Abstract
We developed a simple and fast method to identify temperature-sensitive alleles of essential plant genes. We used primary and tertiary structure information to identify residues in the core of the protein of interest. These residues were mutated and tested for temperature sensitivity, taking advantage of the exceptionally rapid 1-week complementation assay in the moss Physcomitrella patens. As test molecules, we selected the actin-binding proteins profilin and actin-depolymerizing factor, because they are essential and their loss-of-function phenotype can be fully rescued. Screening a small number of candidate mutants, we successfully identified temperature-sensitive alleles of both profilin and actin-depolymerizing factor. Plants harboring these alleles grew well at the permissive temperature of 20°C to 25°C but showed a complete loss of function at the restrictive temperature of 32°C. Notably, the profilin mutation identified in the moss gene can be transferred to profilins from other plant species, also rendering them temperature sensitive. The ability to routinely generate temperature-sensitive alleles of essential plant proteins provides a powerful tool for the study of gene function in plants.
Conditional mutants are powerful genetic tools. In yeast, temperature-sensitive mutations have yielded a wealth of information regarding gene function and have aided immensely in the discovery and elucidation of many molecular pathways (Hartwell, 1967; Bonatti et al., 1972; Pringle, 1975; Novick and Botstein, 1985; Johnston et al., 1991; Balasubramanian et al., 1994; Chang et al., 1996, 1997; Iida and Yahara, 1999). In plants, a number of studies have generated temperature-sensitive alleles to study processes ranging from plant morphology to signal transduction (Lane et al., 2001; Whittington et al., 2001; Wiedemeier et al., 2002; Quint et al., 2005; Bannigan et al., 2006, 2007).
In addition to temperature-dependent function, conditional expression can be generated in a variety of ways. A common strategy in mouse cells is to incorporate lox-p sites flanking the gene of interest (Sauer and Henderson, 1988; Orban et al., 1992; Vidali et al., 2006). Gene function is conditionally lost by the expression of cre recombinase that fuses the lox-p sites, deleting the intervening sequences. This method and others, such as inducible RNA interference (RNAi; Ketelaar et al., 2004), require long incubation times needed for gene expression and protein depletion. Due to the long time course for these studies, loss-of-function effects can be complicated with the development of the organism. In contrast, temperature-sensitive mutants are potentially fast acting, losing their function in some cases within minutes of exposure to the restrictive conditions (Novick and Botstein, 1985; Pruyne et al., 1998).
In most cases, temperature-sensitive mutants are generated randomly and the elucidation of the gene harboring the mutation is uncovered by cloning the mutagenized gene. In plants, this is done by performing a chromosome walk to the mutagenized allele. In yeast, due to the ease of performing complementation, it is also possible to start with a gene of interest, mutagenize that gene, and screen for temperature-sensitive alleles (Shortle et al., 1984; Budd and Campbell, 1987; Mann et al., 1987). In plants, however, this process has not been widely used, presumably due to the time-consuming nature of performing complementation studies in planta.
Here, we show that the moss Physcomitrella patens is an ideal plant suited for screening potential temperature-sensitive alleles of a gene of interest. To screen for a temperature-sensitive mutation, loss of the gene of interest must produce a measurable phenotype that can be rescued by reintroduction of the wild-type allele of the gene. We chose two proteins, profilin and actin-depolymerizing factor (ADF)/cofilin, as test molecules. Profilin and ADF are well-characterized actin-binding proteins that are important for cellular growth in plants (Staiger et al., 1994; Ramachandran et al., 2000; Dong et al., 2001; Vidali et al., 2001, 2007; Chen et al., 2002, 2003; McKenna et al., 2004; Augustine et al., 2008). In the moss P. patens, both profilin and ADF are essential for protonemal filament growth. Loss of profilin or ADF results in severely stunted plants, composed of morphologically abnormal cells (Vidali et al., 2007; Augustine et al., 2008). These phenotypes are fully rescued by expression of wild-type profilin or ADF, respectively.
Moss has emerged as a facile plant system due to its ability to integrate exogenous DNA molecules by homologous recombination at frequencies enabling gene-targeting studies (Cove et al., 2006). In addition, moss is amenable to transient RNAi (Bezanilla et al., 2003, 2005), which enables the study of terminal phenotypes due to loss of essential genes, something that would not be possible if performing only gene knockout experiments. We have previously demonstrated the ability to knock down essential gene families and obtain quantitative rescue of the knockdown phenotypes (Vidali et al., 2007, 2009; Augustine et al., 2008). We have performed these studies using a rapid transient assay, which enables knock down and complementation studies to be performed within 1 week of transformation (Vidali et al., 2007). This is an extremely rapid assay that is unparalleled in other plant systems. Here, we use this complementation assay to screen for temperature-sensitive alleles of both profilin and ADF. Importantly, we show that the residue that confers temperature sensitivity in moss profilin can also render both Arabidopsis (Arabidopsis thaliana) and lily (Lilium longiflorum) profilins temperature sensitive, demonstrating a wider applicability to this rapid in planta complementation system.
RESULTS AND DISCUSSION
Temperature-Sensitive Mutations from Yeast Do Not Transfer to Plant Proteins
P. patens has three functionally redundant profilin genes (PRFa, PRFb, and PRFc) and a single essential ADF/cofilin gene. For profilin, we chose to use PRFa to screen for temperature-sensitive mutants, since it is the most abundant profilin gene in protonemal tissue (Vidali et al., 2007). In fission (Schizosaccharomyces pombe) and budding yeast (Saccharomyces cerevisiae), temperature-sensitive alleles of both profilin and cofilin have been identified (Balasubramanian et al., 1994; Lappalainen et al., 1997; Iida and Yahara, 1999). As an initial approach, we tested whether the moss proteins were rendered temperature sensitive if the analogous yeast mutations were introduced into the moss proteins. For profilin, we generated PRFa-E47K, which is analogous to the E42K mutant profilin cdc3-124 from fission yeast, which encodes a protein known to be highly unstable (Balasubramanian et al., 1994; Lu and Pollard, 2001). Two mutants from budding yeast cofilin were introduced into moss ADF. ADF-D12AD13A and ADF-E139AR140AK142A are analogous to cof1-5 and cof1-22, respectively (Lappalainen et al., 1997).
For our complementation studies, we cotransform moss protoplasts with two plasmids: an RNAi construct that targets the untranslated regions of the genes of interest, and an expression construct of the rescuing gene lacking untranslated regions (Vidali et al., 2007). To ensure that plants are silencing the genes of interest, we use an RNAi system that contains, in tandem, an internal reporter of gene silencing with sequences of the genes of interest (Bezanilla et al., 2005). Briefly, the moss line used for transformation expresses a nuclear-localized GFP-GUS. The RNAi construct simultaneously silences the genes of interest as well as the nuclear localization signal-GFP-GUS reporter. This is due to the presence of sequences of the target gene and GUS in the RNAi construct. One week after transformation, only plants lacking nuclear GFP are analyzed. These plants are derived from single protoplasts and are actively silencing (Bezanilla et al., 2005; Vidali et al., 2007).
Temperature sensitivity of essential genes is evaluated as follows. Transformed protoplasts are regenerated at the permissive temperature in protoplast regeneration medium for 4 d. Protoplast regeneration is inhibited at 32°C. Therefore, allowing it to occur at the permissive temperature enables for efficient regeneration and transformation. During this time, the protoplasts rebuild their cell walls and have very limited protonemal growth. After 4 d, the regenerated plants are transferred to regular growth medium containing antibiotic to select for the plasmids. Duplicate plates for each transformation are incubated at permissive and restrictive temperatures for an additional 3 d to allow for selection and optimal growth that occurs after removal from the protoplast regeneration medium. Seven days after the transformation, plants are analyzed for growth and morphology.
PRFa-E47K did not rescue the profilin RNAi phenotype (Table I). In contrast, the mutants of ADF derived from yeast cofilin temperature-sensitive alleles were able to rescue. However, they were not temperature sensitive (Table I). These results suggest that mutations that render yeast proteins temperature sensitive are not necessarily transferable to plant proteins. This could be due to the fact that yeast grows optimally at different temperatures. Alternatively, the mutation could affect the interaction with molecular partners present in yeast but absent in the plant cell. Thus, the lesions that are not tolerated in one organism may be tolerated in another.
Table I.
PRFa and ADF mutations tested for rescue of the loss-of-function phenotype and for temperature sensitivity in moss
n.d., Not determined.
| Mutation | Rescue | Temperature Sensitive |
|---|---|---|
| PRFa-E47Ka | − | n.d. |
| PRFa-V103A | ++ | Yes |
| PRFa-V103S | − | n.d. |
| PRFa-V103W | − | n.d. |
| PRFa-V103K | − | n.d. |
| ADF-D12AD13Aa | ++ | No |
| ADF-E139AR140A | ++ | No |
| ADF-E139AR140AK142Aa | ++ | No |
| ADF-F29K | − | n.d. |
| ADF-I30A | − | n.d. |
| ADF-I30K | − | n.d. |
| ADF-I30S | −/+ | n.d. |
| ADF-V31A | − | n.d. |
| ADF-V31W | − | n.d. |
| ADF-V69A | ++ | Yes |
| ADF-I91A | − | n.d. |
Analogous mutations in yeast are temperature sensitive.
PRFa-V103A Is Temperature Sensitive
Many temperature-sensitive mutants in yeast have been identified by random mutagenesis (Bonatti et al., 1972; Mann et al., 1987). This approach is likely to yield alleles that are temperature sensitive specifically in yeast. To find a more generalized strategy, we reasoned that mutations in the core of the molecule may affect a protein's stability and activity in a temperature-dependent manner. If the protein is less stable at elevated temperatures, it may result in either unfolding or denaturation at the restrictive temperature. Therefore, one approach is to identify and mutagenize buried residues in the protein of interest. Using the same rationale, Varadarajan et al. (1996) developed an algorithm that identifies buried residues in highly hydrophobic environments without necessarily knowing the three-dimensional structure of the protein. Using this algorithm, studies have identified residues within various proteins that, when mutated, render these proteins temperature sensitive (Varadarajan et al., 1996; Chakshusmathi et al., 2004). Importantly, this approach identified temperature-sensitive lesions in the bacterial ccdB and the yeast Gal4 genes, for which no temperature-sensitive alleles had previously been identified (Chakshusmathi et al., 2004).
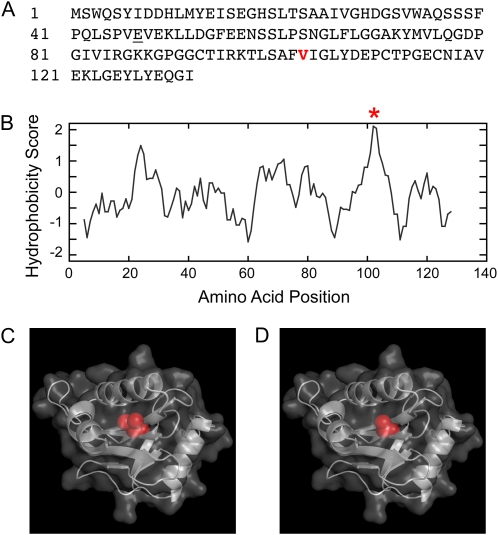
Using the algorithm of Varadarajan et al. (1996), we identified Val-103 (V103) as the most buried residue in PRFa. Hydrophobicity determinations, such as those developed by Kyte and Doolittle (1982), also identify this region in the molecule as the most hydrophobic (Fig. 1B). Additionally, we made a homology model of PRFa using the Arabidopsis PRF1 crystal structure (Thorn et al., 1997). Inspection of the three-dimensional model reveals that V103 is in fact buried in the core of the profilin molecule (Fig. 1C).
Figure 1.
V103 is a buried residue in moss PRFa. A, Primary amino acid sequence of PRFa. The underlined residue is the analogous amino acid that is altered in the fission yeast temperature-sensitive profilin allele. The residue in red is V103, identified by the algorithm of Varadarajan et al. (1996). B, V103 is indicated with a red asterisk in the Kyte-Doolittle hydrophobicity plot. C, PRFa homology model with V103 highlighted. D, PRFa-V103A homology model with Ala highlighted.
Using site-directed mutagenesis, we replaced V103 with four different residues, as suggested by Chakshusmathi et al. (2004). These residues are Ala, Ser, Trp, and Lys. To determine if the profilin mutants were functional, we tested whether the mutants could rescue the profilin RNAi phenotype.
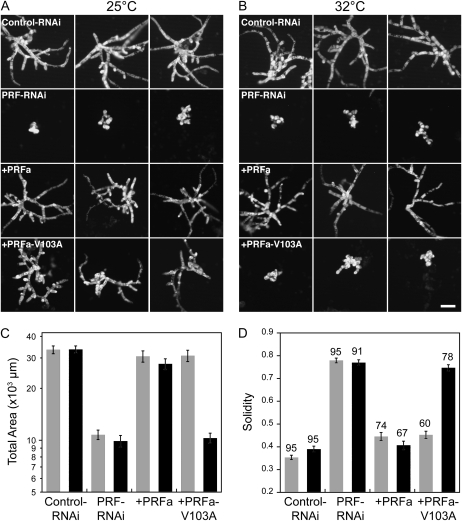
Transformation with PRF-RNAi (untranslated region-RNAi construct from Vidali et al., 2007) results in an inhibition of growth (Fig. 2). Cotransformation of PRF-RNAi with a PRFa coding sequence construct results in plants that form polarized extensions with abundant branches indistinguishable from control plants (Fig. 2). We tested the four profilin mutants to determine which could rescue the PRF-RNAi phenotype at 25°C, the standard temperature for moss growth. PRFa-V103S, PRFa-V103W, and PRFa-V103K were unable to restore wild-type growth in the PRF-RNAi plants (Table I). These plants were very small and contained spherically shaped cells that phenocopied PRF-RNAi plants. In contrast, PRFa-V103A restored wild-type growth to the profilin RNAi plants at 25°C (Fig. 2).
Figure 2.
PRFa-V103A is temperature sensitive. A and B, Three representative micrographs of chlorophyll autofluorescence from 1-week-old moss plants grown at the indicated temperatures and transformed with the indicated constructs. Panels with a plus sign show plants transformed with the indicated construct in addition to the PRF-RNAi construct. Bar = 100 μm. C, Average area of chlorophyll autofluorescence from silenced plants. Area is plotted in a logarithmic scale due to the log normal distribution of the data. D, Average solidity values. Numbers above the bars indicate the total number of plants analyzed for each condition. Gray and black bars in C and D represent data acquired at 25°C and 32°C, respectively. Error bars represent se.
These results indicate that V103 is a critical residue for profilin stability. Only the most modest modification (V103A) functions properly at 25°C. Because our interest is to identify at least one useful mutation, we focused on V103A to investigate its temperature sensitivity.
To test for temperature sensitivity, we analyzed complementation of PRF-RNAi at permissive (25°C) and nonpermissive (32°C) temperatures. The complementation experiments are performed by transforming protoplasts, allowing the protoplasts to recover for 4 d at 25°C and then shifting to the nonpermissive temperature, 32°C, for 3 d. The control RNAi, the PRF-RNAi, and PRFa-complemented plants behave similarly at the permissive and restrictive temperatures. In contrast, cotransformation with PRFa-V103A results in small plants composed of spherical cells at 32°C (Fig. 2). These plants are very similar in area and morphology to the PRF-RNAi plants.
To determine the extent of temperature sensitivity of this temperature-sensitive allele, we used a quantitative analysis of plant area and morphology. Images of individual plants were analyzed by automated morphometry using digital images and custom-made ImageJ macros (Vidali et al., 2007; see “Materials and Methods”). Plant area is determined from the chlorophyll autofluorescence of each plant. The morphology of the plant is represented by solidity, which is the ratio of plant area to the convex hull area. A solidity value of 1 corresponds to a solid object, while decreasing values correspond to objects containing more branched structures. Quantitative analysis of plant area and morphology shows that PRFa-V103A significantly rescues plant growth and morphology at the permissive temperature but not at the restrictive temperature (Fig. 2; Table II).
Table II.
Statistical analyses of moss profilin studies
A plus sign indicates that the construct was expressed together with the RNAi plasmid. Adjusted P values are shown; values in boldface indicate that the difference is statistically significant. The α level was set at 0.05.
| Construct | Area | Solidity |
|---|---|---|
| Control-RNAi 25°C versus control-RNAi 32°C | 1 | 0.4643 |
| PRF-RNAi 25°C versus PRF-RNAi 32°C | 0.8549 | 0.9994 |
| +PRFa 25°C versus +PRFa 32°C | 1 | 0.5754 |
| +PRFa-V103A 25°C versus +PRFa-V103A 32°C | <0.0001 | <0.0001 |
| +PRFa 25°C versus +PRFa-V103A 25°C | 0.999 | 1 |
| +PRFa 32°C versus +PRFa-V103A 32°C | <0.0001 | <0.0001 |
These results clearly demonstrate the ability to generate a temperature-sensitive allele of moss profilin. It is important to note that, in this case, the residue selection was based solely on sequence information (Varadarajan et al., 1996). This makes the presented approach useful for molecules without available three-dimensional structural information. Additionally, since the temperature range used in this study does not hinder wild-type moss growth, it enables future live cell temperature-shift experiments for detailed analysis of moss profilin function.
ADF-V69A Is Temperature Sensitive
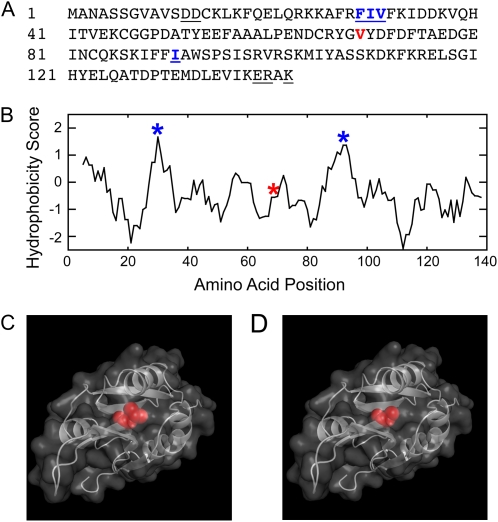
To further investigate the relevance of this method to other molecules, we applied it to another actin-binding protein, ADF. The algorithm of Varadarajan et al. (1996) identified Phe-29, Ile-30, and Val-31 as highly buried residues in ADF (Fig. 3). Using site-directed mutagenesis, we generated ADF-F29K, -I30A, -I30K, -I30S, -V31A, and -V31W. None rescued the ADF RNAi phenotype (Table I). These hydrophobic residues are also recognized by traditional Kyte and Doolittle (1982) hydrophobicity plots (Fig. 3). Another highly hydrophobic and possibly buried region can be identified in this plot but is not detected by the algorithm of Varadarajan et al. (1996). We selected Ile-91 in this region for mutagenesis; a similar mutation in this residue (I91A) also generated a nonfunctional protein (Fig. 3; Table I). The lack of complementation supports the idea that both the algorithm and the hydrophobicity plots can identify residues in critical locations, likely the protein core. We confirmed that the residues identified were indeed in ADF's core by analyzing a three-dimensional model (see below).
Figure 3.
Val-69 is a core residue in moss ADF. A, Primary amino acid sequence of ADF. The underlined residues are the analogous amino acids that are altered in two budding yeast temperature-sensitive cofilin alleles, cof1-5 and cof1-22. The residues underlined and in blue were identified as residing in hydrophobic regions of the protein. The residue in red is Val-69. B, Val-69 is indicated with a red asterisk in the Kyte-Doolittle hydrophobicity plot. The hydrophobic regions are indicated with blue asterisks. C, ADF homology model with Val-69 highlighted. D, ADF-V69A homology model with Ala highlighted.
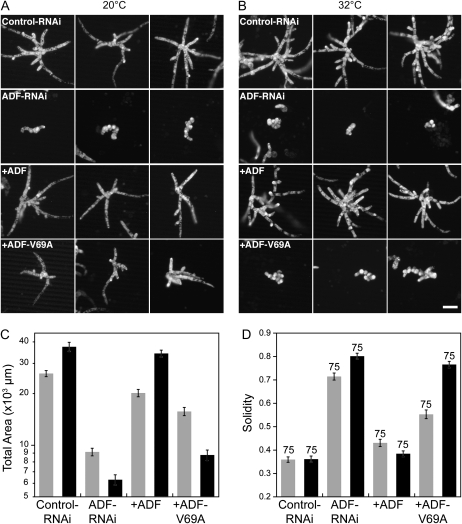
Unfortunately, these results suggest that ADF is more sensitive to changes in its hydrophobic core region than profilin. Therefore, we generated a three-dimensional homology model of moss ADF using the Arabidopsis ADF1 crystal structure (Bowman et al., 2000; Fig. 3C). Inspection of this model with Swiss-PdbViewer (Guex and Peitsch, 1997; Schwede et al., 2003) revealed that Val-69 might also make a good target for mutagenesis, since it is located in a buried region of the molecule. This site had not been identified previously by the algorithm of Varadarajan et al. (1996) or by hydrophobicity plots. We generated and tested ADF-V69A for rescue of the ADF RNAi phenotype. At 25°C, ADF-V69A shows weak rescue of the ADF RNAi phenotype (data not shown). Because of the weak rescue at 25°C, we reasoned that the rescue might improve if we lowered the permissive temperature. Thus, we lowered the permissive temperature to 20°C. Using these conditions, we show that ADF-V69A partially rescues the RNAi phenotype at 20°C and phenocopies the ADF RNAi phenotype at 32°C (Fig. 4). ADF-V69A plants are slightly smaller and less polarized than control and ADF-rescued plants (Fig. 4; Table III). Importantly, they are well polarized when compared with the ADF RNAi plants.
Figure 4.
ADF-V69A is temperature sensitive. A and B, Three representative micrographs of chlorophyll autofluorescence from 1-week-old moss plants grown at the indicated temperatures and transformed with the indicated constructs. Panels with a plus sign show plants transformed with the indicated construct in addition to the ADF-RNAi construct. Bar = 100 μm. C, Average area of chlorophyll autofluorescence from silenced plants. Area is plotted in a logarithmic scale due to the log normal distribution of the data. D, Average solidity values. Numbers above the bars indicate the total number of plants analyzed for each condition. Gray and black bars in C and D represent data acquired at 20°C and 32°C, respectively. Error bars represent se.
Table III.
Statistical analyses of moss ADF studies
A plus sign indicates that the construct was expressed together with the RNAi plasmid. Adjusted P values are shown; values in boldface indicate that the difference is statistically significant. The α level was set at 0.05.
| Construct | Area | Solidity |
|---|---|---|
| Control-RNAi 20°C versus control-RNAi 32°C | 0.045 | 1 |
| ADF-RNAi 20°C versus ADF-RNAi 32°C | <0.0001 | 0.0005 |
| +ADF 20°C versus +ADF 32°C | <0.0001 | 0.2971 |
| +ADF-V69A 20°C versus +ADF-V69A 32°C | <0.0001 | <0.0001 |
| +ADF 20°C versus +ADF-V69A 20°C | 0.1464 | <0.0001 |
| +ADF 32°C versus +ADF-V69A 32°C | <0.0001 | <0.0001 |
We believe that due to its small size and compact structure, ADF is not very tolerant of changes in its core. It is also important to note that using molecular modeling we were able to identify additional buried residues that were not detected by the algorithm of Varadarajan et al. (1996) or by traditional hydrophobicity determinations. We recommend using this approach in combination with the algorithm of Varadarajan et al. (1996) to increase the number of candidate sites for mutagenesis. The original recommendations indicate that four residues should be selected from each protein (Chakshusmathi et al., 2004). Because of the small nature of these proteins, the algorithm of Varadarajan et al. (1996) only produced one and two major sites for profilin and ADF, respectively. Larger proteins should easily yield additional sites for testing. This simple approach, combined with more sophisticated analyses of protein stability and thermodynamics, should further help narrow the required number of permutations needed to identify a temperature-sensitive allele.
Analogous Mutations in Arabidopsis and Lily Profilin Are Temperature Sensitive
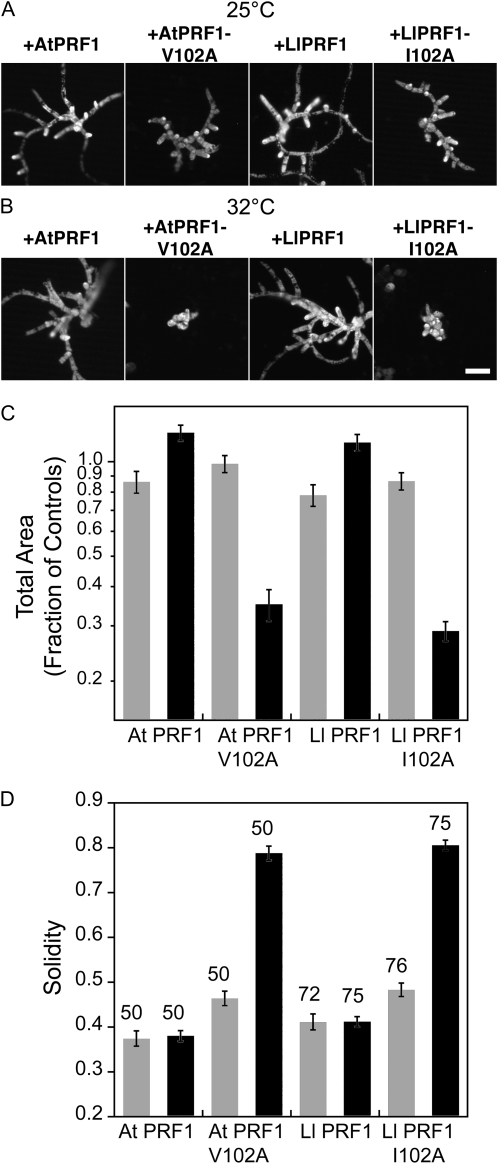
Our data demonstrate the ability to efficiently screen for temperature-sensitive mutations in the moss profilin and ADF proteins based on alterations to their core amino acids. To generalize this approach to other plant proteins, we tested whether other plant profilins could be rendered temperature sensitive by introducing mutations analogous to V103A. Our previous study showed that lily PRF1 (LlPRF1) rescues the profilin RNAi phenotype in moss (Vidali et al., 2007). Here, we show that Arabidopsis PRF1 (AtPRF1) also rescues loss of profilin (Fig. 5).
Figure 5.
Lily and Arabidopsis profilins are rendered temperature sensitive. A and B, Representative micrographs of chlorophyll autofluorescence from 1-week-old moss plants grown at the indicated temperatures and transformed with the indicated constructs. Panels with a plus sign show plants transformed with the indicated construct in addition to the PRF-RNAi construct. Bar = 100 μm. C, Average area of chlorophyll autofluorescence from silenced plants expressed as a fraction of the area of control-RNAi plants. Area is plotted in a logarithmic scale due to the log normal distribution of the data. D, Average solidity values. Numbers above the bars indicate the total number of plants analyzed for each condition. Gray and black bars in C and D represent data acquired at 25°C and 32°C, respectively. Error bars represent se.
We introduced analogous mutations to PRFa-V103A into both the lily and Arabidopsis profilins. In lily, we replaced Ile-102 with Ala (I102A), and in Arabidopsis, we replaced Val-102 with Ala (V102A). Both the lily and Arabidopsis mutants rescued the PRF-RNAi phenotype effectively at the permissive temperature (25°C), generating plants with elongated branched structures (Fig. 5; Table IV). Interestingly, both mutants appeared to have a slight defect in polarization at the permissive temperature, exemplified by an increase in solidity values as compared with rescue with the wild-type proteins (Fig. 5; Table IV). Partial rescue at the permissive temperature is common for temperature-sensitive alleles in many systems. In fact, the ADF-V69A mutant also displays a partial rescue at the permissive temperature.
Table IV.
Statistical analyses of lily and Arabidopsis profilin studies
A plus sign indicates that the construct was expressed together with the RNAi plasmid, in this case PRF-RNAi. Adjusted P values are shown; values in boldface indicate that the difference is statistically significant. The α level was set at 0.05.
| Construct | Area | Solidity |
|---|---|---|
| +AtPRF1 25°C versus +AtPRF1 32°C | 0.0076 | 1 |
| +AtPRF1-V102A 25°C versus +AtPRF1-V102A 32°C | <0.0001 | <0.0001 |
| +LlPRF1 25°C versus +LlPRF1 32°C | <0.0001 | 1 |
| +LlPRF1-I102A 25°C versus +LlPRF1-I102A 32°C | <0.0001 | <0.0001 |
| +AtPRF1 25°C versus +AtPRF1-V102A 25°C | 0.7319 | 0.0034 |
| +AtPRF1 32°C versus +AtPRF1-V102A 32°C | <0.0001 | <0.0001 |
| +LlPRF1 25°C versus +LlPRF-I102A 25°C | 0.4396 | 0.005 |
| +LlPRF1 32°C versus +LlPRF-I102A 32°C | <0.0001 | <0.0001 |
Nevertheless, both the lily and Arabidopsis mutant profilins show a very significant temperature-sensitive phenotype (Fig. 5; Table IV). Neither mutant was able to rescue at the nonpermissive temperature (32°C), resulting in dwarfed plants that phenocopy PRF-RNAi. These results are significant because they demonstrate an effective method to screen for temperature-sensitive alleles in proteins from other plant species.
CONCLUSION
Here, we present a simple and efficient way to identify temperature-sensitive alleles of plant genes. Because of the speed and ease of complementation of moss plants, this identification can be achieved in 1 or 2 months. By simultaneously silencing multiple genes, this approach can overcome the difficulty of working with gene families, a common problem in plants. Most importantly, if further analysis is going to be performed in Physcomitrella, the temperature-sensitive allele can be incorporated into the endogenous locus via homologous recombination. In the case of gene families, the additional family members would need to be disrupted by homologous recombination. This would generate a stable temperature-sensitive line for detailed characterization of phenotype and gene function. For example, in the case of profilin and ADF, their roles in controlling actin dynamics could be directly evaluated. Having temperature-sensitive alleles in hand enables additional studies, such as the identification of genetic suppressors and enhancers.
When working with other plants, the identified alleles can be incorporated into knockout or knockdown lines. For example, in Arabidopsis, we envision two possible strategies. In one case, a line expressing the temperature-sensitive allele is generated and subsequently crossed with a heterozygous knockout line for the essential gene. Subsequent self-crossing would result in a line producing a homozygous knockout plant with a copy of the temperature-sensitive allele. Alternatively, in the case of large gene families and similar to our strategy in moss, an inducible RNAi construct (Ketelaar et al., 2004) targeting the untranslated regions of the mRNAs can be expressed in the presence of the temperature-sensitive allele, rendering the plant temperature sensitive after exposure to the inducer.
MATERIALS AND METHODS
Buried Residue Prediction and Identification
Identification of buried residues was done with the program predbur_dos.exe (Chakshusmathi et al., 2004) running on a personal computer platform. For moss (Physcomitrella patens) profilin (PRFa), V103 was identified as the residue with a 90% burial based on average hydrophobicity values. The following window was found: SAF[V]IGL. For moss ADF, this program identified as 90% buried: Val-29, (AFR[F]IVF), Ile-30 (FRF[I]VFK), and Val-31 (RFI[V]FKI).
Profilin and ADF Models
Moss profilin and ADF were modeled based on the structure of Arabidopsis (Arabidopsis thaliana) profilin 1 (AtPRF1; 1a0k) and ADF1 (AtADF1; 1f7s) using Swiss model (http://swissmodel.expasy.org; Schwede et al., 2003).
Tissue Culture and Protoplast Transformation
All tissue culture and transformations were performed as described previously (Vidali et al., 2007), with the following modification for temperature sensitivity testing. Plants were plated on regeneration medium for 4 d at 25°C or 20°C. At day 4, they were transferred to permissive temperature or 32°C on medium containing hygromycin for selection of transformants. Plants were photographed 3 d after transfer to hygromycin.
DNA Constructs
The PRF-RNAi construct and the PRFa expression constructs were generated as described previously (Vidali et al., 2007). The ADF-RNAi construct and the ADF expression construct were generated as described previously (Augustine et al., 2008). The mutations were introduced into the PRFa, ADF, AtPRF1, and lily (Lilium longiflorum) LlPRF1 entry constructs before transferring them to the expression vector, pTH-Ubi-Gateway (Vidali et al., 2007), using an LR clonase reaction. When possible, the mutation or a closely placed silent mutation were designed to generate a new restriction site for easy identification of mutant clones. All mutations were confirmed by DNA sequencing.
Mutations were introduced by amplifying the plasmid containing the entry clone with a mutagenizing primer using PCR-based site-directed mutagenesis (Weiner et al., 1994). Primers used for amplification are presented in Supplemental Table S1.
Morphometric Analysis
Plants were tested for complementation in the 1-week transient assay. They were photographed as described previously (Vidali et al., 2007). The algorithm used for analysis was modified as follows. Plants undergoing active silencing were manually separated from the background with a selection tool. After digitally separating the red channel, corresponding to the chlorophyll autofluorescence, the resulting eight-bit image was thresholded by the maximum entropy method (ImageJ) and binarized. The total area of the plants was estimated as the number of pixels with an intensity value of 1. The other parameter that was used was solidity. This parameter is defined as the ratio of the convex hull area over the area. For this, the area of the largest object in the selected image was used to calculate both the area and the convex hull area. All image analysis was done using macros written for ImageJ (http://rsb.info.nih.gove.ij/). The code for all macros is available upon request.
Statistical Analyses
Statistical analyses were performed as described previously (Vidali et al., 2007). Briefly, because the distribution of areas was found to be log normal, all comparisons of area were done using the natural logarithm of the values. Solidity values are normally distributed and did not need transformation for comparison. ANOVA was done with the statistical function of Kaleidagraph, using the Tukey post hoc tests for multiple comparisons. The level of α for statistical significance was set at 0.05.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Arabidopsis PRF1, UniProtKB/Swiss-Prot Q42449, PDB 1a0k; Arabidopsis ADF1, UniProtKB/Swiss-Prot Q39250, PDB 1f7s; Physcomitrella PRFa, UniProtKB/TrEMBL A9RDI7; lily PRF1, UniProtKB/Swiss-Prot Q9SNW7; Physcomitrella ADF, UniProtKB/TrEMBL A9TF31.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Peter Hepler and Ming Li for commenting on the manuscript.
This work was supported by the National Science Foundation (grant nos. MCB–0516702, MCB–0640530, and MCB–0747231), by the David and Lucille Packard Foundation (support to M.B.), by the National Science Foundation Integrative Graduate Education and Research Traineeship (grant no. DGE–065412) to the University of Massachusetts Institute for Cellular Engineering (fellowship support to R.C.A.), and by the Howard Hughes Medical Institutes Undergraduate Science Program (support to S.N.F. and K.A.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Magdalena Bezanilla (bezanilla@bio.umass.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Augustine RC, Vidali L, Kleinman KP, Bezanilla M (2008) Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J 54: 863–875 [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL (1994) The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol 125: 1289–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A, Scheible WR, Lukowitz W, Fagerstrom C, Wadsworth P, Somerville C, Baskin TI (2007) A conserved role for kinesin-5 in plant mitosis. J Cell Sci 120: 2819–2827 [DOI] [PubMed] [Google Scholar]
- Bannigan A, Wiedemeier AM, Williamson RE, Overall RL, Baskin TI (2006) Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol 47: 949–958 [DOI] [PubMed] [Google Scholar]
- Bezanilla M, Pan A, Quatrano RS (2003) RNA interference in the moss Physcomitrella patens. Plant Physiol 133: 470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Perroud PF, Pan A, Klueh P, Quatrano RS (2005) An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol (Stuttg) 7: 251–257 [DOI] [PubMed] [Google Scholar]
- Bonatti S, Simili M, Abbondandolo A (1972) Isolation of temperature-sensitive mutants of Schizosaccharomyces pombe. J Bacteriol 109: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, Nodelman IM, Hong Y, Chua NH, Lindberg U, Schutt CE (2000) A comparative structural analysis of the ADF/cofilin family. Proteins 41: 374–384 [DOI] [PubMed] [Google Scholar]
- Budd M, Campbell JL (1987) Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci USA 84: 2838–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakshusmathi G, Mondal K, Lakshmi GS, Singh G, Roy A, Ch RB, Madhusudhanan S, Varadarajan R (2004) Design of temperature-sensitive mutants solely from amino acid sequence. Proc Natl Acad Sci USA 101: 7925–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P (1997) cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol 137: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P (1996) Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci 109: 131–142 [DOI] [PubMed] [Google Scholar]
- Chen CY, Cheung AY, Wu HM (2003) Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wong EI, Vidali L, Estavillo A, Hepler PK, Wu HM, Cheung AY (2002) The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D, Bezanilla M, Harries P, Quatrano R (2006) Mosses as model systems for the study of metabolism and development. Annu Rev Plant Biol 57: 497–520 [DOI] [PubMed] [Google Scholar]
- Dong CH, Xia GX, Hong Y, Ramachandran S, Kost B, Chua NH (2001) ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hartwell LH (1967) Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol 93: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Yahara I (1999) Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells 4: 21–32 [DOI] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA (1991) The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol 113: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Allwood EG, Anthony R, Voigt B, Menzel D, Hussey PJ (2004) The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr Biol 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Hofte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG (1997) Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J 16: 5520–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Pollard TD (2001) Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell 12: 1161–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C, Buhler JM, Treich I, Sentenac A (1987) RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell 48: 627–637 [DOI] [PubMed] [Google Scholar]
- McKenna ST, Vidali L, Hepler PK (2004) Profilin inhibits pollen tube growth through actin-binding, but not poly-L-proline-binding. Planta 218: 906–915 [DOI] [PubMed] [Google Scholar]
- Novick P, Botstein D (1985) Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell 40: 405–416 [DOI] [PubMed] [Google Scholar]
- Orban PC, Chui D, Marth JD (1992) Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci USA 89: 6861–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR (1975) Induction, selection, and experimental uses of temperature-sensitive and other conditional mutants of yeast. Methods Cell Biol 12: 233–272 [DOI] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A (1998) Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol 143: 1931–1945 [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM (2005) Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J 43: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85: 5166–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D, Novick P, Botstein D (1984) Construction and genetic characterization of temperature-sensitive mutant alleles of the yeast actin gene. Proc Natl Acad Sci USA 81: 4889–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Yuan M, Valenta R, Shaw PJ, Warn RM, Lloyd CW (1994) Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr Biol 4: 215–219 [DOI] [PubMed] [Google Scholar]
- Thorn KS, Christensen HE, Shigeta R, Huddler D, Shalaby L, Lindberg U, Chua NH, Schutt CE (1997) The crystal structure of a major allergen from plants. Structure 5: 19–32 [DOI] [PubMed] [Google Scholar]
- Varadarajan R, Nagarajaram HA, Ramakrishnan C (1996) A procedure for the prediction of temperature-sensitive mutants of a globular protein based solely on the amino acid sequence. Proc Natl Acad Sci USA 93: 13908–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Augustine RC, Kleinman KP, Bezanilla M (2007) Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Chen F, Cicchetti G, Ohta Y, Kwiatkowski DJ (2006) Rac1-null mouse embryonic fibroblasts are motile and respond to platelet-derived growth factor. Mol Biol Cell 17: 2377–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, McKenna ST, Hepler PK (2001) Actin polymerization is essential for pollen tube growth. Mol Biol Cell 12: 2534–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen P, Guérin C, Franco P, Lee M, Burkart GM, Augustine RC, Blanchoin L, Bezanilla M (2009) Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC (1994) Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151: 119–123 [DOI] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Wiedemeier AM, Judy-March JE, Hocart CH, Wasteneys GO, Williamson RE, Baskin TI (2002) Mutant alleles of Arabidopsis RADIALLY SWOLLEN 4 and 7 reduce growth anisotropy without altering the transverse orientation of cortical microtubules or cellulose microfibrils. Development 129: 4821–4830 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.