Abstract
Selenium (Se) is an essential micronutrient for animals and humans but becomes toxic at high dosage. Biologically based Se volatilization, which converts Se into volatile compounds, provides an important means for cleanup of Se-polluted environments. To identify novel genes whose products are involved in Se volatilization from plants, a broccoli (Brassica oleracea var italica) cDNA encoding COQ5 methyltransferase (BoCOQ5-2) in the ubiquinone biosynthetic pathway was isolated. Its function was authenticated by complementing a yeast coq5 mutant and by detecting increased cellular ubiquinone levels in the BoCOQ5-2-transformed bacteria. BoCOQ5-2 was found to promote Se volatilization in both bacteria and transgenic Arabidopsis (Arabidopsis thaliana) plants. Bacteria expressing BoCOQ5-2 produced an over 160-fold increase in volatile Se compounds when they were exposed to selenate. Consequently, the BoCOQ5-2-transformed bacteria had dramatically enhanced tolerance to selenate and a reduced level of Se accumulation. Transgenic Arabidopsis expressing BoCOQ5-2 volatilized three times more Se than the vector-only control plants when treated with selenite and exhibited an increased tolerance to Se. In addition, the BoCOQ5-2 transgenic plants suppressed the generation of reactive oxygen species induced by selenite. BoCOQ5-2 represents, to our knowledge, the first plant enzyme that is not known to be directly involved in sulfur/Se metabolism yet was found to mediate Se volatilization. This discovery opens up new prospects regarding our understanding of the complete metabolism of Se and may lead to ways to modify Se-accumulator plants with increased efficiency for phytoremediation of Se-contaminated environments.
Selenium (Se) has been studied extensively because of its essentiality for animals and humans and because of its toxicity at high dosage. Like a double-edged sword, Se is essential for the function of selenoenzymes but becomes toxic due to the nonspecific replacement of sulfur in sulfur-containing proteins (Stadtman, 1974; Brown and Shrift, 1982). The difference between beneficial and toxic levels of Se is quite narrow, making both Se deficiency and Se pollution common problems in different regions (Terry et al., 2000).
Plants appear to be a promising solution for both sides of the Se problem (Pilon-Smits and LeDuc, 2009). Some crops have the ability to accumulate Se in health-beneficial chemical forms (Whanger, 2002; Dumont et al., 2006). Wheat (Triticum aestivum) grain grown in seleniferous soils accumulates selenomethionine (SeMet) and is one of the main dietary sources for Se (Lyons et al., 2005). Broccoli (Brassica oleracea var italica) has the ability to accumulate high level of Se-methylselenocysteine (SeMCys) and SeMet when grown on seleniferous soil (Cai et al., 1995). These selenoamino acids have been shown to be potent chemoprotective agents against cancer (Ip et al., 2000; Whanger, 2002). Other plant foods, such as garlic (Allium sativum) and Brazil nut (Bertholletia excelsa), have been enriched with Se and marketed as dietary Se supplements (Dumont et al., 2006). On the other hand, Se-hyperaccumulating plant species such as Astragalus bisulcatus and secondary accumulators such as Indian mustard (Brassica juncea) have attracted great interest for their ability to accumulate and volatilize Se for phytoremediation of Se-contaminated soils (Banuelos et al., 2007). Se volatilization converts highly toxic selenate and selenite into volatile dimethyl selenide (DMSe) and dimethyl diselenide (DMDSe), which are 500 to 700 times less toxic (Wilber, 1980). This process provides a low-cost, environmentally friendly, and highly efficient approach for cleanup of Se-contaminated environments (Banuelos et al., 2002; Pilon-Smits, 2005).
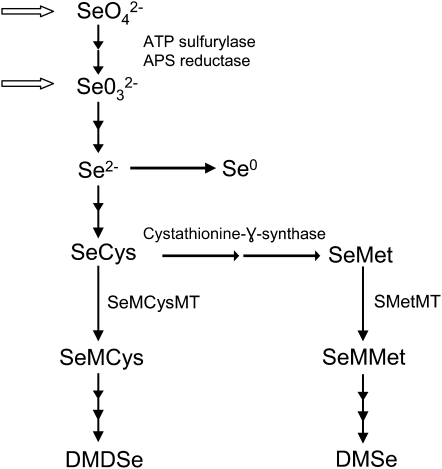
The conversion of inorganic forms of Se into volatile Se in plants is believed to occur via the sulfur metabolic pathway, as outlined in Figure 1 (Terry et al., 2000; Sors et al., 2005). Se is present in soils predominantly as selenate (SeO42−) and selenite (SeO32−). While selenate is actively taken up into plants through sulfur transporters, selenite enters plant cells passively. The reduction of these oxidized forms of Se results in the production of selenoamino acids, such as selenocysteine (SeCys) and SeMet (Fig. 1). In Se-nonaccumulator plants, SeCys and SeMet are readily incorporated into proteins nonspecifically. In Se-accumulating plants, they are metabolized primarily into various nonproteinogenic selenoamino acids. These selenoamino acids can be further metabolized into the volatile Se compounds DMSe and DMDSe. While Se nonaccumulators mainly volatilize DMSe, accumulators primarily emit DMDSe (Terry et al., 2000; Ellis and Salt, 2003). Although Se volatilization is an important step in the Se cycle and provides a protective mechanism for plants and microorganisms to avoid toxicity in seleniferous environments, this metabolic process is not well understood.
Figure 1.
Outline of Se metabolism in plants. The open arrows indicate that both selenate (SeO42−) and selenite (SeO32−) in soil are taken up into plants. Se metabolism from selenate involves a series of reduction steps to form selenide (Se2−), which is assimilated into the selenoamino acids SeCys and SeMet. These selenoamino acids can be methylated and further metabolized into the volatile Se compounds DMDSe and DMSe. Abbreviations not defined in the text: SeMMet, Se-methylmethionine; SeMCysMT, Se-methylselenocysteine methyltransferase; SMetMT, S-adenosyl-l-Met:l-Met S-methyltransferase.
Several sulfur metabolic pathway enzymes have been evaluated for their roles in stimulating Se volatilization (Pilon-Smits and LeDuc, 2009). Cystathionine-γ-synthase is believed to be involved in the formation of SeMet. Overexpression of this enzyme resulted in a 2- to 3-fold increased rate of Se volatilization in transgenic Indian mustard (Van Huysen et al., 2003). S-Adenosyl-l-Met:l-Met S-methyltransferase is responsible for the methylation of SeMet to Se-methylselenomethionine. Overexpression of this enzyme in Escherichia coli produced a 10-fold increase in the rate of Se volatilization when the bacteria were supplied with SeMet (Tagmount et al., 2002). Similarly, expression of a Se-methylselenocysteine methyltransferase to methylate SeCys to SeMCys was shown to stimulate a 2- to 3-fold increase of Se volatilization in transgenic Indian mustard (LeDuc et al., 2004). Although increasing the activities of these known sulfur metabolism enzymes causes increased Se volatilization, additional proteins may be involved in this process (Van Hoewyk et al., 2008).
Microorganisms adapted to high-Se-contaminated environments develop mechanisms to convert inorganic Se compounds into volatile forms. Several methyltransferases from these bacteria were reported to stimulate the emission of DMSe and DMDSe by unknown mechanisms (Ranjard et al., 2002, 2004; Swearingen et al., 2006). To identify novel plant genes whose products promote the production of volatile Se and to gain a better understanding of the metabolic processes associated with Se volatilization, we used a genomics-based approach to isolate genes from broccoli, a plant species known to have high capacity to volatilize Se (Duckart and Waldron, 1992; Terry et al., 1992). Using this approach, a broccoli COQ5 methyltransferase gene designated BoCOQ5-2 was isolated. Functional complementation of a yeast coq5 mutant by BoCOQ5-2 confirmed its identity. BoCOQ5-2 was found to promote Se volatilization when it was expressed in both bacteria and transgenic Arabidopsis (Arabidopsis thaliana).
COQ5 genes encode C-methyltransferases involved in the biosynthesis of ubiquinone or coenzyme Q (Dibrov et al., 1997; Lee et al., 1997). Ubiquinone is an important lipid-soluble compound found in membranes of almost all living species. Ubiquinone is well known for its function as the electron carrier in the mitochondrial respiratory chain for energy production. Moreover, it is widely accepted that ubiquinone also participates in other cellular processes, such as control of cellular redox status and detoxification of harmful reactive oxygen species (ROS; Kawamukai, 2002; Turunen et al., 2004). Indeed, plants with high ubiquinone levels have been demonstrated to be able to suppress ROS generation (Ohara et al., 2004). Increased ubiquinone biosynthesis was found to be associated with increases in tolerance to a variety of stresses in both plants and other organisms (Ohara et al., 2004; Zhang et al., 2007). Se has been shown to induce the production of ROS in Arabidopsis (Tamaoki et al., 2008). Ubiquinone functioning as an antioxidant may protect cells against the oxidative stress to facilitate Se metabolism.
BoCOQ5 methyltransferase represents, to our knowledge, the first plant enzyme that is not known to be involved in sulfur/Se metabolism yet mediates Se volatilization. The cloning and characterization of the methyltransferase from the economically important vegetable crop broccoli extends our understanding of factors affecting Se metabolism. Such information may lead to ways to generate modified Se-accumulator plants with increased efficiency in the phytoremediation of Se-contaminated soils.
RESULTS
Genomics-Based Cloning of Methyltransferase cDNAs from Broccoli
Although it is a well-established phenomenon that plants such as broccoli possess the ability to volatilize Se, many of the specific genes and enzymes catalyzing or facilitating the volatilization process have not been isolated and characterized. Three microorganism methyltransferases, thiopurine methyltransferase from Pseudomonas, UbiE methyltransferase from Geobacillus stearothermophilus, and metalloid methyltransferase (MmtA1) from Hydrogenophaga sp. Esa.33, have been reported to promote Se volatilization in bacteria (Ranjard et al., 2002, 2004; Swearingen et al., 2006). To isolate potential proteins that promote Se volatilization from broccoli, we first BLAST searched the Arabidopsis database (The Arabidopsis Information Resource) using the amino acid sequences of these bacterial methyltransferases to identify Arabidopsis proteins that share high sequence similarity with their bacterial counterparts. They represented the annotated thiol methyltransferase (At2g43940), COQ5 methyltransferase (At1g23360), and sterol methyltransferase (At1g20330). Arabidopsis and B. oleracea genes typically share high nucleotide sequence identity in their coding regions. To identify Brassica sequences that were most closely related to the Arabidopsis homologs, the nucleotide sequences of these Arabidopsis homologs were then used to BLAST search against the Green Plant GB database (The Arabidopsis Information Resource) and the Brassica oleracea Genome Project database (The Institute for Genomic Research). Primers were designed based on the Brassica sequences that aligned to the 5′ and 3′ ends of the Arabidopsis gene sequences and used to amplify and clone the broccoli homologs. These clones were designated as BoTMT, BoCOQ5-1, and BoMmtA1.
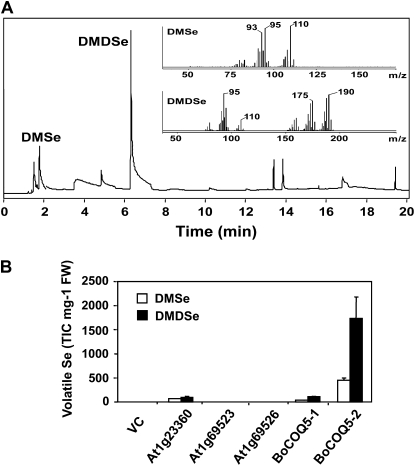
DMSe and DMDSe are the major volatile Se compounds found in nature. To examine whether any of these broccoli clones encoded a protein that promotes Se volatilization, the function of these proteins was examined following heterologous expression in E. coli. Volatile Se compounds in the head space of bacterium cultures treated with 200 μm Na2SeO4 were analyzed by gas chromatography-mass spectrometry (GC-MS). DMSe and DMDSe emerged at 1.91 and 6.41 min, respectively, and were identified by MS with the characteristic ion masses (Fig. 2A, inset). Bacteria carrying the vector-only control produced trace amounts of these volatile Se compounds. Similarly, bacteria expressing the BoMmtA1 or BoTMT construct produced trace or low levels of DMSe and DMDSe (data now shown). In contrast, bacteria expressing the BoCOQ5-1 construct emitted more Se in comparison with vector-only control (Fig. 2B).
Figure 2.
GC-MS screening of broccoli and Arabidopsis homologs of bacterial UbiE methyltransferase. A, A typical gas chromatogram of head space samples from XL1-Blue cells expressing BoCOQ5-2. Insets show the mass spectra of DMSe and DMDSe. B, Relative levels of DMSe and DMDSe in the head space of bacterial cells transformed with broccoli or Arabidopsis homologs of bacterial UbiE methyltransferase. Bacteria were treated with 200 μm Na2SeO4. FW, Fresh weight; TIC, total ion count; VC, vector-only control. Error bars represent sd.
COQ5 methyltransferase belongs to a protein family that contains both predicted chloroplast and mitochondrial forms in Arabidopsis. The BoCOQ5-1 cDNA encoded a predicted chloroplast-targeted protein. To examine whether the mitochondrial form also had the ability to stimulate Se volatilization, a full-length broccoli cDNA was cloned by screening a broccoli cDNA library (Lyi et al., 2005) using a fragment of the putative Arabidopsis mitochondrial COQ5 gene (At5g57300) probe. In addition, three annotated chloroplast COQ5 genes from Arabidopsis (At1g23360, At1g69523, and At1g69526) were also cloned and inserted into pBluescript KS vector to evaluate their abilities to volatilize Se. At1g23360, the BoCOQ5-1 homolog, was previously shown to be involved in phylloquionone (vitamin K) methylation in Arabidopsis (Lohmann et al., 2006). It shared a similar capacity in producing volatile Se compounds as BoCOQ5-1. However, the other two putative chloroplast COQ5 gene products exhibited no activity in promoting Se volatilization (Fig. 2B). Interestingly, the predicted mitochondrial COQ5 gene product from broccoli was by far the most effective one in promoting Se volatilization, with DMDSe as the major form of volatile Se (Fig. 2B). This clone, designated BoCOQ5-2, was subjected to further studies.
Structure Analysis of BoCOQ5-2
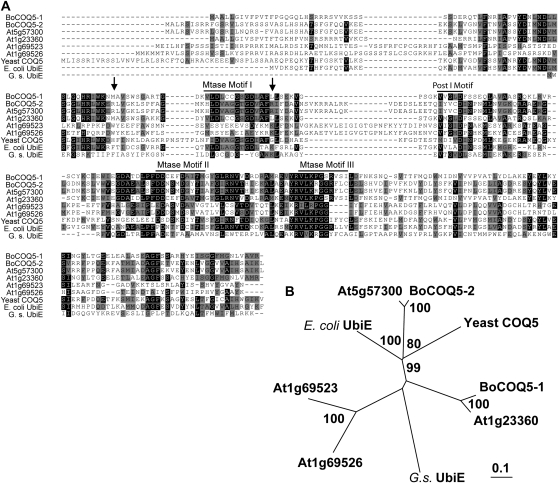
The BoCOQ5-2 cDNA (GenBank accession no. EU879952) contains an open reading frame of 870 nucleotides. It encodes a polypeptide of 289 amino acids with a calculated molecular mass of approximately 32.2 kD. Like the characterized bacterial UbiE methyltransferase and yeast COQ5 (Barkovich et al., 1997; Lee et al., 1997), BoCOQ5-2 as well as BoCOQ5-1 contain four conserved methyltransferase domains: Mtase motifs I, II, III, and Post I motif (Fig. 3A), which are characteristic of the group of methyltransferases using S-adenosyl-Met as the methyl donor (Baba et al., 2004). Interestingly, although BoCOQ5-1 (homolog of At1g23360) shared the same conserved domains, it showed dramatically less activity in stimulating Se volatilization in comparison with BoCOQ5-2. Protein structures for UbiE/COQ5 are not available in the Protein Data Bank (http:/www.rcsb.org/pdb) at this time. Examination of the primary sequences of BoCOQ5-2 and BoCOQ5-1 revealed that there was a 15-amino acid insertion in BoCOQ5-2. Structural modeling of BoCOQ5-2 by threading methods revealed that this insertion did not appear to pack near the active site. Several positively charged Arg residues (i.e. Arg-75 and Arg-101) were present and close to the active site in BoCOQ5-2 but not in BoCOQ5-1. They may represent the characteristic amino acids associated with the pronounced activity of BoCOQ5-2. Future experiments will clarify this hypothesis. Examination of the two predicted chloroplast-targeted Arabidopsis COQ5 proteins (At1g69523 and At1g69526) revealed that they did not have an intact Mtase motif III, which may contribute to the inability of these two proteins to promote Se volatilization. There is also the possibility that these annotated COQ5 genes might have different abilities to produce ubiquinone and/or affect other cellular processes in influencing Se volatilization.
Figure 3.
Comparison of the BoCOQ5-2 sequence with the sequences of related COQ5/UbiE homologs. A, Sequence alignment of the deduced amino acids of BoCOQ5-2 with related COQ5/UbiE proteins from other organisms. The bars indicate the conserved methyltransferase (Mtase) motifs and the Post I motif. At5g57310, At1g23360, At1g69523, and At1g69526 are annotated Arabidopsis COQ5 methyltransferases; yeast COQ5 (P49017), E. coli UbiE (YP_671909), and G. stearothermophilus UbiE (AAR04820) are also shown. Arrows point to two Arg residues in BoCOQ5-2 that may be potentially important for its catalytic activity. B, A phylogenetic tree produced based on the amino acid sequences shown in A. The percentage bootstrap values determined from 1,000 trials are indicated along the branches. The length of the branch lines indicates the extent of divergence according to the scale at bottom.
Results of the phylogenetic analysis of BoCOQ5-2 and related COQ5/UbiE homologs are shown in Figure 3B. BoCOQ5-2 shares 19.5% amino acid sequence identity with G. stearothermophilus UbiE and 46.0% with yeast COQ5.
BoCOQ5-2 Partially Complemented the Yeast coq5 Mutant
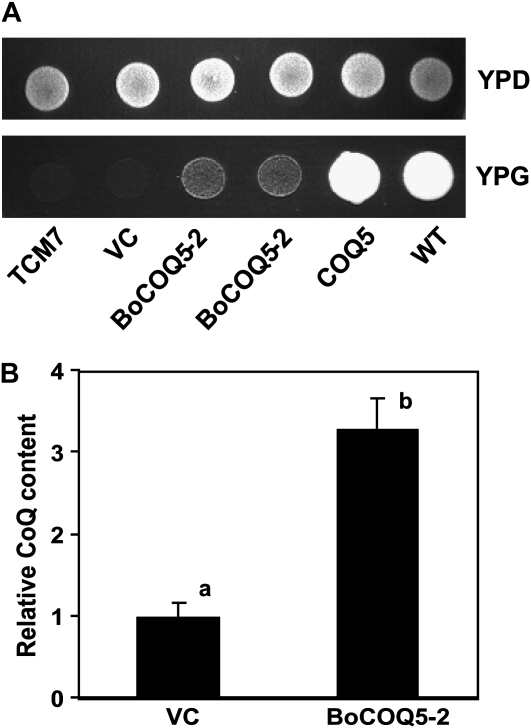
COQ5/UbiE methyltransferases are required for ubiquinone biosynthesis (Dibrov et al., 1997; Lee et al., 1997). The yeast strain TCM7 is a coq5 null mutant that can not grow on YPG medium with glycerol as the sole carbon source (Dibrov et al., 1997). To examine whether the BoCOQ5-2 cDNA encoded a protein with COQ5 methyltransferase function, the coding region of BoCOQ5-2 was cloned into the yeast expression vector pFL61 (Minet et al., 1992). Yeast COQ5 as a positive control was also introduced into the yeast expression vector. These constructs along with the vector-only control were transformed into the yeast strain TCM7. As shown in Figure 4A, all of the yeast strains grew on yeast rich medium (YPD) containing Glc. The presence of BoCOQ5-2 enabled TCM7 to grow on YPG medium with glycerol, although the growth rate was lower than in wild-type yeast grown on glycerol (Fig. 4A). In contrast, no complementation was observed with the vector-only control. These results suggest that BoCOQ5-2 encodes a protein with COQ5 methyltransferase function. Ubiquinone biosynthetic enzymes are known to form a protein complex, and the COQ5 methyltransferase is also required for proper interaction of this complex (Baba et al., 2004). The slow growth of the BoCOQ5-2 yeast strain on YPG medium could be explained by the possibility that BoCOQ5-2 was not able to properly interact with other yeast COQ proteins to restore the full activity of the yeast COQ5 enzyme.
Figure 4.
Functional analysis of BoCOQ5-2. A, Complementation of the yeast coq5 mutant. From left to right, the yeast strains are: TCM7 mutant, the TCM7 mutant expressing empty vector (VC), BoCOQ5-2 (two different lines), yeast COQ5, and wild-type (WT) yeast. Yeast strains were grown on YPD medium containing Glc (top) and YPG medium containing glycerol (bottom). B, Relative ubiquinone contents in bacteria transformed with empty vector or BoCOQ5-2. Ubiquinone levels represent average values of three replicates. Lowercase letters above the bars denote significant differences (P < 0.05).
To further confirm that the BoCOQ5-2 gene product possessed COQ5 methyltransferase activity, ubiquinone was extracted from bacteria cells containing BoCOQ5-2 or the empty vector and quantified by HPLC. As shown in Figure 4B, E. coli cells expressing BoCOQ5-2 exhibited a greater than 3-fold increase in total ubiquinone content in comparison with the vector-only control. This result supports the contention that BoCOQ5-2 indeed encodes a COQ5 methyltransferase.
Expression of BoCOQ5-2 in Bacteria Resulted in a Dramatic Enhancement in Se Volatilization and Tolerance
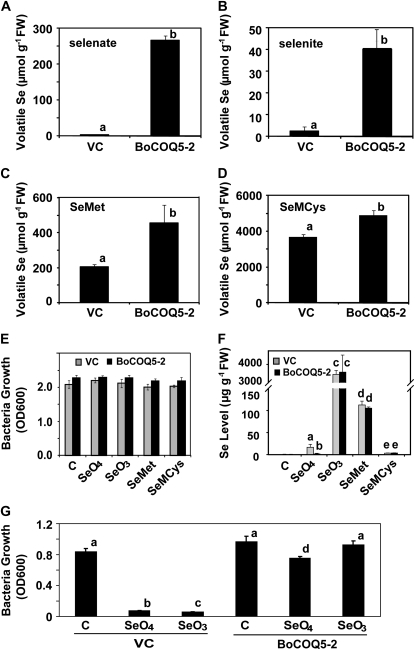
To examine the effect of BoCOQ5-2 on the production of volatile Se in the presence of various forms of Se compounds, E. coli cells carrying BoCOQ5-2 or vector-only control were grown in Luria-Bertani (LB) medium without or with Na2SeO4, Na2SeO3, SeMet, or SeMCys. When bacteria were not exposed to Se, there were no detectable amounts of volatile Se compounds. However, when they were treated with different forms of Se, various levels of total volatile Se were detected (Fig. 5, A–D). The biggest difference in the levels of Se volatilization occurred when the bacteria were exposed to 200 μm Na2SeO4. While the control cells produced trace amounts of volatile Se, as reported in other studies (Ranjard et al., 2002; Swearingen et al., 2006), bacteria expressing BoCOQ5-2 produced an over 160-fold increase in total volatile Se (Fig. 5A). When treated with 200 μm Na2SeO3, the BoCOQ5-2-transformed cells exhibited relatively low levels of volatilization, with an 11-fold increase in comparison with the vector-only control (Fig. 5B). When the cells were exposed to 200 μm SeMet or SeMCys, the control bacteria produced high levels of volatile Se, especially with the SeMCys treatment (Fig. 5, C and D). Expression of BoCOQ5-2 promoted a 2.2-fold (Fig. 5C) or 1.4-fold (Fig. 5D) increase of total volatile Se when bacteria were exposed to SeMet or SeMCys, respectively.
Figure 5.
Total levels of volatile Se in bacteria cells treated with various Se compounds (A–D), bacteria growth (E), Se accumulation (F), and Se tolerance (G). A to D, GC-MS analysis of total levels of volatile Se in the head space of bacterial cultures supplemented with 200 μm Na2SeO4 (A), Na2SeO3 (B), SeMet (C), or SeMCys (D) in LB medium. VC, Vector-only control. E, Effect of various forms of Se compounds on bacteria growth. Bacteria strains were grown in LB medium without (C) or with 200 μm Na2SeO4 (SeO4), Na2SeO3 (SeO3), SeMet, or SeMCys at 37°C for 24 h. Bacteria growth was measured at OD600. F, Total Se accumulation. Bacteria were grown in LB medium without (C) or with 200 μm Na2SeO4 (SeO4), Na2SeO3 (SeO3), SeMet, or SeMCys at 37°C for 24 h. Cell were pelleted, washed extensively, and subjected to inductively coupled plasma analysis. G, Effects of BoCOQ5-2 expression on bacteria Se tolerance. Cell densities of bacteria grown in the absence (C) and presence of 100 μm Na2SeO4 (SeO4) or Na2SeO3 (SeO3) in minimal M9 medium were measured at OD600. Data are means of three repeats. Lowercase letters above the bars indicate significant differences (P < 0.05). FW, Fresh weight.
When bacterial cells were grown on the rich LB medium, the growth of both vector-only control and BoCOQ5-2-transformed cells was not affected by exposure to 200 μm Se (Fig. 5E). To see if the increased Se volatilization from BoCOQ5-2-transformed cells under these treatments affected Se accumulation, total Se contents were determined. Analysis of total Se levels revealed that the bacteria cells had dramatically different abilities to take up different forms of Se (Fig. 5F). The BoCOQ5-2-transformed cells accumulated significantly less total Se than the vector control cells when exposed to selenate, the form of Se that gave the biggest difference in volatilization rate. No significant difference in total Se accumulation between the vector-only control and BoCOQ5-2-transformed cells was observed when the bacteria were exposed to other forms of Se (Fig. 5F). Noticeably, when bacteria were supplied with SeMCys, the control cells appeared to volatilize almost all of the Se taken up and caused very low-level accumulation of Se (Fig. 5F).
To examine whether expression of BoCOQ5-2 increased bacterial tolerance to Se, the growth of the bacterial cells in M9 medium in the presence or absence of 100 μm Na2SeO4 or Na2SeO3 was compared as reported previously (Neuhierl et al., 1999; Lyi et al., 2005). The minimal M9 medium was used for the tolerance assay because bacteria are able to tolerate extremely high concentrations of Se in LB-rich medium. As shown in Figure 5G, after 24 h of incubation, the growth of vector control cells in M9 medium was reduced to less than 10% of that of the untreated samples (Fig. 5G). In contrast, the BoCOQ5-2-transformed bacteria treated with selenate or selenite exhibited 72% and 88% growth, respectively, in comparison with untreated cells. These results establish that expression of BoCOQ5-2 significantly enhanced Se tolerance in E. coli.
Expression of the BoCOQ5-2 Gene in Broccoli, and Subcellular Localization of the Protein
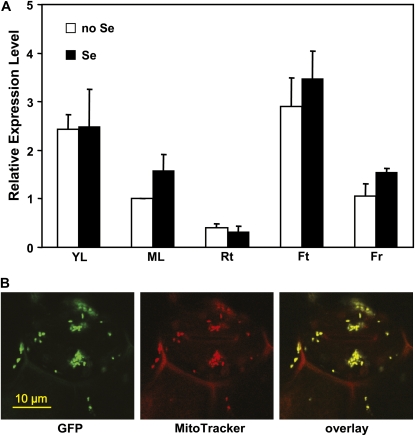
BoCOQ5-2 represents a single-copy sequence in the broccoli genome and showed no cross-hybridization with other COQ5 methyltransferase homologs (data not shown). To examine the expression pattern of BoCOQ5-2 in broccoli, BoCOQ5-2 transcript from different tissues was determined by quantitative reverse transcription-PCR analysis. BoCOQ5-2 was expressed highly in young leaves and florets, and showed relatively low expression in mature leaves, flowers, and roots (Fig. 6A). When the broccoli plants were treated with 20 μm Na2SeO4, a concentration imposing no negative effect on broccoli growth (Lyi et al., 2005), a similar expression pattern for BoCOQ5-2 was observed. No significant difference between Se-treated and nontreated samples was observed. BoCOQ5-2 expression appears not to be affected by Se treatment in broccoli plants.
Figure 6.
Expression profile of BoCOQ5-2 in broccoli, and subcellular localization of BoCOQ5-2 protein in transgenic Arabidopsis. A, Real-time PCR determination of the relative expression levels of BoCOQ5-2 in various tissues of broccoli treated with or without 20 μm Na2SeO4. The expression of BoCOQ5-2 in mature leaf without Se treatment was set to 1. YL, Young leaf; ML, mature leaf; Rt, root; Ft, floret; Fr, flower. Data are means of three repeats with two biological replicates. B, Subcellular localization of BoCOQ5-2-GFP. GFP signal in leaf epidermal cells of Arabidopsis plants expressing the BoCOQ5-2-GFP fusion (left), red fluorescence from MitoTracker Red (middle), and overlay of both GFP and MitoTracker Red signals (right) are shown. These images represent multiple analyses of three independent transgenic lines. Bar = 10 mm.
BoCOQ5-2 contains a 23-amino acid N-terminal target sequence, which was predicted to be a mitochondrial protein by the MitoProt II-v1.101 program (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html). To experimentally determine the subcellular localization of BoCOQ5-2 in plants, a BoCOQ5-2-GFP fusion construct was transformed into Arabidopsis plants. Stable transgenic lines were examined by laser scanning confocal microscopy. Strong, punctate, green fluorescence was evident in leaf cells of BoCOQ5-2-GFP transformants (Fig. 6B). The size, shape, and subcellular distribution of the punctate green fluorescence were consistent with those reported for mitochondria (Logan and Leaver, 2000). To confirm that the BoCOQ5-2 targeted to mitochondria, the mitochondrion-specific fluorescent dye MitoTracker Red was used to highlight the mitochondria. Colocalization of green (BoCOQ5-2-GFP) and red punctuate fluorescence was observed (Fig. 6B). The overlaid GFP and MitoTracker images clearly demonstrate that BoCOQ5-2 was expressed in mitochondria.
Overexpression of BoCOQ5-2 Increased Se Volatilization and Tolerance in Transgenic Arabidopsis
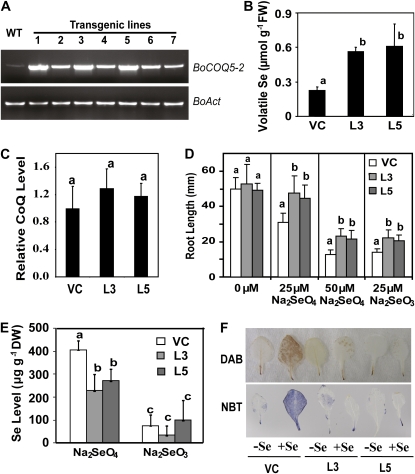
To test whether BoCOQ5-2 promoted Se volatilization in plants, BoCOQ5-2 under the control of the cauliflower mosaic virus 35S promoter was introduced into Arabidopsis. Several Arabidopsis transgenic lines overexpressing BoCOQ5-2 were produced (Fig. 7A). Two homozygous lines (L3 and L5) were obtained and used for Se volatilization test. These BoCOQ5-2-overexpressing plants were grown individually in vials to the same developmental stage and size. Volatile Se compounds from these transgenic plants treated with 50 μm Na2SeO3 or Na2SeO4 for 3 d were analyzed by GC-MS. Unlike the bacterial cultures that volatilized both DMSe and DMDSe when exposed to inorganic Se compounds, the only species of volatile Se detected in the Arabidopsis plants was DMSe. This is in agreement with previous reports that Se-nonaccumulating plant species emit mainly DMSe (Terry et al., 2000). When the transgenic Arabidopsis plants expressing BoCOQ5-2 or empty vector were treated with Na2SeO3, the BoCOQ5-2 transformants produced three times more DMSe than the vector control plants (Fig. 7B). When the transgenic plants were exposed to Na2SeO4, no significant difference of DMSe emission levels was observed between the vector-only control and BoCOQ5-2 transgenic lines (data not shown).
Figure 7.
Analysis of BoCOQ5-2 transgene levels, volatile Se content, ubiquinone level, and Se tolerance in transgenic Arabidopsis. A, Semiquantitative reverse transcription-PCR assay of BoCOQ5-2 transcript level in independent transgenic Arabidopsis lines. Expression of ACTIN (BoAct) was used as an internal control. B, GC-MS analysis of DMSe contents in transgenic Arabidopsis exposed to 50 μm Na2SeO3 for 3 d. C, Relative ubiquinone levels in transgenic Arabidopsis. D, Root length of transgenic Arabidopsis seedlings grown for 12 d on MS medium supplied with or without Se as indicated. E, Total Se accumulation. Transgenic Arabidopsis was treated with 50 μm Na2SeO3 for 3 d. Data represent means (n = 10) ± sd. F, In situ detection of ROS by DAB and NBT staining of representative leaves after exposing the transgenic plant to 25 μm Na2SeO3. The brown and purple precipitations indicate the production of ROS. Data are means of three repeats. Lowercase letters above the bars indicate significant differences (P < 0.05). DW, Dry weight; FW fresh weight; L3 and L5, transgenic line 3 and line 5; VC, vector-only control; WT, wild type. [See online article for color version of this figure.]
To examine whether overexpression of BoCOQ5-2 in Arabidopsis also resulted in enhanced accumulation of ubiquinone, ubiquinone was extracted from leaf tissue of Arabidopsis. The amount of ubiquinone present in plants is usually low. Comparison of the ubiquinone levels in these two overexpressing lines with that in the vector-only control revealed a slight but not statistically significantly increased level of ubiquinone accumulation (Fig. 7C). Such a slight increase may suggest that the ubiquinone metabolism in plant mitochondria is under tight control. Indeed, a previous study in overexpression of the yeast coq2 gene in transgenic tobacco (Nicotiana tabacum) showed that it is not very effective in increasing mitochondrial ubiquinone content (Ohara et al., 2004).
To examine if the increased Se volatilization in transgenic Arabidopsis plants overexpressing BoCOQ5-2 led to an enhanced tolerance to inorganic Se, the root length as a parameter for Se tolerance (Van Hoewyk et al., 2005) was measured. As shown in Figure 7D, in the absence of Se, there was no significant difference in root growth of 12-d-old seedlings between empty vector control and BoCOQ5-2 transgenic lines. In the presence of 25 μm Na2SeO3, the BoCOQ5-2 transgenic lines had 1.3- to 1.5-fold longer roots than controls. Interestingly, although BoCOQ5-2 transformants did not show increased levels of Se volatilization when treated with Na2SeO4, the two transgenic lines exhibited enhanced selenate tolerance, with 1.4- to 1.8-fold longer roots compared with empty vector controls when exposed to 25 or 50 μm Na2SeO4 (Fig. 7D). These results suggest that expression of BoCOQ5-2 enhanced plant tolerance to inorganic Se.
Furthermore, the total Se concentration in leaves of these Arabidopsis transformants exposed to 50 μm Na2SeO4 or Na2SeO3 was determined. When plants were treated with Na2SeO4, the two overexpressing lines accumulated significantly less total Se than the vector-only control (Fig. 7E). In contrast, when these transformants were exposed to Na2SeO3, no significant difference in total Se contents was observed between the vector-only control and the BoCOQ5-2-overexpressing lines.
Exposing Arabidopsis plants to selenite has been shown to induce the production of hydrogen peroxide and superoxide in leaves (Tamaoki et al., 2008). To test whether overexpression of BoCOQ5-2 helped plants detoxify ROS, 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining were performed for in situ detection of ROS production. In agreement with a previous report (Tamaoki et al., 2008), increased staining was observed in leaves of the vector-only control Arabidopsis treated with Na2SeO3. In contrast, the BoCOQ5-2-overexpressing lines showed less staining than the vector-only control when treated with selenite (Fig. 7F), indicating that ROS production was reduced in the BOCOQ5-2 overexpressing plants.
DISCUSSION
Previous research on Se volatilization has been mostly focused on sulfur/Se assimilation pathway enzymes, especially those thought to catalyze rate-limiting steps (Tagmount et al., 2002; Van Huysen et al., 2003; LeDuc et al., 2004). In this study, BoCOQ5-2 was found to represent, to our knowledge, the first plant enzyme that is not known to be part of the sulfur/Se assimilation/volatilization pathway and yet able to significantly stimulate Se volatilization in both bacteria and plants.
The assimilation of Se consists of a series of reduction reactions that change selenate and selenite into Se-containing amino acids. It is believed that the two prevailing volatile forms of Se, DMSe and DMDSe, are derived from the methylation of SeMet and SeMCys, respectively, in plants (Terry et al., 2000; Sors et al., 2005). BoCOQ5-2 was able to stimulate Se volatilization into both DMSe and DMDSe in E. coli and into DMSe in transgenic Arabidopsis. To pinpoint whether BoCOQ5-2 facilitates reduction and/or late-step assimilation of Se compounds, we examined the capacity of BoCOQ5-2 to mediate Se volatilization in response to different chemical forms of Se in a bacterial system. While bacteria containing the vector-only control produced trace amounts of volatile Se when exposed to inorganic Se, they emitted much more DMSe and DMDSe when treated with selenoamino acids. This observation indicates that bacteria exhibit a limited ability to assimilate inorganic Se for volatilization but are naturally able to metabolize SeMet and SeMCys to produce volatile Se compounds (Turner et al., 1998). If BoCOQ5-2 promotes one or more early steps in Se metabolism, it is expected that bacteria expressing BoCOQ5-2 would produce a significant amount of volatile Se following the reduction of inorganic Se and the formation of selenoamino acids. If it stimulates late-step assimilation of selenoamino acids, expression of this gene is expected to dramatically enhance Se volatilization following supplementation with selenoamino acids, as shown in the case for S-adenosyl-l-Met:l-Met S-methyltransferase (Tagmount et al., 2002). The BoCOQ5-2-transformed bacteria showed a 160-fold increase in the level of Se volatilization when exposed to selenate but exhibited only a 1.4- to 2.2-fold increase when treated with selenoamino acids. The finding that BoCOQ5-2 had dramatically different responses to inorganic Se salts compared with selenoamino acids suggests that BoCOQ5-2 likely exerts strong effects on early steps of the Se assimilation/volatilization process. The fact that the BoCOQ5-2-transformed bacteria also exhibited a greater than 1.4-fold increase in volatile Se when they were exposed to selenoamino acids suggests that BoCOQ5-2 may also be involved in the late step of Se metabolism.
While bacteria expressing BoCOQ5-2 volatilized both DMSe and DMDSe, the Arabidopsis BoCOQ5-2 transformants produced DMSe as the only form of volatile Se. DMSe represents the major form of volatile Se compounds in Se-nonaccumulator plants, whereas DMDSe is the main volatile form of Se in Se-accumulator plants (Terry et al., 2000). Arabidopsis as a Se nonaccumulator has been previously shown to be incapable of converting inorganic Se into SeMCys, the precursor for DMDSe formation (Ellis et al., 2004). Thus, the lack of DMDSe production in the BoCOQ5-2-transformed Arabidopsis most likely was due to the absence of the SeMCys precursor. These results also suggest that BoCOQ5-2 functions to enhance Se assimilation processes instead of altering the metabolic fate of Se in vivo.
Although BoCOQ5-2 in E. coli dramatically promoted Se volatilization when exposed to selenate, the BoCOQ5-2 transgenic Arabidopsis did not exhibit an increased rate of Se volatilization in comparison with the control plants when supplied with selenate. One explanation for this response is the limited ability to reduce selenate to selenite in Arabidopsis. This reduction step is known to be rate limiting in Se assimilation and volatilization in plants (de Souza et al., 1998; Pilon-Smits et al., 1999) and is not highly operational in Arabidopsis plants (Ellis et al., 2004; Van Hoewyk et al., 2005). The limited ability to reduce selenate to selenite in Arabidopsis might also limit the effect of BoCOQ5-2 to stimulate Se volatilization when the transgenic plants are treated with selenate.
There are differences in the ability of bacteria or plants to take up and metabolize different forms of Se. Here, we showed that bacteria took up and accumulated extremely high levels of Se when grown in medium containing selenite but relatively low levels when exposed to SeMet, selenate, and SeMCys. When bacteria were treated with selenate, the increased rate of Se volatilization in the BoCOQ5-2-transformed cells resulted in a lower level of Se accumulation. However, when bacteria were treated with selenite, SeMet, or SeMCys, the increased levels of Se volatilization were not linked to reduced Se accumulation, which indicates that the levels of Se accumulation do not necessarily reflect only uptake and volatilization.
As reported previously (Zhang et al., 2006; Tamaoki et al., 2008), Arabidopsis plants accumulated more Se when exposed to selenate than selenite. In higher plants, selenate and selenite are taken up, transported, and metabolized differently (Terry et al., 2000). Selenate is actively taken up and transported easily to the shoot, where some of it is reduced and assimilated into selenoamino acids in chloroplasts. Selenite enters plant cells passively and the bulk of it is nonenzymatically reduced and metabolized primarily in roots (Zayed et al., 1998; Afton et al., 2009). Notably, when the transgenic plants were treated with selenate, the BoCOQ5-2 transformants accumulated significantly less total Se despite the observation of no increased rate of Se volatilization in comparison with the vector-only control. As in the case in bacteria, when plants were treated with selenite, the increased Se volatilization in the BoCOQ5-2 transformants did not affect the levels of total Se accumulation.
The enhanced Se volatilization in selenite-treated BoCOQ5-2 transgenic Arabidopsis was accompanied by an increased resistance to selenite, as demonstrated in other studies (Van Huysen et al., 2003; LeDuc et al., 2004). Interestingly, although the enhanced Se volatilization was only observed in selenite-treated plants, an increased resistance to selenate was also observed. Such an increased tolerance to selenate in the overexpressing lines could be due to reduced levels of total Se accumulation.
Se is essential for bacteria but not for higher plants, although Se has been suggested to be crucial for Se-hyperaccumulator plants (Pilon-Smits and LeDuc, 2009). While bacteria contain the specific SeCys insertion machinery for controlled incorporation of SeCys into selenoproteins, the higher plants are believed to have lost such machinery during evolution and only nonspecifically incorporate selenoamino acids into proteins (Terry et al., 2000). Furthermore, Se shares the sulfur metabolic pathway (Sors et al., 2005) and competes with sulfur for assimilation. The alteration of sulfur metabolites could indirectly affect Se metabolism. The relative amounts of Se and sulfur in the bacterial growth medium and in plant hydroponic solution might also affect Se volatilization in bacteria and plants.
It is intriguing to speculate whether the increased levels or capacity of ubiquinone biosynthesis are responsible for the BoCOQ5-2-mediated Se volatilization. As a redox-active molecule, ubiquinone may indirectly participate in the reduction of inorganic Se for the production of selenoamino acids in stimulating Se volatilization. Indeed, when the bacteria were exposed to selenite, both the vector control and the BoCOQ5-2-transformed cells produced orange elemental Se. While the control cells remained colored for an extended period of growth, the BoCOQ5-2-transformed cells facilitated the reduction and further metabolization of Se and quickly became uncolored. However, in plants, the role of ubiquinone as a redox-active molecule participating in the reduction of Se may be limited due to the compartmentalization within the cell. Ubiquinone in plants is synthesized in the mitochondria, which is separated from chloroplast and cytosol, the major sites for Se assimilation and volatilization reactions (Terry et al., 2000).
Ubiquinone as an antioxidant plays an important role in oxidative stress tolerance. Cellular ubiquinone level has been demonstrated to be involved in tolerance to a variety of stresses in both plants and other organisms. Ubiquinone-overexpressing tobacco exhibits high tolerance to methyl viologen and salt (Ohara et al., 2004). Similarly, ubiquinone-overproducing yeast shows more resistance to hydrogen peroxide, Cu2+, and salt (Zhang et al., 2007). In contrast, ubiquinone-deficient mutants in E. coli and yeast are hypersensitive to hydrogen peroxide and Cu2+ (Soballe and Poole, 2000; Saiki et al., 2003). Also, the yeast ubiquinone biosynthesis mutants are sensitive to cadmium (Kennedy et al., 2008). These studies establish a strong causal relationship between ubiquinone levels and oxidative stress tolerance.
Ubiquinone acting as a lipid-soluble antioxidant may help the cells deal with the oxidative stress caused by Se toxicity and to facilitate the metabolism of Se in the Se assimilation pathway. This might be the primary mechanism for BoCOQ5-2-mediated Se tolerance and volatilization in plants. Se compounds are proposed to be directly toxic to plants (Ellis et al., 2004). Transcriptome analysis of Se-responsive genes provides evidence in support of this, since nearly 50% of up-regulated genes were categorized as responsive to biotic/abiotic stimuli and stress (Van Hoewyk et al., 2008). Such toxicity is associated with the generation of ROS in plants (Tamaoki et al., 2008). Although there was no statistically significant increase in total ubiquinone levels in the BoCOQ5-2-overexpressing plants, the expression of BoCOQ5-2 may enhance the capacity of the plants to synthesize ubiquinone. The increased capacity to synthesize ubiquinone could aid the cells to detoxify the harmful ROS. Indeed, there was a clear suppression of ROS production in Se-treated BoCOQ5-2 transformants as assessed by DAB and NBT staining, which suggests an increased radical-scavenging capacity of the overexpressing lines. Furthermore, our preliminary proteomic analysis of the protein expression profile between the Se-treated and nontreated BoCOQ5-2 transgenic Arabidopsis samples appears also to suggest that this is the case. Among the 21 differentially expressed proteins (≥1.5-fold changes), there were only two proteins involved in general stress responses (Y. Yang, T.W. Thannhauser, and L. Li, unpublished data).
The exact mechanism of BoCOQ5-2 in stimulating Se volatilization remains to be revealed. The finding of BoCOQ5-2 as a facilitator of Se volatilization in bacteria and plants has clearly demonstrated that proteins outside of the Se metabolism pathway also have significant effects on Se volatilization. The cloning and characterization of BoCOQ5-2 have opened up new avenues for understanding the complete metabolism of Se.
The enhancement of Se volatilization by BoCOQ5-2 in bacteria and plants may also imply a potentially practical application for manipulation of this process. Selenate represents one of the most common forms of environmentally toxic Se. Arabidopsis as a Se nonaccumulator has a limited ability to reduce selenate. As a result, overexpression of BoCOQ5-2 in transgenic Arabidopsis exhibited a limited effect on Se volatilization when exposed to selenate. In contrast, Se secondary accumulators, such as broccoli and Indian mustard, volatilize significantly larger amounts of Se when exposed to selenate than selenite (de Souza et al., 2000). Broccoli and Indian mustard are able to volatilize approximately 2% of selenate taken up (de Souza et al., 2000). Increase in Se volatilization rate in these plants could substantially improve the efficiency of Se removal. Therefore, manipulation of this metabolic process in natural Se accumulators may greatly improve their efficiency and effectiveness in decontamination of Se-polluted environments.
MATERIALS AND METHODS
RNA Analysis
Total RNA from broccoli (Brassica oleracea var italica) tissues grown in a growth room at 22°C with a 14-h daylength was extracted and reverse transcribed into cDNAs as described by Lu et al. (2006). Real-time PCR analysis was performed essentially as described by Lyi et al. (2007). The gene-specific primers used were BoCOQ5-2RTF and BoCOQ5-2RTR (Table I).
Table I.
List of primers used in this study
The underlined nucleotides represent the added restriction enzyme sequences.
| Primer | Sequence (5′–3′) |
|---|---|
| BoTMT2F | GAATTCGATGGCTGAGGTACAAC |
| BoTMT2R | TCTAGATCAATTGATCTTCTTCCACCTAG |
| BoCOQ5-1F | CTCGAGATGGCGGCTCTACTCGGTATC |
| BoCOQ5-1R | CCATGGACCTCACAGCGACC |
| BoMmtA1F | GAATTCTATGGACTCGGTGACG |
| BoMmtA1R | TCTAGATCATTCAGAAGAAGTTTTCTCTGG |
| BoCOQ5-2RTF | ATTGCGGAAAGTTGATGAGG |
| BoCOQ5-2RTR | TCTCAGCAGCACGTTGTTTC |
| BoActinRTF | GTCTGTGACAATGGTACCGGAAT |
| BoActinRTR | ACAGCCCTGGGAGCATCA |
| BoCOQ5-2F | GCGGCCGCATGGCACTTCGAT |
| BoCOQ5-2R | GCGGCCGCTCAGAGCTTAATGGC |
| YCOQ5F | ATGTTGATTTCTTCACGGATCGTTC |
| YCOQ5R | TTAAACTTTAATGCCCCAATGGATGG |
| BoCOQ5-2KpnIF | TAGGTACCAATGGCACTTCGATCG |
| BoCOQ5-2BamHIR | TTGGATCCTCAGAGCTTAATGGCAG |
cDNA Isolation and Plasmid Construction
To isolate the broccoli homologs, the protein sequences of bacterial methyltransferases of bTMP (AAP12368), UbiE/COQ5 (AAR04820), and MmtA1 (AAT78751) were used to BLAST search against the Arabidopsis (Arabidopsis thaliana) database (http://www.arabidopsis.org). The nucleotide sequences of the Arabidopsis homologs were used to BLAST search against The Institute for Genomic Research B. oleracea Genome Project database (http://www.tigr.org/tdb/e2k1/bog1/) or other databases (www.arabidopsis.org and http://brassica.bbsrc.ac.uk/). Gene-specific primers based on the B. oleracea sequences were designed and employed to amplify the broccoli homologs from broccoli cDNA using Pfu DNA Polymerase (Stratagene). The amplified fragments were cloned and sequenced. Screening a broccoli cDNA library using Arabidopsis gene probes followed the procedure as described previously (Lyi et al., 2005).
To make expression constructs in bacteria, the open reading frames of full-length clones were amplified using primers with added restriction enzyme sites (Table I) and subcloned in-frame into pBluescript SK vector. The constructs and the empty vector were transformed into the Escherichia coli strain XL1-Blue. All of the constructs were sequenced to verify the inserts and the nucleotide sequences at the cloning sites.
Yeast Expression Constructs and Complementation
The coding sequences of BoCOQ5-2 and yeast COQ5 were amplified with the primer pairs BoCOQ5-2F/BoCOQ5-2R and YCOQ5F/YCOQ5R, respectively (Table I) and subcloned directionally into yeast expression vector pFL61 (Minet et al., 1992). The resulting constructs, termed pFL-BoCOQ5-2 and pFL-COQ5, were mobilized into yeast coq5 mutant strain TCM7, kindly provided by Dr. Bernard D. Lemire (University of Alberta, Canada; Dibrov et al., 1997). Transformants were selected on the dropout medium depleted of uracil and then plated on YPD medium (containing 2% Glc) and YPG (containing 2% glycerol), respectively, for complementation of the mutant phenotype.
HPLC Analysis of Ubiquinone Content
Extraction of ubiquinone in bacteria was performed following the published method with slight modifications (Johnson et al., 2005). Overnight cultures (50 mL) of XL1-Blue cells were harvested and disrupted with a French press. The cell lysates were first extracted with methanol (2 mL) and petroleum ether (6 mL) and then with 6 mL of petroleum ether. The upper organic extracts were combined, dried under a stream of N2, and resuspended in 500 μL of ethanol.
Extraction of ubiquinone from leaf tissue of Arabidopsis was carried out following the procedure as described (Norris et al., 1995). Briefly, 1.0 g of leaves was extracted in 3 mL of chloroform and 6 mL of methanol. Water and chloroform (1.5 mL each) were added, and the chloroform phase was collected. Following one more extraction with 2 mL of chloroform, the two chloroform phases were combined and dried under a stream of N2. The extract was then redissolved in methanol.
Aliquots (100 μL) of samples were injected onto a Symmetry C-18 column (250 × 4.6 mm; Waters) and separated by reverse-phase HPLC at a flow rate of 1 mL min−1 with ethanol:methanol (70:30, v/v). Ubiquinone was identified and quantified using ubiquinone standard from Sigma.
Plant Transformation
To generate the construct for overexpression of BoCOQ5-2 in Arabidopsis, the coding region of BoCOQ5-2 was amplified using primers BoCOQ5-2KpnIF and BoCOQ5-2BamHIR (Table I) and subcloned into pCAMBIA1300S binary vector containing cauliflower mosaic virus 35S promoter. To make the BoCOQ5-2-GFP translational fusion construct, the BoCOQ5-2 coding region was inserted in front of the GFP5 coding sequence (Siemering et al., 1996) and then subcloned into pCAMBIA1300S vector. Both constructs were sequenced to verify the insertions and the nucleotide sequences at the cloning sites.
The constructs along with the empty vector were electroporated into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis using the floral dipping method (Clough and Bent, 1998). T1 seeds were surface sterilized and germinated on Murashige and Skoog (MS) medium containing 25 mg L−1 hygromycin. Hygromycin-resistant plants were transferred into soil and verified by PCR for the presence of transgene. Positive transformants were allowed to grow to produce T2 seeds. Homozygous transgenic plants were selected in the T3 generation.
GC-MS Analysis of Volatile Se Compounds
Overnight E. coli culture (1 mL) was mixed with 4 mL of fresh LB medium in a screw-capped vial (Supelco) with 50 μg mL−1 carbenicillin, 1 mm isopropylthio-β-galactoside, and 200 μm Se compound. The vial was then capped with septa and shaken at 250 rpm in a 37°C incubator for 24 h. Transgenic Arabidopsis plants were grown on 10 mL of solid MS medium with 1% Suc in the vials covered with Millipore tape for 3 weeks under 14 h of light at 22°C. Na2SeO4 or Na2SeO3 was then added to a final concentration of 50 μm and screw capped. The plants were allowed to grow for 3 d.
The volatile Se compounds in the head space were exposed to solid-phase microextraction fiber for 10 min and immediately inserted into the GC injection port using a 0.75-mm inlet liner (Supelco). The inlet liner was subsequently left in the injection port for 20 min to thermally desorb the analytes. GC was performed using an Agilent 6890N network GC system coupled with a 30-m × 0.25-mm column (J&W Scientific). Splitless injection mode was used with helium carrier gas at a flow rate of 1 mL min−1. The injector temperature was set at 230°C. The column oven temperature was started at 40°C and immediately ramped to 100°C at 7.5°C min−1, held for 1 min, and then ramped at 25°C min−1 to 270°C.
MS using a Joel JMS-GCMateII GC-MS system and selected ion monitoring mode was employed for the identification of volatile Se compounds. The ion masses 93, 94, 95, 109, 110, 175, and 190 were chosen for the identification of DMSe and DMDSe peaks. The chromatograms and mass spectra were analyzed using the GCmatePro-3.0 program. Quantification of volatile Se compounds was performed by comparing with standard curves derived from pure DMSe and DMDSe (Sigma).
Analysis of Total Se Level
To determine the level of Se accumulation, bacteria cells grown in 150 mL of LB medium containing 50 μg mL−1 carbenicillin, 1 mm isopropylthio-β-galactoside, and 200 μm Se compound for 24 h were harvested and washed five times with distilled water. For plant tissues, transgenic Arabidopsis plants were grown in Magenta boxes containing MS medium with 1% Suc for 3 weeks and treated with 50 μm Na2SeO4 or Na2SeO3 for 3 d. The whole plants were harvested, washed, and dried. The samples were weighed and analyzed for total Se using an inductively coupled plasma trace analyzer emission spectrometer (model ICAP 61E trace analyzer; Thermo Electron) as described by Lyi et al. (2005).
Analysis of Se Tolerance
To determine bacterial Se tolerance, XL1-Blue cells transformed with BoCOQ5-2 or pBluescript empty vector were grown overnight in 5 mL of M9 medium containing 2% Glc and 100 μg mL−1 ampicillin. The overnight cultures were used to inoculate 3 mL of the same medium, adjusted to an optical density at 600 nm (OD600) of 0.05, and grown in the presence or absence of 100 μm Na2SeO4 or Na2SeO3 at 37°C for 40 h. Cell density at OD600 was then determined.
To assess Se tolerance in plants, seeds of transgenic Arabidopsis were germinated on MS medium containing 1% Suc in the absence or presence of 25 or 50 μm Na2SeO4 or 25 μm Na2SeO3. Plates were left vertically in a growth room at 22°C with a 14-h light photoperiod. Root length of 12-d-old seedlings was measured.
Confocal Microscopy Analysis
Young leaves of Arabidopsis transformants expressing a BoCOQ5-2-GFP fusion were stained with 500 nm MitoTracker Red 580 (Molecular Probes) for 30 min, destained in distilled water, and analyzed with a Leica Upright DMRE-7 confocal microscope (Leica Microsystems). GFP5 was excited by 489-nm light and observed using a detection window from 498 to 520 nm. MitoTracker Red was observed using a 578-nm excitation light and detection window from 588 to 608 nm.
In Situ ROS Detection
Surface-sterilized transgenic Arabidopsis seeds were grown on MS salt plates with or without 25 μm Na2SeO3 for 10 d. Young leaves were cut and vacuum infiltrated either in 1 mg mL−1 DAB-HCl (Sigma) in 50 mm Tris (pH 7.6) or in 0.1% NBT in 10 mm KPO4 buffer (pH 7.8). The samples were incubated overnight for DAB staining and for 20 min for NBT staining at room temperature. Stained leaves were cleared in boiling ethanol (96%) for 10 min prior to photography.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU879952.
Acknowledgments
We are very grateful to Dr. Bernard D. Lemire for kindly providing the yeast COQ5 construct and the yeast coq5 mutant strain and to Dr. Daniel Ripoll for modeling BoCOQ5 proteins. We thank Dr. Ivan Keresztes for technical advice with GC-MS analysis, Dr. Xiangjun Zhou and Ms. Li-Wei Chiu for helpful discussions, and Mr. Laurence Heller for help with HPLC analysis.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Li Li (LL37@cornell.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Afton SE, Catron B, Caruso JA (2009) Elucidating the selenium and arsenic metabolic pathways following exposure to the non-hyperaccumulating Chlorophytum comosum, spider plant. J Exp Bot 60: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba SW, Belogrudov GI, Lee JC, Lee PT, Strahan J, Shepherd JN, Clarke CF (2004) Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J Biol Chem 279: 10052–10059 [DOI] [PubMed] [Google Scholar]
- Banuelos G, LeDuc DL, Pilon-Smits EA, Terry N (2007) Transgenic Indian mustard overexpressing selenocysteine lyase or selenocysteine methyltransferase exhibit enhanced potential for selenium phytoremediation under field conditions. Environ Sci Technol 41: 599–605 [DOI] [PubMed] [Google Scholar]
- Banuelos GS, Lin ZQ, Wu L, Terry N (2002) Phytoremediation of selenium-contaminated soils and waters: fundamentals and future prospects. Rev Environ Health 17: 291–306 [DOI] [PubMed] [Google Scholar]
- Barkovich RJ, Shtanko A, Shepherd JA, Lee PT, Myles DC, Tzagoloff A, Clarke CF (1997) Characterization of the COQ5 gene from Saccharomyces cerevisiae: evidence for a C-methyltransferase in ubiquinone biosynthesis. J Biol Chem 272: 9182–9188 [DOI] [PubMed] [Google Scholar]
- Brown TA, Shrift A (1982) Selenium: toxicity and tolerance in higher plants. Biol Rev Camb Philos Soc 57: 59–84 [Google Scholar]
- Cai XJ, Block E, Uden PC, Zhang X, Quimby BD, Sullivan JJ (1995) Allium chemistry: identification of selenoamino acids in ordinary and selenium-enriched garlic, onion, and broccoli using gas-chromatography with atomic-emission detection. J Agric Food Chem 43: 1754–1757 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Souza MP, Lytle CM, Mulholland MM, Otte ML, Terry N (2000) Selenium assimilation and volatilization from dimethylselenoniopropionate by Indian mustard. Plant Physiol 122: 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza MP, Pilon-Smits EA, Lytle CM, Hwang S, Tai J, Honma TS, Yeh L, Terry N (1998) Rate-limiting steps in selenium assimilation and volatilization by Indian mustard. Plant Physiol 117: 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov E, Robinson KM, Lemire BD (1997) The COQ5 gene encodes a yeast mitochondrial protein necessary for ubiquinone biosynthesis and the assembly of the respiratory chain. J Biol Chem 272: 9175–9181 [DOI] [PubMed] [Google Scholar]
- Duckart EC, Waldron LJ (1992) Selenium uptake and volatilization from plants growing in soil. Soil Sci 153: 94–99 [Google Scholar]
- Dumont E, Vanhaecke F, Cornelis R (2006) Selenium speciation from food source to metabolites: a critical review. Anal Bioanal Chem 385: 1304–1323 [DOI] [PubMed] [Google Scholar]
- Ellis DR, Salt DE (2003) Plants, selenium and human health. Curr Opin Plant Biol 6: 273–279 [DOI] [PubMed] [Google Scholar]
- Ellis DR, Sors TG, Brunk DG, Albrecht C, Orser C, Lahner B, Wood KV, Harris HH, Pickering IJ, Salt DE (2004) Production of Se-methylselenocysteine in transgenic plants expressing selenocysteine methyltransferase. BMC Plant Biol 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip C, Thompson HJ, Zhu Z, Ganther HE (2000) In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res 60: 2882–2886 [PubMed] [Google Scholar]
- Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A (2005) COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem 280: 31397–31404 [DOI] [PubMed] [Google Scholar]
- Kawamukai M (2002) Biosynthesis, bioproduction and novel roles of ubiquinone. J Biosci Bioeng 94: 511–517 [DOI] [PubMed] [Google Scholar]
- Kennedy PJ, Vashisht AA, Hoe KL, Kim DU, Park HO, Hayles J, Russell P (2008) A genome-wide screen of genes involved in cadmium tolerance in Schizosaccharomyces pombe. Toxicol Sci 106: 124–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDuc DL, Tarun AS, Montes-Bayon M, Meija J, Malit MF, Wu CP, AbdelSamie M, Chiang CY, Tagmount A, deSouza M, et al (2004) Overexpression of selenocysteine methyltransferase in Arabidopsis and Indian mustard increases selenium tolerance and accumulation. Plant Physiol 135: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Hsu AY, Ha HT, Clarke CF (1997) A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J Bacteriol 179: 1748–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Lohmann A, Schottler MA, Brehelin C, Kessler F, Bock R, Cahoon EB, Dormann P (2006) Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J Biol Chem 281: 40461–40472 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O'Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyi SM, Heller LI, Rutzke M, Welch RM, Kochian LV, Li L (2005) Molecular and biochemical characterization of the selenocysteine Se-methyltransferase gene and Se-methylselenocysteine synthesis in broccoli. Plant Physiol 138: 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyi SM, Zhou X, Kochian LV, Li L (2007) Biochemical and molecular characterization of the homocysteine S-methyltransferase from broccoli (Brassica oleracea var. italica). Phytochemistry 68: 1112–1119 [DOI] [PubMed] [Google Scholar]
- Lyons GH, Genc Y, Stangoulis JC, Palmer LT, Graham RD (2005) Selenium distribution in wheat grain, and the effect of postharvest processing on wheat selenium content. Biol Trace Elem Res 103: 155–168 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Neuhierl B, Thanbichler M, Lottspeich F, Böck A (1999) A family of S-methylmethionine-dependent thiol/selenol methyltransferases: role in selenium tolerance and evolutionary relation. J Biol Chem 274: 5407–5414 [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, Dellapenna D (1995) Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara K, Kokad Y, Yamamoto H, Sato F, Yazaki K (2004) Engineering of ubiquinone biosynthesis using the yeast coq2 gene confers oxidative stress tolerance in transgenic tobacco. Plant J 40: 734–743 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits E (2005) Phytoremediation. Annu Rev Plant Biol 56: 15–39 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EA, Hwang S, Mel LC, Zhu Y, Tai JC, Bravo RC, Chen Y, Leustek T, Terry N (1999) Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol 119: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EA, LeDuc DL (2009) Phytoremediation of selenium using transgenic plants. Curr Opin Biotechnol 20: 207–212 [DOI] [PubMed] [Google Scholar]
- Ranjard L, Prigent-Combaret C, Favre-Bonte S, Monnez C, Nazaret S, Cournoyer B (2004) Characterization of a novel selenium methyltransferase from freshwater bacteria showing strong similarities with the calicheamicin methyltransferase. Biochim Biophys Acta 1679: 80–85 [DOI] [PubMed] [Google Scholar]
- Ranjard L, Prigent-Combaret C, Nazaret S, Cournoyer B (2002) Methylation of inorganic and organic selenium by the bacterial thiopurine methyltransferase. J Bacteriol 184: 3146–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R, Ogiyama Y, Kainou T, Nishi T, Matsuda H, Kawamukai M (2003) Pleiotropic phenotypes of fission yeast defective in ubiquinone-10 production: a study from the abc1Sp (coq8Sp) mutant. Biofactors 18: 229–235 [DOI] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol 6: 1653–1663 [DOI] [PubMed] [Google Scholar]
- Soballe B, Poole RK (2000) Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146: 787–796 [DOI] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Salt DE (2005) Selenium uptake, translocation, assimilation and metabolic fate in plants. Photosynth Res 86: 373–389 [DOI] [PubMed] [Google Scholar]
- Stadtman TC (1974) Selenium biochemistry. Science 183: 915–922 [DOI] [PubMed] [Google Scholar]
- Swearingen JW Jr, Fuentes DE, Araya MA, Plishker MF, Saavedra CP, Chasteen TG, Vasquez CC (2006) Expression of the ubiE gene of Geobacillus stearothermophilus V in Escherichia coli K-12 mediates the evolution of selenium compounds into the headspace of selenite- and selenate-amended cultures. Appl Environ Microbiol 72: 963–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagmount A, Berken A, Terry N (2002) An essential role of S-adenosyl-L-methionine:L-methionine S-methyltransferase in selenium volatilization by plants: methylation of selenomethionine to selenium-methyl-L-selenium-methionine, the precursor of volatile selenium. Plant Physiol 130: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki M, Freeman JL, Pilon-Smits EAH (2008) Cooperative ethylene and jasmonic acid signaling regulates selenite resistance in Arabidopsis. Plant Physiol 146: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N, Carlson C, Raab TK, Zayed AM (1992) Rates of selenium volatilization among crop species. J Environ Qual 21: 341–344 [Google Scholar]
- Terry N, Zayed AM, De Souza MP, Tarun AS (2000) Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol 51: 401–432 [DOI] [PubMed] [Google Scholar]
- Turner RJ, Weiner JH, Taylor DE (1998) Selenium metabolism in Escherichia coli. Biometals 11: 223–227 [DOI] [PubMed] [Google Scholar]
- Turunen M, Olsson J, Dallner G (2004) Metabolism and function of coenzyme Q. Biochim Biophys Acta 1660: 171–199 [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D, Garifullina GF, Ackley AR, Abdel-Ghany SE, Marcus MA, Fakra S, Ishiyama K, Inoue E, Pilon M, Takahashi H, et al (2005) Overexpression of AtCpNifS enhances selenium tolerance and accumulation in Arabidopsis. Plant Physiol 139: 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Takahashi H, Inoue E, Hess A, Tamaoki M, Pilon-Smits EA (2008) Transcriptome analyses give insights into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiol Plant 132: 236–253 [DOI] [PubMed] [Google Scholar]
- Van Huysen T, Abdel-Ghany S, Hale KL, LeDuc D, Terry N, Pilon-Smits EA (2003) Overexpression of cystathionine-gamma-synthase enhances selenium volatilization in Brassica juncea. Planta 218: 71–78 [DOI] [PubMed] [Google Scholar]
- Whanger PD (2002) Selenocompounds in plants and animals and their biological significance. J Am Coll Nutr 21: 223–232 [DOI] [PubMed] [Google Scholar]
- Wilber CG (1980) Toxicology of selenium: a review. Clin Toxicol 17: 171–230 [DOI] [PubMed] [Google Scholar]
- Zayed A, Lytle CM, Terry N (1998) Accumulation and volatilization of different chemical species of selenium by plants. Planta 206: 284–292 [Google Scholar]
- Zhang D, Shrestha B, Niu W, Tian P, Tan T (2007) Phenotypes and fed-batch fermentation of ubiquinone-overproducing fission yeast using ppt1 gene. J Biotechnol 128: 120–131 [DOI] [PubMed] [Google Scholar]
- Zhang L, Byrne PF, Pilon-Smits AH (2006) Mapping quantitative trait loci associated with selenate tolerance in Arabidopsis thaliana. New Phytol 170: 33–42 [DOI] [PubMed] [Google Scholar]