Abstract
Exposure of cells to DNA-damaging agents triggers a complex biological response involving cell cycle arrest and modulation of gene expression. Genomic sequencing has revealed the presence of archaeal genes homologous to components of the eucaryal nucleotide excision repair (NER) pathway, which is involved in the repair of ultraviolet (UV) light-induced DNA damage. However, the events involved in the cell response to UV irradiation and their regulation have not been studied in Archaea. We show here that UV radiation induces the formation of cyclobutane pyrimidine dimers (CPDs) in the hyperthermophilic archaeon Sulfolobus solfataricus, and that these lesions are efficiently repaired in vivo in the dark, suggesting that a NER pathway is active. DNA damage is a signal for concomitant growth arrest and transcriptional induction of the NER genes XPF, XPG and XPB. The cell response to UV irradiation includes transcriptional regulation of genes encoding two DNA binding proteins involved in chromosome dynamics. Moreover, several of these genes are also strongly induced by the intercalating agent actinomycin D. Thus, response to DNA damage in S.solfataricus has features essentially conserved in all three domains of life.
INTRODUCTION
All cells and organisms respond to DNA damage via a complex network of pathways of DNA repair and damage tolerance mechanisms, cell cycle checkpoints, recombination and programmed cell death (1). Ultraviolet (UV) light is the most common environmental damaging agent and all organisms are equipped with pathways for repair of UV-induced lesions. One of the best understood UV damage repair systems is the nucleotide excision repair (NER) pathway. In both Eucarya and Bacteria, NER is based on a cut-and-paste mechanism requiring protein complexes that recognise the damage, helicases and nucleases that remove the damaged strand, and DNA polymerases and ligases that restore the correct sequence (2). However, NER proteins of Bacteria and Eucarya are not evolutionary related. Whereas in Bacteria, lesion recognition and removal are completed by only three proteins (UvrA, UvrB and UvrC), in Eucarya, the NER system is considerably more complex and requires 15–18 polypeptides.
Despite the relevance of the DNA damage response in cell biology, the mechanisms, regulation and intricate connections of the events involved in this process are well understood only in the case of Escherichia coli. Variation of the pattern of gene expression is part of the cellular reaction to DNA damage-induced stress and operates as a defence mechanism. In E.coli, DNA damage induces transcriptional derepression of approximately 20 genes of the SOS regulon (3). In Eucarya, the regulatory network underlying this response is considerably more complex and not completely elucidated. Proteins sensing the DNA damage initiate signal transduction chains that result in activation of numerous genes and proteins. Expression profiling in yeast and keratinocytes demonstrated the complexity of the transcriptional response to DNA damage, with up- and down-regulation of several dozens of genes involved in any cellular process (4,5).
In mammals, UV irradiation affects the expression of a large number of genes, including transcription factors such as the proto-oncogenes AP-1 and NF-kB and the anti-oncogene p53. These genes control the cell response to UV irradiation, switching from cell cycle arrest to apoptosis. A p53-mediated SOS response has been suggested on the basis of the up-regulation of several genes associated with enhancement of repair capacity, including the NER gene XPB (6,7).
Genome sequencing has revealed that almost all Archaea encode homologues to the eucaryal NER proteins (8), adding to the hypothesis of a common evolutionary ancestry of Eucarya and Archaea, suggested by the conservation of the transcription and replication apparatus (reviewed in 9,10). However, the biochemical and molecular events involved in the cellular response to UV irradiation have not been studied in the third kingdom. Here we investigate the effect of UV exposure in the crenarchaeon Sulfolobus solfataricus. We show that UV-induced DNA damage is repaired in vivo in the absence of light. Exposure to UV induces transcriptional regulation of genes encoding NER factors and DNA binding proteins involved in DNA dynamics. Moreover, some, but not all, such genes are also induced by the intercalating agent actinomycin D.
MATERIALS AND METHODS
In silico analysis
The genome databases of S.solfataricus P2 (11) and other Archaea (http://www-archbac.u-psud.fr/projects/sulfolobus/) were searched for homologues of Saccharomyces cerevisiae and human NER proteins using BLAST and PSI-BLAST. GenBank accession numbers of proteins used as baits were the following: human XPB, P19447; S.cerevisiae Rad25, Q00578; human XPF, Q92889; S.cerevisiae Rad1, NP_015303; human XPG, P28715; S.cerevisiae Rad2, A29839. Homologous sequences were aligned using ClustalW and Multalign (12).
Gene cloning and labelling
All enzymatic DNA reactions were performed according to standard techniques. Gene-specific probes were designed using the sequences of the S.solfataricus P2 genome database. Oligonucleotide sequences are available on request. Appro priate DNA fragments were amplified from S.solfataricus P2 genomic DNA and cloned either in pCRII or pCR2.1 vector (Invitrogen); amplified fragments were always checked by DNA sequencing. DNA probes were labelled with 32P-dCTP using the RediPrime II kit (Amersham); RNA probes were synthesised using 32P-UTP and T7 RNA polymerase (New England Biolabs) as reported (13). Probes spanned the following regions (numbers relative to the predicted first codon): Alba, +1/+273; SsXpg, +1/+600; SsXpf, +19/+595; β-prefoldin, +8/+377; Sav, +573/+999; Smj12, +1/+348; Lrs14, +1/+375; Sul7, +1/+192; 23S rRNA, +1088/+1458.
Cell growth and irradiation
Sulfolobus solfataricus cultures were grown in complete medium at 80°C as described (14) until the OD600 reached 0.3–0.4. Fixed volumes of culture were poured in 110 mm plastic Petri dishes (10 ml/plate) and were irradiated with a germicide UV lamp (254 nm, 30 W) at room temperature for different durations. UV energy (irradiance) was measured using a Quantum-photo-radiometer HD9021 equipped with a LP9021 UVC probe (Delta Ohm), and used to calculate the actual UV dose received by the samples (fluence). Cultures were immediately incubated at 80°C and aliquots were taken after different time spans and analysed. For mock-treated control cultures, exactly the same procedure was followed without irradiation. Escherichia coli BL21 strain was treated in the same way, except for the different growth temperature (37°C). If needed, drugs were added to exponentially growing cells at indicated times before UV irradiation.
Analysis of cyclobutane pyrimidine dimers (CPDs) in genomic DNA and kinetics of repair
Sulfolobus solfataricus cultures were irradiated with 200 J/m2 or mock treated, then immediately incubated at 80°C in the dark. Genomic DNA was extracted after 0–20 h with the Nucleospin Tissue Kit (Macherey-Nagel) following the manufacturer’s protocol. All manipulations were carried out in rooms equipped with a safety red light. Formation and repair of CPDs were followed as reported (15) with modifications. Briefly, samples were digested with T4 endonuclease V (TEV; Epicentre) for 2 h at 37°C in 50 mM Tris–HCl, pH 7.5 and 5 mM EDTA. Twenty units of enzyme were used for 400 ng of DNA. Formamide (80%) was added and samples were denatured for 1 h at 37°C and separated by electrophoresis in 0.8% agarose gel in Tris-acetate buffer containing 1 µg/ml ethidium bromide. Gels were photographed under UV light using the Chemidoc system (Bio-Rad).
RNA preparation and analysis
Aliquots of irradiated and control cultures were taken after different durations of recovery at 80°C; total RNA was extracted using the RNAeasy kit (Qiagen) following the manufacturer’s instructions. Samples were overdigested with DNAse I (Boehringer) and the absence of residual contaminant DNA was checked by PCR. RNA samples were subjected to non-denaturing agarose gel electrophoresis to control the integrity of ribosomal RNA bands and were quantified spectrophotometrically. Northern blots were performed using 10–15 µg of total RNA and hybridised under stringent conditions with strand-specific RNA probes (1–5 × 104 c.p.m./ml) as reported (13). All experiments were repeated independently at least three times. Each filter was stripped and hybridised sequentially with several probes. Radioactivity was determined by autoradiography with a Storm PhosphoImager and quantified with the IQ-Mac software (Molecular Dynamics). Values were normalised to the level of ribosomal RNA in each lane and were expressed as a fraction of the level of the mRNAs in unirradiated samples C1, which was designated 1.
Primer extensions were performed as reported (16) with the following variations. End-labelled primer (0.5 pmol, 1.5 × 106 c.p.m.) was precipitated with 5–10 µg of RNA, resuspended in 3 µl of 2× RT buffer (100 mM Tris–HCl, pH 8.3, 100 mM KCl, 6 mM MgCl2, 10 mM dithiothreitol), denatured for 5 min at 70°C, frozen in dry ice and defrosted on ice. dNTP (final concentration of 1.6 mM each) and 20 units of anti-RNAse (Ambion) were added. Reaction mixtures were incubated for 1 h at 42°C, then 80 units of the M-MuLv reverse transcriptase (Ambion) were added and incubated for 1 h at 42°C. Five microlitres of stop solution were added and samples were denatured and immediately run on 6% sequencing gels. The primers used were the following: SsXpb-I, 5′-GAAGTCATCCTCATCTAACCA; SsXpb-II, 5′-AACTTCAACGTGAGACCTC.
RT–PCRs were performed using the Superscript One-step Kit (Invitrogen) according to the manufacturer’s protocols. The primers used were the following (see Figs 2 and 3): a1, AGAAATTACCTCCAGAAG; a2, GCTTTTATACCTGTTTGG; b1, GCTGATGATAGGGAAAAGG; b2, AAGGAAATCAAATAAAGAGGTAG; c, ACCTAGTGCTTTTTCCA GTTCTG; d1, GACTGAAGGCTACACCG; d2, CCACTTAGATGACTAGTTAC.
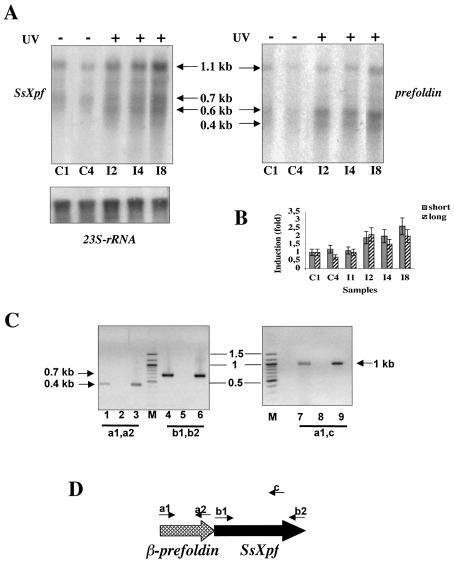
Figure 2.
UV-induced regulation of SsXpf. (A) Total RNA was extracted from controls and UV-irradiated (200 J/m2) cultures and hybridised with the SsXpf riboprobe (left), with the prefoldin riboprobe (right) and with ribosomal RNA for normalisation (bottom). Each lane contained 10 µg of total RNA. C1 and C4, unirradiated cultures, incubated at 80°C for 1 or 4 h, respectively, after mock treatment; I2–I8, irradiated cultures incubated at 80°C for 2, 4 and 8 h, respectively, after irradiation. (B) PhosphoImager quantitation of SsXpf transcripts; values (mean of four independent experiments) were normalised to the level of ribosomal RNA in each lane and were expressed as a fraction of the level of the RNA sample C1, that was designated 1. Short and long refer to the two main transcripts (1.1 and 0.7 kb). (C) RT–PCR analysis of the gene cluster prefoldin-SsXpf. Lanes 1, 2 and 3 contained the primers a1 and a2 shown in (D); lanes 4, 5 and 6 the primers b1 and b2; lanes 7, 8 and 9 the primers a1 and c. Lanes 1, 4 and 7 contained 200 ng of RNA and complete components; in lanes 2, 5 and 8 reverse transcriptase was omitted; lanes 3, 6 and 9 contained 100 ng of genomic DNA instead of RNA. Molecular weight markers (kb) are indicated by bars.
Figure 3.
UV-induced regulation of SsXpg. (A) Northern blot of total RNA prepared as in Figure 2A, hybridised with the SsXpg riboprobe (left), with the Sav riboprobe (right) and with ribosomal RNA for normalisation (bottom). (B) Quantitation of SsXpg transcripts; values (mean of three independent experiments) were normalised and expressed as in Figure 2C. Symbols as in Figure 2. (C) The gene cluster Sav-SsXpg showing the length of the two ORFs. (D) RT–PCR experiment performed using the primers d1 and d2 shown in Figure 3C. Lane 1 contained 200 ng of RNA and complete components; in lane 2 reverse transcriptase was omitted; lane 3 contained 100 ng of genomic DNA instead of RNA. Molecular weight markers (kb) are indicated by bars.
RESULTS
Induction of CPDs by UV irradiation and their repair in vivo
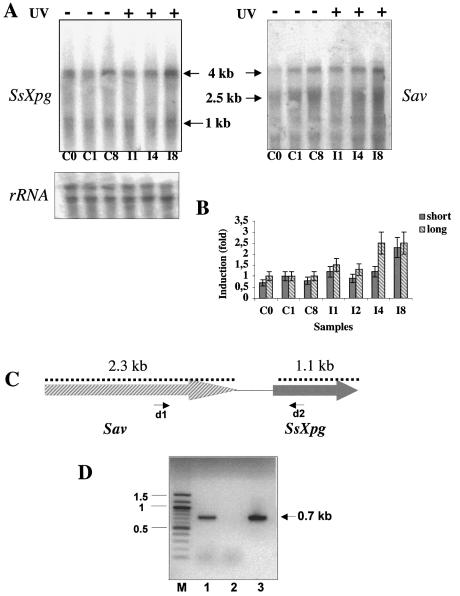
To set up the conditions for UV irradiation of S.solfataricus cultures, we first analysed the effect of different UV doses on cell growth (Fig. 1A). Cultures were grown at 80°C until the exponential phase (0.3–0.4 OD600), then irradiated at room temperature in thin layers and immediately incubated at 80°C. Aliquots were taken after different times and analysed for the presence of CPDs. To rule out from the analysis the effects of the double temperature shock suffered by the cells during these manipulations, control cultures were subjected exactly to the same procedure, but were not irradiated. All UV doses used (200–3300 J/m2) delayed the growth, and the maximal fluence also induced significant reduction of absorbance, indicating cell lysis. A Rec+ strain of E.coli irradiated under the same conditions gave a similar response, with growth delay at all the fluences used (Fig. 1B). In particular, when irradiated at 3300 J/m2 both Sulfolobus and E.coli resumed normal growth only after approximately seven generation times.
Figure 1.
Effects of UV irradiation in S.solfataricus and E.coli. (A) Sulfolobus solfataricus P2 cultures were grown at 80°C, irradiated with UV light at the indicated fluence (J/m2) at room temperature, or mock treated, and immediately incubated at 80°C; absorbance at 600 nm was measured at indicated time points. (B) Escherichia coli BL21 cultures were grown at 37°C, irradiated with UV light at the indicated fluence (J/m2) at room temperature, or mock treated, and immediately incubated at 37°C; absorbance at 600 nm was measured at indicated time points. (C) UV-induced damage and repair in Sulfolobus. Cells were grown, irradiated with UV light (200 J/m2) and immediately incubated at 80°C. The genomic DNA isolated from controls (C0), irradiated cells at each time point (I0, I2, I8 and I20, where numbers indicate hours after irradiation), and naked DNA irradiated at 200 J/m2 (N) were cut with T4 endoV (lanes +), or mock treated (lanes –). DNAs were then denatured and separated by electrophoresis in a 1% agarose gel stained with ethidium bromide. Arrows indicate molecular weight markers (kb).
We chose the lowest dose of irradiation to analyse the molecular events occurring in the cell because it does not induce massive cell death. UV light induces two prevalent types of DNA lesions, CPDs, characterised by covalent linkage between adjacent pyrimidines, and 6–4 photoproducts. These lesions can be repaired by both NER and photolyase; however, whereas photolyase can only act in the presence of light (photoreactivation), NER acts independently of light. Previous work has shown that UV-induced mutations can be reversed by photoreactivation in Sulfolobus acidocaldarius (17). To monitor DNA damage and follow the kinetics of repair of genomic DNA by NER in vivo, S.solfataricus cultures were exposed to UV (200 J/m2) and immediately incubated at 80°C in the dark to avoid photoreactivation by photolyase. Cells were harvested at various times after UV irradiation, genomic DNA was isolated and treated with TEV, an enzyme which specifically cuts CPD sites (15). Denatured DNA was separated by agarose gel electrophoresis (Fig. 1C). Digestion with TEV of purified DNA exposed to UV and processed as described above produced a low molecular weight smear, with the bulk ∼0.5 kb (lane N+). DNA extracted from unirradiated cells was not cleaved (lane C0+). In contrast, DNA extracted immediately after irradiation was completely digested by TEV, producing a smear between 2 and 0.5 kb, with the bulk ∼1 kb (lane I0+). Two hours after irradiation, the TEV digested DNA had a higher average length (lane I2+), suggesting initial repair. After 8 h, the band corresponding to the high molecular weight genomic DNA reappeared (lane I8+), and after 20 h, the pattern remained unchanged (lane I20+). Under the conditions used in our experiments, one CPD every 1000 bases was induced on average, based on the mean size of the DNA smear obtained by TEV digestion. This high number of lesions could be completely removed within 8 h, corresponding to approximately one generation time for this organism. To the best of our knowledge this is the first direct demonstration of CPD induction in Archaea and their removal in vivo using a pathway operating in the dark.
Effects of UV irradiation on transcription of S.solfataricus NER genes
In all organisms studied, gene activation is a critical initial step of the cell response to DNA damage. Having set up the conditions for induction and removal of UV-induced damage, we used the same conditions to analyse the transcriptional response to UV irradiation in S.solfataricus. Total RNA was extracted from irradiated and control cultures after increasing time spans of recovery, and RNA was analysed by northern blotting. Archaeal genes implicated in repair of UV lesions were obvious candidates for UV-dependent regulation. Whereas a small subset of Archaea contains orthologues of the bacterial UvrABC system (clearly a result of lateral gene transfer), orthologues of the eucaryal NER genes XPF/RAD1, XPG/RAD2, XPB/RAD25 and XPD/RAD3 are conserved in all Archaea (crenarchaea and euryarchaea). Therefore, we tested whether S.solfataricus homologues of the first three genes might be transcriptionally regulated by DNA damage.
Eucaryal XPF is a modular protein in which a C-terminal nuclease domain is fused to a non-functional N-terminal helicase domain (8). The Sulfolobus XPF (SsXPF) is a shorter protein, containing only the C-terminal nuclease domain. It has a flap endonuclease activity in vitro in association with the sliding clamp PCNA that is strikingly similar to the eucaryal XPF/Rad1 proteins (18,19). The SsXpf antisense probe detected multiple transcripts, with a sharp and most abundant one of ∼1.1 kb, and shorter smeared transcripts of 0.7–0.6 kb (Fig. 2A). All transcripts were induced by UV irradiation by ∼2-fold starting from 2 h after exposure and were maintained at the same levels for at least 8 h. Since the full-length SsXpf ORF is 702 bp long, we suspected a polycistronic transcription. Mapping of RNA start sites by primer extension showed two main initiation sites located at –3 and more than 350 bases upstream of the first ATG, respectively (data not shown). The analysis of the corresponding genomic sequence showed the presence, directly upstream of SsXpf, of a 0.4 kb ORF (SSO0730) homologous to the β subunit of prefoldin (also called Gim), a molecular chaperone shared by Eucarya and Archaea (20). Hybridisation of the same filter with a probe for the prefoldin gene showed a similar, but not identical, pattern: two major transcripts, of ∼1.1 kb and 600 bases, respectively, and a shorter (0.4 kb) smeared one (Fig. 2A). The 0.6 kb band was most induced by UV irradiation. A transcript spanning the two genes could be amplified by RT–PCRs (Fig. 2C). The different relative intensity of the bands hybridising with the SsXpf and prefoldin probes suggests that the two genes are present in both monocistronic and bicistronic transcripts of different relative abundance (Fig. 2D).
The eucaryal XPG protein belongs to the Fen1/Rad2 family of nucleases, including the 3′ nuclease of NER (Rad2/XPG) as well as the flap endonuclease (Fen-1/Rad27) responsible for the excision of the Okazaki fragments (8). In contrast, Archaea contain a single protein of this family, most similar to Fen-1, that shows nuclease activity in vitro (21). Recently, the Sulfolobus protein has been suggested to participate in the lagging-strand replication in association with PCNA (22); however, its involvement in NER has not been tested. It is probable that the archaeal nuclease functions in both replication and repair, whereas in Eucarya each paralogue has acquired more specialised functions.
Figure 3A shows the analysis of the steady-state levels of SsXpg RNA. The SsXpg antisense probe detected two main transcripts, of ∼4 and ∼1 kb, respectively. The level of both transcripts was unchanged until 8 h after UV irradiation; at this point they showed a 2-fold increase (Fig. 3B). The size of the smaller transcript fits with the length of the ORF, which is ∼1.1 kb (S. D. Bell, personal communication) suggesting that the 4 kb transcript is again the result of a polycistronic transcription. Analysis of the genomic environment showed an ORF coding for a conserved putative ATPase of the AAA family (SSO0176), of ∼2.3 kb, mapping upstream of SsXpg; the two ORFs are separated by a 240 bp intergenic region (Fig. 3C). This protein is homologous to the CDC48/VCP/p97 family, highly conserved in Eucarya and Archaea, involved in a variety of biological functions, ranging from membrane fusion to spindle pole formation, protein trafficking through the ER and protein degradation and regulation of the cell cycle; in all cases they have been suggested to act as molecular chaperones (23). The homologue from S.acidocaldarius (SAV) has been characterised biochemically (24), however, its function in vivo is unknown. Hybridisation of the same filter with a probe from the Sav gene indeed showed two transcripts, of ∼4 and ∼2.5 kb, respectively (Fig. 3A). The filter also showed weaker bands of lower molecular weight, that are likely due to cross-hybridisation of the Sav probe with other transcripts, since the Sulfolobus genome contains several genes of the same ATPase family. A transcript spanning SsXpg and Sav could be amplified by RT–PCRs (Fig. 3D).
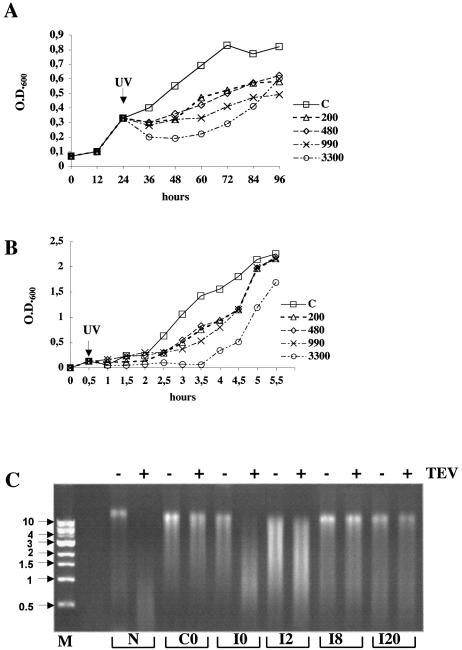
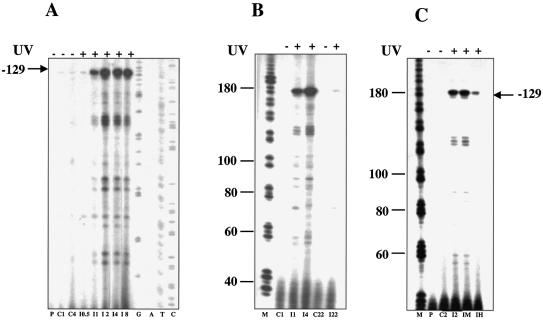
The eucaryal helicase XPB, in complex with the other NER helicase XPD and other proteins, forms the factor TFIIH, which is essential for both transcription and repair. Sulfolobus solfataricus encodes two paralogues of the helicase XPB/Rad25 (SSO0959, hereby designed SsXpb-I, and SSO0473 named SsXpb-II). Neither of these has been characterised biochemically, however, they contain all the domains and signatures typical of this family of proteins (Fig. 4A), and share high sequence similarity with each other, with 58% amino acid identity and long stretches of nucleotide identity. Northern blots hybridised with RNA probes complementary to either genes had a smeared appearance even at high stringency (data not shown). To discriminate between the two genes we performed primer extension using primers annealing to short regions in which the sequences diverge. SsXpb-II showed a single start site mapping at –4 from the first ATG of the ORF, and a site internal to the ORF (+10), likely due to a spurious stop of reverse transcriptase. The transcript level did not show significant variations upon UV exposure (Fig. 4B). The SsXpb-I transcript was barely detectable in the controls and was induced by more than 10-fold after UV irradiation (Fig. 5). Transcript levels reached a maximum at 2 h, were maintained at a high level for at least 8 h, and returned to control levels after 22 h. Surprisingly, the main start site was located 129 bases upstream of the first ATG of the ORF. This situation is highly unusual in archaeal genes that generally show very short, or even absent, 5′ leaders; however, an even longer leader has been found in the RadA gene of Methanococcus voltae which is also UV induced (25). Looking at the P2 genome database, no other gene is annotated within 4 kb upstream and 2 kb downstream of SsXpb-I. A short (369 bp) ORF hypothetically coding for a 123 amino acid polypeptide maps directly upstream of SsXpb-I; the location and sequence of this short ORF is conserved in Sulfolobus tokodai but not in other Archaea. The distal (–129) start site of SsXpb-I maps within this ORF.
Figure 4.
(A) The Sulfolobus Xpb homologues. The boxes indicate conserved motives detected by using the Psi-Blast program. DEXDc, DEAD, conserved helicase superfamily ATP-binding domain. SFN2-N, SNF2 family N-terminal domain. This domain is found in proteins involved in transcription regulation (e.g. SNF2, STH1, brahma, MOT1), DNA repair (e.g. ERCC6, RAD16, RAD5), DNA recombination (e.g. RAD54) and chromatin unwinding (e.g. ISWI). HELICc, helicase superfamily C-terminal domain. Arrows indicate the primers used in Figures 4B and 5. (B) Primer extension of SsXpb-II. Representative gel chosen among three independent experiments is shown. Each lane contained 10 µg of total RNA. Initiation sites are indicated. G, A, T, sequence ladder. Other symbols as in the caption to Figure 2.
Figure 5.
UV-induced regulation of SsXpb-I. (A) Time course primer extension. Each lane contained 10 µg of total RNA prepared as in the caption to Figure 2; a representative gel chosen among four independent experiments is shown. I0.5–I8, RNA extracted after 0.5–8 h of incubation at 80°C following irradiation (200 J/m2). (B) Time course showing later times; C22 and I22, RNA extracted from controls and irradiated cultures, respectively, after 22 h of incubation at 80°C. (C) Dose–response experiment. C2, RNA from control cultures; I2, RNA from irradiated cultures (200 J/m2) after 2 h of incubation at 80°C following irradiation; IM, as I2 but UV fluence was 480 J/m2; IH, as I2 but UV fluence was 1500 J/m2. P, primer without RNA incubated under reaction conditions; G, A, T, C, sequence ladder; M, molecular weight markers (nt), other symbols as in the caption to Figure 2.
In a dose–response experiment, we found that at a very high intensity of irradiation (1500 J/m2), the SsXpb-I induction was reduced, probably reflecting the fact that a considerable fraction of cells were too severely damaged, or dead (Fig. 5C).
Effects of UV irradiation on transcription of genes coding for DNA-binding proteins
Packaging of DNA into higher order chromatin structures affects the accessibility of DNA to enzymes and regulatory complexes involved in transcription, replication and repair. In Eucarya, chromatin modelling is needed to facilitate access of the repair machinery to DNA lesions (26,27). Such modelling is achieved through transcriptional and post-transcriptional control.
Whereas Archaea belonging to subdomain Euryarchaeota possess true eucaryal-like histones, this is not the case for Crenarchaeota. Instead, Sulfolobus chromatin contains several DNA binding proteins (reviewed in 10,28). The most abundant is Sul7d (formerly named Sso7d, Sac7d or Ssh7), a 7 kDa architectural protein specific to the Sulfolobus genus. This protein affects DNA conformation introducing DNA bending, compaction and negative supercoiling (29,30). The other main component of the Sulfolobus chromatin is Alba, a 10 kDa protein conserved in several archaea and eukaryotes, whose DNA binding activity is regulated by acetylation (31).
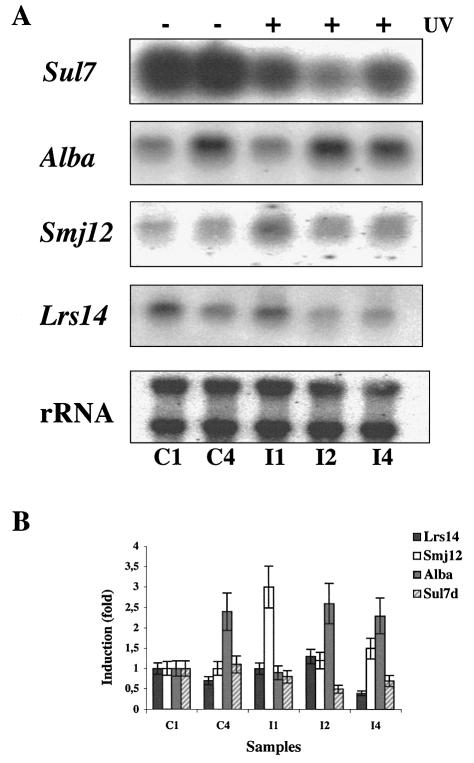
We have investigated the effects of UV irradiation on transcription regulation of Sulfolobus chromatin components. We found that the Alba steady-state RNA level increased with time but was not regulated by UV (Fig. 6). In contrast, the Sul7d transcript was reduced 1 h after exposure to UV, reached the minimum after 2 h, increased after 4 h and returned to normal levels after 8 h (data not shown). Analysis of protein extracts from the same samples showed a parallel transient down-regulation of the Sul7d protein (data not shown).
Figure 6.
UV-induced regulation of chromatin genes. (A) Northern blot of total RNA from control and irradiated cultures (200 J/m2) hybridised with indicated probes. (B) Quantitation of the data (mean of three independent experiments) normalised and expressed as in Figure 2B. Symbols as in the caption to Figure 2.
We have also analysed the effects of UV irradiation on transcription of Smj12 and Lrs14. They are members of a large family of helix–turn–helix DNA-binding proteins found in Archaea and Bacteria and share significant sequence similarity, but have different properties. Whereas Lrs14 is a sequence-specific transcription repressor (32,33), Smj12 binds DNA non-specifically and induces positive supercoiling (34). Interestingly, we found that whereas Lrs14 transcription was not affected by UV irradiation, Smj12 was up-regulated (Fig. 6). The increase of Smj12 RNA (3-fold) was transient and peaked at 1 h after irradiation. Lastly, the single-strand binding protein SSB (35) RNA was not regulated by UV (data not shown).
All these transcripts were sharp and single-gene sized. Primer extension experiments showed a single transcription start site for Sul7d and Smj12, mapping 10 and five bases upstream of the first codon, respectively (data not shown).
Transcription regulation by actinomycin D
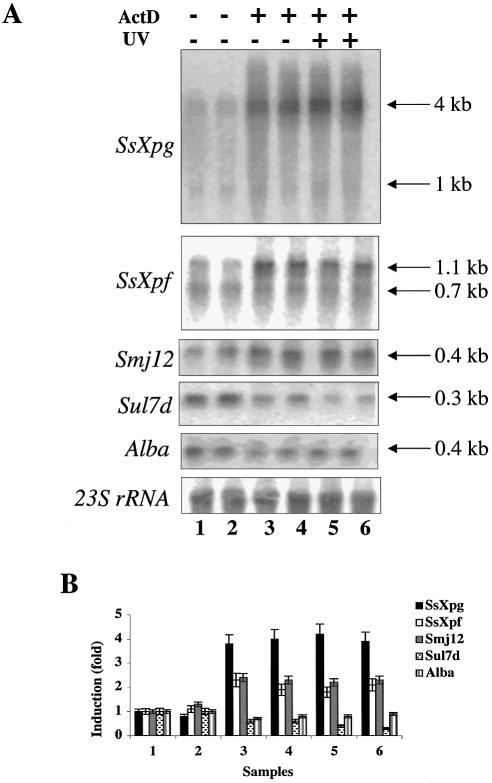
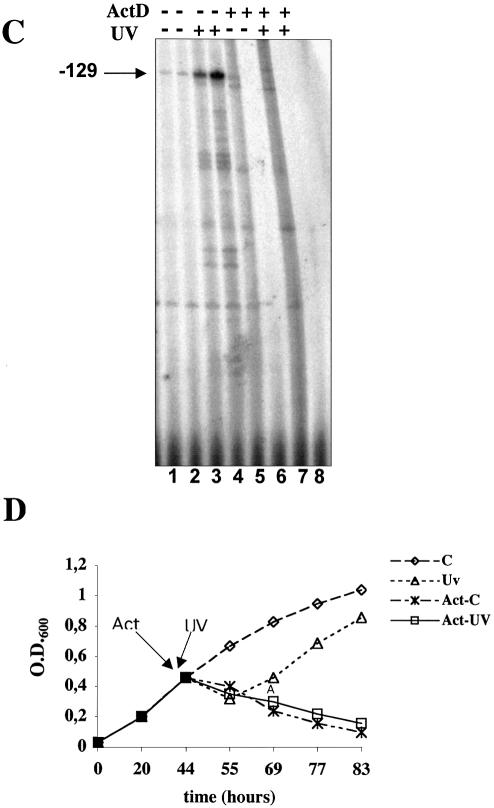
Actinomycin D is commonly used to inhibit transcription because it intercalates into DNA, blocking the progression of RNA polymerase. However, due to its intercalating activity, it may also induce DNA damage and trigger events recalling the UV response. For instance, in human cells both UV and actinomycin D induce transcriptional activation of NFkB, of the HIV-1 promoter (36) and that (p53 dependent) of p21 (37). Likewise, actinomycin D and UV activate plant genes involved in defence against pathogens (38). Sulfolobus solfataricus is sensitive to actinomycin D (growth is arrested 20 min after the addition of 5 µg/ml, and transcription is arrested 40 min after the addition of 10 µg/ml actinomycin D) (39). Therefore, we tested whether the UV-regulated genes in Sulfolobus were also affected by actinomycin D. Cultures were treated with 10 µg/ml actinomycin D, which completely arrested cell growth (Fig. 7D), and RNA was extracted after 140 and 260 min (Fig. 7A). Actinomycin D strongly activated SsXpg, SsXpf and Smj12. In contrast, Sul7d and Alba transcripts were reduced. When cultures pretreated with actinomycin D were exposed to UV, no significant change in the level of transcripts was observed (Fig. 7A and B).
Figure 7.
Actinomycin D-induced regulation of NER and chromatin genes. (A) Exponentially growing cells were treated with actinomycin D (10 µg/ml) as indicated, and after 20 min were UV irradiated (200 J/m2) or mock treated. RNA was extracted after 2 h (lanes 1, 3 and 5) or 4 h (lanes 2, 4 and 6) after UV irradiation, and hybridised with the indicated probes. (B) Quantitation of the data (mean of three independent experiments) normalised and expressed as in Figure 2B. (C) Primer extension of SsXpb-I; cultures were treated with actinomycin D (10 µg/ml), UV, or both, as indicated, and RNA was extracted after 2 h (lanes 1, 3, 5 and 7) or 4 h (lanes 2, 4, 6 and 8). (D) Effect of actinomycin D (10 µg/ml) on S.solfataricus growth. C, control culture; UV, irradiated culture; Act-C, cultures treated with actinomycin D; Act-UV, cultures treated with actinomycin and UV irradiated.
SsXpb-I RNA was not induced after 2 h of treatment with actinomycin D (Fig. 7C, lane 5) and was absent after 4 h (Fig. 7C, lane 6); when cultures were pre-treated with actinomycin D and then were UV irradiated, SsXpb-I RNA was not induced after 2 h (Fig. 7C, lane 7) and was absent after 4 h (Fig. 7C, lane 8). Thus, SsXpb-I was not activated by actinomycin D and pre-treatment with actinomycin D prevented UV-dependent up-regulation.
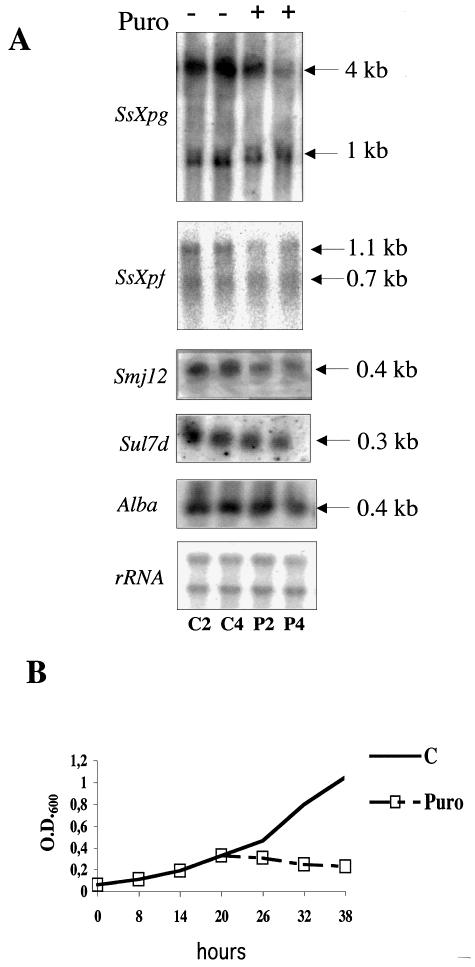
Interestingly, for both SsXpg and SsXpf, actinomycin D induced mainly longer transcripts. One hypothesis to explain this finding is that active transcription is required for the synthesis of a factor responsible for the degradation or processing of longer transcripts. To test whether protein synthesis may have the same effect, we used puromycin, an antibiotic that interferes with protein synthesis without damaging DNA. Puromycin completely blocked S.solfataricus growth at 50 µg/ml (Fig. 8B); however, it did not induce gene activation, but, as expected, the level of all transcripts was slightly reduced after 2 or 4 h (Fig. 8A). This experiment suggests that the actinomycin-induced transcription induction is not due to the block of translation, but is either a consequence of DNA damage or of transcription arrest.
Figure 8.
Effect of puromycin on gene transcription. (A) Exponentially growing cells were treated with puromycin (50 µg/ml) and RNA was extracted after 2 h (C2, P2) or 4 h (C4, P4) of incubation at 80°C. C, RNA from control cultures; P, RNA from cultures treated with puromycin. (B) Effect of puromycin (50 µg/ml) on S.solfataricus growth.
DISCUSSION
Since S.solfataricus contains very well conserved homologues of eucaryal NER genes, as with all other Archaea whose genomes are available, these genes were chosen as likely candidates for UV-dependent regulation. Indeed the genes encoding the two NER nucleases SsXpg and SsXpf were induced moderately by UV, and of the two Xpb paralogues, SsXpb-I was highly induced (∼10-fold) by UV, whereas SsXpb-II was constitutively expressed.
We have also analysed the effect of actinomycin D, an intercalating drug that induces a dual effect: DNA damage and a block of transcription. SsXpf and SsXpg were strongly induced by actinomycin D, whereas neither of the SsXpb paralogues was affected. Actinomycin D has been reported to induce gene expression of some UV responsive genes in eukaryotes, although it is not clear whether this induction is related to its effect on RNA synthesis or its DNA-damaging activity (36). UV radiation also represses general transcription, because the RNA polymerase elongation complex is blocked at sites of UV-induced DNA damage, and is a signal for the so called transcription coupled repair (TCR) pathway that is responsible for the preferential removal of DNA lesions from transcribed strands. The existence of TCR in Archaea has not been reported; our results suggest that both UV- and actinomycin-induced DNA damage may elicit a similar response and therefore be transduced by the same mechanism.
The differential regulation of the Sulfolobus NER genes is difficult to rationalise because they show different timing, duration and extent of induction in response to the same stimulus. However, this is a common feature of the transcriptional response to DNA damage in both Bacteria and Eucarya. Some genes may be partially induced in response to even endogenous levels of DNA damage, while other genes of the same pathway appear to be induced only when high or persistent DNA damage is present. Moreover, repair genes can be induced by several different (and unrelated) damaging agents. It is possible that the different regulation for Sulfolobus repair genes might reflect the function of some NER genes in other pathways, as XPG proteins are involved in the removal of Okazaki fragments during replication, whereas XPF plays a role in recombination, and XPB, in complex with other proteins, constitutes the factor TFIIH, which is essential for both transcription and repair. Although there are no experimental data, an attractive hypothesis that should be tested is that the two different XPB paralogues in Sulfolobus may play different roles in these two processes.
A striking feature of SsXpf and SsXpg is the complexity of their transcription pattern. They show both monocistronic and polycistronic transcripts. Interestingly, in both cases the larger transcripts span a chaperone gene. At this stage it is not clear whether the two transcripts result from different initiation or RNA processing. Polycistronic transcription units are not infrequent in Archaea. As in Bacteria, often co-transcription reflects a functional association. Molecular chaperones have been suggested to participate in NER in E.coli by maintaining repair proteins in their properly folded forms (40). In Eucarya, the 26S proteasome regulates DNA repair by controlling degradation of repair proteins, but also assisting the assembly of the NER complex at UV-induced CPDs (41). Further experiments are required to test whether the observed co-transcription in Sulfolobus of two nuclease genes with two chaperone genes has any functional meaning. It is possible that, like other molecular chaperones, the prefoldin and SAV genes can be up-regulated by a number of different stresses, including DNA damage.
Among the genes coding for DNA binding proteins, we found three different patterns: Alba, Lrs14 and Ssb were not affected by DNA damage, Sul7d was repressed, and Smj12 was activated by both UV and actinomycin D. Intriguingly, Sul7d and Smj12 have opposite effects on DNA conformation: the former is a very abundant protein which induces negative supercoiling and compacts positive or relaxed DNA (30). Chromosomal DNA in hyperthermophiles is positively supercoiled or relaxed (reviewed in 42). In contrast, Smj12 is not highly abundant and induces positive supercoiling; however, free regions surrounding the sites of protein binding might be relaxed to compensate the torsional stress (34). Damage formation and repair are coupled intimately with structural and dynamic properties of chromatin, because compaction of DNA into higher order structures and the presence of chromatin components hinder detection and repair of DNA damage. It has been recently shown that in human cells UV induces global chromatin relaxation mediated by p53 (43). Analogously, it could be hypothesised that in Sulfolobus, access of the repair machinery to lesions can be facilitated by a reduction in Sul7d, which compacts DNA, and an increase in Smj12, which relaxes DNA.
Taken together, our data provide the first evidence that UV exposure in S.solfataricus (and presumably Archaea in general) results in both gene activation and repression. DNA damage-regulated genes may serve as a good model for the study of transcription regulation in Archaea. Genome sequencing and biochemical studies suggested that the composition and function of the transcriptional machinery are fundamentally conserved between Eucarya and Archaea (9). However, available data suggest that, as in Bacteria, most regulation occurs by transcriptional repression. It will be interesting to elucidate the mechanisms of up- and down-regulation and identify elements responsible for this regulation.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Giovanni R. Giusto for his contribution to some experiments, to Derek Richard for critically reading the manuscript and to the IIGB-Tigem DNA sequencing core for the excellent assistance in DNA sequencing. M.F.W. is a Royal Society URF. This work was partially supported by Agenzia Spaziale Italiana, contract no. N.I/R/356/02, by the CNR-MIUR Project ‘Legge 95/95’ and the CNR-MIUR Project ‘Legge 449/97-DM 30/10/2000’.
REFERENCES
- 1.Eckardt-Schupp F. and Klaus,C. (1999) Radiation inducible DNA repair processes in eukaryotes. Biochimie, 81, 161–171. [DOI] [PubMed] [Google Scholar]
- 2.deLaat W.L., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- 3.Walker G.C. (1985) Inducible DNA repair systems. Annu. Rev. Biochem., 54, 425–457. [DOI] [PubMed] [Google Scholar]
- 4.Birrell G.W., Brown,J.A., Wu,H.I., Giaever,G., Chu,A.M., Davis,R.W. and Brown,J.M. (2002) Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl Acad. Sci. USA, 99, 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sesto A., Navarro,M., Burslem,F. and Jorcano,J.L.(2002) Analysis of the ultraviolet B response in primary human keratinocytes using oligonucleotide microarrays. Proc. Natl Acad. Sci. USA, 99, 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eller M.S., Maeda,T., Magnoni,C., Atwal,D. and Gilchrest,B.A. (1997) Enhancement of DNA repair in human skin cells by thymidine dinucleotides: evidence for a p53-mediated mammalian SOS response. Proc. Natl Acad. Sci. USA, 94, 12627–12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu M.H. and Yung,B.Y. (2002) UV stimulation of nucleophosmin/B23 expression is an immediate-early gene response induced by damaged DNA. J. Biol. Chem., 277, 48234–48240. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell S.D. and Jackson,S.P. (2001) Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol., 4, 208–213. [DOI] [PubMed] [Google Scholar]
- 10.Ciaramella M., Pisani,F.M. and Rossi,M. (2002) Molecular biology of extremophiles: recent progress on the hyperthermophilic archaeon Sulfolobus. Antonie Van Leeuwenhoek, 81, 85–97. [DOI] [PubMed] [Google Scholar]
- 11.She Q., Singh.,R.K., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C., Clausen,I.G., Curtis,B.A., De Moors,A., Erauso,G., Fletcher,C., Gordon,P.M., Heikamp-de Jong,I., Jeffries,A.C., Kozera,C.J., Medina,N., Peng,X., Thi-Ngoc,H.P., Redder,P., Schenk,M.E., Theriault,C., Tolstrup,N., Charlebois,R.L., Doolittle,W.F., Duguet,M., Gaasterland,T., Garrett,R.A., Ragan,M.A., Sensen,C.W. and Van der Oost,J. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA, 98, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belin D. (1997) The use of RNA probes for the analysis of gene expression. Mol. Biotechnol., 7, 153–163. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Garcia P. and Forterre,P. (1997) DNA topology in hyperthermophilic archaea: reference states and their variation with growth phase, growth temperature and temperature stresses. Mol. Microbiol., 23, 1267–1279. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Y., Pao,A., Adair,G.M. and Tang,M. (2001) Cyclobutane pyrimidine dimers and bulky chemical DNA adducts are efficiently repaired in both strands of either transcriptionally active or promoter-deleted APRT gene. J. Biol. Chem., 276, 16786–16796. [DOI] [PubMed] [Google Scholar]
- 16.Prisco A., Moracci,M., Rossi,M. and Ciaramella,M. (1995) A gene encoding a putative membrane protein homologous to the major facilitator superfamily of transporters maps upstream of the beta-glycosidase gene in the archaeon Sulfolobus solfataricus. J. Bacteriol., 177, 1614–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood E.R., Ghane,F. and Grogan,D.W. (1997) Genetic responses of the thermophilic archaeon Sulfolobus acidocaldarius to short-wavelength UV light. J. Bacteriol., 179, 5693–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts J.A., Bell,S.D. and White,M.F. (2003) An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol., 48, 361–371. [DOI] [PubMed] [Google Scholar]
- 19.White M.F. (2003) Archaeal DNA repair: paradigms and puzzles. Biochem. Soc. Trans., 31, 690–693. [DOI] [PubMed] [Google Scholar]
- 20.Hartl F.U. and Hayer-Hartl,M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science, 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- 21.Hosfield D.J., Frank,G., Weng,Y., Tainer,J.A. and Shen,B. (1998) Newly discovered archaebacterial flap endonucleases show a structure-specific mechanism for DNA substrate binding and catalysis resembling human flap endonuclease-1. J. Biol. Chem., 273, 27154–27361. [DOI] [PubMed] [Google Scholar]
- 22.Dionne I., Nookala,R.K., Jackson,S.P., Doherty,A.J. and Bell,S.D. (2003) A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell, 11, 275–282. [DOI] [PubMed] [Google Scholar]
- 23.Lord J.M., Ceriotti,A. and Roberts,L.M. (2002) ER dislocation: Cdc48p/p97 gets into the AAAct. Curr. Biol., 12, R182–R184. [DOI] [PubMed] [Google Scholar]
- 24.Confalonieri F., Marsault,J. and Duguet,M. (1994) SAV, an archaebacterial gene with extensive homology to a family of highly conserved eukaryotic ATPases. J. Mol. Biol., 235, 396–401. [DOI] [PubMed] [Google Scholar]
- 25.Reich C.I., McNeil,L.K., Brace,J.L., Brucker,J.K. and Olsen,G.J. (2001) Archaeal RecA homologues: different response to DNA-damaging agents in mesophilic and thermophilic Archaea. Extremophiles, 5, 265–275. [DOI] [PubMed] [Google Scholar]
- 26.Green C.M. and Almouzni,G. (2002)When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep., 3, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ura K. and Hayes,J.J. (2002) Nucleotide excision repair and chromatin remodeling. Eur. J. Biochem., 269, 2288–2293. [DOI] [PubMed] [Google Scholar]
- 28.White M.F. and Bell,S.D. (2002) Holding it together: chromatin in the Archaea. Trends Genet., 18, 621–626. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Garcia P., Knapp,S. Ladenstein,R. and Forterre,P. (1998) In vitro DNA binding of the archaeal protein Sso7d induces negative supercoiling at temperatures typical for thermophilic growth. Nucleic Acids Res., 26, 2322–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Napoli A., Zivanovic,Y., Bocs,C., Buhler,C., Rossi,M., Forterre,P. and Ciaramella,M. (2002) DNA bending, compaction and negative supercoiling by the architectural protein Sso7d of Sulfolobus solfataricus.Nucleic Acids Res., 30, 2656–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell S.D., Botting,C.H., Wardleworth,B.N., Jackson,S.P. and White,M.F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science, 296, 148–151. [DOI] [PubMed] [Google Scholar]
- 32.Napoli A., van der Oost,J., Sensen,C.W., Charlebois,R.L., Rossi,M. and Ciaramella,M. (1999) An Lrp-like protein of the hyperthermophilic archaeon Sulfolobus solfataricus which binds to its own promoter. J. Bacteriol., 181, 1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell S.D. and Jackson,S.P. (2000) Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem., 275, 31624–31629. [DOI] [PubMed] [Google Scholar]
- 34.Napoli A., Kvaratskelia,M., White,M.F., Rossi,M. and Ciaramella,M. (2001) A novel member of the bacterial-archaeal regulator family is a nonspecific DNA-binding protein and induces positive supercoiling. J. Biol. Chem., 276, 10745–10752. [DOI] [PubMed] [Google Scholar]
- 35.Wadsworth R.I. and White,M.F. (2001) Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res., 29, 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassè C., Giannoni,F., Nguyen,V.T., Dubois,M.F. and Bensaude,O. (1999) The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem., 274, 16097–16106. [DOI] [PubMed] [Google Scholar]
- 37.Ljungman M., Zhang,F., Chen,F., Rainbow,A.J. and McKay,B. (1999) Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene, 18, 583–592. [DOI] [PubMed] [Google Scholar]
- 38.Junghae Choi J., Klosterman,S.J. and Hadwiger,L.A. (2001) A comparison of the effects of DNA-damaging agents and biotic elicitors on the induction of plant defense genes, nuclear distortion and cell death. Plant Physiol., 125, 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bini E., Dikshit,V., Dirksen,K., Drozda,M. and Blum,P. (2002) Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA, 8, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou Y., David,J.D.J. and Van Houten,B. (1998) Involvement of molecular chaperonins in nucleotide excision repair. Dnak leads to increased thermal stability of UvrA, catalytic UvrB loading, enhanced repair and increased UV resistance. J. Biol. Chem., 273, 12887–12892. [DOI] [PubMed] [Google Scholar]
- 41.Lommel L., Ortolan,T., Chen,L., Madura,K. and Sweder,K.S. (2002) Proteolysis of a nucleotide excision repair protein by the 26 S proteasome. Curr. Genet., 42, 9–20. [DOI] [PubMed] [Google Scholar]
- 42.Forterre P., Bergerat,A. and Lopez-Garcia,P. (1996) The unique DNA topology and DNA topoisomerases of hyperthermophilic archaea. FEMS Microbiol. Rev., 18, 237–248. [DOI] [PubMed] [Google Scholar]
- 43.Rubbi C. and Milner,P. (2003) p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J., 22, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]