Abstract
CXXC finger protein 1 (CFP1) is a component of the Setd1A and Setd1B methyltransferase complexes, localizes to euchromatic regions of the genome, and specifically binds unmethylated CpG dinucleotides in DNA. Murine embryos lacking CFP1 exhibit peri-implantation lethality, a developmental time that correlates with global epigenetic reprogramming. CFP1-deficient embryonic stem (ES) cells exhibit a 70% reduction in global cytosine methylation and a 60% decrease in maintenance DNA methyltransferase (DNMT1) activity. DNMT1 protein level is reduced 50% in CFP1-deficient ES cells. Experiments were performed to investigate the role of CFP1 in regulating maintenance cytosine methylation. Coimmunoprecipitation experiments reveal that endogenous DNMT1 and CFP1 interact in vivo. Protein regions required for the interaction between DNMT1 and CFP1 were mapped. Amino acids 169–493 and 970–1617 of DNMT1 are each sufficient for interaction with CFP1. Three regions spanning the CFP1 protein, amino acids 1–123, 103–367, and 361–656, are each sufficient for interaction with DNMT1. Interestingly, a single-point mutation (C375A) within CFP1 that abolishes the interaction with the Setd1A and Setd1B histone H3K4 methyltransferase complexes does not disrupt the interaction between CFP1 and DNMT1. This result indicates that CFP1 intersects the cytosine methylation machinery independently of its association with the Setd1 complexes.
Introduction
Epigenetics refers to heritable patterns of gene expression that occur without alteration of nucleotide sequence. Epigenetic modifications that facilitate changes in chromatin structure and thus influence transcription include posttranslational covalent modifications of histone tails and cytosine methylation.
CXXC finger protein 1 (CFP1) is a unique DNA-binding protein that contains a conserved CXXC domain both necessary and sufficient for specific binding to unmethylated CpG dinucleotides (Voo et al., 2000; Lee et al., 2001). CFP1 also contains two plant homeodomain (PHD) fingers. Recently, a global proteomic screen of Saccharomyces cerevisiae proteins containing PHD fingers revealed that the PHD of Spp1, the yeast homolog of CFP1, binds histone H3K4me2/3 (Shi et al., 2007). Additionally, the PHD within the NURF nucleosome remodeling factor binds histone H3K4me3 and targets the NURF complex to Hox promoters during development to activate transcription (Wysocka et al., 2006), thus demonstrating a biological effect of PHD finger interaction with methylated histone H3.
CFP1 is a component of the mammalian Setd1A (Lee and Skalnik, 2005) and Setd1B (Lee et al., 2007) histone H3K4 methyltransferase complexes, which are analogous to the Set1/COMPASS complex in S. cerevisiae (Miller et al., 2001; Nagy et al., 2002; Roguev et al., 2003). Spp1 is required for Set1/COMPASS histone methyltransferase activity in Schizosaccharomyces pombe, but only for appropriate histone H3K4me3 in S. cerevisiae (Roguev et al., 2003; Schneider et al., 2005). While the mammalian Setd1A and Setd1B complexes both localize to euchromatic regions of the genome, they exhibit nonoverlapping subnuclear localization patterns (Lee et al., 2007). This suggests that the Setd1A and Setd1B complexes function at distinct genomic targets.
Mice lacking CFP1 exhibit peri-implantation lethality (4.5–6.5 days postcoitus) (Carlone and Skalnik, 2001). CFP1 is additionally required for postgastrulation development and survival, because zebrafish embryos injected with antisense morpholino oligonucleotides directed against CFP1 exhibit defective primitive hematopoiesis and decreased survival (Young et al., 2006). In addition, siRNA-mediated depletion of CFP1 in human leukemia cell lines leads to defects in cell proliferation and terminal myeloid differentiation (Young and Skalnik, 2007). Lastly, CFP1-deficient murine embryonic stem (ES) cells are viable; however, they are unable to differentiate upon removal of leukemia inhibitory factor from the culture medium and exhibit a threefold increase in apoptosis under normal culture conditions (Carlone et al., 2005).
CFP1-deficient ES cells exhibit a ∼43% increase in histone H3K4me2 and ∼28% reduction in histone H3K9me2 under normal conditions, and an approximately fourfold induction of histone H3K4me3 upon induction of differentiation (Lee and Skalnik, 2005). CFP1-deficient ES cells also exhibit a 70% deficiency in global cytosine methylation that affects single-copy genes, imprinted loci, and repetitive elements (Carlone et al., 2005). Taken together, these data indicate that ES cells lacking CFP1 contain reduced heterochromatin. A 60% reduction in maintenance DNA methyltransferase (DNMT1) activity appears to contribute to the global cytosine methylation deficiency, because de novo DNMT activity is not decreased in ES cells lacking CFP1 (Carlone et al., 2005). DNMT1 protein expression is reduced 50% in the absence of CFP1 (Carlone et al., 2005) due to decreased DNMT1 protein stability and reduced translation (Jill Butler, unpublished data).
Recent reports reveal intricate interrelationships linking cytosine methylation and histone modifications, thus providing a unifying framework for the control of chromatin structure and gene regulation (Burgers et al., 2002). For example, DNMT proteins associate with histone deacetylase (HDAC) complexes (Fuks et al., 2000, 2001; Robertson et al., 2000); cytosine methylation in Neurospora is dependent on methylation of histone H3 and the presence of heterochromatin protein 1 (HP1) (Freitag et al., 2004); human HP1 recruits DNMT1 to methylate DNA and silence gene expression (Smallwood et al., 2007), and inhibition of HDAC activity by trichostatin A results in a loss of cytosine methylation (Selker, 1998; Tamaru and Selker, 2001; Tamaru et al., 2003). Further, the chromatin remodeling protein DDM1 in Arabidopsis and the related factor LSH in mammals are required for normal cytosine methylation (Jeddeloh et al., 1998, 1999; Dennis et al., 2001). Disruption of the Suv39h1 histone H3K9 methyltransferase gene in murine ES cells leads to altered localization of DNMT3b and decreased cytosine methylation at peri-centric satellite repeats (Lehnertz et al., 2003), and loss of DNMT1 leads to perturbations in the histone code consistent with reduced heterochromatin (Espada et al., 2004). Hence, DNA methylation and covalent histone modifications are integrated and mutually reinforcing mechanisms to establish and maintain heterochromatin.
However, the molecular mechanisms linking cytosine methylation and histone modifications are not well understood. Interestingly, organisms that lack cytosine methylation, such as Caenorhabditis elegans and S. cerevisiae, contain CFP1 orthologs that lack the CXXC DNA-binding domain (Voo et al., 2000). This suggests that CFP1 served an ancestral role in the regulation of covalent histone modifications, and acquired DNA-binding capacity in organisms that utilize cytosine methylation as an additional epigenetic modification. The influence of CFP1 on both cytosine methylation and histone methylation suggests a role in mediating cross talk between these epigenetic modifications.
Studies were performed to gain insight into the molecular mechanism of how CFP1 affects cytosine methylation. The results presented in this report demonstrate that CFP1 and DNMT1 physically interact. The minimal regions sufficient for the interaction between DNMT1 and CFP1 were mapped and include conserved domains involved in chromatin targeting. Experiments were also performed to map the minimal region required for CFP1 interaction with Setd1A and Setd1B. Interestingly, CFP1 interaction with Setd1A or Setd1B is not required for its interaction with DNMT1, strongly suggesting that CFP1 directly intersects with both histone methylation and cytosine methylation.
Materials and Methods
Cell culture and transient transfections
Human embryonic kidney (HEK-293) cells were cultured as previously described (Lee and Skalnik, 2002; Carlone et al., 2005). Transient cotransfections in HEK-293 cells were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) per the manufacturer's protocol. Briefly, cells were grown to 90–95% confluence as a monolayer in 10 cm tissue culture dishes. Ten micrograms of plasmid DNA was added to Lipofectamine 2000 reagent in OPTI-MEM serum-free medium (Invitrogen). The mixture was added drop-wise to the cells, which were then incubated for ∼40 h at 37°C with 5% CO2.
Construction of DNMT1 and CFP1 expression vectors
The human DNMT1 cDNA (generously provided by Dr. Sriharsa Pradhan; New England Biolabs, Ipswich, MA) or human CFP1 cDNA was ligated to the pcDNA3-FLAG vector that encodes an amino terminal FLAG epitope, as previously described (Lee et al., 2000). The CFP1 cDNA was ligated to the pcDNA3-Myc vector that encodes an amino-terminal Myc epitope. Constructs carrying truncations of the DNMT1 or CFP1 cDNAs were generated using a combination of polymerase chain reaction (PCR) and restriction enzyme digestion methods, and then subcloned into the pcDNA3-FLAG or pcDNA3-Myc expression vectors. Primer sequences for PCR amplification are available upon request. An exogenous nuclear localization signal (NLS) was made by annealing complementary oligonucleotides corresponding to the endogenous DNMT1 NLS that extends from amino acids 193–213. These oligonucleotides were designed to create a 5′ EcoRI restriction enzyme site and a 3′ ClaI restriction enzyme site when annealed. DNMT1 N-terminal deletion constructs lacking the endogenous NLS were subcloned downstream of the exogenous NLS in the pcDNA3-FLAG vector using the EcoRI and ClaI restriction enzyme sites. Site-directed mutagenesis was carried out using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) per the manufacturer's protocol. The sequences of the oligonucleotide primers used to generate amino acid substitutions within the human CFP1 cDNA are as follows: C375A, 5′-cctgcgtcactgccccaggcgctggggcccggctgtgtg-3′; Y390A, C391A, S392A, 5′-ccgcccagcccagctccaaggcagccgccgatgactgtggcatgaagct-3′; and C580A, 5′-gagctcacgggtgacttcgcccgcctgcccaagcgccag-3′. The nucleotide substitutions within the CFP1 primer sequences are underlined. The nucleotide sequences of all constructs were confirmed by automated DNA sequencing.
Coimmunoprecipitation and western blot analysis
Nuclear extracts were prepared from HEK-293 cells transiently expressing Myc and FLAG epitope-tagged proteins. Cells were washed with 1× PBS, and then with hypotonic buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [pH 7.9 at 4°C], 1.5 mM MgCl2, and 10 mM KCl). Cells were then swollen on ice for 10 min in hypotonic buffer before being lysed by Dounce homogenization 10 times. Nuclei were pelleted by centrifugation at 4°C and resuspended in lysis buffer (20 mM Tris HCl [pH 7.4], 400 mM NaCl, 25% glycerol, 5 mM ethylenediaminetetraacetic acid, 0.1% NP-40, 1 mM dithiothreitol, and 3% protease inhibitor cocktail [Sigma, St. Louis, MO]). Nuclei were lysed by Dounce homogenization 30 times on ice, and debris was pelleted by centrifugation at 4°C.
Either anti-Myc–conjugated agarose (Sigma) or FLAG M2–conjugated agarose (Sigma) was incubated with soluble nuclear extracts for 3 h at 4°C. Alternatively, the soluble nuclear extracts were precleared with protein G agarose (Millipore, Billerica, MA), and then incubated with CFP1 antiserum (Lee et al., 2007) or a custom DNMT1 antiserum directed against the peptide encoded by the 349–825 bp region of the murine cDNA for 1 h at 4°C, followed by incubation with protein G agarose for 3 h at 4°C. Beads were washed four times with buffer containing 300 mM NaCl, 0.1% NP-40, and then boiled in Laemmli sample buffer. The proteins were separated on PAGEr-Gold precast 4–12% or 4–20% Tris-glycine gradient gels (Lonza Group, Basel, Switzerland) by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Amersham, GE Healthcare, Piscataway, NJ). Western blot analysis was performed using primary antisera directed against the Myc epitope (Santa Cruz Biotechnology, Santa Cruz, CA), FLAG epitope (Sigma), or the Setd1 complex components Setd1A (Lee et al., 2007), Setd1B (Lee et al., 2007), Wdr5 (Lee et al., 2007), Wdr82 (Lee et al., 2007), Ash2 (Bethyl Laboratories, Montgomery, TX), and Rbbp5 (Bethyl Laboratories), followed by horseradish peroxidase–conjugated secondary antibodies. Proteins were detected using an ECL detection kit (Amersham, GE Healthcare, Montgomery, TX).
Results
CFP1 and DNMT1 interact in vivo
To determine if DNMT1 and CFP1 interact in vivo, full-length Myc-CFP1 and full-length FLAG-DNMT1 were transiently expressed in HEK-293 cells, and Myc-CFP1 was immunoprecipitated from nuclear extracts using anti-Myc–conjugated agarose. The immunoprecipitated material was subjected to western blot analysis using antiserum against the FLAG epitope. These experiments demonstrate that FLAG-DNMT1 coimmunoprecipitates with Myc-CFP1 (Fig. 1A). Empty vector was cotransfected individually and with each construct to serve as a negative control for Myc immunoprecipitation reactions. The reciprocal experiment demonstrated that Myc-CFP1 coimmunoprecipitates with FLAG-DNMT1 upon immunoprecipitation with FLAG M2–conjugated agarose, thus confirming the interaction (Fig. 1B). These results indicate that full length CFP1 and DNMT1 interact in vivo.
FIG. 1.
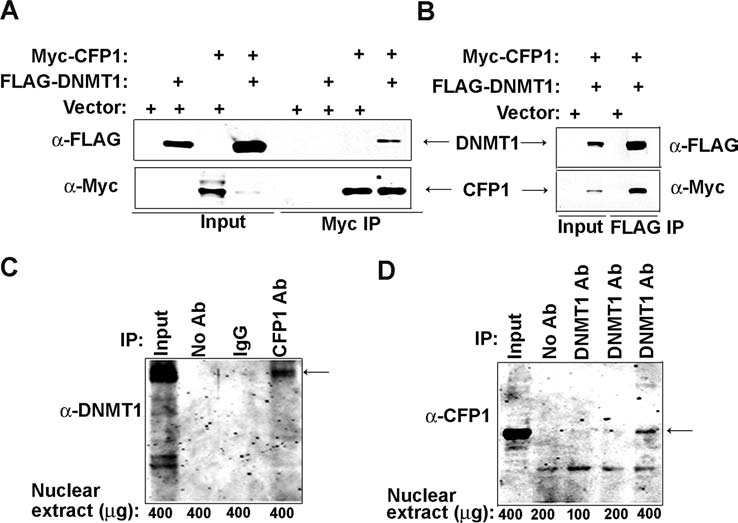
CFP1 and DNMT1 physically interact in vivo. (A) Nuclear extracts were prepared from HEK-293 cells transiently expressing Myc-CFP1 (full length) and FLAG-DNMT1 (full length), as indicated above the lanes, and were subjected to immunoprecipitation using anti-Myc–conjugated agarose. Western blot analysis with anti-FLAG antiserum was used to detect FLAG-DNMT1. The membrane was re-probed with anti-Myc antiserum to verify Myc-CFP1 protein expression and Myc immunoprecipitation efficiency. Transfection and immunoprecipitation of the empty vector serves as a negative control for the Myc immunoprecipitation. (B) Nuclear extracts were prepared from HEK-293 cells transiently expressing Myc-CFP1 (full length) and FLAG-DNMT1 (full length), as indicated above each lane, and were subjected to immunoprecipitation using FLAG M2–conjugated agarose. Western blot analysis with anti-Myc antiserum was used to detect Myc-CFP1. The membrane was re-probed with anti-FLAG antiserum to verify FLAG-DNMT1 protein expression and FLAG immunoprecipitation efficiency. Transfection and immunoprecipitation of the empty vector serves as a negative control for the FLAG immunoprecipitation. (C) Endogenous CFP1 was immunoprecipitated from HEK-293 nuclear extracts using CFP1 antiserum and protein G agarose. Western blot analysis was performed using anti-DNMT1 antiserum. The signal for DNMT1 is marked with an arrow. Immunoprecipitation reactions containing control serum (IgG control) or lacking primary antibody (No Ab) serve as negative controls for the CFP1 immunoprecipitation. (D) Endogenous DNMT1 was immunoprecipitated from HEK-293 nuclear extracts using antiserum against DNMT1 and protein G agarose. Western blot analysis was performed using anti-CFP1 antiserum. The signal for CFP1 is marked with an arrow. An immunoprecipitation reaction lacking primary antibody (No Ab) serves as a negative control for the DNMT1 immunoprecipitation.
To examine whether CFP1 and DNMT1 interact at physiologic concentrations in vivo, endogenous CFP1 was immunoprecipitated from HEK-293 nuclear extracts using CFP1 antiserum. The immunoprecipitated material was subjected to western blot analysis using antiserum against DNMT1. Endogenous DNMT1 immunoprecipitates with CFP1 in HEK-293 nuclear extracts (Fig. 1C). Addition of protein G agarose to nuclear extracts without antibody or with control serum served as negative controls. The reciprocal experiment was performed using DNMT1 antiserum to coimmunoprecipitate CFP1 from HEK-293 nuclear extracts. Western blot analysis using CFP1 antiserum confirmed the interaction with endogenous DNMT1 (Fig. 1D). These results demonstrate a physical interaction between endogenous DNMT1 and CFP1.
Independent domains within the amino and carboxyl termini of DNMT1 are each sufficient for interaction with CFP1
The amino terminus of DNMT1 interacts with a number of chromatin-associated proteins, such as DNMT3a and 3b (Kim et al., 2002), HDAC1 (Fuks et al., 2000; Robertson et al., 2000) and HDAC2 (Rountree et al., 2000), the methyl CpG-binding proteins, MeCP2 (Kimura and Shiota, 2003) and MBD2 and MBD3 (Tatematsu et al., 2000), HP1 (Fuks et al., 2003; Smallwood et al., 2007), and the histone H3K9 methyltransferases, SUV39H1 (Fuks et al., 2003) and G9a (Esteve et al., 2006). A diagram illustrating conserved domains within DNMT1 is shown in Figure 2A. To further characterize the minimal region of DNMT1 required for interaction with CFP1, FLAG-tagged, truncated DNMT1 protein fragments were transiently expressed in HEK-293 cells along with Myc-CFP1 (aa 1–367), followed by Myc immunoprecipitation. The C-terminal truncated proteins, FLAG-DNMT1 (aa 1–650) and (aa 1–489), interact with Myc-CFP1 (Fig. 2B). Analysis of the N-terminus of DNMT1 revealed that the first 173 amino acids, encoding the DNMT1-associated protein 1 (DMAP) and proliferating cell nuclear antigen (PCNA)–binding regions, are not sufficient for interaction with Myc-CFP1 (Fig. 2B). The FLAG-DNMT1 (aa 169–493) protein fragment encodes the NLS and a large portion of the targeting sequence (TS) that directs DNMT1 to late-replicating heterochromatin (Easwaran et al., 2004). This portion of DNMT1 is sufficient for the interaction with Myc-CFP1 (Fig. 2B).
FIG. 2.
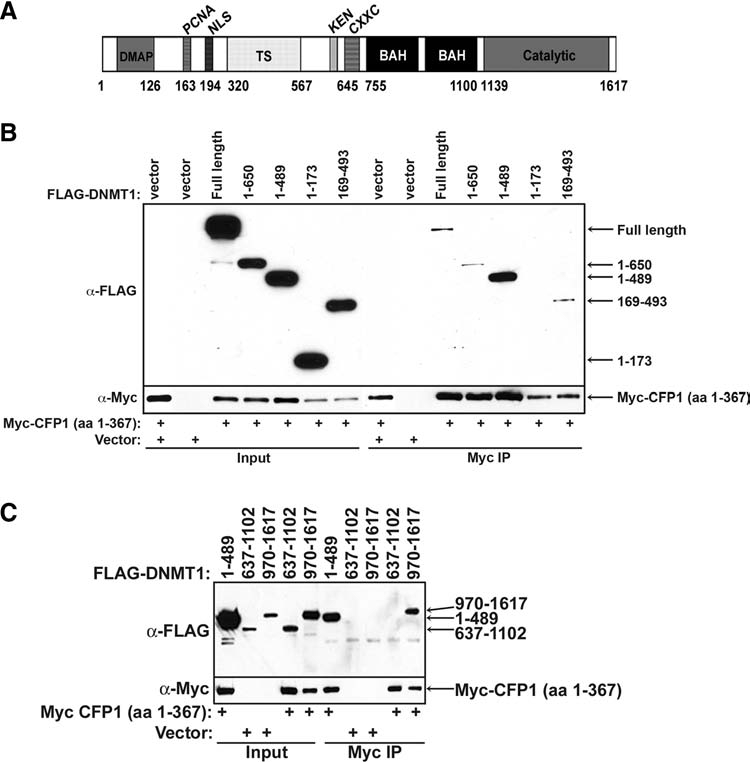
Two independent domains of DNMT1 are each sufficient for interaction with CFP1. (A) A schematic representation of the conserved domains within DNMT1. DMAP, DNMT1-associated protein 1 binding region; PCNA, proliferating cell nuclear antigen–binding region; NLS, nuclear localization signal; TS, region required to target DNMT1 to chromatin independently of the replication fork; CXXC, conserved cysteine-rich DNA-binding domain; BAH, bromo-adjacent homology domain. The amino acid position of each domain is denoted below the diagram. (B) A series of FLAG-DNMT1 C-terminal deletion mutants encoding amino acids 1–650, 1–489, 1–173, and 169–493 were coexpressed with Myc-CFP1 (aa 1–367) in HEK-293 cells. Nuclear extracts were prepared and subjected to Myc immunoprecipitation, followed by western blot analysis using anti-FLAG antiserum. The FLAG-DNMT1 truncation mutants analyzed for interaction are denoted above each lane. The membranes were re-probed with Myc antiserum to verify Myc-CFP1 (aa 1–367) protein expression and Myc immunoprecipitation efficiency. The empty vector was transfected alone or with Myc-CFP1 (aa 1–367) into HEK-293 cells to serve as negative controls for the Myc immunoprecipitation. (C) A series of FLAG-DNMT1 deletion mutants encoding amino acids 1–489, 637–1102, or 970–1617 were tested for interaction with Myc-CFP1 (aa 1–367). FLAG-DNMT1 proteins were immunoprecipitated with Myc-conjugated agarose and subjected to western blot analysis with anti-FLAG antiserum. The FLAG-DNMT1 truncation mutants analyzed are denoted above each lane. The membranes were re-probed with Myc antiserum to verify Myc-CFP1 (aa 1–367) protein expression and Myc immunoprecipitation efficiency. The empty vector was cotransfected with FLAG-DNMT1 (aa 637–1102) or FLAG-DNMT1 (aa 970–1617) to serve as negative controls for the Myc immunoprecipitation.
It was also necessary to analyze the C-terminus of DNMT1 for possible interaction with CFP1. The highly conserved C-terminus of DNMT1 has previously been found to interact with chaperone 23 (p23) (Zhang and Verdine, 1996), the tumor suppressor p53 (Esteve et al., 2005), and the histone H3K27 methyltransferase, EZH2 (Vire et al., 2006). The highly conserved C-terminal catalytic domain contained within the FLAG-DNMT1 (aa 970–1617) protein fragment is sufficient to interact with Myc-CFP1 (aa 1–367) (Fig. 2C). Bromo-adjacent homology (BAH) domains are found in transcriptional regulatory proteins and often mediate protein–protein interactions (Callebaut et al., 1999). However, the BAH domains of DNMT1 are not sufficient for interaction with Myc-CFP1 (Fig. 2C), as the FLAG-DNMT1 (aa 637–1102) protein fragment fails to interact with Myc-CFP1 (aa 1–367). Lastly, central regions of DNMT1, including amino acids 392–661, 169–330, and 169–392, failed to interact with CFP1 (aa 1–367) (data not shown). These results demonstrate a complex pattern of interactions, in which either of two nonoverlapping domains of DNMT1 is sufficient for interaction with CFP1.
CFP1 interacts with DNMT1 through several conserved domains
CFP1 contains multiple highly conserved domains that contribute to CFP1 subcellular localization and function (Voo et al., 2000). For example, the acidic, basic, and coiled-coil domains are necessary and sufficient to target CFP1 to the nuclear matrix (Lee and Skalnik, 2002), while the CXXC domain is required for DNA binding (Lee et al., 2001). To determine if any of these conserved domains are required for the interaction with DNMT1, various CFP1 protein fragments were expressed in HEK-293 cells and tested for interaction with DNMT1. A schematic diagram representing the organization of conserved domains within the CFP1 protein is shown in Figure 3A. Truncations encoding Myc-tagged C-terminal deletion mutants of CFP1 were transiently expressed in HEK-293 cells along with FLAG-DNMT1 (aa 1–489). Immunoprecipitation was carried out using FLAG M2-conjugated agarose. Western blots probed with anti-Myc antiserum demonstrate that Myc-CFP1 (aa 1–123), which contains the PHD1 domain, is sufficient for interaction with FLAG-DNMT1 (Fig. 3B). The middle portion of CFP1 was also analyzed for interaction with DNMT1. Western blot analysis demonstrates that Myc-CFP1 (aa 103–367), which contains the CXXC, acidic, and basic domains, is sufficient for interaction with FLAG-DNMT1 (Fig. 3C). However, Myc-CFP1 (aa 213–367), which lacks the CXXC domain, fails to interact with FLAG-DNMT1, indicating that the acidic and basic domains of CFP1 are not sufficient for interaction with DNMT1 (Fig. 3C). Taken together, these results indicate that multiple regions within CFP1 are sufficient, but no individual domain is necessary, for interaction with DNMT1.
FIG. 3.
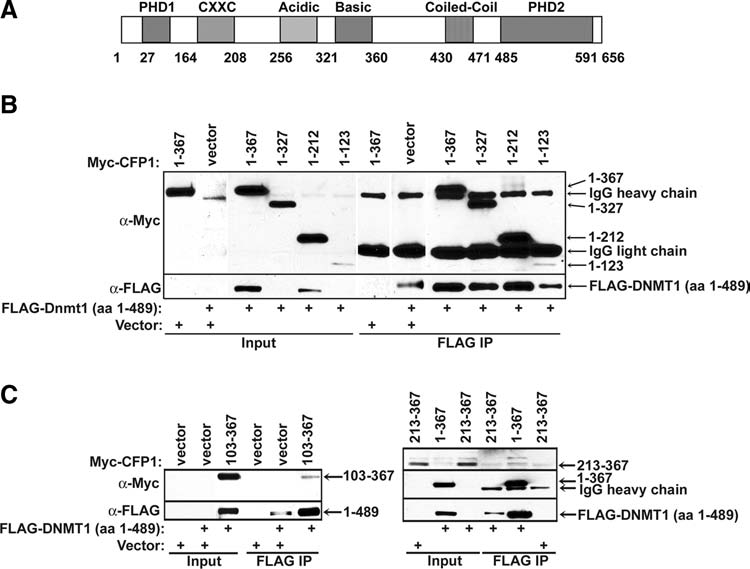
Identification of CFP1 domains that interact with DNMT1. (A) A schematic representation of conserved domains within CFP1. The amino acid position of each domain is denoted below the schematic. (B) A series of C-terminal Myc-CFP1 deletion mutants encoding amino acids 1–367, 1–327, 1–212, or 1–123 were coexpressed with FLAG-DNMT1 (aa 1–489) in HEK-293 cells. Nuclear extracts were prepared and subjected to FLAG immunoprecipitation. Western blot analysis was performed using anti-Myc antiserum. The membrane was re-probed with anti-FLAG antiserum to verify FLAG-DNMT1 (aa 1–489) protein expression and FLAG immunoprecipitation efficiency. (C) Myc-CFP1 truncation mutants encoding amino acids 103–367 or 213–367 were coexpressed with FLAG-DNMT1 (aa 1–489) in HEK-293 cells. Nuclear extracts were prepared and subjected to FLAG immunoprecipitation followed by western blot analysis with anti-Myc antiserum. Membranes were re-probed with anti-FLAG antiserum to verify FLAG-DNMT1 (aa 1–489) protein expression and FLAG immunoprecipitation efficiency. Empty vector was transfected alone or with FLAG-DNMT1 (aa 1–489) or Myc-CFP1 (aa 213–367) to serve as negative controls for the FLAG immunoprecipitation.
The carboxyl terminus of CFP1 mediates the interaction with the Setd1A and Setd1B histone methyltransferase complexes
CFP1 is a component of the Setd1A and Setd1B histone H3K4 methyltransferase complexes (Lee and Skalnik, 2005; Lee et al., 2007). The Setd1A and Setd1B complex–interacting region within CFP1 was mapped using truncation analysis to determine whether CFP1 interacts with the cytosine methylation machinery while associated with the Setd1 complexes. A series of FLAG-epitope–tagged CFP1 truncation mutants were expressed in HEK-293 cells. Nuclear extracts were prepared and subjected to immunoprecipitation using FLAG M2–conjugated agarose. Western blot analysis was then performed using antisera directed against components of the Setd1A and Setd1B histone methyltransferase complexes—Setd1A, Setd1B, Ash2, Rbbp5, Wdr5, and Wdr82. The Setd1A and Setd1B complexes consist of identical components with exception of the catalytic (Setd1) subunits (Lee and Skalnik, 2005; Lee et al., 2007). Coimmunoprecipitation studies revealed that a Setd1-interacting domain (SID) lies between the conserved basic and coiled-coil domains of CFP1 (Fig. 4A, B). The SID of CFP1 is required for CFP1 interaction with the Setd1A and Setd1B complexes, because the FLAG-CFP1 (aa 423–656) mutant that lacks this region fails to interact with any components of the Setd1A or Setd1B complexes (Fig. 4B). Further, the C-terminus of CFP1, which contains the PHD2 domain, is also required for interaction with the Setd1A and Setd1B complexes, because the FLAG-CFP1 (aa 1–481) mutant that contains the SID, but lacks the C-terminus, also fails to interact with any Setd1A or Setd1B complex components (Fig. 4). Finally, FLAG-CFP1 (aa 360–597) fails to interact with the Setd1 complexes, demonstrating that the SID and PHD2 domains are not sufficient for this interaction (Fig. 4). A multispecies alignment of CFP1 revealed several highly conserved amino acids within the SID and PHD2 domains (Fig. 4C). Single– or triple–amino acid substitutions were made within the SID or PHD2 domains in the context of full-length CFP1 to determine the functional significance of these amino acids in mediating the interaction between CFP1 and the Setd1A and Setd1B complexes. Full-length FLAG-CFP1 encoding single-alanine substitutions at cysteine 375 (C375A) within the SID, or 580 (C580A) within the PHD2 domain, or a triple mutant encoding alanine substitutions at tyrosine 390, cysteine 391, and serine 392 (YCS→AAA) within the SID all failed to interact with any of the components of the Setd1A and Setd1B complexes (Fig. 4D). These results indicate that the C-terminus of CFP1 is important for the interaction with the Setd1A and Setd1B complexes, and identify subtle mutations within CFP1 that ablate interaction with the Setd1 histone methyltransferase complexes.
FIG. 4.
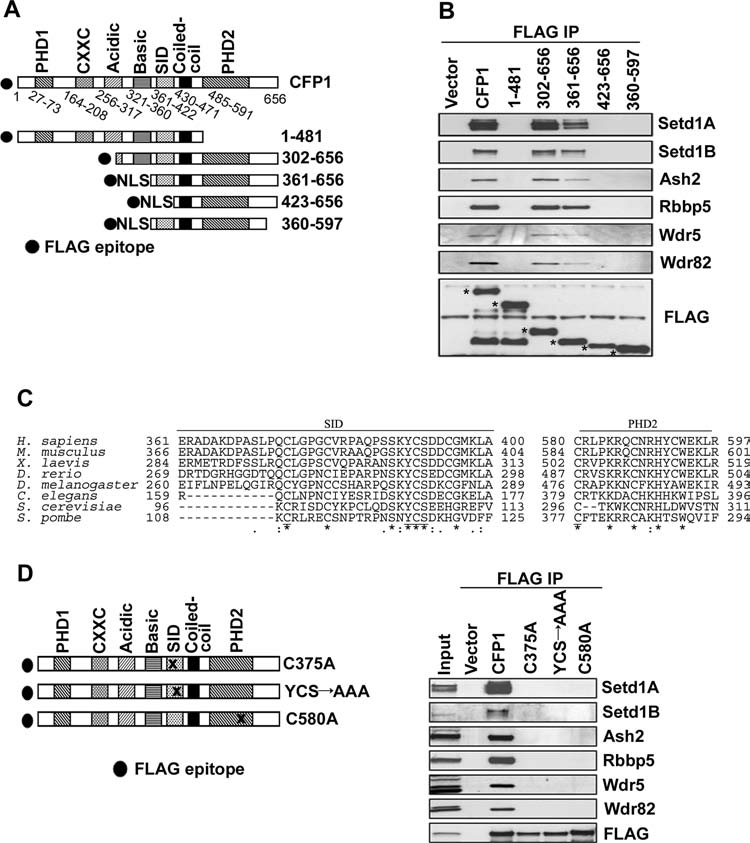
Several conserved amino acids within the C-terminus of CFP1 are required for interaction with the Setd1A and Setd1B histone methyltransferase complexes. (A) A schematic representation of CFP1 is shown. Various CFP1 protein fragments were generated and are shown in schematic form. The filled circle at the N-terminus of each CFP1 truncation mutant represents the FLAG epitope. NLS, nuclear localization signal derived from the CFP1 protein (Lee and Skalnik, 2002). (B) A series of FLAG-CFP1 truncation mutants were expressed in HEK-293 cells and analyzed for interaction with endogenous components of the Setd1A and Setd1B histone H3K4 methyltransferase complexes. The FLAG-CFP1 truncation mutants were immunoprecipitated from nuclear extracts using FLAG M2–conjugated agarose. Western blot analysis was performed using antisera directed against the following Setd1A and B complex components: Setd1A, Setd1B, Ash2, Rbbp5, Wdr5, and Wdr82. The membrane was re-probed with anti-FLAG antiserum to verify FLAG-CFP1 mutant protein expression and FLAG immunoprecipitation efficiency. Asterisks denote the position of each FLAG-tagged fusion protein. (C) A multispecies alignment of the C-terminus of CFP1 is shown. Asterisks mark identical amino acid residues conserved between species. A double filled circle marks a conserved amino acid substitution, while a single filled circle denotes a semi-conserved amino acid substitution. Amino acids chosen for site-directed mutagenesis are underlined. (D) Site-directed mutagenesis was performed to change cysteine 375 within the SID of CFP1 to alanine (C375A). A triple mutant within the SID of FLAG-CFP1 was generated that encoded the following substitutions: tyrosine 390 to alanine, cysteine 391 to alanine, and serine 392 to alanine (YCS→AAA). Finally, cysteine 580 within the PHD2 domain of CFP1 was mutated to alanine (C580A). The full-length FLAG-CFP1 mutants encoding the single– or triple–amino acid point mutations were expressed in HEK-293 cells and analyzed for interaction with the Setd1A and Setd1B complexes, as described above for (A).
CFP1 interaction with the Setd1A complex is not required for interaction with DNMT1
The C-terminus of CFP1 (aa 361–656) is necessary and sufficient for the interaction with the Setd1A and Setd1B complexes (Fig. 4B). To determine if the C-terminus of CFP1 is also sufficient for the interaction with DNMT1, the Myc-CFP1 (aa 361–656) truncation mutant was coexpressed with FLAG-DNMT1 (aa 1–489) in HEK-293 cells and subjected to FLAG immunoprecipitation analysis. Myc-CFP1 (aa 361–656) interacts with FLAG-DNMT1 (aa 1–489) (Fig. 5), thus revealing another DNMT1 interaction domain with CFP1. Importantly, introduction of the C375A point mutation in Myc-CFP1 (aa 361–656), which ablates interaction with the Setd1 histone methyltransferase complexes (Fig. 4D), does not disrupt the interaction with FLAG-DNMT1 (Fig. 5). Thus, CFP1 association with the Setd1A and Setd1B complexes is not required for its interaction with DNMT1.
FIG. 5.
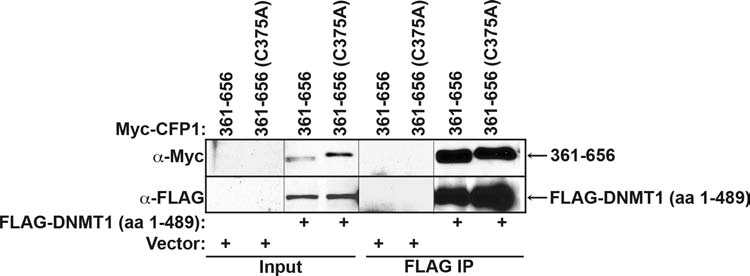
Interaction with Setd1A or Setd1B is not required for CFP1 interaction with DNMT1. FLAG-DNMT1 (aa 1–489) was coexpressed with Myc-CFP1 (aa 361–656) or Myc-CFP1 (aa 361–656; C375A) in HEK-293 cells. Nuclear extracts were prepared, and FLAG M2–conjugated agarose was used to immunoprecipitate FLAG-DNMT1 (aa 1–489). Western blot analysis using anti-Myc antiserum was used to detect Myc-CFP1 (aa 361–656) or Myc-CFP1 (aa 361–656; C375A). Membranes were re-probed with anti-FLAG antiserum to verify FLAG-DNMT1 (aa 1–489) protein expression and FLAG immunoprecipitation efficiency. Transfection and immunoprecipitation of Myc-CFP1 (aa 361–656) or Myc-CFP1 (aa 361–656; C375A) with empty vector served as negative controls for the FLAG immunoprecipitation.
Discussion
The results reported in this study demonstrate that CFP1 and DNMT1 physically interact in vivo. Immunoprecipitation analysis of Myc-CFP1 protein fragments revealed three regions, amino acids 1–123, 103–367, and 361–656, that are sufficient for the interaction with FLAG-DNMT1. These regions all contain conserved domains involved in Zn2+ binding and/or targeting to euchromatic regions of the genome—namely, the PHD1, CXXC, and PHD2 domains. The CXXC domain of CFP1 is the sole DNA-binding domain within the protein and specifically recognizes unmethylated CpG dinucleotides (Voo et al., 2000; Lee et al., 2001). PHDs within several chromatin-associated proteins have recently been identified as binding modules for methylated histone H3K4 or histone H3K36 (Martin et al., 2006; Shi et al., 2006; Taverna et al., 2006; Shi et al., 2007). Further, the PHD within the yeast homolog of CFP1, Spp1, binds histone H3K4me2/3 (Shi et al., 2007). It will be of interest to determine if the PHDs within mammalian CFP1 bind methylated histone H3K4. Importantly, a point mutation (C375A) that abolishes the interaction between CFP1 and the Setd1A and Setd1B complexes does not ablate the interaction between CFP1 and DNMT1, thus illustrating that CFP1 interacts independently with enzymes responsible for cytosine methylation and histone methylation. These results are consistent with previously published studies that failed to detect DNMT1 interaction with either the Setd1A or Setd1B methyltransferase complexes (Lee and Skalnik, 2005; Lee et al., 2007).
Immunoprecipitation analysis of truncated FLAG-DNMT1 proteins revealed two regions (amino acids 169–493 and 970–1617) sufficient for interaction with Myc-CFP1. A similar result was recently observed for the interaction between DNMT1 and the EZH2 histone H3K27 methyltransferase (Vire et al., 2006). Two regions within the N-terminus of DNMT1, amino acids 1–343 and 305–609, as well as the C-terminal region encoding amino acids 1124–1620 are each sufficient for interaction with EZH2 (Vire et al., 2006). The C-terminus of DNMT1 requires interaction with the N-terminus for catalytic activity (Fatemi et al., 2001; Margot et al., 2003). Perhaps CFP1 binding to both regions stabilizes the intramolecular interaction within DNMT1 to enhance enzymatic activity.
Direct interaction between CFP1 and DNMT1 fragments was not observed using an in vitro histidine pull-down assay (data not shown). However, these studies utilized protein fragments isolated from Escherichia coli. Therefore, the interaction would not have occurred if posttranslational modifications of DNMT1 or CFP1 play a role in mediating their interaction. DNMT1 interacts with a number of chromatin-associated proteins (Fuks et al., 2000, 2003; Robertson et al., 2000; Rountree et al., 2000; Tatematsu et al., 2000; Kim et al., 2002; Kimura and Shiota, 2003; Esteve et al., 2006; Vire et al., 2006; Smallwood et al., 2007), and CFP1 is a component of the ∼450 kDa Setd1A and Setd1B complexes (Lee and Skalnik, 2005; Lee et al., 2007). Therefore, it is also possible that additional molecules or proteins mediate the interaction between DNMT1 and CFP1.
While cytosine methylation is critical for embryonic development and survival, the differentiation and survival defects observed in ES cells lacking CFP1 are more severe than those observed in cells deficient solely in global cytosine methylation. Mouse embryos lacking DNMT1 exhibit a 70% reduction in global cytosine methylation and exhibit embryonic lethality between embryonic days 9.5–11 (Li et al., 1992). CFP1-deficient ES cells exhibit a 70% reduction in global cytosine methylation, yet mouse embryos lacking CFP1 die prior to implantation (E4.5–6.5) (Carlone and Skalnik, 2001; Carlone et al., 2005). ES cells lacking CFP1 exhibit a threefold increase in apoptosis in culture, whereas DNMT1−/− ES cells do not undergo apoptosis until induction of differentiation (Lei et al., 1996). Lastly, DNMT1+/− ES cells express DNMT1 at 50% of wild-type levels, similar to what is observed in ES cells lacking CFP1, yet do not exhibit a deficiency in global cytosine methylation or methyltransferase activity (Li et al., 1992, 1996). These observations suggest that a mechanism in addition to a 50% reduction in DNMT1 protein levels contributes to the epigenetic and differentiation defects observed in ES cells lacking CFP1.
DNMT1 maintenance methylation activity at the DNA replication fork is well characterized. However, the kinetics of the DNMT1 methylation reaction are too slow to efficiently methylate all sites while moving at the speed of the replication fork (Pradhan et al., 1999; Easwaran et al., 2004). DNMT1 additionally methylates nucleosomal DNA (Okuwaki and Verreault, 2004), and maintenance methyltransferase activity of DNMT1 is independent of its association with PCNA (Spada et al., 2007). These results imply that additional mechanisms exist to target DNMT1 to chromatin independently of protein interactions at the DNA replication fork. The TS region of DNMT1 (amino acids 324–628) is responsible for targeting DNMT1 to chromatin independent of DNA replication (Easwaran et al., 2004). A portion of the DNMT1 N-terminal CFP1-interacting region, amino acids 169–493, is contained within the TS region. This raises the possibility that CFP1 could be involved in targeting DNMT1 to chromatin independently of DNA replication. It will be important to identify CFP1 target genes and determine if DNMT1 cooccupies these loci.
Why does CFP1, a protein associated with euchromatic regions of the genome, interact with DNMT1, a protein involved in modifying heterochromatic regions? It is possible that the interaction between these proteins occurs at times of transition from euchromatin to heterochromatin states. For example, after transcription the open chromatin template requires repackaging into a transcriptionally inactive chromatin context to suppress aberrant transcription reinitiation (Kaplan et al., 2003; Mason and Struhl, 2003; Carrozza et al., 2005). The presence of DNA methylation at the junction of transcription initiation and coding regions is sufficient for exclusion of histone H3K4me2, and may play a role in protecting transcriptionally repressed regions from aberrant transcription initiation (Okitsu and Hsieh, 2007). An alternative explanation would be a mechanism to specifically silence euchromatic genes. For example, the PHD contained in the inhibitor of growth 2 protein binds histone H3K4me3 within the cyclin D1 and c-Myc promoters and directly leads to transcriptional repression via recruitment of the mSin3a–HDAC1 complex upon DNA damage (Shi et al., 2006). Perhaps CFP1 recruits DNMT1 to specific euchromatic targets in a similar manner to initiate specific gene repression.
A recent study provides evidence that DNA methylation by DNMT1 is required for G9a and HP1 recruitment in order to initiate heterochromatin formation required to silence euchromatic genes (Smallwood et al., 2007). Perhaps decreased heterochromatin formation via reduced cytosine methylation, combined with increased histone H3K4 methylation activity of the Setd1A and Setd1B complexes, results in the global euchromatin architecture observed in CFP1-deficient ES cells. These observations suggest that the function of CFP1 within the Setd1A and Setd1B histone H3K4 methyltransferase complexes in conjunction with its role in regulating cytosine methylation may explain the differentiation and survival defects in CFP1-deficient ES cells.
Acknowledgments
This work was supported by the Riley Children's Foundation, the Lilly Endowment, a Showalter Trust award (to J.-H.L.), and National Science Foundation Grants MCB-0344870 and MCB-0641851 (to D.G.S.). J.S.B. was supported by a predoctoral fellowship from National Institutes of Health Grant T32 CA111198. We thank Dr. Sriharsa Pradhan (New England Biolabs) for generously providing the human DNMT1 cDNA.
References
- Burgers W.A. Fuks F. Kouzarides T. DNA methyltransferases get connected to chromatin. Trends Genet. 2002;18:275–277. doi: 10.1016/S0168-9525(02)02667-7. [DOI] [PubMed] [Google Scholar]
- Callebaut I. Courvalin J.-C. Mornon J.-P. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication, and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- Carlone D.L. Lee J.-H. Young S.R.L. Dobrota E. Butler J.S. Ruiz J. Skalnik D.G. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol Cell Biol. 2005;25:4881–4891. doi: 10.1128/MCB.25.12.4881-4891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlone D.L. Skalnik D.G. CpG binding protein is crucial for early embryonic development. Mol Cell Biol. 2001;21:7601–7606. doi: 10.1128/MCB.21.22.7601-7606.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M.J. Li B. Florens L. Suganuma T. Swanson S.K. Lee K.K. Shia W.J. Anderson S. Yates J. Washburn M.P. Workman J.L. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Dennis K. Fan T. Geiman T. Yan Q. Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H.P. Schermelleh L. Leonhardt H. Cardoso M.C. Replication-independent chromatin loading of DNMT1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J. Ballestar E. Fraga M.F. Villar-Garea A. Juarranz A. Stockert J.C. Robertson K.D. Fuks F. Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- Esteve P.-O. Chin H.G. Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci USA. 2005;102:1000–1005. doi: 10.1073/pnas.0407729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P.-O. Chin H.G. Smallwood A. Feehery G.R. Gangisetty O. Karpf A.R. Carey M.F. Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi M. Hermann A. Pradhan S. Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- Freitag M. Hickey P.C. Khlafallah T.K. Read N.D. Selker E.U. HP1 is essential for DNA methylation in Neurospora. Mol Cell. 2004;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- Fuks F. Burgers W.A. Brehm A. Hughes-Davies L. Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F. Burgers W.A. Godin N. Kasai M. Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F. Hurd P.J. Deplus R. Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A. Bender J. Richards E.J. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A. Stokes T.L. Richards E.J. Maintenance of genomic methylation requires a SWI2/SNF-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Kaplan C.D. Laprade L. Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;310:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kim G.-D. Ni J. Kelesoglu N. Roberts R.J. Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. Shiota K. Methyl-CpG binding protein, MeCP1, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- Lee J.-H. Shimojo M. Chai Y.-G. Hersh L.B. Studies on the interaction of REST4 with the cholinergic repressor element-1/neuron restrictive silencer element. Mol Brain Res. 2000;80:88–98. doi: 10.1016/s0169-328x(00)00129-7. [DOI] [PubMed] [Google Scholar]
- Lee J.-H. Skalnik D.G. CpG binding protein is a nuclear matrix- and euchromatin-associated protein localized to nuclear speckles containing human trithorax: identification of nuclear matrix targeting signals. J Biol Chem. 2002;277:42259–42267. doi: 10.1074/jbc.M205054200. [DOI] [PubMed] [Google Scholar]
- Lee J.-H. Skalnik D.G. CpG binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lee J.-H. Tate C.M. You J.-S. Skalnik D.G. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- Lee J.-H. Voo K.S. Skalnik D.G. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- Lehnertz B. Ueda Y. Derijck A.A.H.A. Braunschweig U. Perez-Burgos L. Kubicek S. Chen T. Li E. Jenuwein T. Peters A. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lei H. Oh S.P. Okano M. Juttermann R. Goss K.A. Jaenisch R. Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E. Bestor T.H. Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Margot J.G. Ehrenhofer-Murray A.E. Leonhardt H. Interactions within the mammalian DNA methyltransferase family. BMC Mol Biol. 2003;4:7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.G. Baetx K. Shi X. Walter K.L. MacDonald V.E. Wlodarski M.J. Gozani O. Hieter P. Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.B. Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. Krogan N.J. Dover J. Erdjument-Bromage H. Tempst P. Johnston M. Greenblatt J.F. Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.L. Griesenbeck J. Kornberg R.D. Cleary M.L. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu C.Y. Hsieh C.-L. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M. Verreault A. Maintenance DNA methylation of nucleosome core particles. J Biol Chem. 2004;279:2904–2912. doi: 10.1074/jbc.M310111200. [DOI] [PubMed] [Google Scholar]
- Pradhan S. Bacolla A. Wells R.D. Roberts R.J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- Robertson K.D. Ait-Si-Ali S. Yokochi T. Wade P.A. Jones P.L. Wolffe A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Roguev A. Schaft D. Shevchenko A. Aasland R. Shevchenko A. Stewart A.F. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J Biol Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- Rountree M.R. Bachman K.E. Baylin S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Schneider J. Wood A. Lee J.S. Schuster R. Dueker J. Maguire C. Swanson S.K. Florens L. Washburn M.P. Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Selker E.U. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc Natl Acad Sci USA. 1998;95:9430–9435. doi: 10.1073/pnas.95.16.9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. Hong T. Walter K.L. Ewalt M. Michishita E. Hung T. Carney D. Pena P. Lan F. Kaadige M.R. Lacoste N. Cayrou C. Davrazou F. Saha A. Cairns B.R. Ayer D.E. Kutateladze T.G. Shi Y. Cote J. Chua K.F. Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. Kachirskaia I. Walter K.L. Kui J.-H.A. Lake A. Davrazou F. Chan S.M. Martin D.G.E. Fingerman I.M. Briggs S.D. Howe L. Utz P.J. Kutateladze T.G. Lugovskoy A.A. Bedford M.T. Gozani O. Proteome-wide analysis in S. cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A. Esteve P.-O. Pradhan S. Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada F. Haemmer A. Kuch D. Rothbauer U. Schermelleh L. Kremmer E. Carell T. Langst G. Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H. Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamaru H. Zhang X. McMillen D. Singh P.B. Nakayama J.I. Grewal S.I. Allis C.D. Cheng X. Selker E.U. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Tatematsu K.-I. Yamazaki T. Ishikawa F. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication fork in late S phase. Genes Cells. 2000;5:677–688. doi: 10.1046/j.1365-2443.2000.00359.x. [DOI] [PubMed] [Google Scholar]
- Taverna S.D. Ilin S. Rogers R.S. Tanny J.C. Lavender H. Li H. Baker L. Boyle J. Blair L.P. Chait B.T. Patel D.J. Aitchison J.D. Tackett A.J. Allis C.D. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E. Brenner C. Deplus R. Blanchon L. Fraga M.F. Didelot C. Morey L. van Eynde A. Bernard D. Vanderwinden J.M. Bollen M. Esteller M. DiCroce L. de Launoit Y. Fuks F. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Voo K.S. Carlone D.L. Jacobsen B.M. Flodin A. Skalnik D.G. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J. Swigut T. Xiao H. Milne T.A. Kwon S.Y. Landry J. Kauer M. Tackett A.J. Chait B.T. Badenhorst P. Wu C.-L. Allis C.D. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Young S.R.L. Mumaw C. Marrs J.A. Skalnik D.G. Antisense targeting of CXXC finger protein 1 inhibits genomic cytosine methylation and primitive hematopoiesis in zebrafish. J Biol Chem. 2006;281:37034–37044. doi: 10.1074/jbc.M604546200. [DOI] [PubMed] [Google Scholar]
- Young S.R.L. Skalnik D.G. CXXC-finger protein 1 is required for normal proliferation and differentiation of the PLB-985 myeloid cell line. DNA Cell Biol. 2007;26:80–90. doi: 10.1089/dna.2006.0535. [DOI] [PubMed] [Google Scholar]
- Zhang X. Verdine G.L. Mammalian DNA cytosine-5-methyltransferase interacts with p23 protein. FEBS Lett. 1996;392:179–183. doi: 10.1016/0014-5793(96)00810-1. [DOI] [PubMed] [Google Scholar]


