Abstract
The architecture of grass genomes varies on multiple levels. Large long terminal repeat retrotransposon clusters occupy significant portions of the intergenic regions, and islands of protein-encoding genes are interspersed among the repeat clusters. Hence, advanced assembly techniques are required to obtain completely finished genomes as well as to investigate gene and transposable element distributions. To characterize the organization and distribution of repeat clusters and gene islands across large grass genomes, we present 961- and 594-kb contiguous sequence contigs associated with the rf1 (for restorer of fertility1) locus in the near-centromeric region of maize (Zea mays) chromosome 3. We present two methods for computational finishing of highly repetitive bacterial artificial chromosome clones that have proved successful to close all sequence gaps caused by transposable element insertions. Sixteen repeat clusters were observed, ranging in length from 23 to 155 kb. These repeat clusters are almost exclusively long terminal repeat retrotransposons, of which the paleontology of insertion varies throughout the cluster. Gene islands contain from one to four predicted genes, resulting in a gene density of one gene per 16 kb in gene islands and one gene per 111 kb over the entire sequenced region. The two sequence contigs, when compared with the rice (Oryza sativa) and sorghum (Sorghum bicolor) genomes, retain gene colinearity of 50% and 71%, respectively, and 70% and 100%, respectively, for high-confidence gene models. Collinear genes on single gene islands show that while most expansion of the maize genome has occurred in the repeat clusters, gene islands are not immune and have experienced growth in both intragene and intergene locations.
Genome sequencing of the maize (Zea mays) genome is nearing completion (Bennetzen et al., 2001; Chandler and Brendel, 2002; Wessler, 2006); it is the largest and most difficult-to-assemble plant genome sequenced to date. Maize is an important economic, agricultural, industrial, and research crop; however, with a genome close to the size of the human genome (2.8 Gb) and its high percentage of repetitive elements, acquiring the maize genome seemed a daunting task. Approximately 67% of the genome is made up of transposable elements (TEs; Haberer et al., 2005; Kronmiller and Wise, 2008), increasing the difficulty of assembly (Rabinowicz and Bennetzen, 2006). Much exploratory work has gone into isolating and sequencing just the gene areas and ignoring the repetitive regions, both by methylation filtration (Rabinowicz et al., 1999; Palmer et al., 2003; Whitelaw et al., 2003) and high-C0t (Whitelaw et al., 2003; Yuan et al., 2003) systems, which have assisted researchers with selecting only genic regions to sequence. These methods have captured a majority of the maize genic sequence (Fu et al., 2005), but they still have the potential to miss important regions. The current genome-sequencing project aims to capture the entire gene set of maize, including regulatory regions. However, the current strategy will not provide a fully assembled genome but rather assembled bacterial artificial chromosome (BAC) contigs ordered and orientated to provide complete gene regions that are adjacent to potentially incomplete TE clusters.
The landscape of the maize genome provides an interesting challenge for both sequencing and subsequent annotation. A high density of long terminal repeat (LTR) retrotransposons has had a direct effect on the genome size of many plant genomes, including maize (SanMiguel et al., 1996; Bennetzen et al., 2005; Hawkins et al., 2006; Piegu et al., 2006). Besides expanding genome size, LTR retrotransposons can have an impact on evolution of the species (Kidwell and Lisch, 2000). LTR retrotransposon insertions tend to form nested clusters (SanMiguel and Bennetzen, 1998), which are separated by small regions of several genes. Large nested repeat clusters consist of TE insertions inside TE sequences, expanding the repeat cluster and breaking up the sequence of the TEs found within, hindering repeat and gene annotation and increasing the difficulty of assembly. However, full sequence completion of the repetitive regions can be of great benefit to understanding the evolutionary history of the maize genome. LTR retrotransposons can provide an estimated time since insertion by calculating the divergence of their LTRs (Kimura, 1980; Ma and Bennetzen, 2004), and carefully sequenced assemblies of nested repeat clusters can help to illustrate their expansion, proliferation, and evolution across the genome (Kronmiller and Wise, 2008).
Previous studies of large contiguous regions of maize have provided a general view of the landscape of the genome. Unfinished sequence totaling 7.8 Mb from chromosome 1 and 6.6 Mb from chromosome 9 shows a gene density of one gene per 33 and 27 kb, respectively (Bruggmann et al., 2006). BAC contigs ranging in size from 126 to 405 kb show a gene density of one gene per 19 kb and genes found in small groups between large repeat clusters (Brunner et al., 2005). Genome-wide analysis of maize BACs has painted a different picture: while gene density of 100 random BACs at one gene per 44 kb was similar to the above results, genes were not observed in tight clusters (Haberer et al., 2005). When investigating gene-specific areas of maize, this dichotomy of gene density is also seen. Analysis of gene-rich regions such as the 22-kd α-zein gene family on maize chromosome 4 reveals a high density of genes, with one gene observed per 10 kb over 346 kb (Song et al., 2001). The Adh1 locus on maize chromosome 1 contains two genes across 280 kb, or one gene per 140 kb. Perhaps the only message learned here is that the gene density across the maize genome varies to a great degree, and large contiguous sequenced regions can begin to capture the true diversity of maize chromosome architecture.
In order to characterize large contiguous regions of maize sequence, we identified and sequenced two B73 BAC contigs from the centromeric region of chromosome 3. These contigs of 961 and 594 kb correspond to contigs 117 and 119, respectively, on maize WebFPC (Wei et al., 2007) and span regions associated with the rf1 (for restorer of fertility1) locus for Texas (T) cytoplasmic male sterility (cmsT; Duvick et al., 1961; Wise et al., 1996). As a foundation for the isolation of the Rf1 locus, four rf1 male-sterile mutants were recovered from a screen of 123,500 flowering plants (Wise et al., 1996). A 5.5-kb Mu1-hybridizing EcoRI restriction fragment was identified that cosegregated with the rf1-m3207 allele. Sequences from this fragment were hybridized to a Rf1 cDNA library, and probes designed from the identified cDNA, p6140-1 (Wise et al., 1999), were found to cosegregate with the rf1 locus in a recombinant population selected from over 10,000 progeny.
Using probes designed off the 5.5-kb cosegregating restriction fragment and the p6140-1 cDNA, we have identified two BAC contigs spanning the rf1 locus. Sixteen BACs were sequenced to completion to provide high-quality finished sequence. Here, we present two methods for computational finishing of highly repetitive grass genomes, which were successfully utilized to close 11 TE-induced gaps. Sixteen nested repeat clusters were found, each spanning as much as 155 kb and containing a variety of LTR retrotransposon types and ages of insertion. Genes are found tightly clustered, showing a density rate of one gene per 16 kb within gene islands. Finally, comparative analysis with rice (Oryza sativa) and sorghum (Sorghum bicolor) shows that while many genes are retained across all three species, genes have both been lost and translocated across the genomes.
RESULTS AND DISCUSSION
Mapping, Sequence, and Assembly of Maize rf1 Contigs
Analysis of Multiple B73 Maize BAC Libraries Leads to Two Separated rf1-Associated Contigs
Six tightly linked and cosegregating low-copy amplification of insertion mutagenized sites (AIMS) fragments (Frey et al., 1998) were identified from three rf1-m families (Wise et al., 1996) and, along with sequences selected from the cosegregating 6140-1 cDNA (Wise et al., 1999), were used as probes against the first B73 library filters (ZMMBBa; Clemson University Genomics Institute). From each of the resulting short nonoverlapping contigs, overgo probes were designed off sequenced BAC ends. Low-copy hybridizing probes were used for the next round of hybridization to the B73 BAC library; identified BACs were used to extend the length of the existing rf1 contigs.
After the National Science Foundation-sponsored maize physical mapping project was under way (NSF-PGR no. 9872655), additional BAC clones were identified by hybridization to the ZMMBBb and ZMMBBc libraries and subsequently via in silico overlaps from the maize WebFPC database (Coe et al., 2002; Wei et al., 2007), and a minimal tiling path was constructed from a total of 796 BACs from the three B73 BAC libraries. The minimal tiling path formed two contigs both located on chromosome 3. rf1-associated contig 1 (rf1-C1) and rf1-associated contig 2 (rf1-C2) correspond to contigs 117 and 119, respectively, in maize WebFPC and are located in maize bin 3.04.
Sequencing and Initial Assembly of Maize BACs
Sixteen BAC clones were fully shotgun sequenced to provide the most accurate representation of this region. Initial sequencing produced 8- to 9-fold coverage; however, after initial assembly, additional plates were produced if the BAC was deemed highly repetitive. Once the draft sequence was completed, BACs averaged 10× coverage, depending on their repeat content (Table I). BACs were finished via standard gap-closing techniques (see “Materials and Methods”).
Table I.
Sequenced BACs across the rf1 locus
| BAC (ZMMBB) | Length | Sequence Deptha | BAC Percentage Repetitive | BAC Percentage Genic | Post-Finish Gaps | Computationally Closed Gaps | Gaps Remaining | Reasons for Gaps |
|---|---|---|---|---|---|---|---|---|
| rf1-C1 | ||||||||
| b0322P15 | 158,174 | 9.17 | 80.23 | 0.00 | 2 | 2 | 0 | |
| b0452F11 | 127,944 | 12.90 | 84.47 | 0.00 | 0 | 0 | 0 | |
| b0153H24 | 132,164 | 9.38 | 74.09 | 0.00 | 1 | 1 | 0 | |
| c0080G23 | 167,218 | 8.90 | 66.66 | 5.37 | 0 | 0 | 0 | |
| b0211C05 | 142,755 | 9.54 | 74.97 | 4.05 | 0 | 0 | 0 | |
| b0331I02 | 149,683 | 11.48 | 85.60 | 0.00 | 2 | 1 | 1 | Dinucleotide gapb |
| c0360P05 | 178,651 | 8.16 | 82.62 | 1.75 | 1 | 1 | 0 | |
| b0348F20 | 129,989 | 11.86 | 75.49 | 0.00 | 1 | 2 | 0 | |
| rf1-C2 | ||||||||
| b0345O22 | 112,741 | 10.52 | 93.01 | 0.00 | 3 | 3 | 0 | |
| c0294D02 | 171,602 | 7.38 | 78.56 | 4.43 | 2 | 0 | 0 | |
| b0103L15 | 154,263 | 8.41 | 85.58 | 0.00 | 1 | 1 | 0 | |
| b0622H01 | 156,570 | 10.39 | 80.46 | 1.24 | 0 | 0 | 0 | |
| b0335C07 | 132,421 | 12.36 | 84.13 | 0.78 | 0 | 0 | 0 |
To not count sequences from overlapping BACs twice, information presented here is calculated from the start of a BAC sequence to the start of the next overlapping BAC sequence.
Four areas of small nucleotide repeats causing the gap: GA × 300 bp, unresolved sequence gap, GA × 400 bp, TTAGGG × 620 bp, AT × 50 bp.
At this stage, BAC assemblies were as close to best possible condition that finished sequencing could bring (Table I, Post-Finish Gaps). Remaining gaps were closed with computational methods. BAC sequences were assembled with two programs, CAP3 (Huang and Madan, 1999) and phrap (Ewing and Green, 1998; Ewing et al., 1998; http://www.phrap.org). Incomplete regions were examined in order to present and submit completely finished BACs. Twelve gaps made it to this stage of computational finishing. Eleven of these gaps were caused by LTR retrotransposon misassemblies, one was caused by long strings of dinucleotide/gap/dinucleotide/hexanucleotide/dinucleotide simple polymer repeats. By careful analysis of the repeats, identification the retrotransposons, their associated LTRs and their nested structure, and mapping of paired end sequences of plasmid subclones, we were able to determine the correct sequence of all of the retrotransposon-caused sequence gaps; however, the correct sequence for the lone simple repeat gap remains elusive (Table I, Gaps Remaining).
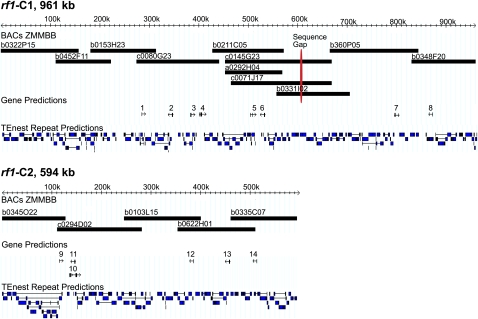
Finished BAC clones were verified with restriction digest analysis. For nongap regions, base pair quality is well within sequencing standards, with less than one error in 1 × 105 per BAC assembly. In the minimal tiling path, BACs average 32 kb of overlap, although the areas of overlap between ZMMBBb0211C05 and ZMMBBb0331I02 multiple BACs were sequenced to resolve mapping discrepancies. Fully assembled, the rf1-C1 is 961 kb, the rf1-C2 is 594 kb, and they have been submitted to GenBank (Benson et al., 2006) as EF517601 and EF517600, respectively (Fig. 1).
Figure 1.
Combined genetic and physical map of maize sequence contigs. GBrowse display of the rf1 BAC contigs of maize chromosome 3 showing BAC path, predicted genes, and annotated TEs. rf1-C1 is 961 kb and contains 11 repeat clusters and eight predicted genes. rf1-C2 is 594 kb and contains five repeat clusters and six predicted genes. The two BAC contigs are separated by approximately 30 Mb. One gap remains, caused by dinucleotide and hexanucleotide polymer repeats; this is shown on BAC ZMMBBb0331I02, found at approximately 607 kb on rf1-C1.
Characterization of Repetitive Gaps in Maize Sequence Assembly
In particular, two methods proved very useful to resolve maize sequence gaps that were unclosable with traditional laboratory-based finishing methods. Eleven gaps in the BAC assemblies were closed with purely computational methods. Two cases of a gap causing misassembly were found to be common in maize BACs, both involving the duplicated regions of LTRs of retrotransposons. The first misassembly type is much like any misassembly caused by a duplicated area within a BAC; the traces for one LTR all assemble into the second copy, breaking the sequence of the first LTR and causing a gap. This was seen most often in TEs with long LTRs where the whole sequence trace or even both end sequences from an entire subclone were within the LTR boundaries. This was also commonly seen on LTR retrotransposons with a recent age of insertion, and fewer polymorphisms introduced over the time since insertion between the two LTRs caused more assembly confusion.
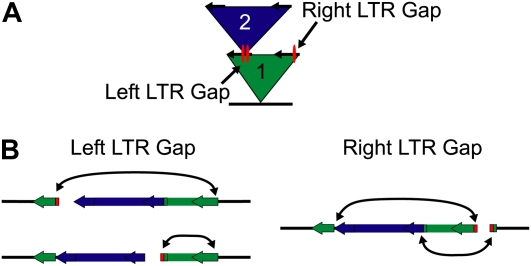
The second common case of misassembly was also caused by the LTRs of retrotransposons, seen when a LTR retrotransposon nested into one of the LTRs of an existing LTR retrotransposon. In this type, the gap can be found in either of the two LTRs of the first retrotransposon (Fig. 2A). Once this insertion occurs, the sequence of one LTR is interrupted with the sequence of the nested transposon. To cause a gap, during assembly the sequence from the complete LTR incorrectly aligns to both LTR locations, removing the join between the interrupted LTR and the nested TE. This recruitment causes a gap; now one or both of the contig ends that point into the gap have assembled traces belonging to the other LTR (Fig. 2B) and can cause one of two gaps in the nested LTR or one gap in the unnested LTR of the original LTR retrotransposon.
Figure 2.
Nested LTR retrotransposons cause sequence assembly gaps. Diagram of the commonly seen type of gap caused by nested LTR retrotransposons. A, Nested TE insertion view of the gap region. The blue TE (labeled 2) is found nested within the LTR of the green LTR retrotransposon (labeled 1). This can cause an assembly gap in one of three locations: at the insertion point of the blue TE on either the left or the right of the insertion, or on the other LTR of the green TE at the corresponding location of the insertion point. B, A sequence view of the three gap locations caused by insertion of the blue TE into the LTR of the green LTR retrotransposon. The blue TE has inserted into the left LTR of the green TE, and an assembly gap can be found on the left LTR to either the left or the right of the blue TE insertion. In either case, the sequence of the left LTR of the green TE has been split apart, and sequences belonging to the right LTR have incorrectly assembled at this split location (shown as the arrow pointing to the red sequence) and cause the gap assembly. The assembly gap can also occur on the right LTR of the green TE. Here, the join sequences between the left LTR and the blue TE, found on both sides of the blue TE insertion, can assemble incorrectly into the sequence of the right LTR and prevent the sequence from aligning. Successful closing of these types of gaps is crucial to characterization of maize nested repeat clusters.
The closure of the final unfinished gap, found in ZMMBBb0331I02 (Table I), has been hindered by long strings of simple repeat sequences. Simple repeats, such as homonucleotide polymers (AAAA), dinucleotide polymers (GAGAGA), or even larger repeated segments, inhibit thorough sequencing by allowing the DNA polymerase to slip on the DNA template or sequencing product, resulting in either a loss of polymerase or unreadable sequence beyond the difficult region. On one contig end this gap has a 305-bp string of GAs. The other side, starting from the gap and traveling into the contig, has approximately 700 bp of unique sequence, followed by 396 bp of GA repeated, followed by 620 bp of TTAGGG repeated, followed by 50 bp of ATs. Plasmid subclones surrounding the gap have not been able to close the gap when sequenced with the transposon-bombing method, and primers designed from the surrounding area have been unable to amplify PCR products. Sequencing off of primers designed in the most internal unique regions provides less than 100 bp of sequence. All of these results suggest a strongly bound hairpin across this area preventing complete sequence, with a possible fifth simple repeat section still within the gap.
Computational Methods for Closing Difficult Gaps: Genome-Based Approach
Two computational methods were designed to combat the misassemblies caused by repetitive sequences. The first method is termed the genome-based approach because it uses the biological or genomic information present in the BAC sequence to determine the correct assembly configuration. As explained above, many assembly gaps occurred when similar sequences are found in multiple locations in the BAC. In maize, this occurs frequently with the long LTRs of retrotransposons, when the traces for one or more location collapse their assembly into a single copy. Our genome-based approach uses the structure of the nested TEs to suggest the gap-filling sequence.
The first step in the genome-based approach was to run both contigs surrounding the gap with TEnest (Kronmiller and Wise, 2008). This gave two nested structure pictures of the contigs, and the TE insertions leading into the gap were examined for any gap-split TE insertions. For example, for the gap presented in Figure 2, one contig end would contain a partial LTR (and possibly some internal TE sequence) of one nested retrotransposon near the end of the gap and the other contig would contain the other sections of this partial retrotransposon along with the complete sequence of the nested TE. Other TE insertions, more than presented in the simple example of Figure 2, could confound the identification of the split TE, but they could also be of assistance. If the two TEs shown in the example were both nested in an older TE insertion, the older TE would also be split around the gap even farther from the problem region, providing more evidence for the nesting pattern.
Once the nesting structure of the TEs was identified using the above process, a string of DNA sequence could be filled in to span the gap. Sequence surrounding the gap was built to resemble the predicted nested TE structure. This built sequence contains three sections. The split LTR is formed by identifying its missing sequence donated by the corresponding full LTR. The join point between the split LTR and the nested TE exactly identified the nested location on the other side of the split LTR. Finally, the sequence of the nested TE is added to complete the sequence spanning the gap. A low-quality backbone phd file (Ewing et al., 1998) was created from the proposed gap-spanning sequence and used to drive the phrap assembly. From here, the correct sequence traces were found either during the assembly or by the user in Consed (Gordon et al., 1998). Several iterations were generally required to add or remove any sequence differences between the proposed backbone and the true sequence. Ultimately, sequences were found to span across the gaps, and custom sequencing primers were designed to help span low-quality regions if necessary.
Computational Methods for Closing Difficult Gaps: Sequence-Based Approach
The second computational method used for difficult gap closure used the sequence information from paired end plasmids. Essentially mimicking a localized constrained assembly, the sequence-based approach would back out of the gap into the contig looking for unique sequence unduplicated in the BAC assembly. This process backed up on both contigs for at least 4 kb (the largest plasmid clone length) but often much longer to find unique sequence. At the unique locations, all of the traces found in this area and the plasmid end pairs for these traces were built into separate assemblies. The phd file backbone sequences would be made from these small localized assemblies, and again overlapping sequences and their mate pairs would be added and assembled to the localized assemblies, continuing until the contigs identified and correctly assembled missing sequences or sequences incorrectly placed and walked into the gap.
This sequence-based approach was most useful on the simpler gaps caused by duplicated regions in the BAC that condensed the sequence into one region. In these misassemblies, the collapsed traces were identified by their plasmid mate pairs anchored in unique sequence and forced to assemble into the duplicated copy. This process also proved to be helpful to build a backbone phd sequence when closing gaps by the genome-based approach explained above. Often, the sequences that were needed to span the gap were hard to identify or did not match the predicted backbone sequence well enough to find by assembly or by hand, and this sequence-based method was useful to draw them to the correct location.
TE Annotation Reveals Large Repeat Clusters
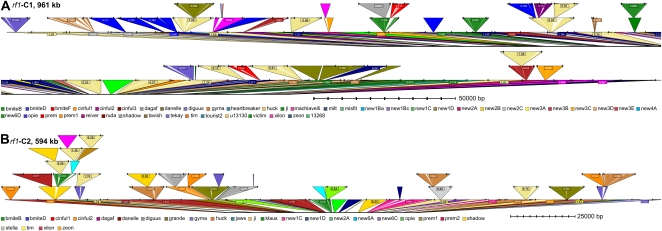
The two sequence contigs were repeat annotated with TEnest (Kronmiller and Wise, 2008) using the maize repeat database. For the rf1-C1 961-kb contig (EF517601), TEnest identified 60 whole LTR retrotransposons, three solo LTR sequences, six whole DNA transposons, and 42 partial TEs. For the rf1-C2 594-kb contig (EF517600), TEnest identified 41 whole LTR retrotransposons, four solo LTR sequences, two whole DNA transposons, and 18 partial TEs (Figs. 1 and 3). The ratios of solo LTR to whole LTR retrotransposon to DNA transposon to partial TE insertion were consistent with the genome-wide analysis of maize TEs we presented earlier (Kronmiller and Wise, 2008). The families of TEs identified, the abundance of solo LTR sequences, and the estimated age of insertion for LTR retrotransposons in these two sequence contigs were also found to be consistent with the previous results. The overall TE content is greater in these contigs, 78% compared with the previously reported 67% across the sampling of the finished maize BACs (both studies using the same repeat databases), possibly showing the bias of BACs selected for sequencing with high gene and low repetitive content in previously sequenced BACs.
Figure 3.
TEnest graphical display of maize sequence contigs. A, TEnest insertion display output of the rf1-C1 961-kb maize contig, split into two sections. B, TEnest insertion display output of the rf1-C2 594-kb maize contig. TEs are shown as triangles inserted into the black DNA line. The TE families are shown below (for detailed display, see Supplemental Fig. S1).
Definite separation between gene areas and repeat areas can be seen when large sections of the maize genome are evaluated. In maize, this phenomenon is known as oceans and islands, where islands of genes are found within oceans of repetitive clusters (SanMiguel et al., 1998). For this analysis of repeat clusters, we defined a cluster or ocean as a group of nested or closely inserted TEs. TEs found inserted less than 5 kb from each other and not separated by a predicted non-transposon-related gene were grouped together as a repeat cluster. Groups of TEs identified by this definition that contained less than three TE insertions were designated as TE insertions within a gene island and so were left out of repeat clusters. In total, 16 TE clusters were identified in the two rf1-associated contigs. Repeat clusters range in size from 23 to 155 kb, ranging from three to 18 TE insertions (Table II). While sizes of TE clusters are generally evenly distributed across the contigs, the two largest clusters are found on the smaller 594-kb rf1-C2 contig. This corresponds to its higher repeat percentage, 82% versus 75% of rf1-C1.
Table II.
TEs identified with TEnest by repeat cluster
| Repeat Cluster | Start | End | Size | TE Insertions | LTR Retrotransposon | Solo LTR | DNA Transposon | MITE | Partial |
|---|---|---|---|---|---|---|---|---|---|
| rf1-C1 | |||||||||
| RC1 | 1 | 31,288 | 31,287 | 3 | 3 | ||||
| RC2 | 34,881 | 82,158 | 47,277 | 4 | 4 | ||||
| RC3 | 101,703 | 191,706 | 90,003 | 14 | 7 | 3 | 4 | ||
| RC4 | 289,569 | 341,973 | 52,404 | 6 | 4 | 1 | 1 | ||
| RC5 | 366,102 | 389,224 | 23,122 | 5 | 2 | 1 | 2 | ||
| RC6 | 407,934 | 509,832 | 101,898 | 13 | 11 | 2 | |||
| RC7 | 536,833 | 573,823 | 36,990 | 6 | 3 | 1 | 2 | ||
| RC8 | 615,187 | 691,808 | 76,621 | 9 | 6 | 1 | 2 | ||
| RC9 | 699,169 | 792,216 | 93,047 | 12 | 7 | 5 | |||
| RC10 | 876,233 | 918,911 | 42,678 | 4 | 4 | ||||
| RC11 | 924,464 | 960,629 | 36,165 | 8 | 2 | 1 | 5 | ||
| rf1-C2 | |||||||||
| RC12 | 1 | 120,813 | 120,812 | 14 | 10 | 2 | 2 | ||
| RC13 | 147,102 | 302,699 | 155,597 | 18 | 12 | 2 | 2 | 2 | |
| RC14 | 313,302 | 367,983 | 54,681 | 8 | 4 | 1 | 3 | ||
| RC15 | 401,526 | 456,413 | 54,887 | 6 | 4 | 2 | |||
| RC16 | 515,062 | 593,635 | 78,573 | 12 | 6 | 1 | 5 |
TEnest displays clusters of TE insertions, with multiple layers of chronologically inserted TEs nested into one another. As repeat clusters become more dense and complex, the heights or levels of these TE insertion clusters increase. The level heights of TEnest-displayed repeat clusters observed here correspond to the lengths of repeat clusters; large repeat clusters contain more TE insertions, which have higher levels of nested TEs. The largest repeat group, repeat cluster 13 (RC13) at 155 kb, has 18 TE insertions, 12 of which are full LTR retrotransposons (Table II). This cluster has a height of six nested TEs. Estimated times since TE insertion are spread evenly throughout the repeat clusters; larger clusters do not have younger or older LTR retrotransposon insertions when compared with smaller clusters. As expected, TE nested clusters are seen with older insertions found lower in the cluster and younger insertions found at higher levels. Partial TE insertions, resulting from whole TEs that have either undergone a deletion or rearrangement at the sequence location or that have mutated significantly so that characterization becomes increasingly difficult, are most often found at the lowest levels of nested TE clusters and so correspond to the oldest TE insertions. This is expected, as after enough time for mutations to accumulate the identified TE fragments cannot be reconstructed.
LTR retrotransposons were examined for differing insertion patterns between repeat clusters. Of the three most abundant retrotransposons found in maize (Meyers et al., 2001), Huck, a Gypsy element, was found to be distributed evenly across the contigs and repeat clusters. Opie, a Copia element, was found nested almost exclusively in one location. Four full and two partial Opie retrotransposons were identified in RC2 and RC3, while only three full and one partial Opie elements were found across the rest of the contigs. Ji, a Copia element with sequence identity similar to Opie, was found to have a scattered distribution across rf1-C1 but to have three full and two partial insertions on rf1-C2 in RC12 and RC13. Xilon, a Gypsy element, has two full-length insertions found in RC11 out of three total Xilon elements found. Six MITE DNA transposons were identified, three in RC3 and two in RC13, inserted close to each other within each cluster. This is in contrast to other results that show that MITEs preferentially insert into the 3′ upstream regulatory regions of genes (Mao et al., 2000; Zhang et al., 2000). Six solo LTRs were identified across the two rf1-associated contigs. Four Gyma, one Ruda, and one Danelle solo LTR are seen scattered evenly across the repeat clusters. An interesting finding is the high observation of full-length Gyma solo LTRs, all seen in the 594-kb rf1-C2.
Distances between clusters, which can also be characterized as length of gene islands, range in size from 4 to 98 kb, averaging 33 kb long. These sizes heavily rely on the definition of repeat clusters and would significantly change with modifications to this rule that would separate or combine the repeat cluster sets. Gene islands are not devoid of TE insertions, as described by the definition for repeat clusters. We also attempted to characterize the differences between TEs found inserted within TE clusters versus those found in gene islands. Many of the TE insertions within gene islands are partial LTR retrotransposons: 18 TEs out of 36 total gene island TEs. This suggests that ancient TE insertions have occurred in these areas and have since been mutated beyond recognition. Eleven whole LTR retrotransposons were found in gene islands. These are not younger, recently integrated LTR retrotransposons but rather older yet complete insertions. Instead, the recently inserted LTR retrotransposons are seen almost exclusively at the top levels of repeat clusters. There is one observed exception: a Shadowspawn LTR retrotransposon inserted into rf1-C2 at 389 kb has an estimated time since insertion of 0.231 million years ago. Also seen is nested Ji retrotransposon (0.154 million years ago) inserted within an older Huck (0.654 million years ago) found in a gene island between 456 and 507 kb on rf1-C2. The Huck TE follows the observed pattern of older LTR retrotransposons inserted into gene islands, the younger Ji does not, but because it is inserted within the Huck element, the selective pressures against its insertion may not be as strong than if it was to insert directly within the gene island; thus, it has less chance to disrupt nearby gene functions.
Predicted Maize Genes Are Found Clustered in Islands
Sequence file repeats masked by TEnest were used for gene prediction. These masked files were analyzed with three programs: GeneSeqer (Schlueter et al., 2003), FGENESH (Salamov and Solovyev, 2000), and GeneMark.hmm (Lukashin and Borodovsky, 1998). Each of these programs has a monocot- or maize-specific model. EST and protein sequences from Arabidopsis (Arabidopsis thaliana), Avena sativa, Brachypodium distachyon, Hordeum vulgare, O. sativa, Saccharum officinalis, S. bicolor, Secale cereale, Triticum aestivum, and maize were aligned to the two repeat masked contigs. Results from repeat masking, gene prediction, and sequence alignments were visually displayed with the Generic Model Organism Database package Generic Genome Browser (GBrowse; Stein et al., 2002; Fig. 1). Exon structure for each gene model identified by the three prediction programs was plotted on GBrowse and compared with the evidence-based sequence alignments. A consensus approach between the results of the three gene prediction programs and the EST and protein alignments was used to pick candidate genes and build gene models. Eight predicted genes were identified in rf1-C1 and six were identified in rf1-C2 (Table III). The sequences for these predicted gene exons were exported and examined for sequence similarity to the characterized genes in GenBank (Benson et al., 2006) to determine possible gene functions. PCR primers were designed in predicted gene exons for high-resolution genetic mapping.
Table III.
Predicted genes identified across the two maize sequence contigs of chromosome 3
| Gene | Start–Stop Coordinates | Collinear Comparative Alignments
|
Predicted Functionc | |||
|---|---|---|---|---|---|---|
| Sorghum Genea | Sorghum Locationa | Rice Geneb | Rice Locationb | |||
| rf1-C1 | ||||||
| 1 | 287,591–288,464 | Sb03g005160 Sb03g005163 Sb03g009440 Sb03g009450 Sb03g009460 | 3: 5,388,191–5,390,991 3: 5,396,753–5,397,361 3: 10,169,571–10,170,223 3: 10,181,483–10,182,246 3: 10,186,387–10,187,099 | Os01g14700 Os01g14710 | 1: 8,228,201–8,228,903 1: 8,231,933–8,232,871 | Heavy-metal-associated domain-containing protein, Arabidopsis, NP_850876 |
| 2 | 344,659–345,858 | Sb03g005180 Sb03g009480 | 3: 5,406,762–5,408,141 3: 10,196,462–10,197,835 | Os01g14720 | 1: 8,231,697–8,232,599 | Transcription regulator, Arabidopsis, NP_198156 |
| 3 | 396,751–397,645 | Hypothetical gene | ||||
| 4 | 399,956–405,975 | Sb03g005190 Sb03g009490 | 3: 5,416,176–5,420,210 3: 10,205,042–10,209,131 | T-complex protein 1 subunit β, maize, ACG33558 | ||
| 5 | 516,699–521,496 | Sb03g009540 | 3: 10,260,909–10,265,042 | Os01g14820 | 1: 8,281,320–8,285,398 | Pigment-defective 320, Arabidopsis, NP_566296 |
| 6 | 527,928–528,314 | Sb03g009560 | 3: 10,275,285–10,337,556 | MFS18 protein, maize, ACG25280 | ||
| 7 | 797,020–799,513 | Sb03g009580 | 3: 10,333,816–10,338,112 | Os01g14860 | 1: 8,327,829–8,330,335 | Glycogen synthase kinase-3 MsK-3, maize, NP_001150105 |
| 8 | 874,156–875,402 | Sb03g009600 | 3: 10,346,011–10,347,106 | Os01g14890 | 1: 8,341,448–8,342,475 | Predicted gene, maize, NP_001130618 |
| rf1-C2 | ||||||
| 9 | 120,996–121,223 | Hypothetical gene | ||||
| 10 | 127,687–144,158 | Sb03g013600 | 3: 17,162,324–17,171,223 | Os01g23640 | 1: 13,278,985–13,287,453 | mov34/MPN/PAD-1 family protein, maize, NP_001149862 |
| 11 | 129,898–130,077 | Hypothetical gene | ||||
| 12 | 382,024–383,247 | Sb03g013615 | 3: 17,230,890–17,232,099 | Ubiquitin-protein ligase, Ricinus, EEF52805 | ||
| 13 | 458,175–460,116 | Sb03g013620 | 3: 17,234,336–17,236,738 | Os01g24780 | 1: 13,923,887–13,926,971 | Cytochrome P450, Triticum, AAR11387 |
| 14 | 511,265–512,304 | Hypothetical gene | ||||
Predicted maize genes aligning to predicted sorghum genes of sorghum genome assembly version 1 are shown. Sorghum genome location is displayed as chromosome: start location–end location.
Predicted maize genes aligning to predicted rice genes of rice genome assembly version 5 are shown. Rice genome location is displayed as chromosome: start location–end location.
Predicted functions are for proteins found to be similar to predicted rf1-associated genes by BLASTX. “Predicted gene” refers to predicted genes that align to uncharacterized proteins in GenBank, and “hypothetical gene” refers to predicted genes that were identified by gene prediction software and align to sequenced ESTs found in GenBank but do not align to uncharacterized proteins in GenBank.
Complete gene models were identified for all 14 predicted genes. Gene model and exon coordinates are given in Supplemental Table S1. Predicted functions were assigned to nine of the identified genes (Table III). Genes that we were unable to assign function were given one of two notations: predicted, if the predicted protein has a full-length alignment to other submitted nonfunctionally characterized proteins; or hypothetical, if the predicted protein has a less than full alignment to submitted proteins. Hypothetical predicted genes, while having complete gene model predictions, are suspect due to their incomplete alignments and may be pseudogenes or false gene predictions. This corresponds to one predicted gene and four hypothetical genes. A gene density of one gene per 111 kb is much less than other observed rates of gene densities over long distances of the maize genome: for example, one gene per 19 kb over 2.8 Mb (Brunner et al., 2005), one gene per 33 kb in 7.8 Mb (Bruggmann et al., 2006), and one gene per 27 kb over 6.5 Mb (Bruggmann et al., 2006). However, high-confidence gene models of Haberer et al. (2005) show gene density of one gene per 83 kb. Our presented gene densities are in line with the near-centromeric region of these chromosome 3 contigs and also with the increase of maize sequence resources that has become available since these previous studies, allowing us to cull questionable gene predictions.
The lengths of predicted genes range from 180 to 1,578 bp (with introns removed), having a median of 719 bp and a mean of 798 bp. Full genes (including introns) range from 180 to 16,472 bp in length, giving a median of 1,212 bp and a mean of 2,786 bp. Exons have a median of 117 bp and a mean of 205 bp in length. The number of exons per gene ranges from one to 14. Introns have a median of 151 and a mean of 529 bp in length. In one example, a TE inserted within the intron of a gene has increased the length of the intron. Gene 10 on rf1-C2, a mov/MPN/PAD-1 family protein, has an almost complete Jaws retrotransposon found within intron 5.
We identified 14 gene islands as a result of characterization of 16 nested TE clusters. Because our repeat cluster definition (explained above) did not allow repeat clusters to contain predicted non-transposon-related genes, all of the predicted genes are found in these 14 gene islands. While genes found within gene islands or between islands do not seem to form any tight clusters, there is obvious clustering of genes when observed on a contig-wide scale. Gene islands have just one or a few predicted gene annotations, and no gene islands contain large clusters of genes.
Collinearity between Orthologous Regions in Maize, Rice, and Sorghum
To examine sequence collinearity between grass genomes, the 14 gene islands were aligned to the rice assembly (International Rice Genome Sequencing Project, 2005) version 5 and the sorghum assembly version 1 (sbi1; http://www.phytozome.net/sorghum; Paterson et al., 2009). The maize gene islands were compared with the rice genome directly in the VISTA (Dubchak et al., 2000; Mayor et al., 2000; Bray et al., 2003; Brudno et al., 2003; Couronne et al., 2003; Frazer et al., 2004) comparative genome browser. Sorghum was compared with the maize gene islands by first using WU-BLASTN (http://blast.wustl.edu) to align TEnest repeat masked gene island sequences to the sorghum genome assembly sbi1. Each identified region of similarity was compared in the VISTA malign browser.
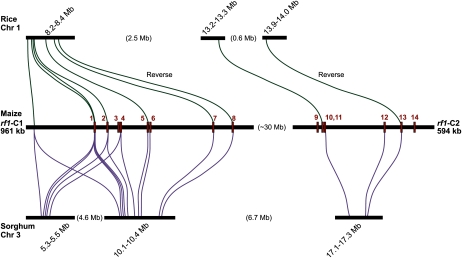
Seven out of the 14 predicted maize genes align when compared to the rice genome, all seven seen in a syntenic location on rice chromosome 1. As illustrated in Figure 4 and Table III, predicted maize genes 1, 2, 5, 7, and 8 on rf1-C1 correspond to gene exons of rice chromosome 1 between 8.2 and 8.4 Mb with a conserved order. rf1-C2 genes 10 and 13 also align to gene exons of rice chromosome 1 in a conserved order and orientation, approximately 5 Mb farther along the rice chromosome at 13.2 Mb. Of the seven genes found in conserved collinear locations, two genes, 8 and 13, are found in a reverse orientation relative to maize. One nonpredicted region on the maize contigs, a region near 85 kb on rf1-C1, aligns to rice gene Os01g14670 on rice chromosome 1 also in this conserved location, near 8.2 Mb. These conserved gene regions show expanded intragene distance in maize as compared with rice, as expected by the increased density of repeat clusters surrounding gene islands.
Figure 4.
Comparative analysis of maize sequence contigs with rice and sorghum. The two sequenced rf1-associated BAC contigs are shown in the center; predicted genes are shown as red rectangles on the black sequence contig lines, with gene identification numbers found in red above. Comparative sequence analysis with rice is shown at the top, and shared sequence regions between maize and rice are shown as green connecting lines. Comparative sequence analysis with sorghum is shown at the bottom, and shared sequence regions between maize and sorghum are shown as blue connecting lines. Collinear regions are seen between maize chromosome 3, rice chromosome 1, and sorghum chromosome 3. Seven out of 14 predicted genes are found in collinear order and in orientation between maize and rice. Ten out of 14 predicted genes are found in collinear order and in orientation between maize and sorghum; three of these genes are found duplicated in a second location on sorghum chromosome 3. One nonpredicted gene region at the left end of rf1-C1 aligns to collinear regions in both rice and sorghum. This is probably a maize pseudogene.
Ten of the 14 predicted maize genes align to the sorghum genome. On rf1-C1, predicted genes 1, 2, and 4 align with a conserved order and orientation to a 50-kb region on sorghum chromosome 3 near 5.4 Mb. This same set of predicted maize genes, along with genes 5, 6, 7, and 8, are found also on sorghum chromosome 3 near 10.2 Mb (Table III; Fig. 4). This shows that at least 500 kb of the maize sequence is duplicated in the sorghum genome on the same chromosome, while only one copy of this region is found in rice, and only one copy of this region is found in the currently sequenced maize genome. Similar to the rice genome comparison, the nonpredicted region near 85 kb on rf1-C1 aligns to sorghum chromosome 3 at both 5.4 and 10.2 Mb. rf1-C2 gene predictions show that genes 10, 12, and 13 are shared between maize and sorghum over the sequence of this contig in similar order and orientation. The four maize genes that did not have sorghum counterparts correspond to the four hypothetical gene predictions, further suggesting that these may not be real genes.
The set of seven predicted maize genes found on rice chromosome 1 in a conserved order are found in the set of 10 genes found conserved when compared with the sorghum genome. The two genes in conserved order and location but found in a reverse direction in rice are seen in the same orientation in maize and sorghum, suggesting that the direction change for these genes occurred either in rice after the split to maize/sorghum or in the maize/sorghum ancestor. Three maize genes are found in two locations on sorghum chromosome 3, and these genes are not found duplicated in the rice genome. These three genes are not seen duplicated in the initial maize genome sequence, either on chromosome 3 or elsewhere.
CONCLUSION
Based on the sequence length, 78% of the rf1-associated contigs consist of repetitive sequences (Table I). For an extremely repetitive organism, maize BAC clones are not overly difficult to assemble. Compared with the assembly of much less repetitive genomes, such as rice (35% repetitive; International Rice Genome Sequencing Project, 2005), Arabidopsis (10% repetitive; Arabidopsis Genome Initiative, 2000), human (44% repetitive; Lander et al., 2001), mouse (37.5% repetitive; Waterston et al., 2002), and Drosophila (3.9% repetitive; Kaminker et al., 2002), this near-centromeric region of maize chromosome 3 is not proportionally more difficult (Celniker et al., 2002) to bring to sequence completion. This is due to a number of reasons. First, the maize genome has many families of TEs; therefore, within a given BAC, there is less of a chance to contain multiple copies of a type of element. Second, the average size of TEs in maize is larger than those of other sequenced organisms, again decreasing the chance of obtaining multiple copies in a single BAC. Third, simple repeats are much less common in maize than in some other sequenced organisms. Simple repeats are generally small (usually less than 500 bp, similar in length to sequence traces) and tandemly duplicated, causing havoc with assembly algorithms. Fourth, the phenomenon of nesting TEs in maize is only seen on a small scale in previously sequenced genomes (Quesneville et al., 2005). Nesting within a TE will break up the repetitive sequence into smaller sections. Once broken up, these segments are flanked by unique sequences in relation to other similar elements, so they are actually easier to assemble. Unfortunately for sequence assemblies, LTRs of maize TEs are in general much longer than those of other sequenced genomes. LTRs are very similar to each other, and they cause many of the gaps seen in initial draft assemblies.
Seven of the predicted maize genes are found conserved in the rice genome, and 10 of the predicted maize genes are found in the sorghum genome. One nonpredicted gene region is found conserved in both rice and sorghum; this is not near any predicted maize genes and suggests that it is a pseudogene. Fifty percent of predicted maize genes are found in collinear locations on rice chromosome 1, and 71% of predicted maize genes are found collinear to sorghum chromosome 3. For high-confidence gene models (the set of predicted maize genes excluding those termed hypothetical), 70% are found in collinear locations on rice chromosome 1 and 100% are found in collinear locations on sorghum chromosome 3. Of genes found conserved across both compared organisms, 27% of shared genes are not seen collinear between maize and rice and 23% of shared genes are not seen in collinear locations between maize and sorghum. Gene islands are not found conserved in their entirety in their orthologous locations. Rather, gene islands are made up of one to two collinear genes, with additional genes found on other chromosome locations or not found in the comparison organism. In the maize-to-rice comparison, one gene island is found containing at least two genes in the collinear region. The distance between these two genes expanded by almost 7-fold in maize. In the maize-to-sorghum comparison, three sets of genes are found with two genes in a gene island in the collinear region. One set of genes is seen with a similar distance between the genes in maize and sorghum, one set has had an approximately 3-fold expansion in maize relative to sorghum, and the final set of genes, the same set observed in the maize-to-rice comparison, has experienced an almost 9-fold increase of intergene distance in the maize genome. While the most common increase of intergene distance has occurred between gene islands, increase in genome sequence is not limited to repeat clusters. In several instances, genes found on the ends of collinear regions of rice and sorghum did not have a maize counterpart; however, due to the increased intergene distances, these genes may be found off the ends of our sequenced contigs.
Sixteen repeat clusters were identified across the two sequenced contigs. These clusters are 23 to 155 kb long and contain a variety of TEs and LTR retrotransposons with a range of insertion ages. In a few cases, several LTR retrotransposon families are seen highly clustered in tight groupings within one to two repeat clusters and may indicate preferential nesting of TEs. Recent insertions of LTR retrotransposons, those that can be considered as the currently active replicating and transposing elements, are seen almost exclusively in the top levels of nested repeat clusters. Insertions into these locations are farther away from genes; therefore, mutations in these regions have a less detrimental effect on the organism.
Gene islands, located between each repeat cluster, range from 4 to 98 kb long and contain from one to four gene predictions. The average gene density across islands is one gene per 16 kb for islands that contain genes. This density is not consistent across islands; larger gene islands do not necessarily contain more genes. While it may be an artifact of our definition of repeat oceans and gene islands, TEs found inserted in gene islands are seen on a very small scale as opposed to the large nested repeat clusters. In all but one case, LTR retrotransposon insertions in gene islands are estimated to have older ages of insertion when compared with the younger TE insertions on upper levels of repeat clusters. This suggests that TEs integrated near genes are rare or not selected for, possibly due to their potential to cause plant-altering mutations. One LTR retrotransposon is seen within the intron of predicted gene 10, increasing the size of the intron by 4.5 kb. The rice and sorghum ortholog counterparts to maize predicted gene 10 do not share this observed increase of intron length due to TE insertion.
The architecture of maize varies across its expanse. From comparative sequence analysis of related grass genomes to the clustering of genes or repeats, diversity is observed at different sequence scales and across various sequence lengths. We hope the assembly techniques presented here will assist the community, ultimately providing long contiguous grass genome assemblies that facilitate examination of the genome as a whole.
MATERIALS AND METHODS
Identification of BACs in the rf1 Region
Three maize (Zea mays) rf1-m allele families (rf1-m3207, rf1-m7323, and rf1-m7212; Wise et al., 1996) were analyzed by a modification of the AIMS method (Frey et al., 1998). DNA was extracted from each individual plant of a segregating population. Ten μL of DNA (10 μg μL−1) was digested with the 4-bp recognition restriction enzyme (MseI or BfaI) in a 40-μL reaction volume. Adaptors with ligase and ligase buffer were added, incubated at room temperature, precipitated, and rehydrated into 30 μL of double-distilled water. This preamplification product was amplified by PCR with the preamplification template, Mu-specific primer (AIM-Mu1, 5′-GAGAAGCCAACGCCAACGCCTCC-3′) and adaptor primer (AIM-AdF, 5′-GCACACGCGATTCGATGTCGAC-3′). Five microliters of a diluted (1:500) preamplification product was used as a template in the exponential amplification using 5 μL of [δ-33P]ATP (Perkin-Elmer), Mu-selective primer (AIMS-Mu4, 5′-GCGCTCTTCGTCCATAATGGCAATTATCTC-3′), and 5 μL of unlabeled adaptor primer (AIMS-Ads, 5′-GACCACGCGTATCGATGTCGACGAG-3′) in a 50-μL reaction. Amplified products were analyzed on acrylamide sequencing gels, and specific fragments were cloned.
Two different BAC genomic library filters were obtained from the Clemson University Genomics Institute, ZMMBBa and ZMMBBb. After probing, additional ZMMBBb and ZMMBBc (Children's Hospital of Oakland Research Institute) BACs were computationally identified using maize WebFPC (http://www.genome.arizona.edu/fpc/maize/).
Genomic DNA, cDNA, AIMS, and RFLP probe fragments were labeled by random priming with [α-32P]dCTP (Feinberg and Vogelstein, 1983). All fragments used as probes were screened to verify copy number by hybridizing to Southern blots of genomic DNA digested with HindIII. Hybridization reactions were performed in Church hybridization buffer (EDTA, pH 8.0, 7% SDS, 0.5 m sodium phosphate buffer, pH 7.2, and 1% bovine serum albumin) at 65°C. High-stringency washes consisted of two 30-min washes in 1× SSPE (0.2 m monobasic sodium phosphate, 3.6 m sodium chloride, and 20 mm EDTA) and 0.1% sodium dodecyl lauryl sulfate, a 60-min wash in 1× SSPE and 0.1% SDS, and a 15-min wash in 0.1× SSPE and 0.1% SDS at 65°C.
BAC DNA was extracted by a modified alkaline lysis protocol obtained from the Clemson University Genomics Institute. BACs were digested with HindIII and run on a 0.9% LE agarose gel for fingerprint analysis. TIFF images were edited for lane tracking, individual band calling, and size fractionation with IMAGE software (Sulston et al., 1989). BAC restriction digest fingerprint data were transferred to the Finger Print Contig Program (FPC; Soderlund et al., 1997, 2000; Pampanwar et al., 2005) for contig analysis. Preliminary contigs were generated with a tolerance of 7 and a cutoff of 1e−12. The agarose gels were bidirectionally transferred to Hybond N (Amersham Pharmacia Biotech) for marker confirmation via Southern hybridization. Final contig assemblies were achieved by reciprocal [α-32P]dCTP random priming reactions with HindIII-digested BAC DNA as the template.
Sequenced BAC ends from each of the original putative contigs were used to make low-copy overgo probes, which were designed using Overgo Maker software (http://genome.wustl.edu/tools/software/overgo.cgi). The set lengths of the overgos are paired 24-mer oligonucleotides that contain an 8-bp complementary overlap with a GC range of 40% to 60%. The oligonucleotides were annealed to each other, and a fill-in reaction was performed using [α-32P]dCTP and dATP. The BAC-end overgos were labeled by a revision of the random priming technique with [α-32P]dCTP and dATP. The hybridization protocol for overgos was similar to those explained above for AIMS probes, except overgos were hybridized at 58°C and were washed for two 15-min washes in 1× SSPE and 0.1% SDS and a 15-min wash in 0.5× SSPE and 0.1% SDS at 58°C.
BAC Sequencing and Assembly
BAC clones were sequenced by MWG Biotech. BACs were sheered and cloned into 3-kb subclone libraries, and subclones were end sequenced to a coverage of 8× to 10×. The BAC sequences were initially assembled with the phred/phrap package (Ewing and Green, 1998; Ewing et al., 1998; http://www.phrap.org) to determine the coverage condition. If the BAC assembly was highly repetitive or assembled into many separate contigs, additional plates of sequence were produced to increase the sequence depth.
Finishing assembly was conducted with phrap and CAP3 (Huang and Madan, 1999). To increase the quality of poor regions, low-quality and failed subclone sequences were identified for resequencing. If low quality was due to the DNA structure (hairpin folding) or difficult sequence (mononucleotide/dinucleotide strings), subclone sequences were identified and resequenced with alternate sequencing chemistries. To close gaps, sequencing primers were designed and sequenced off the subclone and BAC template in order to walk in the direction of the gap. For larger gaps, PCR primers were designed surrounding the area and amplified to make templates for sequencing into the gap. Entire plasmid subclones and PCR products were identified that spanned the gap regions and other unsequenceable areas and were fully sequenced with transposon-bombing insertion methods (Kimmel et al., 1997).
Assembly of repetitive gap regions was aided with the use of TEnest (Kronmiller and Wise, 2008). Individual BACs and combined BAC contigs were run with TEnest using default parameters on the provided maize repeat database. Collapsed repeat spanning assemblies were manipulated with Consed (Gordon et al., 1998). HindIII restriction digests were compared with in silico digestion of finished sequence files (Marra et al., 1997). Any discrepancies found between the two digestions were reexamined for sequence misassemblies.
Annotation of BAC Contigs
Sequence files masked with TEnest were used for gene predictions. Three programs were used: GeneSeqer (Schlueter et al., 2003), FGENESH (Salamov and Solovyev, 2000), and GeneMark.hmm (Lukashin and Borodovsky, 1998). Predicted gene models were compared across the three prediction programs to determine a consensus for predicted genes. Protein and EST databases for Arabidopsis thaliana, Avena sativa, Brachypodium distachyon, Hordeum vulgare, rice (Oryza sativa), Saccharum officinalis, Secale cereale, sorghum (Sorghum bicolor), Triticum aestivum, and maize were downloaded from GenBank (Benson et al., 2006) and aligned to determine gene models. Predicted gene exons were exported and examined for similarity to the plant protein, EST, and predicted gene sets of GenBank to determine possible functions. Output from gene prediction programs, alignments to protein and EST sequences, and predicted genes were displayed with the Generic Model Organism Database package GBrowse (Stein et al., 2002).
Comparative Analysis of Sequence Contigs
Orthologous regions were identified using the VISTA comparative genomics tools (Dubchak et al., 2000; Mayor et al., 2000; Bray et al., 2003; Brudno et al., 2003; Frazer et al., 2004). Identified maize gene islands were compared with the rice genome (International Rice Genome Sequencing Project, 2005) using GenomeVISTA (Couronne et al., 2003). Sorghum genome assembly sbi1 (http://www.phytozome.net/sorghum) was downloaded and aligned with BLASTN and TBLASTX to maize gene islands to locate genes exhibiting similarity; these regions were compared using mVISTA. Coordinates of aligned regions were pulled out of the table of conserved regions from the VISTA output.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF517601 (rf1-C1) and EF517600 (rf1-C2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. TEnest graphical display of maize sequence contigs.
Supplemental Table S1. Summary of gene model and exon coordinates for rf1-C1 and rf1-C2.
Supplementary Material
Acknowledgments
We thank Karin Gobelman-Werner for expert technical assistance in construction of the sequence-ready BAC contigs.
This work was supported by the U.S. Department of Agriculture-National Research Initiative (grant no. 2002–35301–12064).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Roger P. Wise (rpwise@iastate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Chandler VL, Schnable P (2001) National Science Foundation-sponsored workshop report: maize genome sequencing project. Plant Physiol 127: 1572–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot (Lond) 95: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2006) GenBank. Nucleic Acids Res 34: D16–D20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Dubchak I, Pachter L (2003) AVID: a global alignment program. Genome Res 13: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, Pontaroli AC, Wei F, Haberer G, Fuks G, Du C, et al (2006) Uneven chromosome contraction and expansion in the maize genome. Genome Res 16: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: RESEARCH0079 [DOI] [PMC free article] [PubMed]
- Chandler VL, Brendel V (2002) The maize genome sequencing project. Plant Physiol 130: 1594–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E, Cone K, McMullen M, Chen SS, Davis G, Gardiner J, Liscum E, Polacco M, Paterson A, Sanchez-Villeda H, et al (2002) Access to the maize genome: an integrated physical and genetic map. Plant Physiol 128: 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I (2003) Strategies and tools for whole-genome alignments. Genome Res 13: 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Brudno M, Loots GG, Pachter L, Mayor C, Rubin EM, Frazer KA (2000) Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res 10: 1304–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN, Snyder RJ, Anderson EG (1961) The chromosomal location of Rfl, a restorer gene for cytoplasmic pollen sterile maize. Genetics 46: 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279 [DOI] [PMC free article] [PubMed]
- Frey M, Stettner C, Gierl A (1998) A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS). Plant J 13: 717–721 [Google Scholar]
- Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, Schnable PS (2005) Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci USA 102: 12282–12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G, Butler E, Wing RA, Rounsley S, Birren B, et al (2005) Structure and architecture of the maize genome. Plant Physiol 139: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16: 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3: RESEARCH0084 [DOI] [PMC free article] [PubMed]
- Kidwell MG, Lisch DR (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15: 95–99 [DOI] [PubMed] [Google Scholar]
- Kimmel B, Palozzolo M, Martin C, Boeke JD, Devine SE (1997) Transposon-Mediated DNA Sequencing. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Kronmiller BA, Wise RP (2008) TEnest: automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol 146: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo S, Sasinowski M, Presting G, Frisch D, Goff S, et al (2000) Rice transposable elements: a survey of 73,000 sequence-tagged-connectors. Genome Res 10: 982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH (1997) High throughput fingerprint analysis of large-insert clones. Genome Res 7: 1072–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11: 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, Dike S, de la Bastide M, Martienssen RA, McCombie WR (2003) Maize genome sequencing by methylation filtration. Science 302: 2115–2117 [DOI] [PubMed] [Google Scholar]
- Pampanwar V, Engler F, Hatfield J, Blundy S, Gupta G, Soderlund C (2005) FPC Web tools for rice, maize, and distribution. Plant Physiol 138: 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16: 1262–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, Ashburner M, Anxolabehere D (2005) Combined evidence annotation of transposable elements in genome sequences. PLOS Comput Biol 1: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Bennetzen JL (2006) The maize genome as a model for efficient sequence analysis of large plant genomes. Curr Opin Plant Biol 9: 149–156 [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, Stein L, McCombie WR, Martienssen RA (1999) Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet 23: 305–308 [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot (Lond) 82: 37–44 [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20: 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Schlueter SD, Dong Q, Brendel V (2003) GeneSeqer@PlantGDB: gene structure prediction in plant genomes. Nucleic Acids Res 31: 3597–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Humphray S, Dunham A, French L (2000) Contigs built with fingerprints, markers, and FPC V4.7. Genome Res 10: 1772–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R (1997) FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Song R, Llaca V, Linton E, Messing J (2001) Sequence, regulation, and evolution of the maize 22-kD alpha zein gene family. Genome Res 11: 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al (2002) The generic genome browser: a building block for a model organism system database. Genome Res 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Mallett F, Durbin R, Horsnell T (1989) Image analysis of restriction enzyme fingerprint autoradiograms. Comput Appl Biosci 5: 101–106 [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, et al (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR (2006) The maize community welcomes the maize genome sequencing project. Curr Opin Plant Biol 9: 147–148 [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118–2120 [DOI] [PubMed] [Google Scholar]
- Wise RP, Dill CL, Schnable PS (1996) Mutator-induced mutations of the rf1 nuclear fertility restorer of T-cytoplasm maize alter the accumulation of T-urf13 mitochondrial transcripts. Genetics 143: 1383–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Gobelman-Werner K, Pei D, Dill CL, Schnable PS (1999) Mitochondrial transcript processing and restoration of male fertility in T-cytoplasm maize. J Hered 90: 380–385 [DOI] [PubMed] [Google Scholar]
- Yuan Y, SanMiguel PJ, Bennetzen JL (2003) High-Cot sequence analysis of the maize genome. Plant J 34: 249–255 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Arbuckle J, Wessler SR (2000) Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc Natl Acad Sci USA 97: 1160–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.