Abstract
A prominent enzyme in organellar RNA metabolism is the exoribonuclease polynucleotide phosphorylase (PNPase), whose reversible activity is governed by the nucleotide diphosphate-inorganic phosphate ratio. In Chlamydomonas reinhardtii, PNPase regulates chloroplast transcript accumulation in response to phosphorus (P) starvation, and PNPase expression is repressed by the response regulator PSR1 (for PHOSPHORUS STARVATION RESPONSE1) under these conditions. Here, we investigated the role of PNPase in the Arabidopsis (Arabidopsis thaliana) P deprivation response by comparing wild-type and pnp mutant plants with respect to their morphology, metabolite profiles, and transcriptomes. We found that P-deprived pnp mutants develop aborted clusters of lateral roots, which are characterized by decreased auxin responsiveness and cell division, and exhibit cell death at the root tips. Electron microscopy revealed that the collapse of root organelles is enhanced in the pnp mutant under P deprivation and occurred with low frequency under P-replete conditions. Global analyses of metabolites and transcripts were carried out to understand the molecular bases of these altered P deprivation responses. We found that the pnp mutant expresses some elements of the deprivation response even when grown on a full nutrient medium, including altered transcript accumulation, although its total and inorganic P contents are not reduced. The pnp mutation also confers P status-independent responses, including but not limited to stress responses. Taken together, our data support the hypothesis that the activity of the chloroplast PNPase is involved in plant acclimation to P availability and that it may help maintain an appropriate balance of P metabolites even under normal growth conditions.
Organisms require phosphorus (P) continually and in relatively high amounts, and in photosynthetic systems a major use is the regeneration of ribulose-1,6-bisphosphate, the acceptor for CO2 fixation by Rubisco. Chloroplast inorganic phosphate (Pi) pools are also affected by starch biosynthesis, since the conversion of Glc-1-P to ADP-Glc, the penultimate step in the pathway, releases Pi through ATP hydrolysis. Starch is primarily synthesized during the day from excess triose phosphates and broken down at night, a step that consumes Pi through the action of starch phosphorylase and other enzymes (Zeeman et al., 2007). Thus, P is a major player in chloroplast metabolism and is intimately integrated into the carbon budget of plant cells.
Chloroplast P is also required for processes not directly related to photosynthesis, such as gene expression. In particular, the chloroplast ribonuclease polynucleotide phosphorylase (PNPase) both consumes and liberates P. PNPase in bacteria (Soreq and Littauer, 1977) and chloroplasts (Baginsky et al., 2001) is a homotrimer that degrades RNA through phosphorylytic attack, but it also readily synthesizes polynucleotides using nucleotide diphosphate or nucleotide triphosphate precursors, a reaction that generates Pi or inorganic pyrophosphate, respectively. The purified chloroplast enzyme, like its bacterial counterpart, is readily reversible in vitro (Yehudai-Resheff et al., 2001). Several lines of work have clearly placed PNPase, in both prokaryotes and eukaryotic organelles (for review, see Slomovic et al., 2006), as a key player in a polyadenylation-stimulated RNA degradation pathway found in plant mitochondria and chloroplasts.
While the role of PNPase in RNA metabolism is established if incompletely understood, newer evidence has linked PNPase to somewhat unrelated functions. In human cells, for example, a cytosolic fraction of mitochondrial PNPase appears to influence cell differentiation and senescence (Sarkar and Fisher, 2006), and in the mitochondrion itself, PNPase is located in the intermembrane space while mitochondrial RNA is in the matrix, suggesting a metabolic rather than an RNA catalytic role for PNPase (Chen et al., 2006), through which it can nonetheless influence the accumulation of certain mitochondrial RNAs (Slomovic and Schuster, 2008). More closely related to this study, a genetic screen carried out in Arabidopsis (Arabidopsis thaliana) for resistance to fosmidomycin, which inhibits the plastid methylerythritol phosphate (MEP) pathway, identified a chloroplast (cp)PNPase null mutant that was named rif10 (Sauret-Gueto et al., 2006). This led to the suggestion that the MEP pathway might be regulated by plastid metabolic cues, which in turn could be influenced by PNPase.
This study was stimulated by our finding that in the green alga Chlamydomonas reinhardtii, reduced expression of cpPNPase rendered cells unable to acclimate to P deprivation, whereas the same strains had wild-type responses to other nutrient or environmental stresses (Yehudai-Resheff et al., 2007). We also found that both the PNPase transcript and protein were repressed under P deprivation, suggesting that reduced PNPase activity is part of the overall metabolic adjustment to P limitation. Furthermore, repression of PNPase expression required the general P deprivation response regulator Psr1, a likely transcription factor (Wykoff et al., 1999), demonstrating that PNPase regulation is integrated into the global P limitation response. The ortholog of Psr1 in Arabidopsis is PHR1 (Rubio et al., 2001), among whose functions is inducing microRNAs of the miR399 family, which through long-distance signaling (Pant et al., 2008) derepress a suite of genes including some encoding P transporters, through a ubiquitination pathway involving PHO2 (Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). This pathway in turn is modulated by the noncoding RNA IPS1 (Franco-Zorrilla et al., 2007) and possibly others.
Here, we examine the role of cpPNPase in Arabidopsis in the context of the P deprivation response using null mutant alleles. The most obvious growth defect under P limitation is in the elaboration of lateral roots, where the pnp mutant phenocopies pdr2, a mutant thought to define a signal needed for lateral root proliferation (Ticconi et al., 2004). When gene expression was examined using array technology, we found that the pnp mutants already induced, under normal growth conditions, genes normally only induced upon P deprivation. These and other data suggest that pnp mutants exhibit elements of P starvation even when grown under P-replete conditions. Therefore, we propose that cpPNPase in plants, as in unicellular algae, has a key role in P metabolism.
RESULTS
PNP T-DNA Insertion Mutants
cpPNPase is encoded by the locus At3g03710, which specifies a 922-residue protein, consistent with the approximately 100-kD migration of PNPase previously described from spinach (Spinacia oleracea) and pea (Pisum sativum) chloroplasts (Hayes et al., 1996; Li et al., 1998). Limited data have been obtained on Arabidopsis lines reduced or totally deficient for cpPNPase (Walter et al., 2002; Sauret-Gueto et al., 2006). To pursue the analysis of Arabidopsis cpPNPase in the context of the nutrient stress response, we selected two T-DNA insertion alleles, naming them pnp1-1 and pnp1-2 (Fig. 1A). Homozygous mutants for each line were isolated following three outcrosses of the original T3 plants. UV cross-linking was performed to confirm that the pnp1-1 mutant lacked cpPNPase, shown in Figure 1B as an approximately 100-kD RNA-binding activity. The mutants also lack complete PNP transcripts, as revealed by reverse transcription (RT)-PCR using primers flanking the respective T-DNA insertions (data not shown).
Figure 1.
Characterization of pnp1-1 and pnp1-2 T-DNA insertion mutants. A, Diagram of the two T-DNA insertions in the PNP gene corresponding to the lines named pnp1-1 and pnp1-2 (SALK_013306 and SALK_070705, respectively). B, Total (T) or chloroplast (cp) proteins extracted from the wild type (WT) and the pnp1-1 mutant were analyzed for RNA binding by the UV cross-linking assay as described in “Materials and Methods.” The bottom band is an unknown RNA-binding protein whose signal is similar in the different samples. C, The wild type, pnp1-1, and pnp1-2 homozygous mutants were grown on soil for 21 d. Note that the 1-cm bar size is different for the mutants. D, cpRNA patterns in the pnp1-1 and pnp1-2 mutants. Total RNA was isolated from the wild type, pnp1-1, and pnp1-2 and analyzed by ethidium bromide staining to reveal 23S rRNA anomalies (top) or by gel blot using the chloroplast gene probes indicated at right. [See online article for color version of this figure.]
Figure 1C compares wild-type and pnp mutant plants after 21 d of growth on soil. The rosette leaves of pnp mutants emerged and remained pale green compared with those of wild-type plants, as was observed previously (Sauret-Gueto et al., 2006). In addition, the mutant plants were retarded in silique production, and both silique number and seed production were reduced (data not shown). Thus, cpPNPase is required for fully normal plant development, although in its absence a full life cycle is achieved. Another feature of cpPNPase deficiency is incomplete 3′ end processing of certain chloroplast transcripts, including 23S rRNA and various mRNAs. These are exemplified in Figure 1D, and in the cases of 23S rRNA, rbcL, and psbA, the presence of 3′ extensions had been confirmed previously (Walter et al., 2002).
Lateral Root Development under P Limitation
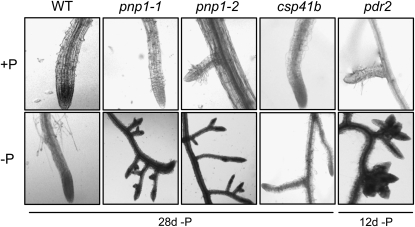
It is well established that when faced with nutrient limitation, plants seek additional sources through altered root system architecture. In the case of P limitation, both root hair density and lateral root proliferation are observed (for review, see Lopez-Bucio et al., 2003). To see whether PNPase deficiency affected this process, we compared the root architecture of wild-type and mutant plants grown under +P and −P conditions. As exemplified in Figure 2, there were no substantive differences observed when several genotypes were compared under +P conditions, although the pnp mutants had a slower overall root elongation rate (data not shown). We used two other mutants as controls. The first was csp41b, a nuclear mutant lacking two related chloroplast endoribonucleases (Bollenbach et al., 2009). We reasoned that if abnormal chloroplast RNA (cpRNA) metabolism had any pleiotropic effect on root architecture, that would be seen for both pnp1-1 and csp41b. A second control was pdr2, a mutant that is known to exhibit abortive lateral root initiation under −P conditions (Ticconi et al., 2004). This control was useful because, as we discovered (Fig. 2, bottom row), the pnp1-1 mutants phenocopied pdr2. In particular, when grown under −P conditions, the pnp mutants were unable to elaborate lateral roots, instead exhibiting a proteoid root-like phenotype after 4 weeks of P starvation. While the same effect was observed earlier for pdr2, it should be borne in mind that pnp mutants have an overall slower growth rate (Fig. 1).
Figure 2.
Lateral root phenotypes under P-replete conditions (top row) or under P deprivation (bottom row). Additional mutants used as controls were csp41b, which lacks two related chloroplast endoribonucleases (Bollenbach et al., 2009), and pdr2, which was previously demonstrated to exhibit the observed phenotype (Ticconi et al., 2004). For the experiment shown here, plants were grown on a full nutrient medium for 2 weeks, then transferred to +P or −P medium for the times indicated. WT, Wild type.
Because the csp41b mutant was indistinguishable from the wild type under −P conditions, we concluded that the aberrant root architecture phenotype of pnp1-1/pnp1-2 was not related to altered cpRNA metabolism per se. Instead, we suspected that some function of PNPase was essential to lateral root elaboration. To investigate this phenomenon in more detail, we crossed the pnp1-1 mutation into two backgrounds expressing GUS reporter genes, under the control of either the auxin-responsive promoter element DR5 (Ulmasov et al., 1997) or the cyclin B1 promoter (Colòn-Carmona et al., 1999), as an indicator of cell division. Both reporters have previously been used in studies of root physiology under P limitation conditions (Ticconi et al., 2004; Nacry et al., 2005; Sanchez-Calderon et al., 2005).
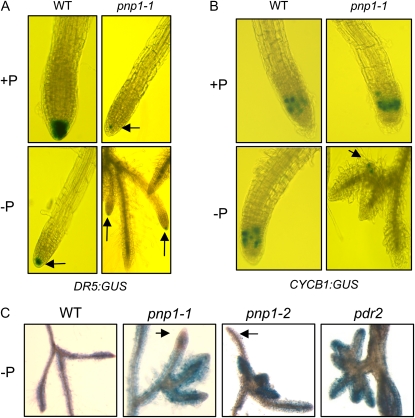
Figure 3, A and B, show results for plants expressing DR5:GUS and CYCB1:GUS, respectively. When grown on +P medium, pnp1-1 exhibited considerably less staining in the primary root tip than did the wild type, which is consistent with its slower overall root elongation rate. When grown on −P medium, the wild type exhibited reduced staining at the root tip, a result consistent with another study showing an age-dependent decrease DR5:GUS expression following P starvation as compared with seedlings grown on +P medium (Sanchez-Calderon et al., 2005). In the case of pnp1-1, we found that only one or two of the lateral root initiates in the proteoid-like clusters stained with GUS under –P conditions (Fig. 3A, arrows). This suggested that while some lateral roots in pnp1-1 had a wild-type-like auxin environment, other roots in the clusters showed no evidence for this marker and were likely auxin deficient.
Figure 3.
Cell division and mortality in lateral roots developing under P deprivation. A, Roots from plants expressing a DR5:GUS transgene were stained histochemically for GUS. Arrows point to smaller stained areas. Plants were germinated and grown for 2 weeks on +P medium, then transferred to +P or −P medium for 4 weeks. B, Roots from plants expressing a CYCB1:GUS transgene were stained histochemically, following growth as for A. C, Plants were grown as for A. Roots were stained with Evans blue. The arrows indicate pnp mutant root tips that are not stained. WT, Wild type. [See online article for color version of this figure.]
Concordant results were obtained with the cyclin marker (Fig. 3B). As observed in another study (Ticconi et al., 2004), a zone of mitotic cells was present in the root meristem under both +P and −P for the wild type, and this was also the case for pnp1-1 when grown under +P conditions. When pnp1-1 was starved for P, however, only a single early-stage initiate in the root clusters stained for GUS (Fig. 3B, arrow). This suggested that cell division had ceased in the majority of lateral root initiates, the same phenomenon that was documented for pdr2 (Ticconi et al., 2004). Since the pnp mutant developed root clusters where the majority of roots appeared to lack both auxin and cell division, we used the dye Evans blue to see if cell death had occurred. As shown in Figure 3C, in the cases of both pnp1-1 and pdr2, the majority of roots in P limitation-stimulated clusters stained with Evans blue, which cannot pass intact membrane barriers. Thus, the majority of these cells are dead, and the affected root tips likely correspond to roots that also failed to stain with GUS driven by DR5 or CYCB1 promoters. In conclusion, pnp1-1 exhibits abortive lateral root initiation under −P conditions, where the aborted roots cease division and undergo cell death.
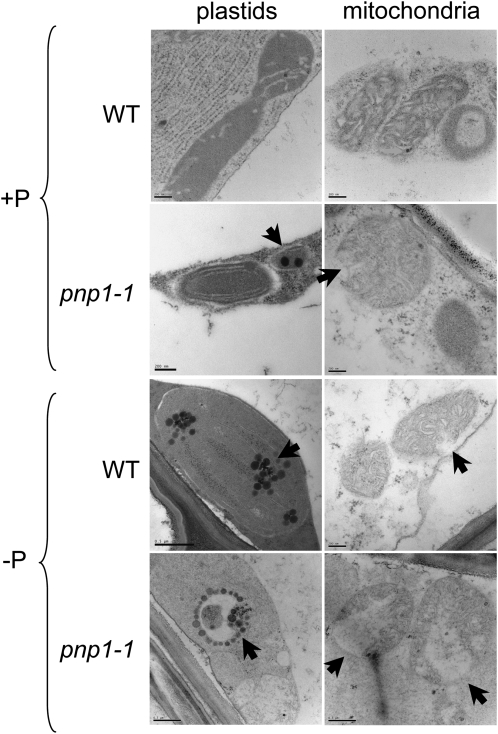
Because PNPase is a plastid protein, we examined organellar ultrastructure in roots under +P and −P conditions, as exemplified in Figure 4. Under +P conditions, we observed an increased frequency of plastoglobules in pnp1-1 as well as apparently ruptured mitochondria. Plastoglobules are associated with senescence and stress conditions and may be a pleiotropic consequence of the pnp mutation. The frequency of plastoglobules increased more dramatically in pnp1-1 than in the wild type under −P conditions, also consistent with an enhanced stress response. In 60% of plastids in the mutant, they were arranged in a circle, as shown in Figure 4. We also noted increased mitochondrial disruption in the mutant, and occasionally in the wild type, under −P conditions. Mitochondrial rupture, as measured by cytochrome c release, has been associated with cell death in male-sterile sunflower (Helianthus annuus) anthers (Balk and Leaver, 2001) and subsequently studied in other contexts where programmed cell death occurs in plants (Reape et al., 2008). Thus, it is possible that the mitochondrial disruption observed in pnp1-1 under −P conditions is symptomatic of the loss of cell integrity ultimately observed (Fig. 3C).
Figure 4.
Ultrastructure of wild-type (WT) and pnp1-1 mutant lateral root cortical cells. Plants were germinated and grown for 2 weeks on +P medium, then transferred to +P or −P medium. Plants were observed after 1, 2, 3, and 4 weeks on −P. Here, only images of root organelles after 1 week on −P are shown. Cortical cells of root tip sections in the longitudinal direction were observed using transmission electron microscopy as described in “Materials and Methods.” Bars = 200 nm, except for pnp1-1 on −P (plastids and mitochondria) and for the wild-type plastid on −P, where bars = 500 nm. Arrows indicate features referred to in the text.
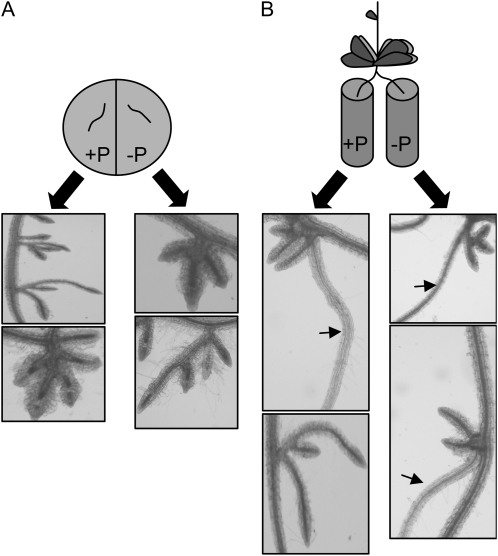
The PNP gene is expressed in both roots and leaves (http://mpss.udel.edu/at/; S. Yehudai-Resheff and D.B. Stern, unpublished RT-PCR data), albeit at a much lower level in roots, raising the question of whether pnp mutants display an altered root architecture under −P conditions because of PNPase deficiency in roots, or in leaves, or both. To address this question, we carried out the experiments shown in Figure 5. We first excised roots from pnp1-1 plants already exposed to −P conditions to observe whether providing them with P would correct the root architecture deficiency. Figure 5A shows that aberrant roots did not regain normal proliferation when placed on +P medium, although some growth occurred, suggesting that the P level encountered by the roots did not alone determine their phenotype. In contrast, when root masses from pnp1-1 plants starved for P were divided and half placed on +P medium and the other half on −P medium, normal root elongation ensued in both samples (Fig. 5B). Thus, although half the pnp roots were in −P medium, P provided from the other roots and cycled through the aboveground tissues rescued the mutant phenotype. We conclude that altered root architecture in pnp1-1 is not a cell-autonomous feature of root cells.
Figure 5.
Split-root experiment using the pnp1-1 mutant. A, Plants were germinated and grown for 2 weeks on +P medium and transferred to −P conditions for 3 weeks. The root segments were excised and placed on −P or +P agar medium. Roots were photographed after 2 additional weeks on these media. B, Plants were germinated and grown for 2 weeks on +P medium and transferred to −P conditions for 3 weeks. The roots were then divided into −P or +P liquid medium while remaining attached to the plants and grown for an additional 2 weeks. The arrows indicate lateral roots that have recovered a wild-type-like phenotype.
Metabolite Comparison of pnp1-1 and the Wild Type
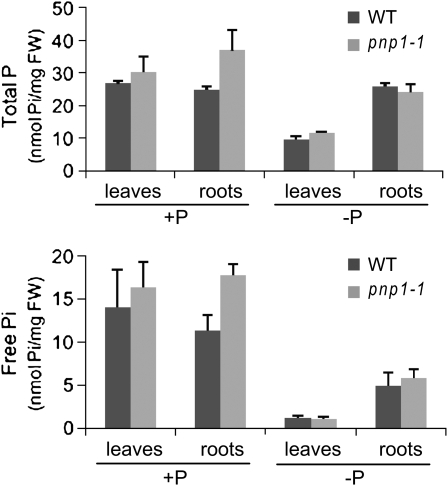
Based on the data described above, we hypothesized that PNPase deficiency either directly or indirectly affected production or transduction of a signal required for normal response to P deficiency. We first measured free or total P in wild-type or pnp mutant leaves and roots grown under +P and −P conditions. As shown in Figure 6, both free and total leaf P declined in both wild-type and mutant plants grown under −P conditions relative to +P, as was expected. For pnp1-1, we found that P levels were slightly higher in leaves, and significantly higher in roots, than those of the wild type when the plants were grown under +P conditions. On the other hand, P levels in pnp1-1 plants grown under −P conditions did not differ from those found in the wild type. We conclude that the pnp1-1 mutation does not have a major effect on P content as related to fresh weight, although both leaf and root P were slightly elevated under +P conditions. We also measured P uptake in wild-type and mutant roots, in case differences in P uptake were masked in the P accumulation data. However, no significant differences were observed (Supplemental Fig. S3).
Figure 6.
P content in leaves and roots. P content was measured in wild-type (WT) and pnp1-1 mutant seedlings. Plants were germinated and grown for 2 weeks on +P medium, then transferred to −P or +P medium for 1 additional week. Free Pi and total P were assessed as described in “Materials and Methods.” Error bars correspond to se. FW, Fresh weight.
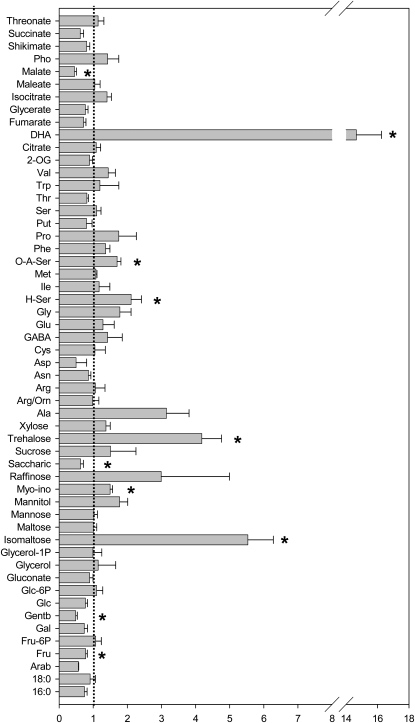
For a broader view of metabolic status, we quantified a panel of soluble, primary metabolites (Roessner et al., 2001) from wild-type and mutant plants grown under +P conditions or starved between 3 h and 3 weeks (Supplemental Tables S1 and S2; Supplemental Figs. S1 and S2). Under +P conditions, the metabolic differences between mutant and the wild type were limited (Fig. 7; Supplemental Table S1). Nonetheless, the sugars trehalose and isomaltose accumulated in the mutant to levels greater than 4-fold higher than in the wild type, whereas saccharate, gentobiose, and Fru were present at significantly lower levels in the mutant. The mutant was additionally characterized by higher levels of O-A-Ser, H-Ser, and Ala. The levels of succinate, malate, and fumarate were considerably lower in the mutant than in the wild type, whereas dehydroascorbate (DHA) accumulated (although it is important to note that the DHA level measured here is not absolutely representative of the in vivo amount, since the extraction conditions used here do not exactly conserve the cellular redox poise of the ascorbate pools).
Figure 7.
Relative metabolite content of pnp1-1 and the wild type grown under +P conditions expressed as mean fold change (±se) to the wild type. Plants were grown for 2 weeks on +P medium, then transferred (at time 0) to +P or −P medium for 3 h, 6 h, 1 week, or 3 weeks. Metabolites were quantified as described in “Materials and Methods.” The complete data set and statistical analysis are presented in Supplemental Table S2. Data are normalized with respect to the internal standard and the fresh weight. Values are presented as means ± se of determinations on six samples of bulked seedlings. Asterisks indicate values that were significantly different from the wild type when assessed by t tests (P < 0.05).
When looking from a global level, the impact of P starvation was fairly similar in both genotypes. The wild type displayed metabolic responses resembling those of many previous studies (Pieters et al., 2001; Uhde-Stone et al., 2003; Liu et al., 2005; Misson et al., 2005; Hernandez et al., 2007; Karthikeyan et al., 2007; Muller et al., 2007), while the pnp mutant generally also displayed many of these characteristic responses. Among observed differences was a far less dramatic change in the pnp mutant for the levels of Fru-6-P and Glc-6-P. Moreover, isomaltose, Man, saccharate, Suc, trehalose, and Xyl all increased in the wild type in response to P starvation, with raffinose being the only sugar that increased in both genotypes. Organic acids displayed mixed behavior in response to P deficiency: tricarboxylic acid (TCA) intermediates downstream of the reaction catalyzed by isocitrate lyase (one of the key enzymes of nitrate assimilation; Hodges et al., 2003) decreased in both genotypes, while those upstream increased. Taken together, these results suggest an altered TCA cycle activity on P stress and, as such, are largely in keeping with those of other recent studies (Morcuende et al., 2007).
With the exception of γ-aminobutyrate (GABA) and Pro, amino acids tend to accumulate with increasing periods of starvation in both genotypes. GABA, a stress-related metabolite (Fait et al., 2008), increased upon P starvation in the mutant, despite decreasing in the wild type, while the increase in the plastidial Asp family was visible following a mere 3 h of starvation in the mutant but only after 1 week in the wild type. Indeed, the changes in a wide range of amino acids were exacerbated in the mutant lines. To assess this statistically, two-way ANOVA tests were conducted (Supplemental Table S2); these tests revealed that the majority of the metabolites that discriminated between the genotypes were amino acids; however, also included in the top 25 discriminating metabolites were the phosphorylated intermediates Glc-6-P, glycerol 1-phosphate, and Fru-6-P, as expected (Morcuende et al., 2007, and refs. therein), and a range of sugars. These results were somewhat surprising, since the pattern of changes in many of these metabolites appears conserved between the genotypes; however, this clearly reflects that the altered metabolic response of pnp1-1 is quite subtle. When studied from a functional rather than a chemical perspective, this list also revealed a high number of stress-related metabolites such as GABA, myoinositol, raffinose, gentiobiose, chlorogenate, DHA, and salicylate, which tended to respond more dramatically in the pnp mutant than in the wild type.
Transcriptome Characterization of pnp1-1
To gain further insight into how the lack of PNPase might affect plant responses to P starvation, we used microarray hybridizations to compare the transcriptomes of pnp1-1 and wild-type plants grown under +P or −P conditions. To be able to relate transcriptome data to the metabolite analysis, we chose two time points for which the two genotypes were most distinct, which were after 3 h and 1 week of P starvation, according to principal component analysis (Supplemental Fig. S2). Total rosette RNA was used with the Affymetrix ATH1 platform to facilitate comparison with previous studies. The threshold of 2-fold change was chosen with a false discovery rate (FDR) < 0.05. Supplemental Tables S3 and S4 include the full data sets of the 3-h and 1-week −P experiments, respectively. Because the 3-h experiment revealed no significant regulated genes in the wild type or pnp1-1 in response to P starvation, we will only discuss the 1-week experiment here.
We first compared the two genotypes grown on +P medium (Table I). A total of 960 genes were found to be differentially expressed, which is perhaps not surprising given the slow growth and partial chlorosis of pnp1-1. What was noteworthy, however, was that the number of differentially expressed genes dramatically decreased when plants were grown on −P medium, to 224, indicating that the wild type and pnp1-1 have more similar gene expression patterns when P is not provided.
Table I.
The number of regulated genes in the wild type and/or pnp1-1 under different growth conditions
| Experimenta | Datab | Inducedc | Repressedc | Total |
|---|---|---|---|---|
| pnp1-1 +P/wild type +P | 16,536 | 601 | 359 | 960 |
| Wild type −P/wild type +P | 16,660 | 409 | 99 | 508 |
| pnp1-1 −P/pnp1−1 +P | 16,585 | 174 | 35 | 209 |
| pnp1-1 −P/wild type −P | 16,751 | 154 | 70 | 224 |
The genotype and the condition listed first correspond to the numerator of the gene expression mean ratio relative to the second, which is the denominator. For example, 601 genes are induced in the pnp1-1 mutant when compared with the wild type under +P conditions.
Data correspond to the genes that were tagged as “present” in at least one replicate as described in “Materials and Methods.”
The induced or repressed genes exhibit at least 2-fold change in expression and FDR < 0.05.
Subsequently, the effect of P deprivation on gene expression was assessed in the wild type. We found that 508 genes were regulated: 80.5% were up-regulated, whereas less than 20% were repressed. We compared our data set with other published experiments and found general consistency, although the experimental protocols were not identical (Misson et al., 2005; Morcuende et al., 2007). A third step of the analysis was to examine the effect of P deprivation in the mutant, and we found that the number of P-regulated genes in pnp1-1 was dramatically reduced with respect to the wild-type (i.e. 209 versus 508; Table I). This is consistent with the data mentioned above, that 960 genes differentiate the two genotypes on +P but only 224 on −P. In terms of transcript regulation, we conclude that P starvation attenuates the differences between pnp1-1 and the wild type.
Classification of Differentially Expressed Genes
We used MapMan to determine which functional categories were most affected in various pairwise comparisons (Supplemental Table S5). When the wild type and pnp1-1 were compared on +P conditions, the categories of photosynthesis, RNA regulation, cell functions, and stress responses were all identified with a P value of 10−10 or less, and various other metabolic functions were also identified with low P values (10−4 or less). Together, these categories likely include pleiotropic effects (e.g. stress responses) but may also include metabolic functions related to a particular role of PNPase. Under −P conditions and using a P value of 10−4 as a cutoff, eight categories rather than 11 (as seen under +P conditions) were identified, consistent with the speculation above that P starvation attenuates differences between the wild type and mutant. Of these eight categories, only two were the same as for +P conditions (photosynthesis and mitochondrial metabolism), consistent with the known large-scale reprogramming of plant gene expression when facing abiotic stress. The eight categories also included three important metabolic networks: major carbohydrate metabolism, oxidative pentose phosphate pathway, and TCA cycle/organic acid transformations. Thus, under P starvation, the pnp mutation affects normal gene expression as related to several major organellar and metabolic functions.
Another form of comparison was done, where we identified the 40 most regulated genes in pnp1-1 versus the wild type when grown under +P conditions (Table II), 15 of which are related to defense or stress responses. Four genes related to photosynthesis are also strongly regulated. Most remarkable among them is petD, which encodes subunit IV of the cytochrome b6/f complex and is induced 100-fold or greater under both +P and −P conditions in the mutant. However, like other chloroplast-localized genes in pnp1-1, altered mRNA processing is likely to account for some of the observed increase, and furthermore, as our cDNA was primed with oligo(dT), we would amplify polyadenylated chloroplast transcripts, which have been reported to hyperaccumulate in PNPase-deficient Arabidopsis plants (Walter et al., 2002). In contrast, two nuclear genes encoding photosynthesis proteins are strongly repressed in pnp1-1 versus the wild type; these are PSBP2 (PSII oxygen-evolving enhancer) and PORA (chlorophyll biosynthesis).
Table II.
The 40 most up- and down-regulated genes in the pnp1-1 mutant relative to the wild type when grown under +P conditions (FDR > 0.05)
ns, The fold change is below the cutoff value of 2. Stress-related gene products are shown in boldface.
| Probe Set Identifier | Arabidopsis Genome Initiative Code | Description |
pnp1-1 versus the Wild Type
|
|
|---|---|---|---|---|
| Fold Change on +P | Fold Change on −Pa | |||
| 244977_at | AtCg00730 | PetD, subunit IV of cytochrome b6/f complex | 102.82 | 151.68 |
| 257673_at | At3g20370 | Meprin and TRAF homology domain-containing protein | 81.11 | 15.88 |
| 250515_at | At5g09570 | Expressed chloroplast protein | 39.15 | 1.61 |
| 263539_at | At2g24850 | Aminotransferase | 32.66 | 9.07b |
| 267472_at | At2g02850 | Plastocyanin-like domain-containing protein | 29.93 | 3.28b |
| 245002_at | AtCg00270 | PsbD, D2 protein of PSII | 28.93 | 34.44 |
| 258941_at | At3g09940 | Monodehydroascorbate reductase, putative | 27.35 | ns |
| 248434_at | At5g51440 | 23.5-kD mitochondrial small HSP | 26.05 | 11.35b |
| 265051_at | At1g52100 | Jacalin-lectin family protein | 26.05 | 17.67 |
| 246888_at | At5g26270 | Expressed protein | 26.02 | 18.81 |
| 261930_at | At1g22440 | Alcohol dehydrogenase | 24.83 | ns |
| 265668_at | At2g32020 | GCN5-related N-acetyltransferase family protein | 22.45 | 20.99 |
| 260522_x_at | At2g41730 | Expressed protein | 22.34 | 9.65 |
| 266752_at | At2g47000 | P-glycoprotein 6 (PGP6), multidrug-resistant transporter | 21.49 | 11.89 |
| 266246_at | At2g27690 | Cytochrome P450 | 20.82 | 5.00b |
| 252921_at | At4g39030 | Enhanced disease susceptibility 5 (EDS5) | 20.69 | 6.70b |
| 256159_at | At1g30135 | JAZ8/TIFY5A (jasmonate-ZIM-domain protein 8) | 20.60 | ns |
| 254385_s_at | At4g21830 | Met sulfoxide reductase domain-containing protein | 20.56 | ns |
| 255110_at | At4g08770 | Peroxidase | 18.24 | ns |
| 263153_s_at | At1g54010 | Myrosinase-associated protein | 18.11 | 6.18 |
| 247718_at | At5g59310 | Lipid transfer protein 4 (LTP4) | 0.01 | 0.03 |
| 258498_at | At3g02480 | ABA-responsive protein-related | 0.03 | ns |
| 262412_at | At1g34760 | 14-3-3 protein GF14 omicron (GRF11) | 0.03 | ns |
| 266516_at | At2g47880 | Glutaredoxin family protein | 0.05 | ns |
| 266098_at | At2g37870 | Protease inhibitor/lipid transfer protein (LTP) family protein | 0.07 | ns |
| 259789_at | At1g29395 | Stress-responsive protein | 0.08 | 0.10b |
| 247095_at | At5g66400 | Dehydrin (RAB18) | 0.08 | 0.04b |
| 248197_at | At5g54190 | NADPH-protochlorophyllide oxidoreductase A (PORA) | 0.1 | 0.08b |
| 263981_at | At2g42870 | Phytochrome rapidly regulated 1 (PAR1) | 0.1 | ns |
| 247717_at | At5g59320 | Lipid transfer protein 3 (LTP3) | 0.1 | 0.13b |
| 260831_at | At1g06830 | Glutaredoxin family protein | 0.11 | 0.24 |
| 245353_at | At4g16000 | Expressed protein | 0.11 | ns |
| 264041_at | At2g03710 | Agamous-like 3 (AGL3) | 0.12 | 0.1 |
| 266873_at | At2g44740 | CYCP4;1, cyclin family protein | 0.13 | ns |
| 250933_at | At5g03170 | Fasciclin-like arabinogalactan protein (FLA11) | 0.13 | ns |
| 252612_at | At3g45160 | Expressed protein | 0.14 | 0.07 |
| 267569_at | At2g30790 | PSII oxygen-evolving complex 23 (PSBP2) | 0.14 | 0.17 |
| 261226_at | At1g20190 | Expansin (EXP11) | 0.14 | ns |
| 251039_at | At5g02020 | Expressed chloroplast protein | 0.14 | 0.15 |
| 251196_at | At3g62950 | Glutaredoxin family protein | 0.15 | ns |
The corresponding data for −P are also indicated (i.e. pnp1-1 on −P versus the wild type on −P). A full data set is given in Supplemental Table S4.
These values are not significant (FDR > 0.05).
P-Independent PNPase-Regulated Genes
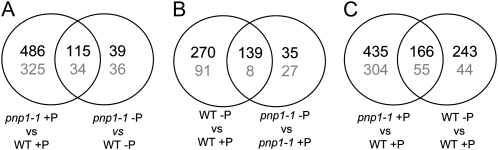
We conducted further analysis to differentiate pleiotropic effects on gene expression related to the slower growth, chlorosis, and possible general stress responses of the pnp1-1 mutant from those that could be directly attributed to the effect of the pnp mutation on chloroplast metabolism as possibly related to P deprivation responses. To do so, we compared sets of regulated genes as shown in Figure 8.
Figure 8.
Venn diagrams showing overlap of significantly induced or repressed genes between the indicated data sets derived from DNA microarray analysis. Significant changes were defined as 2-fold or greater, with FDR < 0.05. A to C represent the overlap between pairs of comparisons. Numbers in black and gray correspond to induced and repressed genes, respectively. WT, Wild type.
Figure 8A shows the overlap between pnp1-1 versus the wild type on +P or −P, revealing 149 genes that are regulated in the same direction. These can be interpreted as nutrient-independent effects of the pnp mutation. Representatives of these 149 genes are listed in Table III, with major categories related to photosynthesis or chloroplast functions. Fourteen genes are chloroplast encoded, and 11 of them are very strongly induced in pnp1-1, which as discussed above likely relates to the RNA maturation function of PNPase. A second cluster of genes included in the overlap in Figure 8A is conspicuous, as it mainly encodes components of the PYK10 complex, which is named after an endoplasmic reticulum body-associated β-glucosidase thought to be involved in plant defense (Nagano et al., 2005). Like the gene encoding PYK10 itself, genes encoding eight jacalin-lectin proteins, three GDSL lipase-like proteins, TSA1-like (DNA topoisomerase), and a meprin and TRAF homology domain-containing protein are all up-regulated in the pnp mutant. All of these proteins except three jacalins (JAL4, -11, and -27) were found to be part of the PYK10 complex (Nagano et al., 2008), and their induction is consistent with a stress situation in the pnp mutant.
Table III.
Selected genes regulated in pnp1-1 relative to the wild type independent of P status
| Probe Set Identifier | Arabidopsis Genome Initiative Code | Description |
pnp1-1 versus the Wild Type
|
|
|---|---|---|---|---|
| Fold Change on +P | Fold Change on −P | |||
| Photosynthesis- and chloroplast-targeted proteins | ||||
| 244977_at | AtCg00730 | PetD | 102.82 | 151.68 |
| 245002_at | AtCg00270 | PsbD | 28.93 | 34.44 |
| 245015_at | AtCg00490 | RbcL | 16.12 | 18.88 |
| 244970_at | AtCg00660 | Rpl20 | 15.03 | 26.41 |
| 245005_at | AtCg00330 | Rps14 | 10.57 | 43.80 |
| 244991_s_at | AtCg00890 | Ndh.B1 | 9.54 | 18.21 |
| 245003_at | AtCg00280 | PsbC | 6.35 | 9.79 |
| 244966_at | AtCg00600 | PetG | 5.57 | 7.03 |
| 245004_at | AtCg00300 | Ycf9 (PsbZ) | 3.24 | 4.99 |
| 245000_at | AtCg00210 | Ycf6 (PetN) | 2.79 | 3.08 |
| 244969_at | AtCg00650 | Rps18 | 2.14 | 2.76 |
| 244988_s_at | AtCg00840 | Rpl23.1 | 0.50 | 0.49 |
| 244964_at | AtCg00580 | PsbE | 0.40 | 0.47 |
| 245024_at | AtCg00120 | AtpA | 0.17 | 0.35 |
| 264868_at | At1g24090 | RNase H domain-containing protein | 7.19 | 5.56 |
| 256459_at | At1g36180 | Acetyl-CoA carboxylase 2 (ACC2) | 6.86 | 13.60 |
| 260648_at | At1g08050 | Zinc finger (C3HC4-type RING finger) family protein | 4.12 | 3.23 |
| 257667_at | At3g20440 | BE1/EMB2729 (branching enzyme 1), α-amylase | 3.58 | 4.98 |
| 245523_at | At4g15910 | Drought-induced protein (Di21) | 3.10 | 2.75 |
| 265479_at | At2g15760 | Calmodulin-binding protein, similar to AR781 | 2.59 | 3.26 |
| 263325_at | At2g04240 | Zinc finger (C3HC4-type RING finger) family protein | 0.47 | 0.47 |
| 248040_at | at5g55970 | Zinc finger (C3HC4-type RING finger) family protein | 0.26 | 0.25 |
| PYK10-associated proteins | ||||
| 265051_at | At1g52100 | JAL11, jacalin-lectin family protein | 26.05 | 17.67 |
| 259384_at | At3g16450 | JAL33, jacalin-lectin family protein, similar to ATMLP-300B (myrosinase-binding protein-like protein 300B) | 13.28 | 4.97 |
| 266988_at | At2g39310 | JAL22, jacalin-lectin family protein | 8.73 | 3.46 |
| 259327_at | At3g16460 | JAL34, jacalin-lectin family protein, similar to MBP1 (myrosinase-binding protein 1) | 7.87 | 4.32 |
| 259381_s_at | At3g16390 | JAL27, jacalin-lectin family protein, similar to ATMLP-470 | 5.25 | 2.71 |
| 262001_at | At1g33790 | JAL4, jacalin-lectin family protein | 3.28 | 4.63 |
| 259382_s_at | At3g16430 At3g16420 | JAL30, PBP1 (PYK10-binding protein 1) JAL31 | 5.60 | 3.10 |
| 263153_s_at | At1g54000 At1g54010 | GLL22, myrosinase-associated protein GLL23, myrosinase-associated protein | 18.11 | 6.18 |
| 263156_at | At1g54030 | GLL25, GDSL-motif lipase similar to ESM1 (epithiospecifier modifier 1) | 3.57 | 2.19 |
| 259009_at | At3g09260 | PYK10 (phosphate starvation response 3.1) | 10.95 | 4.27 |
| 257798_at | At3g15950 | TSA1-like (DNA topoisomerase), similar to TSK-associating protein 1 (TSA1) | 12.46 | 5.78 |
| 249817_at | At5g23820 | MD-2-related lipid recognition domain-containing protein | 3.65 | 3.68 |
| 246855_at | At5g26280 | Meprin and TRAF homology domain-containing protein | 12.51 | 7.61 |
| RNA- and DNA-related proteins | ||||
| 264460_at | At1g10170 | NF-X1-type zinc finger family protein | 4.56 | 3.03 |
| 267140_at | At2g38250 | DNA-binding protein-related, similar to transcription factor | 3.17 | 2.55 |
| 260266_at | At1g68520 | Zinc finger (B-box-type) family protein | 0.40 | 0.46 |
| 262291_at | At1g70790 | C2 domain-containing protein | 0.38 | 0.35 |
| 248764_at | At5g47640 | CCAAT box-binding transcription factor subunit B (NF-YB) (HAP3) | 0.26 | 0.32 |
| 252504_at | At3g46590 | TRFL1 (TRF-LIKE 1) telomere repeat-binding protein | 0.26 | 0.40 |
| 263909_at | At2g36490 | HhH-GPD base excision DNA repair family protein (ROS1) | 0.16 | 0.24 |
| 264041_at | At2g03710 | SEP4 (Sepallata 4); identical to agamous-like MADS-box protein AGL3 (AGL3) | 0.12 | 0.10 |
| Transport | ||||
| 263402_at | At2g04050 | MATE efflux family protein, similar to ATDTX1, antiporter | 13.05 | 7.08 |
| 263401_at | At2g04070 | MATE efflux family protein, similar to ATDTX1, antiporter | 6.49 | 9.09 |
| 253732_at | At4g29140 | MATE efflux protein-related | 2.58 | 2.49 |
| 249188_at | At5g42830 | Transferase family protein, similar to N-hydroxycinnamoyl/benzoyltransferase 6 | 10.47 | 5.68 |
| 255941_at | At1g20350 | ATTIM17-1 (translocase mitochondrial inner membrane subunit 17-1) | 10.44 | 5.51 |
| 253181_at | At4g35180 | LHT7 (Lys/His transporter 7) | 6.44 | 8.49 |
| 265479_at | At2g15760 | Calmodulin-binding protein, similar to AR781 | 2.59 | 3.26 |
| 267483_at | At2g02810 | ATUTR1 (UDP-Gal transporter 1) | 2.45 | 2.34 |
| 265161_at | At1g30900 | Vacuolar sorting receptor 6 precursor (AtVSR6) | 2.33 | 3.60 |
| 254662_at | At4g18270 | ATTRANS11 (translocase 11) | 0.30 | 0.21 |
| 257294_at | At3g15570 | Phototropic-responsive NPH3 family protein | 0.15 | 0.03 |
Effect of the pnp Mutation on the P Starvation Response
In order to explore the apparent attenuation of P starvation-induced transcriptional responses in the pnp1-1 mutant, we analyzed the overlap in regulated genes between pnp1-1 −P versus +P and wild type −P versus +P (Fig. 8B). This revealed 147 genes whose expression changes in the same direction in the two genotypes, representing about 30% and 70% of wild-type and pnp1-1 P-regulated genes, respectively. If one includes data below the 2-fold threshold we had chosen and allows an FDR > 0.5, 92% of the nonoverlapping P-responsive genes in the wild type are also regulated by −P in the same way in pnp1-1. Altogether, these data indicate that the wild type and pnp1-1 possess largely parallel responses to P deprivation, but the average fold change of the 139 overlapping induced genes was globally lower for the pnp1-1 plants (20.7-fold induction for pnp1-1 and 32.4-fold for the wild type), whereas the eight overlapping repressed genes had a similar average regulation level (0.22-fold for pnp1-1 and 0.20-fold for the wild type).
Constitutive P Starvation Responses in pnp1-1
The attenuated transcriptional response of pnp1-1 under P starvation raised the possibility that the plants were experiencing some degree of P stress, even when grown under nominally +P conditions, and thus had constitutive P starvation responses. Therefore, we compared the −P transcriptional responses in the wild type with the pnp versus wild-type effects on gene expression under +P conditions (Fig. 8C). The behaviors of some of the main known P-responsive genes are detailed in Table IV. The global result that emerges is that among the 508 P-responsive genes in the wild type, 43% are also regulated in the same direction in pnp1-1 relative to the wild type under +P conditions. This 43%, or 221 genes, includes 166 that are induced and 55 that are repressed. Among them are several major genes normally induced during P stress, such as those encoding P transporters belonging to the PHT1 family (PHT1;1 and PHT1;2, detected with the same probe; PHT1;4 and PHT1;7, detected with the same probe), the ribonuclease RNS1, and the transcription factor WRKY75, a positive modulator of P starvation responses and root development (Devaiah et al., 2007). Messenger RNAs encoding at least 30 other plastid-targeted proteins are also regulated in pnp1-1 as if it were already P starved (Supplemental Table S6). This strengthens the conclusion that plastid P metabolism is altered in pnp1-1.
Table IV.
Behavior of major genes normally involved in the phosphate deprivation response
Values in bold are those for which the fold change is significant (fold change ≥ 2 and FDR < 0.05); for those in normal type, the fold change is <2 and/or the FDR > 0.05.
| Probe Set Identifier | Arabidopsis Genome Initiative Code | Description |
pnp1-1 +P versus Wild Type +P
|
Wild Type −P versus Wild Type +P
|
pnp1-1 −P versus pnp1-1 +P
|
pnp1-1 −P versus Wild Type −P
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold Change | FDR | Fold Change | FDR | Fold Change | FDR | Fold Change | FDR | |||
| 247629_at | At5g60410 | DNA-binding family protein (SIZ1) | 1.82 | 0.324 | 1.97 | NA | 0.87 | 1.000 | 0.80 | NA |
| 253784_at | At4g28610 | Phosphate starvation response regulator (PHR1) | 0.94 | 0.820 | 0.86 | 0.589 | 0.91 | 1.000 | 1.00 | 0.999 |
| 245976_at | At5g13080 | Transcription factor (WRKY75) | 7.41 | 0.022 | 4.77 | 0.114 | 1.24 | 1.000 | 1.92 | 0.461 |
| 266743_at | At2g02990 | Ribonuclease 1 (RNS1) | 4.04 | 0.024 | 6.66 | 0.008 | 2.60 | 0.316 | 1.58 | 0.634 |
| 264893_at | At1g23140 | C2 domain-containing protein | 3.55 | 0.014 | 48.17 | 0.000 | 11.83 | 0.000 | 0.87 | 0.921 |
| 249152_s_at | At5g43350 At5g43370 | Inorganic phosphate transporter (PT1) (Pht1;1/Pht1;2) | 7.83 | 0.001 | 40.68 | 0.000 | 7.65 | 0.001 | 1.47 | 0.507 |
| 266184_s_at | At2g38940 At3g54700 | Phosphate transporter (PT2) (Pht1;4/Pht1;7) | 9.54 | 0.003 | 367.71 | 0.000 | 51.03 | 0.000 | 1.32 | 0.806 |
| 267646_at | At2g32830 | Inorganic phosphate transporter (PHT5) | 2.37 | 0.085 | 12.00 | 0.001 | 6.39 | 0.011 | 1.26 | 0.968 |
| 249151_at | At5g43360 | Inorganic phosphate transporter (PHT3) | 1.03 | NA | 1.02 | NA | 1.00 | 1.000 | 1.01 | 0.996 |
| 257311_at | At3g26570 | Phosphate transporter family protein PHT2;1 (chloroplast) | 0.74 | 0.782 | 0.74 | 0.876 | 0.75 | 1.000 | 0.75 | 0.902 |
| 258293_at | At3g23430 | Phosphate transporter (PHO1) | 0.74 | 0.656 | 15.88 | 0.012 | 6.43 | 0.186 | 0.30 | 0.223 |
| 267456_at | At2g33770 | Ubiquitin-conjugating enzyme family protein (PHO2/UBC24) | 1.40 | 0.315 | 1.76 | 0.164 | 1.10 | 1.000 | 0.87 | 0.926 |
| 264204_at | At1g22710 | Suc transporter/Suc-proton symporter (PHO3/SUC2) | 0.52 | 0.027 | 0.89 | 0.676 | 1.20 | 1.000 | 0.70 | 0.394 |
| 258158_at | At3g17790 | Acid phosphatase type 5 (ACP5) | 1.78 | 0.176 | 42.25 | 0.000 | 20.71 | 0.002 | 0.87 | 0.954 |
| 263083_at | At2g27190 | Iron(III)-zinc(II) purple acid phosphatase (PAP12) | 1.30 | 0.074 | 5.35 | 0.000 | 4.11 | 0.000 | 1.00 | 1.000 |
| 252006_at | At3g52820 | Purple acid phosphatase (PAP22) | 0.98 | 0.968 | 15.16 | 0.000 | 12.50 | 0.000 | 0.81 | 0.732 |
| 263594_at | At2g01880 | Purple acid phosphatase (PAP7) | 1.03 | NA | 10.00 | 0.307 | 10.88 | 0.597 | 1.12 | 0.950 |
| 255587_at | At4g01480 | Inorganic pyrophosphatase (ATPPA5) | 2.01 | 0.013 | 5.08 | 0.000 | 2.36 | 0.016 | 0.93 | 0.927 |
| 246071_at | At5g20150 | SPX (SYG1/Pho81/XPR1) domain-containing protein (AtSPX1) | 1.83 | 0.055 | 99.05 | 0.000 | 46.20 | 0.000 | 0.85 | 0.766 |
| 267611_at | At2g26660 | SPX (SYG1/Pho81/XPR1) domain-containing protein (AtSPX2) | 0.78 | 0.195 | 5.17 | 0.000 | 4.85 | 0.000 | 0.73 | 0.259 |
| 266132_at | At2g45130 | SPX (SYG1/Pho81/XPR1) domain-containing protein (AtSPX3) | 1.12 | 0.819 | 139.62 | 0.000 | 123.14 | 0.000 | 0.99 | 0.998 |
| 263599_at | At2g01830 | His kinase (CRE1/AHK4/WOL) | 0.40 | 0.004 | 0.55 | 0.033 | 0.98 | 1.000 | 0.71 | 0.273 |
| 258887_at | At3g05630 | Phospholipase D (PLDP2) | 0.89 | NA | 69.58 | 0.000 | 62.57 | 0.000 | 0.80 | 0.613 |
| 258452_at | At3g22370 | Alternative oxidase 1a, mitochondrial (AOX1A) | 4.44 | 0.003 | 1.87 | 0.107 | 0.99 | 1.000 | 2.35 | 0.074 |
| 257642_at | At3g25710 | Basic helix-loop-helix family protein (bHLH32) | 0.47 | 0.134 | 0.70 | 0.561 | 1.03 | 1.000 | 0.69 | 0.662 |
| 264783_at | At1g08650 | Phosphoenolpyruvate carboxylase kinase (PPCK1) | 1.04 | NA | 63.73 | 0.000 | 36.33 | 0.000 | 0.59 | 0.404 |
| 258570_at | At3g04530 | Phosphoenolpyruvate carboxylase kinase (PPCK2) | 1.02 | NA | 31.95 | 0.005 | 9.09 | 0.222 | 0.29 | 0.170 |
| 249846_at | At5g23630 | ATPase E1-E2 type family protein (PDR2) | 0.85 | 0.51 | 1.05 | 0.89 | 1.04 | 1 | 0.85 | 0.63 |
| Plastid carbon transport | ||||||||||
| 248144_at | At5g54800 | Glucose-6-phosphate/phosphate translocator (GPT1) | 1.51 | 0.047 | 1.95 | 0.008 | 1.42 | 0.288 | 1.10 | 0.826 |
| 264400_at | At1g61800 | Glucose-6-phosphate/phosphate translocator (GPT2) | 3.49 | 0.016 | 10.56 | 0.002 | 3.92 | 0.079 | 1.29 | 0.806 |
| 252414_at | At3g47420 | Glycerol-3-phosphate transporter | 1.34 | 0.796 | 33.88 | 0.016 | 24.66 | 0.057 | 0.98 | 0.998 |
| 246445_at | At5g17630 | Glucose-6-phosphate/phosphate translocator | 0.89 | 0.666 | 0.77 | 0.419 | 0.75 | 0.880 | 0.87 | 0.785 |
| 248886_at | At5g46110 | Phosphate/triose-phosphate translocator (TPT) | 0.88 | 0.388 | 0.98 | 0.934 | 0.92 | 1.000 | 0.83 | 0.326 |
| 259185_at | At3g01550 | Triose phosphate/phosphate translocator (PPT2) | 0.80 | 0.831 | 0.35 | 0.856 | 0.36 | 1.000 | 0.82 | 0.954 |
Verification of Transcriptome Results
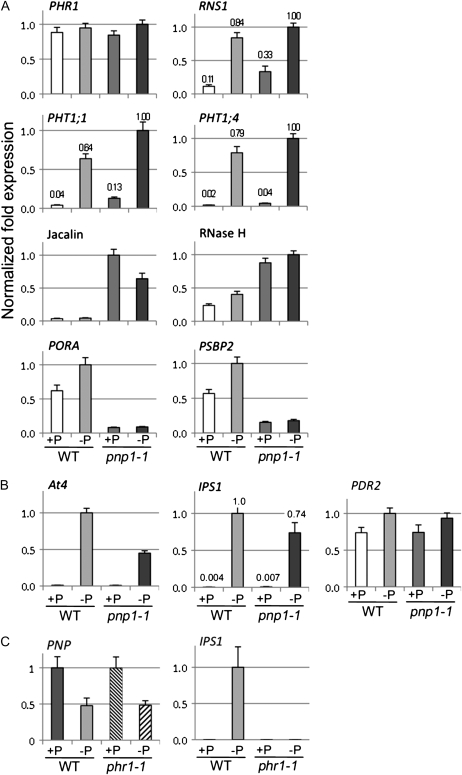
We performed quantitative RT-PCR to validate a portion of the expression data described above, as shown in Figure 9, selecting both P starvation-responsive genes and genes whose expression was independently affected by the pnp1-1 mutation. Figure 9A shows various protein-coding genes. We examined PHR1, which encodes a key P response transcription factor. As shown previously (Rubio et al., 2001), PHR1 expression is not regulated transcriptionally by P starvation in the wild type, and we found the same for the pnp mutant. RNS1, which encodes a P starvation-induced secreted ribonuclease (Bariola et al., 1999), showed the expected induction in the wild type; however, it was also constitutively (under +P conditions) expressed at 3-fold the wild-type level in pnp1-1. The transcripts encoding the transporters Pht1;4 and Pht1;1 are induced in the wild type, as expected (Mudge et al., 2002), and in the mutant. We also verified four genes whose expression was regulated P independently by the pnp1-1 genotype. We confirmed that genes encoding a jacalin-lectin and a potentially chloroplast-targeted RNase H domain-containing protein were induced, in agreement with microarray results (Supplemental Table S7), and that two photosynthetic genes, PORA and PSBP2, were repressed.
Figure 9.
Transcript levels for selected P starvation and pnp1-1-regulated genes as determined by quantitative RT-PCR. The fold change in expression was normalized to the actin gene ACT2 and scaled to the sample with the highest expression level for each tested gene, which was defined as 1.0. Error bars represent se. Data correspond to three biological replicates and at least two technical replicates, as detailed in Supplemental Table S7. For selected genes, relative transcript abundance is shown above the bars. A, Validation of microarray data for selected genes. Gene symbols are used except where there is no gene name but the product is known. Plants were germinated and grown for 2 weeks on +P medium, then transferred to +P or −P medium for 1 additional week. B, Expression levels of At4 and IPS1, two P starvation markers not represented on the ATH1 microarrays, and PDR2. Plants were grown as for A. C, Plants were germinated and grown for 1 week on +P medium, then transferred to +P or −P medium for 10 d, to replicate previously published growth conditions for the phr1 mutant (Rubio et al., 2001). WT, Wild type.
Figure 9B shows expression analysis of two P starvation-induced riboregulators, At4 and IPS1, which are not represented on ATH1 arrays. Both were strongly induced in both genotypes. Although it is not evident because of the scale of the graph, IPS1 was slightly but significantly induced in pnp1-1 under +P conditions, approximately 2-fold relative to the wild type. Also, we examined the expression of PDR2, since mutations in that gene phenocopy pnp1-1 in terms of lateral root abortion. PDR2 expression, however, did not differ in pnp1-1. Finally, we examined the expression of the PNP gene itself, and any dependence on PHR1, because we had previously found that in Chlamydomonas (Yehudai-Resheff et al., 2007) the PNP gene is repressed by P starvation in a Psr1-dependent manner and PHR1 is the ortholog of Psr1. As shown in Figure 9C, PNP transcripts decreased approximately 2-fold under P starvation conditions, which is comparable to the approximately 3-fold decrease observed in Chlamydomonas. This decrease appeared to be PHR1 independent, however. The character of this mutant was verified by measuring IPS1 transcripts, which, as expected (Nilsson et al., 2007), failed to be induced. In summary, our quantitative RT-PCR data supported the conclusions from the microarray approach and yielded additional information regarding the expression of riboregulators and the regulation of the PNP gene.
DISCUSSION
Our previous report illuminated a role for the cpPNPase in P starvation acclimation in Chlamydomonas (Yehudai-Resheff et al., 2007) and stimulated the studies reported here. While we conclude that cpPNPase also is important for a normal P starvation response in Arabidopsis, we used a different set of analytical tools to address issues specific to the multicellular context. In one perspective, the main commonality in the two experimental systems is that cell death occurs, which in the unicellular Chlamydomonas, of course, is lethal, whereas in Arabidopsis it is restricted to lateral root initiates. Both results, however, point to a key role for PNPase apart from RNA metabolism.
Altered cpRNA Maturation in pnp Mutants Is Associated with a Pale-Green Leaf Phenotype
Disruption of the cpPNPase gene in Arabidopsis was previously noted to be associated with 3′ extensions of both mRNAs and 23S rRNA (Walter et al., 2002), which we confirmed and also observed for the atpB mRNA (Fig. 1D). The pale-green phenotype is evident in emerging leaves (Fig. 1C) but was not noted by Walter et al. (2002) because a cosuppressed line rather than T-DNA null mutants was used. However, the phenotype was noted where a pnp mutant allele was obtained in a genetic screen for fosmidomycin resistance (Sauret-Gueto et al., 2006). The pale-green phenotype of young tissues progressively disappears, and cotyledons are indistinguishable from those of the wild type. This contrasts with deficiency in the product of the paralogous At5g14580 locus, which encodes mitochondrial PNPase. In this case, null mutants are embryo lethal and knockdown lines hyperaccumulate antisense and intergenic transcripts, suggesting a perhaps fatal disruption in mitochondrial gene expression (Holec et al., 2006).
cpPNPase Deficiency Affects Root Adaptation to −P Stress
We observed that pnp plants grown under −P conditions were unable to elaborate lateral roots (Figs. 2 and 3), a phenomenon that phenocopies pdr2, albeit on a longer time scale. The pdr2 mutation corresponds to a point mutation in At5g23630, which encodes a P-type ATPase of group V (Ticconi, 2005). Mutations in the same gene have been associated with reduced male fertility (Jakobsen et al., 2005). We also compared the root phenotype of pnp1-1 with that of csp41b-1, the latter of which is deficient in two related chloroplast endoribonucleases and which has pale mature leaves (Bollenbach et al., 2009). Because csp41b-1 resembled the wild type in terms of lateral roots, we conclude that neither the partial chlorosis nor the chloroplast gene expression defect of pnp1-1 is likely to be responsible for its lateral root phenotype under −P conditions.
We used two reporter genes to gain additional insight into the pnp1-1 root phenotype (Fig. 3). Using the DR5 promoter to drive GUS expression, we observed reduced staining in pnp1-1, relative to the wild type, whether grown under +P or −P conditions. Moreover, in most cases only one or two lateral root initiates under −P conditions stained for GUS. Since the DR5 construct essentially measures responsiveness to auxin in that tissue (Ulmasov et al., 1997), we conclude that auxin levels and/or activity are abnormally low. It has been shown that P starvation enhances auxin sensitivity in Arabidopsis roots and helps lead to higher lateral root density and inhibition of primary root elongation (Lopez-Bucio et al., 2002). Thus, our observations are consistent with the conclusion that the failure to elaborate lateral roots in pnp1-1 under low-P conditions is at least in part related to a defective hormonal cue.
A second reporter gene, CYCB1:GUS, revealed no abnormalities under +P conditions but suggested that cell division was only occurring in a single lateral root initiate within the clusters that formed in pnp1-1 under −P conditions. By staining with Evans blue, we concluded that, as in pdr2, mutant lateral root initiates die, followed by initiation of secondary, tertiary, and quaternary lateral roots. This cell death, or at least the loss of membrane integrity, was correlated with organellar abnormalities revealed by electron microscopy (Fig. 4). While we have not investigated whether lateral roots in pnp1-1 undergo programmed cell death, it is worth noting that in plants, mitochondrial abnormalities in particular, but also chloroplast dysfunction or communication, have been associated with programmed cell death (Yao et al., 2004).
P Content and Uptake
To test the hypothesis that PNPase might have a role in P homeostasis, we measured total and free Pi in leaves, where PNPase is predominantly expressed, as well as in roots, where a defective growth phenotype was observed. A slight increase of total P and free Pi was evident under +P conditions for pnp1-1, whereas no significant differences were noted under −P conditions in the wild type (Fig. 6). While most P-containing metabolites that we measured did not exhibit differences between genotypes, phosphorate was slightly elevated in pnp1-1 (Fig. 7). Also, while P uptake did not differ between genotypes, we saw slight induction relative to the wild type of the gene encoding the P transporters Pht1;1 and Pht1;4, under +P conditions (Fig. 9). This is consistent with a degree of P stress in pnp1-1 under +P conditions, as discussed below.
Transcript and Metabolite Profiling
Microarray data revealed an obvious reorientation of the pnp mutant transcriptome toward expression of −P responses when grown on a full nutrient medium, comprising 221 out of the 508 P starvation-regulated genes in the wild type. Taken together with the P transporter data discussed above, we hypothesize that pnp mutant plants adjust their metabolism as if they were already, to some extent, under phosphate stress.
A second set of genes is responding to the absence of PNPase independent of P availability. Overlap analyses (Fig. 8A) between pnp-regulated genes on +P and on −P compared with the wild type revealed 149 genes that fall into two major metabolic classes: regulation of chloroplast activities and oxidative stress responses. Fourteen of these are encoded by the chloroplast genome and are generally strongly up-regulated. As mentioned earlier, the accumulation of these transcripts likely results from perturbation of normal cpRNA degradation pathways. The genotype also affected 12 nucleus-encoded chloroplast proteins. On the other hand, of several genes suggested to be responsible for the communication between chloroplast and nucleus, or retrograde signaling, none was significantly regulated. Altered photosynthesis-related gene expression in pnp1-1 is in agreement with its partial chlorosis and slow-growth phenotype.
We also examined gene expression related to the chloroplast MEP pathway, since the pnp mutant rif10 was identified using fosmidomycin, a strong inhibitor of deoxyxylulose 5-phosphate reductoisomerase (DXR), which catalyzes the second step (Sauret-Gueto et al., 2006). In the case of rif10, the authors reported a similar transcript accumulation for the deoxyxylulose 5-phosphate synthase (DXS), the first enzyme of the MEP pathway, and for DXR but increased accumulation for both at the protein level. Our experiments confirmed the lack of significant regulation for two DXS genes (DXS1 and DXS3) and the DXR gene but revealed 5.8-fold activation of the third DXS gene (DXS2; At3g21500), which is expressed at a low level under normal conditions, based on EST data (Rodriguez-Concepcion and Boronat, 2002), but in our study was also activated in the wild type during P starvation. That regulation of DXS2 in pnp1-1, rather than a posttranslational effect, could explain at least in part the DXS enzyme accumulation described, although it should be noted that the activities of DXS2 and DXS3 remain to be established; indeed, it has been proposed that they do not encode functional proteins and should be renamed DXL (for DXS-like; Phillips et al., 2008).
Overall, in pnp mutants including rif10 and pnp1-1, defects in the MEP pathway lead to less accumulation of chlorophyll and carotenoids. This is not only consistent with the observed chlorosis but may make the plants subject to oxidative stress due to a decrease in photoprotective carotenoids. Finally, because this pathway is also responsible for the biosynthesis of certain hormones, their levels may also be affected in pnp mutants.
Metabolic profiling revealed limited changes between pnp1-1 and the wild type in the presence of P. However, pnp1-1 displayed a different response to P starvation than wild-type Arabidopsis (Morcuende et al., 2007; this work) or common bean (Phaseolus vulgaris; Hernandez et al., 2007). For example, changes in the amino acid profile under −P conditions strongly hint of a perturbation in plastid metabolism, including large changes in the Asp family amino acids. Interestingly, these metabolites are those that cost the most ATP in their production, and their rate of biosynthesis has previously been demonstrated to be highly dependent on the plastidial energy charge (Regierer et al., 2002; Carrari et al., 2005). Importantly, these changes are considerably more dramatic in the pnp mutant than in the wild type, implying a functional role for the PNP protein in this process. Although less easy to rationalize there are also large changes in cytosolic and mitochondrially synthesized metabolites such as Gly, Ser, Ala, and GABA.
Oxidative Stress Responses and Organellar Integrity in pnp1-1
At the transcriptional and metabolic levels, several lines of evidence suggest activation of oxidative stress responses in pnp1-1. Among the metabolites, DHA is the most strongly regulated on +P, with its content increased about 15-fold. Several enzymes involved in the ascorbate cycle, and its biosynthesis, are strongly regulated in pnp1-1. Induced genes include VTC5, an ascorbate biosynthetic enzyme (Dowdle et al., 2007), and two ascorbate oxidases that convert l-ascorbate to mono-DHA (Ishikawa et al., 2006). In contrast, the gene encoding the chloroplast thylakoid enzyme responsible for the same conversion but via hydrogen peroxide rather than oxygen scavenging, tAPX, is repressed. Reduction of mono-DHA, which is a radical, is catalyzed by mono-DHA reductase, which is also strongly induced in pnp1-1. Finally, the initiation and the activity of the ascorbate-glutathione cycle are strongly induced, probably leading to accumulation of DHA. However, the repression of the gene encoding tAPX might lessen the ability of the pnp mutant to detoxify the chloroplast (for review, see Asada, 2006). It should also be noted that a recent study associated oxidative stress resulting from a combination of drought and heat with enhanced malic enzyme activity and a decrease in malate, leading the authors to speculate that malate metabolism plays an important role in the response of Arabidopsis to this stress combination (Koussevitzky et al., 2008). Our findings of 50% lower malate in pnp1-1 accompanying the accumulation of DHA hint of a similar response in this mutant.
The pnp1-1 +P transcriptome revealed induction of additional genes involved in redox homeostasis, including peroxidases, thioredoxins, cytochromes P450, and glutathione S-transferases. We also note the induction of one cytosolic and one chloroplast copper/zinc superoxide dismutase: CSD1 and CSD2, respectively (Kliebenstein et al., 1998). These are involved in superoxide radical detoxification and the positive and negative regulation of genes encoding glutaredoxins. Some of this regulation was also found in the wild type under P starvation.
A general consequence of reactive oxygen species accumulation can be the alteration of membrane lipids, which could lead in turn to organelle disorganization, as we observed in pnp1-1 roots (Fig. 4). This loss of organelle integrity might be linked to the induction of a group of genes related to the PYK10 complex, as discussed in “Results.” The genes encoding PYK10 complex components were found to be repressed in the nai1 mutant (Nagano et al., 2008), which lacks a basic helix-loop-helix transcription factor (Matsushima et al., 2004). Another target of NAI1 is the lipoxygenase LOX3, which is induced more than 12-fold in pnp1-1 on +P; the NAI1 gene itself is induced 1.8- and 1.5-fold in pnp1-1 versus the wild type on +P and −P, respectively. One study of endoplasmic reticulum bodies suggested that the PYK10 complex might form only when subcellular structures are altered or disrupted, as the complex's partners would normally be localized in different compartments (Nagano et al., 2008). The induction of PYK10 complex genes in pnp1-1 could be thus related to the loss of organelle integrity triggered by oxidative stress.
A Systemic Signal May Be Altered in pnp1-1
The split-root experiment (Fig. 5) raised the possibility that a systemic signal was affected in pnp1-1 plants. Such a signal has been hypothesized to arise in the root cap during P starvation (Svistoonoff et al., 2007) and may be integrated in the aerial part of the plant to stimulate adaptation in root architecture. Our results suggest that in the case of pnp1-1, either a major metabolite or hormonal balance may be relevant to this issue.
One metabolite candidate would be sugars, as a tight relationship between sugar metabolism and P deprivation responses is well established and was evident in a transcriptomic comparison between P starvation responses and Suc-regulated metabolism (Muller et al., 2005). It was found that some P-responsive genes are more strongly induced in the presence of Suc and that these enhanced responses were correlated with increased lateral root density (Karthikeyan et al., 2007). It has additionally been proposed that phloem Suc transport may integrate root responses to phosphate deprivation (for review, see Hammond and White, 2008).
In the experiments reported here, pnp1-1 differed from the wild type in particular with respect to early fairly dramatic decreases in Fru-6-P at 3 h and to a delayed increase in raffinose as compared with the wild type. The latter was found to accumulate in the mutant under control conditions. Additional differences in the sugar response to P starvation between the two genotypes are to be found in the opposite behavior of isomaltose and trehalose and the sugar derivative myoinositol, particularly following 1 week of starvation. The differences are reduced in the longest starvation period, suggesting an adjustment of sugar metabolism to P deprivation within the first hours.
Our microarray analysis also revealed gene expression changes in pnp1-1 closely related to Suc metabolism. For example, under +P conditions, several carbohydrate transporters are induced: the plastid Glc-6-P translocator (GPT2), a mannitol transporter, a sugar transporter (STP4), two UDP-Gal transporters, and the Suc transporter SUC1. Also in pnp1-1, glycolysis appears to be globally activated. While it is challenging to link particular metabolite levels to any of these changes in gene expression, it is notable that both metabolite steady-state levels and relevant genes are fluctuating in tandem.
Two other sugars, isomaltose and trehalose, increased approximately 5.5- and 4-fold in pnp1-1 versus the wild type, respectively, when grown on +P (Fig. 7). Their accumulation patterns also differ qualitatively upon P starvation (Supplemental Fig. S1). The role of isomaltose is unclear, as it is apparently not a major form of carbon exported from the chloroplast, at least in wild-type plants (Weise et al., 2004). Nor does isomaltose appear to be accumulating at the expense of maltose, which is the major exported form. However, we cannot exclude a deficiency of carbon export, as we have examined only total metabolites rather than their partitioning into subcellular compartments.
Trehalose is a disaccharide whose phosphorylated form, trehalose-6-phosphate (Tre-6-P), appears to be an important signaling molecule related to sugar metabolism (Paul et al., 2008). Trehalose is produced in a tandem reaction commencing with the synthesis of Tre-6-P from Glc-6-P and UDP-Glc catalyzed by Tre-6-P synthase (TPS); subsequently, Tre-6-P phosphatase (TPP) catalyzes the dephosphorylation of Tre-6-P to trehalose. The accumulation of trehalose in the mutant under +P conditions might suggest an up-regulation of Tre-6-P dephosphorylation, a phenomenon that occurs in the wild type during the short periods of P starvation and might relate to P repartitioning within the cell. On the other hand, trehalose levels could increase in response to stress conditions (redox and/or P starvation). Indeed, although the role of trehalose as an osmoprotectant remains under debate, it is known to accumulate under a variety of environmental stresses, including cold, osmotic imbalance, and salt. Most of its biosynthetic genes respond to plant exposure to a wide range of abiotic and perhaps also biotic stresses (Iordachescu and Imai, 2008).
Consistent with our metabolite data and published gene expression results, transcriptome analyses revealed that five genes related to trehalose biosynthesis are differentially regulated on +P in pnp1-1. Four of them encode TPP (TPPG and TPPD are induced and TPPA and TPPH are repressed), whereas TPS1 is repressed. TPPA, TPPD, and TPPG are predicted to be chloroplast targeted. Because both TPP and TPS are encoded by multigene families whose expression profiles vary widely (Paul et al., 2008), how this suite of expression changes results in increased trehalose content in pnp1-1 is difficult to pinpoint. Furthermore, since Tre-6-P is an important metabolite whose concentration we have not measured directly, its relevance to the pnp root phenotype remains an open question.
Hormone Balance in pnp1-1
Many publications concerning P starvation responses highlight the roles of hormones in signal transduction, particularly auxins and cytokinins (Rubio et al., 2009; for review, see Yuan and Liu, 2008). It is not surprising, then, that many genes involved in hormone biosynthesis or degradation are regulated in the pnp mutant on +P (Supplemental Table S4). Furthermore, expression of DR5:GUS in pnp1-1 (Fig. 3A) suggested a decreased auxin activity in pnp1-1 root tips on +P as well as on −P. However, because we did not carry out detailed analyses of hormone contents, nor create genetic combinations with hormone pathway mutants, we feel it is premature to draw any direct connections between normal PNPase activity and hormone signaling.
In conclusion, the Arabidopsis and Chlamydomonas pnp mutants have few commonalities in their response to P starvation when examined in molecular detail. However, the importance of PNPase, and more generally the chloroplast, in conferring the ability to correctly respond to P starvation is conserved. Given the considerable differences in the survival strategies of a motile, unicellular organism and a sessile, multicellular one, differences in gene regulation and the consequences of PNPase inactivation are perhaps not surprising. Examining additional evolutionarily diverse photosynthetic species in this regard should similarly be interesting. How PNPase activity influences the ability of organisms to respond to P stress remains to be understood in detail, in particular what type of signal it generates and how that signal is integrated into the global response pathway.
MATERIALS AND METHODS
Plant Material
All of the Arabidopsis (Arabidopsis thaliana) plants used in this study are derived from the Columbia-0 ecotype, which was used as the wild type. The two mutant lines, pnp1-1 (SALK_013306) and pnp1-2 (SALK_070705), containing a T-DNA insertion in the PNP gene (At3g03710), were obtained from the SIGnAL collection (Alonso et al., 2003). The precise locations of the T-DNA left borders were confirmed by DNA sequencing. PCR with the left border primer (LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′) and gene-specific primers (pnp1-1, 5′-GCAAAGCTCGCTGTTTAGATG-3′ and 5′-CATAGCCATGTCAACTTTGCC-3′; pnp1-2, 5′-TACGTAGGCGAATTGTTGAGG-3′ and 5′-CCACAAACAGATGCCATACTG-3′) was used to identify T-DNA insertion alleles of pnp1-1 and pnp1-2, respectively. The pnp1-1 line was crossed with the Arabidopsis CYCB1:GUS (Ulmasov et al., 1997) and DR5:GUS (Colòn-Carmona et al., 1999) lines to generate the reporter lines used in Figure 3.
Growth Conditions and Phosphate Starvation Treatment
Seeds were surface sterilized and stratified at 4°C for 3 to 4 d. Unless noted in the figure legends, plants were grown as follows. Seeds were germinated in a full nutrient Murashige and Skoog (MS) liquid medium in a controlled-environment chamber on a shaker at 25°C under fluorescent lights (100 μmol m−2 s−1) with a long-day photoperiod (16 h of light). After 2 weeks, plantlets were transferred into Magenta boxes or onto petri plates with MS medium (1.25 mm KH2PO4) containing 2% (w/v) Suc and 0.75% (w/v) phytagar (Murashige and Skoog, 1962) for the indicated periods. This medium is referred to as +P medium. For −P medium, KH2PO4 was omitted from the nutrient solution and the plantlets were rinsed with the same liquid solution prior to transfer. The phytagar (commercial grade; Gibco-BRL) contributed about 25 μm total P to the final medium.
For the microarray and the quantitative RT-PCR experiments, plants were germinated and grown for 2 weeks on a full nutrient MS medium (+P) containing 0.6% phytagar (w/v) and 0.5% (w/v) Suc (to limit the Suc effect on gene expression) at 22°C in a growth chamber under a 16-h photoperiod (with a fluorescent light intensity of 200 μmol m−2 s−1). Then, they were transferred to fresh +P or −P MS medium. For the −P medium, KH2PO4 was omitted but the potassium was compensated by K2SO4. Plantlets were rinsed with distilled water before transfer. On soil, plants were grown on Metro Mix 360, in a growth chamber, as described above.
RNA Analysis
Total RNA was isolated using Tri Reagent according to the manufacturer's instructions (Molecular Research Center), separated by electrophoresis, and transferred onto a GeneScreen membrane (Perkin-Elmer) as described previously (Bollenbach et al., 2005). Gene-specific probes were labeled by random priming using 100 ng of DNA, 20 μCi of [α-32P]dCTP, and the Klenow fragment (Promega).
UV Cross-Linking
UV cross-linking of proteins to radiolabeled RNA was performed as described previously (Lisitsky et al., 1997). The proteins (10 fmol) were mixed with [α-32P]UTP-RNA (10 fmol) in a buffer containing 10 mm HEPES-NaOH, pH 7.9, 30 mm KCl, 6 mm MgCl2, 0.05 mm EDTA, 2 mm dithiothreitol, and 8% (v/v) glycerol and cross-linked immediately with 1.8 J of UV irradiation (Stratalinker 1800; Stratagene). The RNA was then digested with 10 μg of RNase A and 30 units of RNase T1 at 37°C for 1 h. The proteins were fractionated by SDS-PAGE and analyzed by autoradiography.
Root Analyses
Images of roots were recorded with a stereomicroscope (Olympus SZX12) high-performance CCD camera and imported into Photoshop Image software. Histochemical analysis of the GUS reporter enzyme activity was adapted from Jefferson (1987). Samples were stained using 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Sigma) dissolved in a 100 mm sodium phosphate buffer (pH 7.2) containing 0.2% Triton X-100, 2 mm K4Fe(CN)6·H2O, and 2 mm K3Fe(CN)6·H2O. For cell viability analysis, roots were stained with 10% (w/v) Evans blue (Fisher). After washing with distilled water and mounting in 50% glycerol, root tips were viewed using the Olympus SZX12 microscope.
Transmission Electron Microscopy
Roots were embedded in an epoxy resin (Spurr, 1969). The root tips, from root cap to root hair zone (approximately 2 mm in length), were cut and fixed overnight in 5% (v/v) glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 7.4) at 4°C, postfixed in 1% (w/v) aqueous osmium tetroxide for 3 h at room temperature, and rinsed three times with distilled water. After dehydration through a graded ethanol series, the samples were rinsed in propylene oxide, infiltrated, and embedded in Spurr's resin. Ultrathin sections were cut to a 50- to 70-nm thickness with a diamond knife on a Leica UCT ultramicrotome. The sections were stained with uranyl acetate (2.5%, w/v) and lead citrate. After staining, the sections in the middle longitudinal direction were viewed and photographed using a Philips FEI-Technai 12 microscope.
Determination of Metabolite Levels
Free Pi and total P were assessed as described previously (Versaw and Harrison, 2002). For metabolic profiling, leaf samples were immediately frozen in liquid nitrogen and stored at −80°C. Extraction and quantification of metabolites were carried out using an established gas chromatography-mass spectrometry-based protocol (Roessner et al., 2001), with the exception that the metabolites studied also included subsequent additions to the mass spectral libraries (Schauer et al., 2005).
Principal component analysis was performed with the online tool MetaGeneAlyse (www.metagenealyse.mpimp-golm.mpg.de; Scholz et al., 2004) and TMEV software (Saeed et al., 2003). The data were normalized to the mean of the entire sample set for each metabolite and log10 transformed before the analysis. This transformation reduces the influence of rare high-measurement values but does not change the discrimination in the data set. Statistical analysis of the data was performed by t test and two-way ANOVA.
Microarray Hybridization and Data Analysis
Total RNA was isolated from seedlings, from which roots had been removed, using the RNeasy Plant Minikit (Qiagen), including DNase treatment according to the manufacturer's instructions. RNA quality check, linear amplification, labeling, hybridization, washing, and scanning were performed by the Cornell Microarray Core Facility (http://cores.lifesciences.cornell.edu/brcinfo/?f=10). Affymetrix ATH1 genome array GeneChips were used. Three biological replicates were used for each experimental condition.
Raw array data were normalized at the probe level using gcRMA (Wu et al., 2004). The detection calls (present, marginal, or absent) for each probe set were obtained using the mas5calls function in the Affy package (Gautier et al., 2004). Only genes with at least one present call across all of the compared samples were used to identify differentially expressed genes. Significance of gene expression was determined using the LIMMA test (Smyth, 2004), and raw P values of multiple tests were corrected using FDR (Benjamini and Hochberg, 1995). Genes with FDR < 0.05 and fold change ≥ 2 were identified as differentially expressed. The complete data set was deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE 18071.
Quantitative RT-PCR
One microgram of DNase-treated RNA was reverse transcribed in a 20-μL reaction using SuperScript III (Invitrogen) according to the instructions, including the RNase H treatment. One microliter of this cDNA was amplified using the Fast Sybr Green master mix (Applied Biosystems) and 0.66 μm of each primer in a 15-μL reaction. PCR amplification was performed using the Bio-Rad CFX96 real-time PCR detection system with the following conditions: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The amplification specificity was checked using melting curves. The relative quantification of the samples was determined using the Bio-Rad CFX Manager software, integrating primer efficiencies calculated from a standard curve. For each gene, the sample showing the highest intensity level was used as reference with a value of 1. The final data result from averages of three biological replicates and at least two technical repetitions. Primers are listed in Supplemental Table S8.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Relative metabolite contents of pnp1-1 and wild type grown under -P conditions for different time periods.
Supplemental Figure S2. Principal component analysis of metabolite profiles.
Supplemental Figure S3. Pi uptake in pnp1-1 and wild type.
Supplemental Table S1. Raw data for metabolite contents of pnp1-1 and wild type grown under +P or −P conditions.
Supplemental Table S2. Statistical analysis of metabolite data.
Supplemental Table S3. Microarray data for significantly regulated genes after 3 h of P starvation.
Supplemental Table S4. Microarray data for significantly regulated genes after 1 week of P starvation.
Supplemental Table S5. Statistical significance of the functional categorization of regulated genes using MapMan-defined BINs.
Supplemental Table S6. Genes encoding chloroplast-targeted proteins that are similarly regulated in pnp1-1 +P versus wild type +P and in wild type −P versus wild type +P.
Supplemental Table S7. Normalized expression data for quantitative RT-PCR.
Supplemental Table S8. List of primers used for quantitative RT-PCR.
Supplementary Material
This work was supported by the Triad Foundation, the Binational Agricultural Research and Development Fund (project no. IS–4152–08), and a National Science Foundation Research Experiences for Undergraduates fellowship (to J.J.), by the Alexander von Humboldt Foundation and the Max Planck Society (to A.F. and A.R.F.), and by the National Natural Science Foundation of China (grant no. 30670119).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David B. Stern (ds28@cornell.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage RE, Shabanowitz J, Hunt DF, Schuster G, Gruissem W (2001) Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA 7: 1464–1475 [PMC free article] [PubMed] [Google Scholar]
- Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC, Green PJ (1999) Regulation of S-Like ribonuclease levels in Arabidopsis: antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol 119: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D (2005) RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res 33: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB (2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69: 541–552 [DOI] [PubMed] [Google Scholar]
- Carrari F, Coll-Garcia D, Schauer N, Lytovchenko A, Palacios-Rojas N, Balbo I, Rosso M, Fernie AR (2005) Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth. Plant Physiol 137: 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong J, McBride H, Koehler CM, Teitell MA, et al (2006) Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol 26: 8475–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colòn-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N (2007) Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52: 673–689 [DOI] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13: 14–19 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Hammond JP, White PJ (2008) Sucrose transport in the phloem: integrating root responses to phosphorus starvation. J Exp Bot 59: 93–109 [DOI] [PubMed] [Google Scholar]
- Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W (1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J 15: 1132–1141 [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Ramirez M, Valdes-Lopez O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M, Flesch V, Galvez S, Bismuth E (2003) Higher plant NADP+-dependent isocitrate dehydrogenases, ammonium assimilation and NADPH production. Plant Physiol Biochem 41: 577–585 [Google Scholar]
- Holec S, Lange H, Kuhn K, Alioua M, Borner T, Gagliardi D (2006) Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol 26: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordachescu M, Imai R (2008) Trehalose biosynthesis in response to abiotic stresses. J Integr Plant Biol 50: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Dowdle J, Smirnoff N (2006) Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiol Plant 126: 343–355 [Google Scholar]
- Jakobsen MK, Poulsen LR, Schulz A, Fleurat-Lessard P, Moller A, Husted S, Schiott M, Amtmann A, Palmgren MG (2005) Pollen development and fertilization in Arabidopsis is dependent on the MALE GAMETOGENESIS IMPAIRED ANTHERS gene encoding a type V P-type ATPase. Genes Dev 19: 2757–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG (2007) Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225: 907–918 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Suzuki N, Huntington S, Armijo L, Sha W, Cortes D, Shulaev V, Mittler R (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem 283: 34197–34203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Gupta JD, Hunt AG (1998) Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J Biol Chem 273: 17539–17543 [DOI] [PubMed] [Google Scholar]
- Lisitsky I, Kotler A, Schuster G (1997) The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: the exoribonuclease 100RNP-polynucleotide phosphorylase displays high binding affinity for poly(A) sequences. J Biol Chem 272: 17648–17653 [DOI] [PubMed] [Google Scholar]
- Liu JQ, Samac DA, Bucciarelli B, Allan DL, Vance CP (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41: 257–268 [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I (2004) NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16: 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Muller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Nilsson L, Nielsen LK, Hamborg Nielsen T (2005) Interaction between phosphate starvation signalling and hexokinase-independent sugar sensing in Arabidopsis leaves. Physiol Plant 124: 81–90 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type beta-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol 49: 969–980 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Matsushima R, Hara-Nishimura I (2005) Activation of an ER-body-localized beta-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol 46: 1140–1148 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59: 417–441 [DOI] [PubMed] [Google Scholar]
- Phillips MA, Leon P, Boronat A, Rodriguez-Concepcion M (2008) The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci 13: 619–623 [DOI] [PubMed] [Google Scholar]
- Pieters AJ, Paul MJ, Lawlor DW (2001) Low sink demand limits photosynthesis under Pi deficiency. J Exp Bot 52: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Reape TJ, Molony EM, McCabe PF (2008) Programmed cell death in plants: distinguishing between different modes. J Exp Bot 59: 435–444 [DOI] [PubMed] [Google Scholar]
- Regierer B, Fernie AR, Springer F, Perez-Melis A, Leisse A, Koehl K, Willmitzer L, Geigenberger P, Kossmann J (2002) Starch content and yield increase as a result of altering adenylate pools in transgenic plants. Nat Biotechnol 20: 1256–1260 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids: a metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen ML, Cardona-Lopez X, Rojas-Triana M, Paz-Ares J (2009) Plant hormones and nutrient signaling. Plant Mol Biol 69: 361–373 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon L, Lopez-Bucio J, Chacon-Lopez A, Cruz-Ramirez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- Sarkar D, Fisher PB (2006) Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions. Pharmacol Ther 112: 243–263 [DOI] [PubMed] [Google Scholar]
- Sauret-Gueto S, Botella-Pavia P, Flores-Perez U, Martinez-Garcia JF, San Roman C, Leon P, Boronat A, Rodriguez-Concepcion M (2006) Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis. Plant Physiol 141: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J (2004) Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20: 2447–2454 [DOI] [PubMed] [Google Scholar]
- Slomovic S, Portnoy V, Liveanu V, Schuster G (2006) RNA polyadenylation in prokaryotes and organelles: different tails tell different tales. Crit Rev Plant Sci 25: 65–77 [Google Scholar]
- Slomovic S, Schuster G (2008) Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA 14: 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3. [DOI] [PubMed] [Google Scholar]
- Soreq H, Littauer UZ (1977) Purification and characterization of polynucleotide phosphorylase from Escherichia coli: probe for the analysis of 3′ sequences of RNA. J Biol Chem 252: 6885–6888 [PubMed] [Google Scholar]
- Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Ticconi CA (2005) A genetic analysis of phosphate deficiency responses in Arabidopsis. PhD thesis. University of California, Davis, CA
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37: 801–814 [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Gilbert G, Johnson JMF, Litjens R, Zinn KE, Temple SJ, Vance CP, Allan DL (2003) Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant Soil 248: 99–116 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Kilian J, Kudla J (2002) PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J 21: 6905–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, Weber AP, Sharkey TD (2004) Maltose is the major form of carbon exported from the chloroplast at night. Planta 218: 474–482 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, Spencer F (2004) A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96: 15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Eisfelder BJ, Marvin J, Greenberg JT (2004) The mitochondrion: an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J 40: 596–610 [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Hirsh M, Schuster G (2001) Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol Cell Biol 21: 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB (2007) Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 19: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu D (2008) Signaling components involved in plant responses to phosphate starvation. J Integr Plant Biol 50: 849–859 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2007) The diurnal metabolism of leaf starch. Biochem J 401: 13–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.