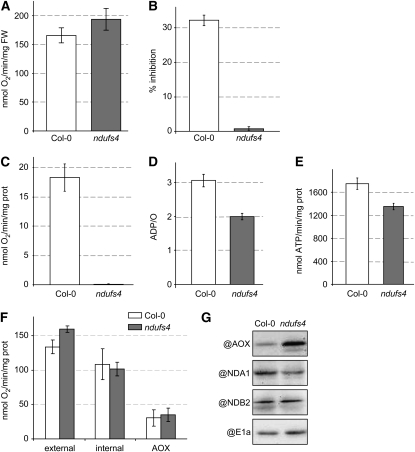
Characterization of the respiration of the ndufs4 mutant A, Total leaf respiration measurement in the dark (n = 4). FW, Fresh weight. B, Effect of rotenone on respiration of isolated mitochondria. The percentage of inhibition was calculated by the difference in respiration rates before and after the addition of rotenone in the medium (n = 3) when mitochondrial respiration is driven by the TCA cycle (state 3). C, Deamino-NADH oxidation capacity (n = 3) of mitochondrial membranes. D, ADP/O ratio calculated from oxygen consumed for a known addition of ADP in transitions between state 3 and state 4 (n = 3). E, ATP synthesis rate measured using luciferase from extracts of respiring isolated mitochondria. F, Capacity of the different alternative pathways (n = 3) in intact mitochondria: external, external NADH dehydrogenase; internal, internal NADH dehydrogenase (rotenone-insensitive TCA cycle substrate-dependent respiration); AOX, antimycin A-insensitive respiration. G, Western blots of mitochondrial fractions purified from Col-0 and ndufs4 plants. PDC-E1α is used as a loading control.
To confirm the absence of complex I function in the ndufs4 plants, we used rotenone as a classical inhibitor of complex I activity (Ayala et al., 2007) and deamino-NADH as a selective substrate for complex I (Rasmusson and Moller, 1991) in mitochondria isolated from Col-0 and ndufs4 plants. The TCA cycle-dependent respiration rate of mitochondria isolated from Col-0 plants was inhibited by 30% by rotenone, indicating that rotenone-insensitive nonphosphorylating bypasses account for about two-thirds of the normal respiratory flux (Fig. 3B). In contrast, rotenone has no effect on the TCA cycle-dependent respiration of mitochondria isolated from ndufs4 plants (Fig. 3B), indicating that this mutant does not need an active complex I to maintain its respiratory rate. To independently confirm the evidence of Figure 1C, showing the lack of complex I activity, we assessed the rate of deamino-NADH oxidation by ruptured mitochondrial samples. Mitochondria from Col-0 plants used deamino-NADH as a substrate for whole chain electron transport, while the rate from ndufs4 mitochondrial membranes was almost undetectable (Fig. 3C). We conclude that complex I activity is absent in the mutant. Hence, fewer protons should be able to be exported to the intermembrane space, and as a consequence, less ATP should be able to be synthesized in the mutant for the same rate of oxygen consumption. We showed this to be true in isolated mitochondria (Fig. 3D), where the mitochondria from ndufs4 showed an approximately 35% decrease in the ADP/O ratio calculated from state 3 to state 4 transitions and produced approximately 20% less ATP in assays of isolated organelles (Fig. 3E).
It is assumed that plants can survive without complex I because of the presence of alternative NADH dehydrogenases in the inner mitochondrial membrane that can complement the loss of complex I by maintaining the oxidation of matrix NADH. We measured the capacity of the internal and external alternative dehydrogenases of purified mitochondria to determine if any compensation in these rates occurred in the mutant. The capacity of the internal dehydrogenases was not affected in ndufs4, whereas the capacity of the external dehydrogenases increased by 15% (Fig. 3F). It has been previously reported in the CMSII mutant that external NADH dehydrogenase activity as well as AOX capacity are increased in isolated mitochondria (Sabar et al., 2000), even though the in vivo flux through the AOX was not increased (Vidal et al., 2007). An increased capacity could be explained by an increased activity or a larger pool of the proteins analyzed. We studied the transcript pools for components of the alternative pathway (Supplemental Table S3), enzyme capacities (Fig. 3F), and protein levels using antibodies (Fig. 3G). There was no induction of any AOX transcripts, but AOX protein is clearly more abundant in the ndufs4 mutant on a mitochondrial protein basis. Among the external NADH dehydrogenase (ND) transcripts, NDA1 was the only one clearly induced by more than 4-fold in ndufs4 (Supplemental Table S3). However, we have recently shown that this protein is dual targeted to mitochondria and peroxisomes in Arabidopsis (Carrie et al., 2008). In isolated mitochondria, NDA1 was in fact less abundant at the protein level in ndufs4 (Fig. 3G). The major external NADH dehydrogenase, NDB2, was not transcriptionally induced (Supplemental Table S3), and there was no change in protein level between Col-0 and ndufs4 (Fig. 3G).
Overall, these data suggest that internal and external dehydrogenases permit considerable flux for the electron transport chain even in the absence of complex I in ndufs4, and even though AOX protein levels are increased, the capacity of this alternative pathway is not significantly altered in ndufs4 (Fig. 3, F and G). Interestingly, the genes encoding components of the mitochondrial respiratory chain (BIN 9) are significantly down-regulated in ndufs4 (Supplemental Table S2), but this down-regulation does not correlate with lower respiratory activity on a mitochondrial protein basis or lowered respiratory rate on a leaf area basis.
Changes in the Mitochondrial Proteome in ndufs4 Are Limited to Complex I
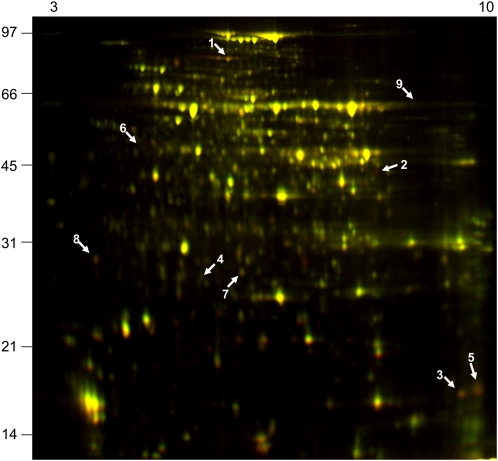
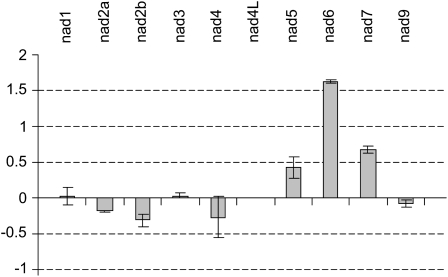
Given that in the ndufs4 mutant complex I was absent and the respiration chain was altered, we performed a differential proteomic analysis using the fluorescence difference gel electrophoresis (DIGE) CyDye fluorophore technology in order to determine if the effects we noted in the mutant could be due to major modifications of other mitochondrial functions rather than simply the loss of complex I. Three independent experiments were run to compare Col-0 and ndufs4 mitochondrial samples, and we identified spots that were consistently decreased more than 2-fold over all experiments in the ndufs4 sample compared with the Col-0 sample (Fig. 4). No spots were consistently increased in abundance. The spots decreasing in abundance were excised and analyzed by liquid chromatography-tandem mass spectrometry. The identities of the corresponding proteins are summarized in Table I. Spot 3 corresponded to NDUFS4, confirming that the protein was highly reduced (below the limits of detection) in the mutant. Three spots identified, namely spots 1, 5, and 8, contained a total of four complex I subunits. Spot 7 corresponds to a chloroplastic carbonic anhydrase that is probably a contaminant of the mitochondrial preparation. We were not able to positively identify the last three proteins decreasing in abundance. These results indicate that, within the detection range of the DIGE technique, the only change in the mitochondrial proteome observed is the reduction in the abundance of complex I subunits. As the DIGE approach involves isoelectric focusing (IEF)-SDS-PAGE, this gel technique is not suitable for the visualization of highly hydrophobic proteins such as mitochondria-encoded complex I subunits. In order to determine if any feedback control of the expression of the mitochondria-encoded complex I subunits was occurring due to the absence of the fully assembled complex, we measured the relative abundance of all mitochondrial transcripts in the ndufs4 mutant by RT-PCR. The only modification of the expression of nad genes in the ndufs4 mutant is the overaccumulation of nad6 transcripts (Fig. 5). Similarly, there was little if any response in the transcripts for nucleus-encoded components for complex I (Supplemental Table S4). Thus, it appears that loss of complex I in ndufs4 does not lead to retrograde signals affecting the expression of nucleus-encoded complex I components and has little systematic impact on the mitochondrial proteome outside of other subunits in complex I, even though there is some measurable response in the transcript pool of a mitochondria-encoded complex I subunit.
Figure 4.
Differential gel electrophoresis of changes in the mitochondrial proteome between the ndufs4 mutant and wild-type plants. Mitochondrial proteins were bound to different fluorescent dyes: Cy2 (red fluorescence) for wild-type samples and Cy5 (green fluorescence) for the ndufs4 samples. Yellow spots, Both dyes are present in equal abundance; green spots, up-regulated spots; red spots, down-regulated spots. Three independent sets of mitochondrial isolates and a set of three gels were run and analyzed using DeCyder quantification software. Protein spots that reproducibly changed in abundance were selected for further analysis (numbered arrows; see Table I for identification). The molecular mass scale (kD) is indicated on the left, and pI values are indicated on the top.
Table I.
Identification of mitochondrial proteins with different abundances between the ndufs4 mutant and wild-type plants
Spot number refers to the annotation in Figure 4. The fold changes and P values were obtained from DeCyder software (GE Healthcare) using three biological replicates. Data from in-gel digestions of the spots were matched using Mascot (Matrix Sciences) to a database of Arabidopsis proteins (The Arabidopsis Information Resource 8). AGI, Arabidopsis Genome Initiative accession number; MOWSE, Molecular Weight Search score derived by Mascot from the MS/MS search; Peptide, number of matching peptides; Coverage, percentage of protein sequence covered by the peptides identified; Localization, subcellular compartment where the protein is located based on MS, GFP, and predictor data according to the SUBA database (Heazlewood et al., 2007).
| Spot No. | Fold Change | P | AGI | Description | Molecular Weight | MOWSE | Peptide | Coverage | Localization |
|---|---|---|---|---|---|---|---|---|---|
| 1 | −4.18 | 0.017 | At5g37510 | EMB1467 NDUFS1 EMB1467 (EMBRYO DEFECTIVE1467); NADH dehydrogenase | 81,130 | 571 | 14 | 21 | Mitochondria |
| 2 | −4.32 | 0.003 | |||||||
| 3 | −4.23 | 0.03 | At5g67590 | NDUFS4 18-kD subunit | 17,124 | 140 | 4 | 22 | Mitochondria |
| 4 | −2.7 | 0.01 | |||||||
| 5 | −2.6 | 0.041 | At3g03100 | NDUFA11 NADH:ubiquinone oxidoreductase family protein | 18,305 | 160 | 3 | 22 | Mitochondria |
| 6 | −2.14 | 0.02 | |||||||
| 7 | −2.11 | 0.018 | At3g01500 | CA1 (CARBONIC ANHYDRASE1); carbonate dehydratase/zinc ion binding | 36,121 | 130 | 5 | 23 | Chloroplast |
| 8 | −2.03 | 0.009 | At1g79010 | NDUFS8-1 NADH-ubiquinone oxidoreductase 23-kD subunit, mitochondrial (TYKY) | 25,487 | 245 | 6 | 30 | Mitochondria |
| At1g16700 | ATMLO14 NDUFS8-2 ATMLO14 (Arabidopsis MILDEW RESISTANCE LOCUS O14); NADH dehydrogenase (ubiquinone) | 25,361 | 211 | 5 | 25 | Mitochondria | |||
| 9 | −2.01 | 0.017 |
Figure 5.
Changes in mitochondrial gene expression. Quantitative RT-PCR was performed to estimate the abundance of mitochondrial transcripts for the nad genes in the ndufs4 mutant and wild-type plants. Data are expressed with a log2 scale as the ratio of ndufs4 to Col-0 signal (n = 3).
The ndufs4 Mutant Causes a Broad Impact on Cellular Metabolite Pools
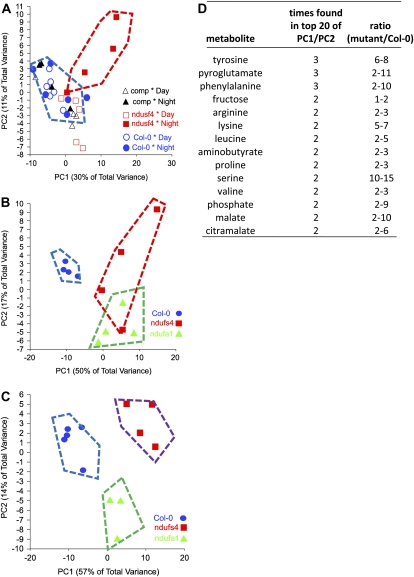
In order to understand the cellular response to the modified respiration in the ndufs4 mutant, we analyzed the abundance of major metabolites using gas chromatography-mass spectrometry (GC-MS). Leaves from 6-week-old plants were harvested at the end of the night and at the end of the day, and the metabolites were extracted and analyzed. Figure 6A shows two-dimensional principal components analyses (2D PCA) of these data for Col-0, ndufs4, and the complemented line (fold changes and P values are given in Supplemental Table S5). This shows that the major variance in the overall metabolome was in the ndufs4 genotype at night and that this variance was eliminated in the complemented line. Two additional experiments were performed using leaves from 4-week-old plants during two successive nights, and the metabolome data were compared during each night for Col-0, ndufs4, and ndufa1. Again, the 2D PCA plots show clear separation of the genotypes (Fig. 6, B and C). The differences between the mutants on the two successive nights were largely the fold differences observed in metabolites (fold changes and P values are given in Supplemental Table S6). As there is some variation between experiments in the fold changes of specific metabolites, we have sought to combine the data by exploring the lists of compounds with the largest loadings in the first and second components that account for 40% to 70% of the variation in each experiment. The tabulated list is presented in Figure 6D. The accumulation of organic acids and amino acids was pronounced and is consistent with an alteration of central metabolism. Notable among the amino acids higher at night are substrates for mitochondrial respiration, and their oxidation bypasses complex I of the respiratory chain. The branched chain amino acids (Leu and Val) accumulate during the night, and their degradation transfers electrons directly to ubiquinone via the electron transfer flavoprotein (Ishizaki et al., 2006). Transcripts for a number of the components of this branched chain amino acid degradation pathway are induced in ndufs4 (Supplemental Table S2). The accumulation of organic and amino acids suggests no shortage of substrate for glycolysis; indeed, a higher flux may operate as glycolysis and the TCA cycle are freed up from adenylate control by the lower OXPHOS efficiency in ndufs4.
Figure 6.
Metabolite changes in the ndufs4 and ndufa1 mutants predominate at night. A, 2D PCA displays of the GC-MS metabolomic data sets (Supplemental Table S5) from Col-0, ndufs4, and the complemented (comp) line during the day and the night (6-week-old plants). B and C, 2D PCA displays of the GC-MS metabolomic data sets (Supplemental Table S6) from Col-0, ndufs4, and ndufa1 during two successive nights (4-week-old plants). Data for four to five plants are shown, and genotype/treatment separations from the wild type are shown by the hatched boxes. D, The 20 metabolites most important for the variance in each data set were identified from PC1/PC2 component weights. Ranges of the ratios of abundance between the wild type and ndufs4/ndufa1 mutants are shown for all metabolites that appeared in these lists in at least two out of three experiments (combined from Supplemental Tables S5 and S6).
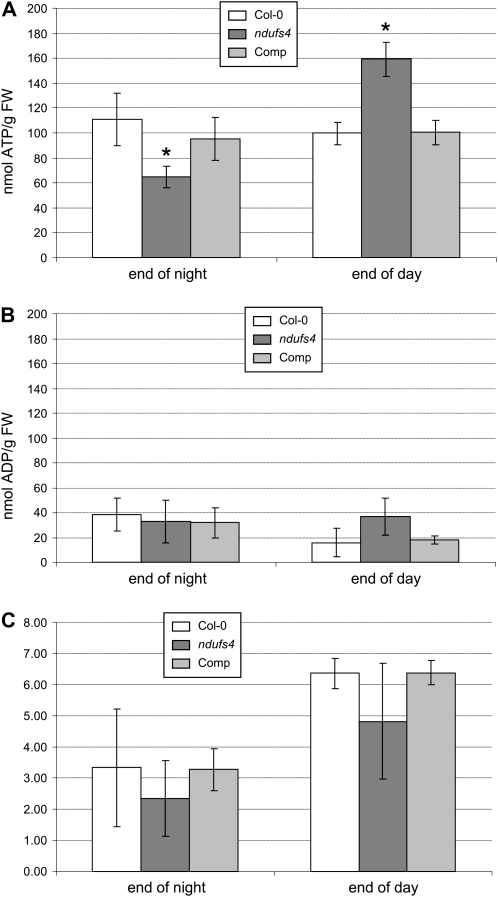
In order to assess this hypothesis, we measured the levels of ATP and ADP in Col-0, ndufs4, and the complemented line. ATP levels in Col-0 and the complemented line are not significantly changed between the end of the night and the end of the day (Fig. 7A). However, in ndufs4, ATP levels are significantly lower in the night and higher in the day (Fig. 7A). ADP levels are relatively stable in all three lines (Fig. 7B). The average ATP/ADP ratio appears lower in ndufs4, but this is not statistically significant given the variations within these data (Fig. 7C).
Figure 7.
Altered adenylate levels in ndufs4. A, ATP content. B, ADP content. C, ATP/ADP ratio. Data are expressed as means of three biological replicates. Asterisks indicate significant differences (P < 0.05) between Col-0 and ndufs4 or the complemented (Comp) line. FW, Fresh weight.
Impact of ndufs4 on Photosynthetic Function
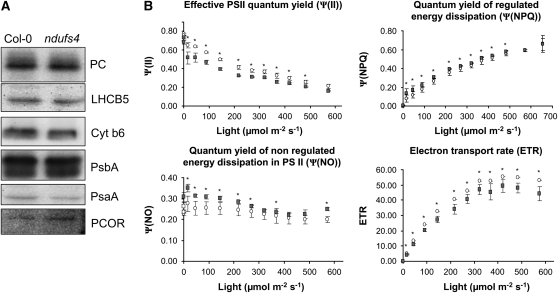
From our microarray data, a large set of transcripts for photosynthesis (BIN 1) are significantly up-regulated (Supplemental Table S2). Modification of photosynthetic activity has been reported in the tobacco (Nicotiana tabacum) complex I mutant, CMSII (Dutilleul et al., 2003a). Thus, loss of mitochondrial complex I does appear to have some retrograde action that influences nuclear gene expression of some plastid functions. Investigating this further in ndufs4, we found that subgroups of genes involved in photosynthesis responded differently: genes involved in the light reactions (BIN1.1) were significantly up-regulated, whereas the expression of genes involved in photorespiration (BIN1.2), the Calvin cycle (BIN1.3), and the reductive pentose phosphate cycle (BIN1.4) was not significantly changed. Analysis of transcription from the chloroplast genome showed no major modification of the expression of chloroplast genes, and overall the plastid transcription seems to be slightly down-regulated in ndufs4 compared with the wild type (Supplemental Fig. S2). We looked at the abundance of some proteins involved in the light reactions of photosynthesis by western-blot analysis of cellular fractions. No visible differences at the protein level were observed between the ndufs4 mutant and the Col-0 control (Fig. 8A).
Figure 8.
Impact of the ndufs4 mutation on photosynthetic proteins and photosynthesis. A, Analysis of the protein abundance of chloroplast proteins by western blot on whole cell fractions. PC, Plastocyanin; LHCB5, light-harvesting complex subunit B5; Cyt b6, cytochrome b6; PsbA, PSII subunit A; PsaA, PSI subunit A; PCOR, protochlorophyllide oxidoreductase. B, Plants were grown for 4 weeks at 22°C, 12 h of light and dark, under 120 μE m−2 s−1 light. Graphs represent the variation of quantum efficiency (Ψ) of photochemical energy dissipation (ΨPSII), light-dependent thermal dissipation (ΨNPQ), light-independent thermal dissipation, and fluorescence energy dissipation (Ψf,D) over a range of light intensities. White circles, Col-0; gray squares, ndufs4. The stars indicate statistically significant differences using Student's t test (P < 0.05).
We also measured the activity of PSII and the electron transport rate (ETR) using chlorophyll a fluorometry from intact leaves. ETR and ΨPSII (photochemical energy dissipation) were significantly lower in ndufs4 than in the wild type, especially at low irradiances (Fig. 8B). The evidence suggests that photochemical efficiency is impaired in the mutant. It is not clear if this modification is a consequence or the cause of the transcriptional modification of nucleus-encoded components for the light reactions of photosynthesis, but it is plausible to suggest that the two phenomena could be linked. It is of interest that superoxide is elevated yet ETR is lower. While there are many processes that can generate superoxide in cells, if the photosynthetic change is responsible for the superoxide elevation, then one scenario that would explain our data is ferridoxin-mediated NADP reduction being lowered, negatively feeding back on ETR and resulting in increased superoxide production at PSI. This could also lead to a more reduced plastoquinone pool that would enhance nonphotochemical quenching (NPQ; ΨNO), as we observed in ndufs4 (Fig. 8B).
Impact of ndufs4 on Stress Tolerance
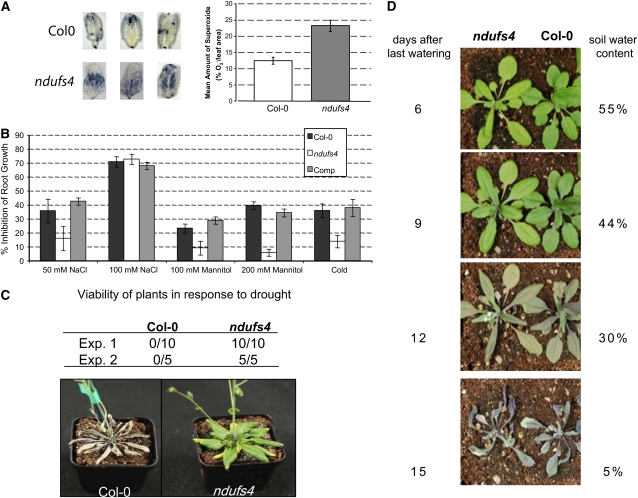
The microarray analysis showed that the expression of the stress-related genes (BIN 20) was significantly increased (Supplemental Table S2), and the metabolite analysis shows elevation of the antioxidant ascorbate and the osmoprotectant compounds putrescine and Pro (Supplemental Tables S5 and S6). The in situ NBT staining technique was used to confirm increased superoxide production in ndufs4 that was observed in other complex I mutants, including CMSII (Dutilleul et al., 2003b), and in a point mutation in the same gene in Arabidopsis, fro1 (Lee et al., 2002; Fig. 9A). This increased NBT staining is indicative of either increased superoxide production or altered antioxidant defense enzymes in the mutant. To determine if the stress tolerance was altered, we performed a series of whole plant stress assays to monitor the behavior of the ndufs4 mutant and the complemented line under three different stresses: cold, salt, and osmotic stress. The root length was measured and compared with control plants grown under nonstressed conditions. The percentage of growth inhibition by the stress treatment was then measured. The growth of the ndufs4 roots was less inhibited under cold, osmotic, or moderate salt stress (Fig. 9B), suggesting some form of stress tolerance. Under high salt concentrations, the ndufs4 mutant behaved like wild-type plants. In all cases, the complemented line was indistinguishable from Col-0.
Figure 9.
Stress tolerance of the ndufs4 mutant. A, NBT stain for superoxide in leaves. Three examples are shown for each ecotype, and the average amount of superoxide was estimated by quantifying the coloration (n = 20). B, In vitro analysis of root growth under stress conditions. Data are expressed as the ratio between the root length of plants grown under control conditions and those of plants grown under stress conditions (n = 20). Comp, Complemented mutant. C, Drought stress assay in individual pots. After 10 d of withholding water from 40-d-old plants, the viability was assessed visually (Exp. 1; a representative image is shown) and by monitoring chlorophyll fluorescence (Exp. 2). D, Drought stress assay with plants staggered (the mutant was sown 10 d before Col-0) and grown in the same pot under normal conditions. The last watering occurred at day 28 after transfer to soil. A photograph was taken every 3 d, and the soil water content was measured.
Osmotic stress has been used in previous studies to mimic a drought stress experiment. Given the osmotic stress tolerance in root growth, we investigated the drought tolerance of the ndufs4 mutant. Control and ndufs4 plants of the same age were grown in separate pots. Watering was stopped and plants were visually inspected over time. This experiment clearly demonstrated superior survival of the ndufs4 mutant after 10 d of nonwatering in multiple experiments (Fig. 9C). Direct comparison of multiple ndufs4 and ndufa1 plants in one experiment showed similar results in both mutants compared with Col-0 (Supplemental Fig. S5). However, when ndufs4 and Col-0 were staggered in planting and grown in the same pot and the watering was stopped after 4 weeks, both ecotypes responded in the same way to the decreasing soil water content (Fig. 8D). The results of these two experiments can be best explained by ndufs4 having a lowered water use rate than Col-0 plants when growing alone and remaining turgid longer, but when the soil water content is the same for both genotypes, ndufs4 does not remain viable any longer than Col-0. Hence, while there appears to be a water deficit/osmotic stress tolerance phenotype, it at least in part reflects differences in water uptake from the soil and/or delayed growth altering transpiration rates, not a markedly enhanced tolerance to a given soil water content. This is despite a clear tolerance to osmotic shock induced by mannitol, which is statistically significant. This is not unexpected, as soil-based water deficit is a different stress than mannitol-induced osmotic shock (Verslues et al., 2006).
DISCUSSION
When the nonphosphorylating pathways of plant respiration were first identified, their most intriguing characteristic was considered to be decreased efficiency of phosphorylation, because this was seen as such a pivotal factor in animal respiration and respiratory disease research (Gellerich et al., 2004). Lowered efficiency was also seen as a wasteful process; hence, elimination of the nonphosphorylating pathways in plants was for a time seen as desirable. The potential for AOX to lower phosphorylation efficiency and enable increased flux through glycolysis and the TCA cycle for providing carbon skeletons for biosynthesis without the stricture of adenylate control was the first general role proposed for this bypass (Lambers, 1982). However, the stress induction of nonphosphorylating pathways and the potential of AOX to limit ROS production from the ubiquinone pool has dominated the study of this nonphosphorylating pathway for many years.
Recently, it was shown that knockout of Aox1a leads to stress sensitivity in high light and drought in Arabidopsis and a stress-induced accumulation of sugars and depletion of organic and amino acids compared with the wild type (Giraud et al., 2008). This suggested a constriction of primary metabolism in the absence of AOX, which is consistent with the concept of nonphosphorylating respiration as a loosener of adenylate restriction on metabolism. In this study, we have characterized an insertion mutant in the ndufs4 gene and have shown that this plant lacks a fully assembled and active complex I. However, it exhibits few other significant changes in mitochondrial composition or respiratory function. Thus, the additional responses observed are likely to be primarily due to the lack of proton translocation by complex I and a concomitant lowering of phosphorylating efficiency rather than a restriction on respiratory rate. In this mutant, the same photosynthesis-derived organic acids will yield approximately one-third less ATP during their oxidation to drive growth and maintenance operations, and consistent with this, there were significant and dynamic alterations of the ATP and ADP pools in ndufs4 (Fig. 7; Supplemental Tables S5 and S6). The mutant exhibits a slower growth rate at all stages of development, and many of the transcripts for housekeeping functions (RNA and protein metabolism, cell cycle) are decreased in abundance, indicating a slower pace of cellular function. This growth delay has also been observed in other characterized plant complex I mutants, such as the CMSII tobacco mutant (Gutierres et al., 1999) and the Arabidopsis ca2 (Perales et al., 2005), otp43 (de Longevialle et al., 2007), and fro1 (Lee et al., 2002) mutants.
Delayed Germination and the Impact of Sugars and GA
We have shown that the germination of the ndufs4 mutant is affected. Germination is the transition from a dormant stage during which the metabolic activity is kept to its minimum to a stage of highly active cell division and expansion, requiring efficient energy production. In a dry seed, mitochondria are barely functional, but during the very early stages of germination, mitochondria are transformed into a mature active form (Howell et al., 2006). During the first few hours, they are mainly powered by external alternative dehydrogenases (Logan et al., 2001) to allow the import of energy-producing components such as the TCA cycle enzymes or the respiratory chain complexes (Howell et al., 2006). It could be predicted that a complex I mutant should not be affected at this stage of germination. Once the mitochondria are functional, the embryonic cells start to rapidly divide and expand. These energy-requiring processes are more likely to be affected in mutants lacking complex I, and this would explain why ndufs4 germination and early seedling growth are delayed. This is confirmed by the facts that mature seeds are not accumulating ATP and that ATP concentration increases during the early stages of germination (Perl, 1986). The mutant's germination is also inhibited at high Suc concentrations. This has also been observed in the fro1 mutant (Lee et al., 2002) and in the css1 mutant, which is affected in the splicing of nad4 (Nakagawa and Sakurai, 2006). There is a strong relationship between sugar and abscisic acid (ABA) signaling, as many Suc-insensitive mutants are unable to synthesize or insensitive to ABA (Dekkers et al., 2008). We have shown that external GA is able to complement the sugar sensitivity of the ndufs4 mutant; thus, we can postulate that this phenotype arises from the mutant being either more sensitive to ABA or less sensitive to GA. Early studies of Brassica napus germination and its control by ABA clearly show the correlation between respiration rate, ATP content, and the inhibitory influence of ABA (Schopfer and Plachy, 1984). While the causative factor in these correlations is not known, lowered ATP synthesis in ndufs4 would be consistent with slower germination and higher ABA (that could be overcome by GA-induced germination, as was observed). Interestingly, a relationship between GA synthesis and impaired respiration has been postulated in the CMSII mutant but in a different context. Under low-nitrogen conditions, which enhanced levels of GA, the CMSII mutant has a similar phenotype to wild-type plants (Pellny et al., 2008), while under normal nitrogen nutrition and with lower GA, a delayed growth phenotype was observed (Gutierres et al., 1997; Pellny et al., 2008). These data suggest that GA signaling or ABA depletion is partially impaired in mutants lacking complex I and that efficient ATP production by respiration is crucial for rapid germination.
Effects on Photosynthesis
In the ndufs4 mutant, the expression of nuclear genes encoding proteins needed for the light reactions of photosynthesis is significantly up-regulated, and while no effect could be shown at the protein level, fluorescence measurements suggest impaired photochemical efficiency. In the CMSII mutant, photosynthesis is reduced by 20% to 30% under normal conditions, but this phenomenon remains unexplained, as photosynthetic capacity, stomatal parameters, and O2 sensitivity are normal in the mutant (Dutilleul et al., 2003a). Interestingly, induction of photosynthetic genes has been shown previously in Chlamydomonas when the mitochondrial respiratory chain is inhibited (Matsuo and Obokata, 2006), whereas we have previously shown that there was an early transcriptional decrease in photosynthetic light reaction gene expression in response to rotenone inhibition (Garmier et al., 2008).
The up-regulation of photosynthetic light reaction gene expression in ndufs4 could be explained by a higher light sensitivity of the photosynthesis apparatus in the ndufs4 mutant. Consistent with this, we noted that genes encoding elements for signaling by light (BIN 30.11) are up-regulated in ndufs4 (Supplemental Table S2) and that the mutant has a “curly leaf” phenotype that is stronger when plants are grown in a long-day photoperiod than in a short-day photoperiod (Supplemental Fig. S3). The overexpression of genes encoding PSII subunits could be a response to a higher light sensitivity. PSII is the protein complex with the highest turnover in plants, and its subunits, especially D1, have to be synthesized continuously to replace photodamaged components. As metabolites known to act in shuttling reducing equivalents from the chloroplast to the mitochondria accumulated in the mutant (e.g. malate used in the malate-oxaloacetate shuttle), perhaps impaired metabolite shuttling from the chloroplast to the mitochondria may be inducing a stress on photosynthetic activity in the mutant. Alternatively, the change in photosynthetic gene transcription may reflect the alterations to mitochondrial function and not any change in the chloroplast. The effect of mitochondrial defects on retrograde signals that can modulate chloroplast gene expression has been observed previously in AOX mutants (Giraud et al., 2008).
There is no apparent photorespiratory blockage at Gly decarboxylase in ndufs4, as Gly levels are not significantly higher than in the wild type in the day (Supplemental Tables S5 and S6). Such a block might be expected given the high rate of NADH production in the mitochondrial matrix and the need for the mitochondrial electron transport chain to oxidize this pool. Notably, deletion of AOX did cause a clear accumulation of Gly and a decrease in Ser (Giraud et al., 2008), showing that loss of the nonphosphorylating components of ETC limits photorespiration but the loss of the classical proton-pumping complex I does not.
Consequences on Respiration at Night
The measurement of respiratory activity in leaves kept in the dark showed no change in oxygen uptake (Fig. 3A), but as the ATP level was lowered in the mutant at the end of the night, this was likely to be occurring with lower phosphorylation efficiency and hence yielding less ATP for growth and maintenance of plants at night. Such conditions may allow a Pasteur effect and elevated glycolytic activity, providing substrate-level phosphorylation to supplement the ATP pool generated by OXPHOS but also leading to higher levels of carbon intermediates requiring respiration. As the rate of respiration did not increase, these would be expected to accumulate; consistent with this scenario, amino acids derived from the convergence of glycolysis and the pentose phosphate pathway in the shikamate pathway (Tyr and Phe) and TCA cycle intermediates were seen to accumulate. Two studies have previously sought to understand the consequences of complex I loss through a broad analysis of metabolite levels: the treatment of Arabidopsis cell culture by rotenone (Garmier et al., 2008) and the characterization of a natural complex I mutation in tobacco (Dutilleul et al., 2005). The results obtained in these studies were starkly different. After inhibition of respiration in Arabidopsis cells by rotenone, the amino acid pools decreased, glycolysis became disconnected from the mitochondria, and there was an activation of cytoplasmic pathways such as fermentation (Garmier et al., 2008). In the CMSII mutant from tobacco, an accumulation of amino acids was observed (Dutilleul et al., 2005).
Our study shows a strong accumulation of amino acids and is in accordance with the CMSII tobacco data, suggesting that the effects of complex I mutations in different species show more similarity to each other than do comparisons of short-term and long-term inhibition studies in the same species. Our metabolomic study shows that the abundance of many more metabolites is significantly changed at night than during the day in both ndufs4 and ndufa1 lines. This observation highlights the important role of mitochondrial respiration in controlling metabolic fluxes at night and the masking of many of the effects of complex I loss during photosynthesis in the day period.
Stress Responses in ndufs4
The T-DNA insertion studied in this report is in the same gene as an ethyl methanesulfonate mutant, fro1, characterized from a screen for plants showing reduced cold induction of stress-responsive genes (Lee et al., 2002). This mutant was named frostbite1 due to a leaf phenotype resembling leaves after freezing (Lee et al., 2002). While we were unable to reproduce this leaf phenotype here in the T-DNA insertion mutant ndufs4, the lack of cold inhibition of root growth we note may be linked to the suggestion by Lee et al. (2002) that cold induction of nuclear gene expression is modulated by mitochondrial function through complex I function. This study and the fro1 mutant thus link mitochondrial respiration, especially changes in complex I activity, with lack of response to environmental changes. Similarly, the CMSII mutant has a higher tolerance to ozone and Tobacco mosaic virus (Dutilleul et al., 2003b), and the ccs1 mutant is more resistant to an inhibitor of cellulose synthesis (Nakagawa and Sakurai, 2006). The ndufs4 mutant has a higher tolerance of cold, osmotic, and moderate salt stress based on root growth assays and shows significant induction of stress-related transcripts (Supplemental Table S1) and metabolites such as Pro and putrescine. The mutant will thus be predisposed to cope with a stress and might not be as affected as the wild type. Taken together, these data clearly indicate that mutants with decreased OXPHOS are more tolerant to moderate stresses.
CONCLUSION
We have shown that the loss of one subunit of complex I can remove complex I function without significant changes in other mitochondrial functions, and we investigated the wider effects of this loss on nuclear and organellar transcript pools and the cellular metabolome. We have described what are, to our knowledge, new phenotypes for complex I mutants such as exogenous GA-stimulated germination and root growth tolerance to abiotic stresses. Overall, we show that the efficiency of ATP production by OXPHOS is lowered in ndufs4 but that this can be tolerated in Arabidopsis through broad rearrangements in cellular metabolism and development, which lead to altered tolerance to moderate abiotic stresses. This constitutive decreased efficiency of phosphorylation gives us insights into the value of the transient modulation of phosphorylation efficiency afforded to plants through the dynamic up-regulation of nonphosphorylating respiratory bypasses during abiotic stress (Clifton et al., 2005) and even during the diurnal cycle (Escobar et al., 2004) that are consistent with the decreased stress tolerance and lowered metabolic pools of knockouts of nonphosphorylating pathways (Giraud et al., 2008).
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized in a solution containing 70% (v/v) ethanol and 0.1% (v/v) Tween 20 for 5 min, washed first in 70% (v/v) ethanol and then in 100% (v/v) ethanol, dried, and plated onto plates of 0.5× Gamborg B5 medium, 1% (w/v) Suc, and 1% (w/v) agar. Plates were cold stratified at 4°C for 2 d. Two-week-old seedlings were transferred on soil and grown in growth chambers under a long-day photoperiod of 16 h of light (150 μE cm−2, 22°C) and 8 h of dark (18°C). Plants used for isolation of mitochondria were grown under a short-day photoperiod (8 h of light and 16 h of dark). Plants used for metabolite extraction were grown under a medium-day photoperiod (12 h of light and 12 h of dark).
Transcript Analysis
Total RNA from leaves harvested in the middle of the light period was extracted using the plant RNeasy kit (Qiagen), and genomic DNA was removed by two DNase treatments using RNase-free DNase (Ambion). Complete removal of both mitochondrial and nuclear DNA was checked by PCR on diluted RNA. Microarray ATH1 chip hybridizations were conducted following Affymetrix instructions. All of the experiments were done on three biological replicates. Microarray data analysis was conducted using AVADIS software (Strands). Arrays were normalized using GC-RMA. A stringent false discovery rate correction was applied to P values when individual fold changes were studied (Nettleton, 2006) but not when genes were studied in functional groups. The latter tests were conducted using MapMan (Thimm et al., 2004; http://gabi.rzpd.de/projects/MapMan/) with the Ath_AFFY_TAIR7 mapping file. A Benjamini-Hochberg correction was applied to the P values calculated by the Wilcoxon rank sum test. Primary data are available at Array Express (www.ebi.ac.uk/microarray/) as E-MEXP-1967 for ndufs4 and E-MEXP-2178 for ndufa1; P < 0.05 data sets are provided in Supplemental Data S1.
Organellar Transcript Analysis
Quantitative RT-PCR was performed using a Roche LightCycler 480 Real Time PCR system as described previously (de Longevialle et al., 2008). The mitochondrial transcriptome analysis was conducted in the same way as the chloroplastic transcriptome analysis, except that primers from Supplemental Table S7 were used.
Isolation of Mitochondria from Arabidopsis Plants
The aerial parts of plants grown under a short-day photoperiod were used as starting material for isolation of mitochondria. Plants were ground at 4°C in extraction buffer (0.3 m Suc, 5 mm tetrasodium pyrophosphate, 10 mm KH2PO4, pH 7.5, 2 mm EDTA, 1% [w/v] polyvinylpyrrolidone 40, 1% [w/v] BSA, 5 mm Cys, and 20 mm ascorbic acid) using a Polytron (40 mL of buffer for 10 g of plants, three bursts of 10 s, speed 4). The homogenate was centrifuged for 5 min at 3,000g, and the supernatant was centrifuged for 10 min at 20,000g. The pellet was resuspended in wash buffer (0.3 m Suc, 1 mm EGTA, and 10 mm MOPS/KOH, pH 7.2) and subjected to the same low-speed (3,000g) and high-speed (20,000g) centrifugations. The pellet was then resuspended in a small volume of wash buffer and loaded on top of a Percoll step gradient (from bottom to top: 1 volume of 50%, 5 volumes of 25%, and 1 volume of 18% Percoll in wash buffer, equivalent to 20 g of plant loaded per gradient). The gradient was centrifuged at 40,000g for 45 min. The fraction located at the interface between the 50% and 25% layers was collected and washed three times in wash buffer. The mitochondrial suspension was divided into aliquots and stored at −80°C.
Measurements of Oxygen Consumption
Oxygen consumption was measured using a Clark-type oxygen electrode (Hansatech Instrument). Respiration on whole Arabidopsis leaves was performed in the dark at 25°C by adding leaf discs (between 15 and 25 mg fresh weight) in the chamber containing 1 mL of distilled water. Respiration rates on isolated mitochondria were determined at 25°C on 100 μg of mitochondrial proteins suspended in 1 mL of respiration buffer (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] BSA, pH 6.8). Different substrates, cofactors, and inhibitors were successively added to the reaction medium to modulate oxygen consumption by mitochondria. Complex I inhibition was measured in the presence of Glu (10 mm), malate (10 mm), CoA (12 μm), TPP (0.2 mm), NAD+ (2 mm), and ADP (0.3 mm) in order to initiate electron transport through the TCA cycle. Rotenone (5 mm) was then added. The fraction of rotenone-insensitive respiration was calculated by the ratio of the rates after and before rotenone addition and expressed as the percentage of respiration inhibition. Complex I activity was measured on mitochondrial membranes obtained after freezing/thawing of the mitochondria, 1 mm deamino-NADH was added to the chamber, and oxygen consumption measured. Rotenone (5 mm) was then added. The capacity of internal NADH dehydrogenases was measured in the presence of Glu (10 mm), malate (10 mm), CoA (12 μm), TPP (0.2 mm), NAD+ (2 mm), ADP (0.3 mm), and rotenone (5 mm). n-Propylgallate (0.5 mm) and KCN (1 mm) were then added to inhibit the terminal oxidases and stop the reaction. The capacity of mitochondria to use external NADH was measured by addition of rotenone (5 mm), NADH (1 mm), CaCl2 (0.1 mm), ADP (0.3 mm), and ATP (0.5 mm). nPG (0.5 mm) and KCN (0.1 mm) were added to stop the reaction. The maximal capacity of AOX was measured in the same buffer but at pH 7.2 in the presence of succinate (10 mm) and ATP (0.5 mm) and after successive additions of the complex III inhibitor myxothiazol (2.5 μm) and the AOX activators pyruvate (10 mm) and dithiothreitol (DTT; 5 mm). Finally, the AOX inhibitor nPG (0.5 mm) was added. The ADP/O ratio was measured when mitochondrial respiration was run through the TCA cycle (state 3) in the presence of Glu (10 mm), malate (10 mm), CoA (12 μm), TPP (0.2 mm), and NAD+ (2 mm). State 4 was reached by the addition of ADP (200 μmol), and the amount of oxygen consumed during state 4 was monitored and used to calculate the ADP/O ratio by dividing 200 (the amount of ADP) by two times the amount of oxygen consumed. Mitochondrial integrity was measured following oxygen uptake after addition of ascorbate (10 mm), cytochrome c (25 μm), and Triton X-100 (0.05%, w/v). In all experiments, it was consistently better than 90%.
Measurements of Adenylate Concentrations
Production of ATP by isolated mitochondria was measured using the ATP Bioluminescent Assay Kit (Sigma). Mitochondria were incubated in respiration buffer containing Glu (10 mm), malate (10 mm), CoA (12 μm), TPP (0.2 mm), NAD+ (2 mm), and ADP (1 mm) for exactly 10 min. Then, TCA was added to a final concentration of 2.3% (v/v). After centrifugation for 15 min at 20,000g, the supernatant was used for ATP production estimation. To extract adenylates, frozen leaves (about 80 mg) were ground and resuspended in 500 μL of 2.3% (v/v) TCA. After centrifugation for 15 min at 20,000g, the supernatant was recovered and neutralized using a few drops of 2.5 m K2CO3. Adenylates were measured according to Ford and Leach (1998) using the ATP Bioluminescent Assay Kit (Sigma).
Protein Overexpression and Antibody Production
A cDNA encoding for the C-terminal part of the NDUFS4 protein (E37-N154) was cloned in the Gateway entry vector pDONR207 (Invitrogen) and transferred to the destination vector pDEST15 (Invitrogen), which allows the expression of the chimeric protein glutathione S-transferase∷NDUFS4. This protein was overexpressed in Escherichia coli BL21 pLysS cells and purified on glutathione S-transferase-Sepharose. The purified protein was injected into a rabbit (four doses of 250 μg). The serum was used unpurified in western-blot experiments.
One-Dimensional SDS-PAGE and Immunodetection
Mitochondrial proteins (20 μg) were solubilized in sample buffer (2% SDS, 125 mm Tris-HCl, 10% glycerol, 10% β-mercaptoethanol, and 0.002% bromphenol blue, pH 6.8) and heated at 95°C for 5 min. The fractions were loaded on 12% (w/v) polyacrylamide/0.1% (w/v) SDS gels. For immunodetection, separated proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and incubated with the primary antibody. A secondary antibody linked to horseradish peroxidase was used, and the signal was detected by chemiluminescence (GE Healthcare). The image was recorded using a Luminescent Image Analyser (LAS 100; Fuji). The list of antibodies used is given in Supplemental Table S1.
Blue Native-PAGE and Complex I Activity Assay
Blue Native-PAGE was performed according to the method described by Jänsch et al. (1996). Mitochondrial proteins (200 μg) were solubilized with dodecyl maltoside (1% [w/v] final) in ACA buffer (750 mm amino caproic acid, 0.5 mm EDTA, and 50 mm Tris-HCl, pH 7.0) and incubated for 20 min at 4°C. The samples were centrifuged for 10 min at 20,000g, and Serva Blue G (0.2% [v/v] final) was added to the supernatant. The samples were loaded onto a 4.5% to 16% gradient gel. Gels were directly used for in-gel complex I activity assay according to Zerbetto et al. (1997). The gel was washed three times for 5 min with distilled water and incubated in the reaction medium (0.14 mm NADH, 1.22 mm NBT, and 0.1 m Tris-HCl, pH 7.4). When the dark blue stain was strong enough, the reaction was stopped by transferring the gel to 40% methanol/10% acetic acid (v/v).
2D DIGE
Mitochondrial proteins from control plants (Col-0) and ndufs4 plants were precipitated overnight at −20°C in 100% (v/v) acetone. After centrifugation at 20,000g and 4°C for 20 min, pellets were resuspended in 10 μL of DIGE lysis buffer (8 m urea, 4% [w/v] CHAPS, and 40 mm Tris, pH 8.5) and centrifuged again at 12,000g and 4°C for 10 min in order to remove insoluble material. Fifty micrograms of proteins from each sample was labeled with a different Cy dye (Cy-3 or Cy-5) by addition of 400 pmol of dye (freshly prepared in dimethylformamide according to the manufacturer's instructions) on ice in the dark for 30 min. The reaction was stopped by addition of Lys (10 mm) for 10 min on ice in the dark. An equal volume (12 μL) of DIGE lysis buffer with 22 mm DTT was added to each sample. Each of the labeled protein samples was mixed, and rehydration buffer (8 m urea, 4% [w/v] CHAPS, 0.5% [v/v] 3–10 nonlinear immobilized pH gradient buffer, 18 mm DTT, and 0.001% [w/v] bromphenol blue) was added to give a final volume of 450 μL. The mix was loaded onto a 24-cm-long strip with immobilized nonlinear pH gradient of 3 to 10 (Immobiline DryStrip; GE Healthcare). Rehydration of the strips and the first IEF dimension electrophoresis were performed on an IPGphor unit (GE Healthcare) using the following settings: 12 h at 30 V (rehydration step), 1 h at 500 V, 1 h gradient from 500 V to 1,000 V, 1 h gradient from 1,000 V to 3,000 V, 2 h gradient from 3,000 V to 8,000 V, and 5 h at 8,000 V. After IEF, strips were incubated for 15 min in an equilibration buffer (6 m urea, 2% [w/v] SDS, 26% [v/v] glycerol, 65 mm DTT, 0.001% [w/v] bromphenol blue, and 50 mm Tris-HCl, pH 8.8) and then for 15 min in an equilibration buffer containing iodoacetamide (6 m urea, 2% [w/v] SDS, 26% (v/v) glycerol, 135 mm DTT, 0.001% [w/v] bromphenol blue, and 50 mm Tris-HCl, pH 8.8). The equilibrated strips were then loaded on top of a 12% (w/v) acrylamide gel. Following separation, gels were scanned using the Typhoon Trio Variable Mode Imager at a resolution of 100 (pixel size) with the photomultiplier tube set to 500 V. Proteins were processed (quantification) using the DeCyder 2-D Differential Analysis software version 6.5 (GE Healthcare). In order to get statistical significance from these experiments, three sets of proteins from three independent experiments were labeled and submitted to electrophoresis. Standard gels were also performed, and precipitation, IEF, and SDS-PAGE were run in parallel with labeled samples. These standard gels were loaded with a mix of 150 μg of each protein sample. After electrophoresis, proteins were visualized by colloidal Coomassie Brilliant Blue (G250) staining. The aim of the standard gel is to allow identification of proteins by excision gel spots followed by mass spectrometry.
Identification of Proteins by Liquid Chromatography-Tandem Mass Spectrometry
Gel spots to be analyzed were cut from colloidal Coomassie Brilliant Blue-stained gels. Proteins were digested with trypsin (trypsin sequencing grade; Roche Diagnostic) according to Sweetlove et al. (2001). Prior to mass spectrometry, peptides were resuspended in 5% acetonitrile/0.1% formic acid. The samples were analyzed with an Agilent 1100 series capillary liquid chromatography system and an Agilent Technologies XCT Ultra IonTrap with an electrospray ionization source equipped with a low-flow nebulizer in positive mode controlled by Chemstation (Rev B.01.03 [204]; Agilent Technologies) and MSD Trap Control version 6.0 (Build 38.15) software (Bruker Daltonik). Samples were loaded with an Agilent 1100 series capillary liquid chromatography system onto a 0.5- × 50-mm C18 (5 μm, 100 Å) reverse-phase column (Higgins Analytical) with a C18 OPTI-GUARD guard column (Optimize Technologies) at 10 μL min−1 equilibrated with 5% acetonitrile and 0.1% formic acid under a regulated temperature of 50°C. Peptides were eluted from the C18 reverse-phase column at 10 μL min−1 into the XCT Ultra by a 9-min acetonitrile gradient (5%–60%) in 0.1% formic acid at 50°C. Initial ion detection utilized a mass range of 200 to 1,400 mass-to-charge ratio (m/z) with a scan mode set to 8,100 m/z per second. Ions reaching an intensity of 80,000 cps were sent to tandem mass spectrometry (MS/MS) analysis, and two precursors were selected for the initial MS scan. MS/MS conditions employed SmartFrag for ion fragmentation (scan range 70–2,200 m/z). The resulting MS/MS spectra were exported from the Data Analysis for LC/MSD Trap version 3.3 (Build 149) software package (Bruker Daltonik) using default parameters for AutoMS(n) and compound Export. For protein identification, data generated from MS/MS spectra were matched against The Arabidopsis Information Resource (release 8) Arabidopsis database using the Mascot search engine (Matrix Sciences). Searches were conducted at parameters set at error tolerances of ±1.2 for MS and ±0.6 for MS/MS; “Max Missed Cleavages” were set to 1. Results were filtered using “Standard scoring,” “Max. number of hits” set to 20, and “Significance threshold” at P < 0.05.
Metabolite Extraction and GC-MS Analysis
Frozen leaves (approximately 100 mg) were ground to a fine powder (without thawing) in a ball mill and extracted with metabolite extraction medium (85% [v/v] HPLC-grade methanol [Sigma], 15% [v/v] untreated MilliQ water, and 100 ng μL−1 ribitol) for 15 min at 65°C with 1,400 rpm shaking. Tubes were centrifuged for 10 min at 20,000g and 4°C in order to pellet cell debris. Supernatants (50 μL) were dried in a speed vac system. Prior to GC-MS analysis, a first derivatization was performed by addition of 20 μL of methoxyamine hydrochloride (stock solution of 20 mg mL−1 in pyridine) on the dry metabolite pellet and incubation for 90 min at 30°C with shaking (1,400 rpm). A second derivatization was performed by addition of 30 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) and incubation for 30 min at 37°C with shaking (1,400 rpm). Ten microliters of a retention time standard mixture (0.29% [v/v] n-dodecane, 0.29% [v/v] n-pentadecane, 0.29% [w/v] n-nonadecane, 0.29% [w/v] n-docosane, 0.29% [w/v] n-octacosane, 0.29% [w/v] n-dotriacontane, and 0.29% [w/v] n-hexatriacontane dissolved in anhydrous pyridine) was then added to each tube, and the samples were transferred to GC-MS vials for analysis. Metabolites were analyzed on an Agilent GC/MSD system composed of an Agilent GC 6890N gas chromatograph fitted with a 7683B Automatic Liquid Sampler and a 5975B Inert MSD Quadrupole MS Detector (Agilent Technologies). The GC apparatus was fitted with a 0.25-mm i.d., 0.25-μm film thickness, 30-m Varian FactorFour VF-5ms capillary column with 10m integrated guard column (Varian). GC-MS run conditions were essentially as described for GC-quadrupole-MS metabolite profiling on the Golm Metabolome Database Web site (http://csbdb.mpimpgolm.mpg.de/csbdb/gmd/analytic/gmd_meth.html; Kopka et al., 2005). Samples were injected into the split/splitless injector operating in splitless mode with an injection volume of 1 μL, purge flow of 50 mL min−1, purge time of 1 min, and a constant inlet temperature of 300°C. Helium carrier gas flow rate was held constant at 1 mL min−1. The GC column oven was held at the initial temperature of 70°C for 1 min before being increased to 76°C at 1°C min−1 and then to 325°C at 6°C min−1 before being held at 325°C for 10 min. Total run time was 58.5 min. Transfer line temperature was 300°C. MS source temperature was 230°C. Quadrupole temperature was 150°C. Electron impact ionization energy was 70 eV, and the MS detector was operated in full-scan mode in the range 50 to 600 m/z with a scan rate of 2.6 Hz. The MSD was pretuned against perfluorotributylamine mass calibrant using the “atune.u” autotune method provided with Agilent GC/MSD Productivity ChemStation software (version D.02.00 SP1; Agilent Technologies). Raw GC-MS data preprocessing and statistical analysis was carried out using in-house MetabolomeExpress software (version 1.0). Peak detection settings were as follows: slope threshold = 200; minimum peak area = 5,000; minimum peak purity factor = 2; minimum peak width (scans) = 5. Peak lists were then selected for library matching against an in-house library of mass spectra and Kovat's retention index information for authentic metabolite standards and “unknown” metabolites observed in biological samples. Library-matching settings were as follows: retention index (RI) window = 2 RI units; mass spectral tag centroid distance = ±1 RI unit; minimum peak area = 25,000 arbitrary peak area units; MS qualifier ion ratio error tolerance = 30%; minimum number of correct ratio qualifier ions = 2; maximum average MS ratio error = 40%. Each positively matched analyte was quantified based on a particular analyte-specific, library-specified quantifier ion. For each run, Kovat's retention indices were internally calibrated against n-alkane internal standard retention times automatically determined using supplied approximate retention times and prominent extracted ion chromatogram peaks at m/z = 85. After library matching, a complete, normalized (analyte × sample) data matrix was constructed from the library-matching report. This matrix construction process included an automated quality-control step (designed to remove technically spurious samples) to ensure that only runs with a positively identified ribitol internal standard peak with a peak area of at least 106 arbitrary peak area units and with an absolute deviation from the median detected ribitol peak area that was less than 50% of that median were used. Missing signal intensity values were automatically filled (prior to normalization) by retrieving, from raw data files, the total signal of the relevant quantifier ion between the average start and stop times of peak integration of the relevant metabolite. Metabolite signal intensities were normalized to fresh tissue mass and internal standard (ribitol) peak area. Metabolite signal intensity ratios were then calculated by dividing the mean normalized signal intensity for each metabolite in ndufs4, complemented, and ndufa1 samples by its corresponding value in Col-0 samples. The statistical significance of each ratio was determined by a two-tailed Welch's t test with P < 0.05 being considered significant (n = 4–5). Tables of all the metabolites identified, their ratios between genotypes, P values, GC-MS retention times, retention indices, and quantifier ion information are given in Supplemental Tables S5 and S6. PCA were carried out on internal standard- and tissue mass-normalized data matrices filtered to exclude analytes of nonbiological origin and include only one GC-MS signal per metabolite. PCA analyses were initiated via the MetabolomeExpress interface, which uses the prcomp() function of the open-source statistical package, R (version 2.5.1), with scaling of all variables to unit variance.
Photosynthesis Measurements
Fluorescence induction kinetics at room temperature (22°C) were measured using a pulse amplitude modulation fluorometer (PAM101; H. Walz) on 32-d-old seedlings as described previously (Giraud et al., 2008). After a 30-min dark acclimation, the Fv/Fm (maximum photochemical efficiency of PSII in the dark-adapted state) was measured as (Fm – F0)/Fm, where Fm is the maximum PSII fluorescence in the dark-adapted state and F0 is the initial (minimum) PSII fluorescence in the dark-adapted state (Butler, 1978). Subsequent to illumination, the utilization of absorbed photons by the PSII antennae in photosynthetic electron transport and thermal dissipation was assessed from the quantum efficiency (Ψ) of photochemical energy dissipation (ΨPSII), light-dependent (ΨNPQ) and light-independent thermal dissipation, and fluorescence energy dissipation (Ψf,D), with Ψf,D + ΨNPQ + ΨPSII = 1 (Hendrickson et al., 2004). Fluorescence measurements were made on four plants for each of the Col-0 and ndufs4 lines.
Root Growth Stress Assays
Surface-sterilized ndufs4 and Col-0 seeds (approximately 200 seeds each) were sown onto a 1% Suc agar plate. When the cotyledons were fully opened, individual plantlets were transferred to 1% Suc agar plates at 22°C (control), 1% Suc agar plates for 48 h at 4°C for cold stress, or 1% Suc agar plates supplemented with 100, 200, 300, or 400 mm mannitol for osmotic stress or 50, 75, 100, 125, or 150 mm NaCl for salt stress (18 plantlets per treatment) at 22°C. Plantlets were kept at long-day growth conditions in a vertical position, and root growth were measured 9 d later.
NBT Detection of Superoxide in Leaves
In accordance with a previously described method (Lee et al., 2002), leaves of 4-week-old wild-type and ndufs4 plants grown under long-day conditions were detached and incubated in a superoxide staining solution (0.1 mg mL−1 NBT in 25 mm HEPES buffer, pH 7.6). As a control, wild-type and ndufs4 leaves were incubated in superoxide staining solution containing 10 mm MnCl2 and 10 units mL−1 superoxide dismutase. Leaves were left in staining solution for 2 h at room temperature in darkness. Chlorophyll was then removed from leaves via transfer to an 80% ethanol solution (70°C) for 10 min, and leaves were mounted onto glass slides in the presence of 40% (v/v) glycerol and scanned. Three leaves per plant, three genotypes per experiment, were assayed for superoxide. Each experiment was repeated three times, and representative images are shown for both experiments. Pixel densitometric analysis for determination of amounts of NBT precipitate in leaves was carried out using ImageJ.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR analysis of ndufs4.
Supplemental Figure S2. Changes in chloroplast gene expression in ndufs4.
Supplemental Figure S3. Phenotype of Col-0 and ndufs4 plants under long- and short-day photoperiod.
Supplemental Figure S4. Characterization of ndufa1.
Supplemental Figure S5. Drought stress assays of Col-0, ndufs4, and ndufa1 in individual pots.
Supplemental Table S1. Antibodies and dilution used in this work.
Supplemental Table S2. Microarray analysis of ndufs4 and ndufa1.
Supplemental Table S3. Expression of the alternative pathway genes in ndufs4 and ndufa1.
Supplemental Table S4. Expression of complex I subunits in ndufs4 and ndufa1.
Supplemental Table S5. Measurements of the metabolite levels in ndufs4 and complemented line versus Col-0.
Supplemental Table S6. Measurements of the metabolite levels in ndufs4 and ndufa1 versus Col-0.
Supplemental Table S7. Primers used for quantitative RT-PCR.
Supplemental Data S1. Microarray data on ndufs4 and ndufa1 versus Col-0.
Supplementary Material
This work was supported by the Australian Research Council Centre of Excellence (grant no. CEO561495) and by an Australian Research Council Australian Professorial Fellowship to A.H.M., a Western Australia State Premier's Fellowship to I.D.S., and a Ph.D. scholarship from the Grains Research and Development Corporation to A.J.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: A. Harvey Millar (harvey.millar@uwa.edu.au).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ayala A, Venero JL, Cano J, Machado A (2007) Mitochondrial toxins and neurodegenerative diseases. Front Biosci 12: 986–1007 [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT (2000) Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci 3: 1301–1306 [DOI] [PubMed] [Google Scholar]
- Brangeon J, Sabar M, Gutierres S, Combettes B, Bove J, Gendy C, Chetrit P, Des Francs-Small CC, Pla M, Vedel F, et al (2000) Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotiana sylvestris NMS1 nuclear mutant. Plant J 21: 269–280 [DOI] [PubMed] [Google Scholar]
- Butler W (1978) Energy distribution in the photochemical apparatus of photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 29: 345–378 [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC (2008) Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Mol Biol 67: 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003. a) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, de Paepe R (2003. b) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Franklin KA, Svensson AS, Salter MG, Whitelam GC, Rasmusson AG (2004) Light regulation of the Arabidopsis respiratory chain: multiple discrete photoreceptor responses contribute to induction of type II NAD(P)H dehydrogenase genes. Plant Physiol 136: 2710–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford SR, Leach FR (1998) Bioluminescent assay of the adenylate energy charge. Methods Mol Biol 102: 69–81 [DOI] [PubMed] [Google Scholar]
- Garmier M, Carroll AJ, Delannoy E, Vallet C, Day DA, Small ID, Millar AH (2008) Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellerich FN, Trumbeckaite S, Muller T, Deschauer M, Chen Y, Gizatullina Z, Zierz S (2004) Energetic depression caused by mitochondrial dysfunction. Mol Cell Biochem 256–257: 391–405 [DOI] [PubMed] [Google Scholar]
- Giegé P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ (2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15: 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Combettes B, De Paepe R, Mirande M, Lelandais C, Vedel F, Chetrit P (1999) In the Nicotiana sylvestris CMSII mutant, a recombination-mediated change 5′ to the first exon of the mitochondrial nad1 gene is associated with lack of the NADH:ubiquinone oxidoreductase (complex I) NAD1 subunit. Eur J Biochem 261: 361–370 [DOI] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, de Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson L, Furbank RT, Chow WS (2004) A simple alternative approach to assessing the fate of absorbed light energy using chlorophyll fluorescence. Photosynth Res 82: 73–81 [DOI] [PubMed] [Google Scholar]
- Hoefs SJ, Dieteren CE, Distelmaier F, Janssen RJ, Epplen A, Swarts HG, Forkink M, Rodenburg RJ, Nijtmans LG, Willems PH, et al (2008) NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet 82: 1306–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA (1988) Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331: 717–719 [DOI] [PubMed] [Google Scholar]
- Howell KA, Millar AH, Whelan J (2006) Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol Biol 60: 201–223 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Jänsch L, Kruft V, Schmitz UK, Braun HP (1996) New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J 9: 357–368 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kussmaul L, Hirst J (2006) The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc Natl Acad Sci USA 103: 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H (1982) Cyanide-resistant respiration: a non-phosphorylating electron transport pathway acting as an energy overflow. Physiol Plant 55: 478–485 [Google Scholar]
- Lee BH, Lee H, Xiong L, Zhu JK (2002) A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Millar AH, Sweetlove LJ, Hill SA, Leaver CJ (2001) Mitochondrial biogenesis during germination in maize embryos. Plant Physiol 125: 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld JR, Newton KJ (1994) The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138: 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Obokata J (2006) Remote control of photosynthetic genes by the mitochondrial respiratory chain. Plant J 47: 873–882 [DOI] [PubMed] [Google Scholar]
- Meyer EH, Giegé P, Gelhaye E, Rayapuram N, Ahuja U, Thony-Meyer L, Grienenberger JM, Bonnard G (2005) AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc Natl Acad Sci USA 102: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Sakurai N (2006) A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 47: 772–783 [DOI] [PubMed] [Google Scholar]
- Nettleton D (2006) A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellny TK, Van Aken O, Dutilleul C, Wolff T, Groten K, Bor M, De Paepe R, Reyss A, Van Breusegem F, Noctor G, et al (2008) Mitochondrial respiratory pathways modulate nitrate sensing and nitrogen-dependent regulation of plant architecture in Nicotiana sylvestris. Plant J 54: 976–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP (2005) Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J Mol Biol 350: 263–277 [DOI] [PubMed] [Google Scholar]
- Perl M (1986) ATP synthesis and utilization in the early stage of seed germination in relation to seed dormancy and quality. Physiol Plant 66: 177–182 [Google Scholar]
- Pla M, Mathieu C, De Paepe R, Chetrit P, Vedel F (1995) Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol Gen Genet 248: 79–88 [DOI] [PubMed] [Google Scholar]
- Priault P, Tcherkez G, Cornic G, De Paepe R, Naik R, Ghashghaie J, Streb P (2006) The lack of mitochondrial complex I in a CMSII mutant of Nicotiana sylvestris increases photorespiration through an increased internal resistance to CO2 diffusion. J Exp Bot 57: 3195–3207 [DOI] [PubMed] [Google Scholar]
- Rasmusson A, Moller I (1991) NAD(P)H dehydrogenases on the inner surface of the inner mitochondrial membrane studied using inside-out submitochondrial particles. Physiol Plant 83: 357–365 [Google Scholar]
- Rasmusson AG, Geisler DA, Moller IM (2008) The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8: 47–60 [DOI] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, de Kouchkovsky Y (2000) Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol 124: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Schopfer P, Plachy C (1984) Control of seed germination by abscisic acid. II. Effect on embryo water uptake in Brassica napus L. Plant Physiol 76: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Mowday B, Hebestreit HF, Leaver CJ, Millar AH (2001) Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett 508: 272–276 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vidal G, Ribas-Carbo M, Garmier M, Dubertret G, Rasmusson AG, Mathieu C, Foyer CH, De Paepe R (2007) Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 19: 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ II, Nikoskelainen EK (1988) Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Zerbetto E, Vergani L, Dabbeni-Sala F (1997) Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18: 2059–2064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.