Abstract
We describe a new magic angle spinning (MAS) NMR experiment for obtaining 15N-15N correlation spectra. The approach yields direct information about the secondary and tertiary structure of proteins, including identification of α-helical stretches and inter-strand connectivity in antiparallel β-sheets, which are of major interest for structural studies of membrane proteins and amyloid fibrils. The method, 15N-15N proton assisted recoupling (PAR), relies on a second order mechanism, third spin assisted recoupling (TSAR), used previously in the context of 15N-13C and 13C-13C polarization transfer schemes. In comparison to 15N-15N proton driven spin diffusion experiments, the PAR technique accelerates polarization transfer between 15N’s by a factor of ~102−103, and is furthermore applicable over the entire range of currently available MAS frequencies (10–70 kHz).
Keywords: recoupling, MAS, SSNMR, solid-state NMR, proteins
1. Introduction
Magic angle spinning (MAS)1 NMR has emerged as the preferred approach for performing detailed studies of the structure and dynamics of insoluble biological systems and systems lacking long range order that are currently not accessible by x-ray diffraction or solution NMR. Specifically, MAS experiments are used to investigate protein folding and misfolding, amyloid aggregation, signal transduction, and molecular transport across biomembranes to name a few of the areas of current research2–13
A number of developments have contributed to the evolving methodology to determine protein structures via MAS NMR. These include access to high field magnets (>15T), improved sample preparation protocols,14 selective isotopic labeling schemes,15–18 adaptation of computational protocols for structure calculations11,19–22 and new methods for assigning spectra and for measuring distances and torsion angles23–43. At present, resonance assignments and structural studies in the solid state rely mainly on multidimensional 13C-13C and 15N-13C-(13C) correlation experiments. In addition, 15N-15N correlation spectra, which were first reported by Reif, et al. almost a decade ago,44 are a valuable tool for estimating 15N-15N distances45 and for measuring the NHi-NHi+1 projection angle θi,,i+1,44,46 To date, however, these experiments have been limited to B0 < 11–13 T and ωr/2π < 12 kHz and therefore have not achieved their full potential.
In this paper, we show that 15N-15N correlation spectroscopy can be extended to MAS frequencies >15 kHz and to magnetic fields >20 T using the 15N-15N proton assisted recoupling (PAR) technique29 that was recently introduced in the context of 13C-13C and 13C-15N recoupling and which relies on a more general third spin assisted recoupling (TSAR) mechanism.29,41
We apply the 15N-15N PAR pulse sequence (see Fig. 1) to a model tripeptide N-f-MLF-OH and to the 56-residue microcrystalline β1 immunoglobulin binding domain of protein G (GB1). The mixing time required for observing structurally relevant 15N-15N contacts (~2.8–4.5 A) in the PAR experiment corresponds to tens of milliseconds, improving on spin-diffusion based techniques (PDSD47, DARR39) by two to three orders of magnitude. In addition, the observed cross peak intensities can be related to the topology of the 15N-15N network in a straightforward manner, thus allowing protein secondary and tertiary structure to be clearly established.
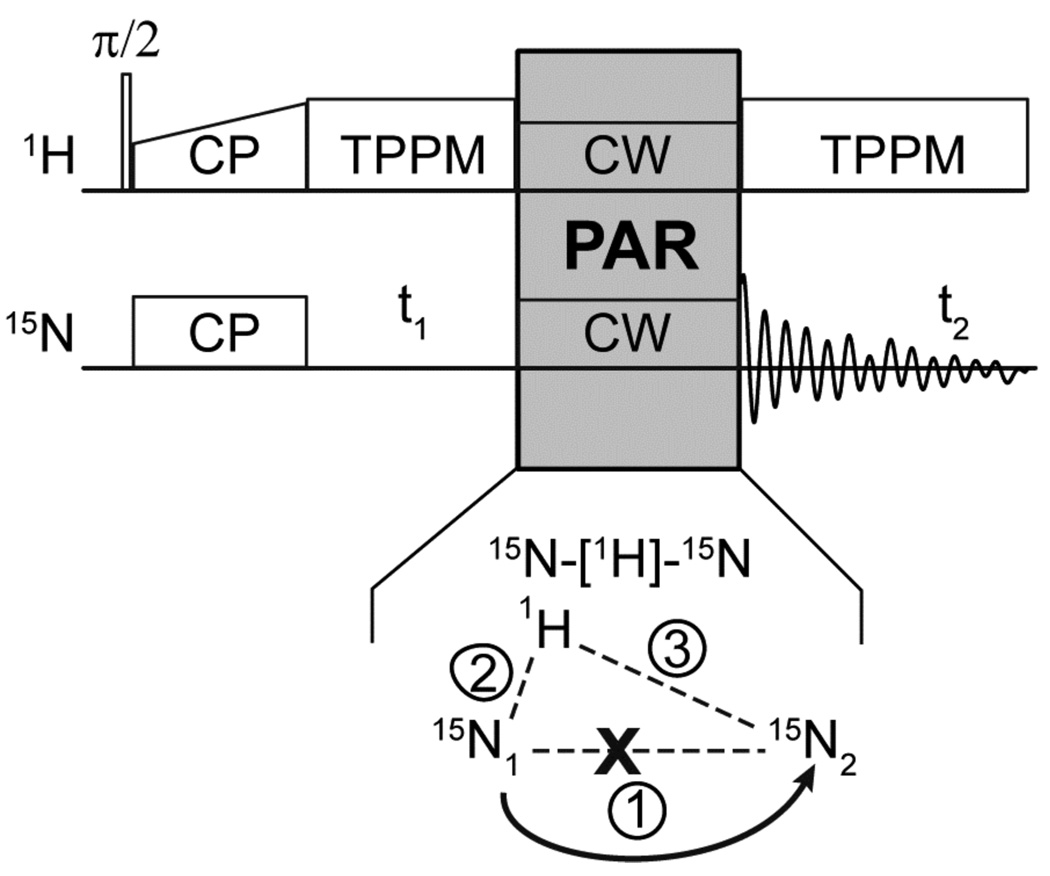
Figure 1.
Pulse sequence for the 2D 15N-15N PAR correlation experiment. The PAR mixing period consists of C.W. irradiations on the 1H and 15N channels. The irradiation strengths are chosen to produce an appreciable second order TSAR mechanism between the 1H-15N1 and 1H-15N2 dipolar couplings (terms 2 and 3 in the spin system graphics), resulting in TSAR terms of the form .
2. 15N-15N correlation spectroscopy
Despite its low gyromagnetic ratio, 15N has been a valuable nucleus for biomolecular MAS SSNMR studies. Metabolic sources of 15N are relatively inexpensive, allowing one, for example, to prepare uniformly 15N labeled proteins to screen sample preparation conditions.14 In addition, an 15N dimension is often incorporated into advanced multidimensional NMR experiments. 15N and 13C labeled samples are routinely used for sequential resonance assignments,25,41,48–61 for measuring torsion angles,62–65 extracting accurate 15N-13C distances,31,32,46,66–72 and finally for locally probing protein backbone dynamics.73–76
The two main challenges for 15N-15N correlation spectroscopy in the solid state have been (1) the poor sensitivity of 15N observed experiments and (2) the relatively restricted range of available methods for transferring magnetization among 15N nuclei. The first issue is currently being addressed by the development of high field dynamic nuclear polarization (DNP)77,78, and the combination of spinning frequencies up to ~70 kHz together with 1H detected experiments.79 The second issue mentioned above is directly related to the small magnitude of 15N-15N couplings, which currently prevents the wide use of advanced first order recoupling techniques developed for 13C-13C polarization transfer and restricts acquisition of 15N-15N correlation experiments primarily to proton driven spin diffusion (PDSD) based experiments.17,46,74,80,81
Although 15N-15N PDSD experiments are relatively straightforward to perform, they are far from ideal for biomolecular systems requiring high resolution conditions available at high magnetic field strengths (B0 > 16 T) and MAS frequencies (ωr/2π > 20 kHz). Such operating conditions require long mixing times which reduces the polarization transfer efficiency (due to the competition with the relaxation), and, more importantly, complicates the interpretation of the 15N-15N polarization transfer buildups in terms of distance restraints.45
3. 15N-[1H]-15N TSAR – 15N-15N PAR experiments
3.1 TSAR mechanism principles
The PAR pulse sequence was recently introduced in the context of 13C-13C recoupling.29 Its underlying mechanism relies on a second-order recoupling process referred to as third spin assisted recoupling (TSAR) that was used to develop the heteronuclear PAINCP41 (proton assisted insensitive nuclei cross polarization) experiment and has lead to an understanding of the beneficial effect of applying a small (< 0.25 ωr) 1H irradiation field to improve the double quantum transfer efficiency of CMpRR (where p ranges from 3.5 to 5).30 The TSAR mechanism, denoted as B-[A]-C, relies on three spin operators that connect spins B and C via a cross term involving dipolar couplings with a third assisting spin A (B-A and C-A dipolar couplings, respectively). In the experiment described here, the 15N-15N PAR pulse sequence relies on a 15N-[1H]-15N TSAR mechanism based on cross terms involving heteronuclear 1H-15N1 and 1H-15N2 dipolar couplings (see inset of Fig. 1) to induce polarization transfer between the nitrogen nuclei. As pointed out in our previous work,29,41 the polarization transfer does not rely on the BC coupling (15N-15N in the experiments described here).
3.2 PAR pulse sequence and effective Hamiltonian
The 15N-15N PAR pulse sequence is illustrated in Fig. 1 and consists of simultaneous C.W. irradiation on the 1H and 15N channels.
The spin dynamics during the TSAR mixing period can be described by the following Hamiltonian:
| (1) |
where denote the shift tensors and resonant offsets of the 15N and 1H nuclei respectively, and the homonuclear and heteronuclear dipolar couplings. The last two terms in Eq. (1) denote the rf fields applied at the 15N and 1H frequencies, respectively. Note that MAS induces a time dependence of the spatial anisotropy of the interactions.
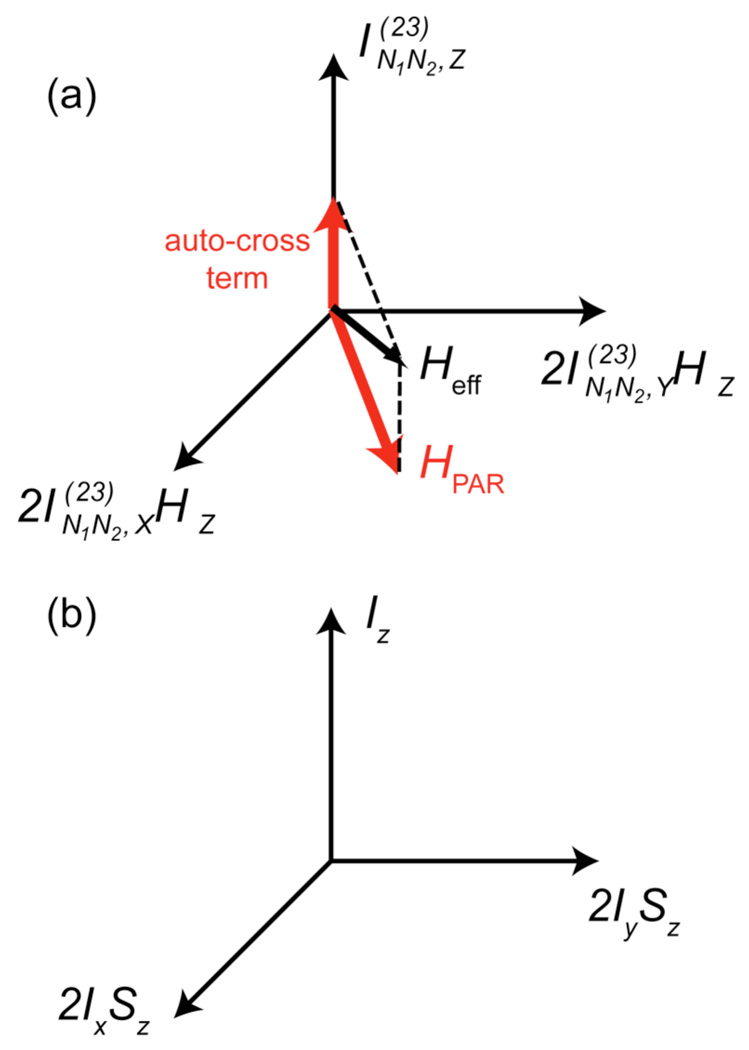
As described in detail in De Paëpe, et al.,29 an effective TSAR Hamiltonian can be derived in the interaction frame described by the two C.W. rf fields of strength ω1N/2π and ω1H/2π for the 15N and 1H channels. The TSAR subspace (see Fig. 2) associated with the polarization transfer is defined by the following operators:, which represent a coupled basis between a fictitious ZQ spin (associated with spins N1 and N2) and a proton spin H. The TSAR cross term resulting from terms 2 and 3 (1H-15N1 and 1H-15N2) in Eq. (1) can be written in the transverse plane defined by the operators and , and leads to polarization transfer between N1 and N2. The other important contribution to the spin dynamics comes from autocross terms created by term 2 with itself (i.e. 1H-15N1 cross 1H-15N1) and term 3 with itself (i.e. 1H-15N1 cross 1H-15N1) respectively. These autocross terms produce an off-resonance contribution along the operator in the TSAR subspace, which leads to a tilting of the effective recoupling axis and reduces the TSAR polarization transfer efficiency. Note that similar longitudinal terms also arise from autocross terms involving the chemical shift tensor with itself.29
Figure 2.
Visualization of the PAR subspace. The space can be seen as a coupled basis between a fictitious ZQ operator involving the two carbons (or nitrogens) and a proton spin. The red arrows indicate PAR recoupling axis and longitudinal tilting field resulting from autocross terms. Panel (b) depicts the coupled basis encountered in solution NMR.
3.3 PAR pulse sequence optimization
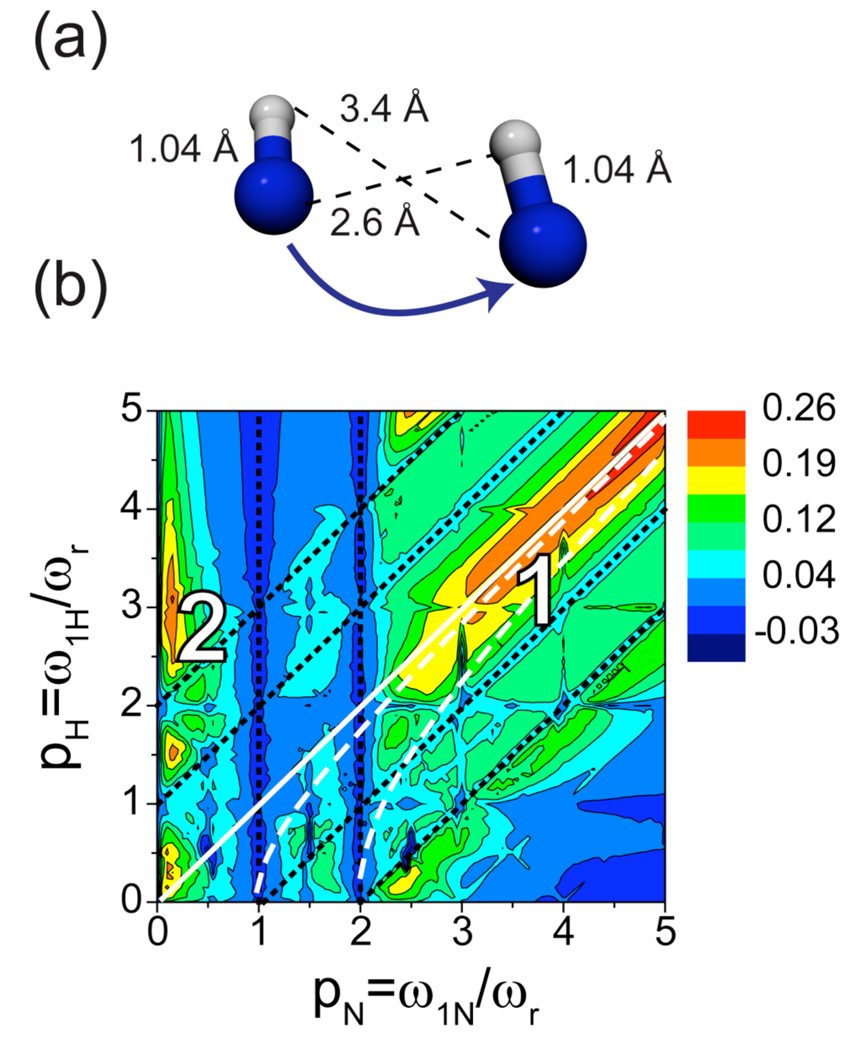
Figure 3b represents a contour plot of the 15N-15N PAR polarization transfer efficiency as a function of the 15N/1H rf field strength in units of the spinning frequency (pN or pH) for a fixed mixing time of 20 ms. The numerical simulations were performed for ω0H/2π= 750 MHz and ωr/2π= 20 kHz with the spin system shown in Fig. 3a (corresponding to backbone nitrogens from neighboring residues in an a-helix with the directly attached protons) and include chemical shifts (the atomic coordinates and chemical shift tensors used in the simulations may be found in Table SI1).
Figure 3.
Numerical simulation of a 15N-15N PAR polarization transfer map for backbone nitrogens in an α-helix. (a) Spin system used in the simulation consisting of the two backbone nitrogens with directly bonded amide protons (see Table SI1). Simulations were performed at ωr/2π=20 kHz and ω0H/2π=750 MHz using 20 ms mixing and include typical isotropic and anisotropic chemical shifts (see Table SI1). (b) Contour plot of the 15N-15N PAR polarization transfer between neighboring nitrogens in an α-helix as a function of the nitrogen and proton irradiation magnitudes in units of spinning frequency: pN and pH. The two main areas used for performing 15N-15N PAR experiments are indicated with numerals 1 and 2. The dashed magenta lines indicate conditions for which the m=1 and m=2 components of the auto cross-term arising from the heteronuclear 15N-1H dipolar coupling are zero respectively. These lines are defined by the following equations: and .
The optimization map in Fig. 3b displays typical features of PAR polarization transfer.29 15N-[1H]-15N TSAR polarization transfer is appreciable for settings that avoid first order recoupling conditions such as 15N rotary resonance (i.e. pN = 1, 2) and 1H-15N Hartmann-Hahn conditions (black dotted lines). Indeed, in these cases the 15N-[1H]-15N TSAR polarization transfer is absent either because of 15N CSA recoupling or because the 15N magnetization is transferred to 1H’s.
The two main regions that lead to appreciable 15N-15N polarization transfer are marked on the map with numbers. Area 1 is located under the pH=pN condition (white solid line) for pN > 2 and area 2 corresponds to settings where pN < 1 and pH > 2. Note that the first of the above conditions leads to more broadband recoupling than the second area as it employs a higher 15N rf field strength. These favorable settings correspond to conditions where the transverse TSAR term dominates the off-resonance longitudinal term originating from autocross terms. More precisely each autocross terms is the sum of two contributions involving the m=1 and the m=2 components of the heteronuclear 15N-1H dipolar interactions associated with the frequencies ωr and 2ωr, respectively. The two white dashed lines displayed in Fig. 3 represent rf settings where each of these contributions is zero.29 These lines are defined by the following equations: and . The contribution to the autocross terms arising from the m=1 component has a higher scaling factor which explains why one set of the optimal rf settings for the TSAR transfer are found along the lines.
4. Experimental PAR experiments: application to peptide and protein
4.1 15N-15N PAR on N-[U-13C,15N]-/-MLF-OH
Figure 4a shows a 2D 15N-15N PAR correlation spectra obtained at ω0H/2π = 900 MHz on the tripeptide N-[U-13C,15N]-f-MLF-OH using the rf power levels corresponding to area 2 -- ω1N/2π ~ 4 kHz and ω1H/2π~ 53 kHz -- with ωr/2π = 20 kHz and τmix = 20 ms. Note that the low 15N rf power is sufficient to cover the backbone nitrogen bandwidth (~2.7 kHz at ω0H/2π = 900 MHz). Such low power rf settings minimize the rf sample heating, reducing the danger of compromising the sample integrity during the experiment because of rf heating.
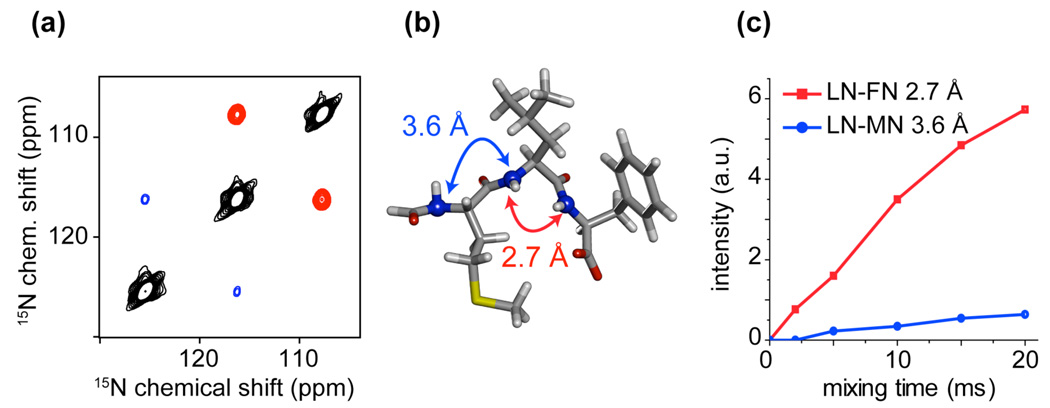
Figure 4.
(a) Low power 2D 15N-15N PAR correlation spectrum obtained on [U-13C,15N]-f-MLF-OH72 at ωr/2π = 20 kHz and ω0H/2π = 900 MHz using 20 ms of mixing time. The red cross-peaks correspond to a short LN-FN sequential contact (rNN = 2.7 Å, ~10 % efficiency at 20 ms) and the blue cross-peaks correspond to the long sequential LN-MN contact (rNN = 3.6 Å, ~5% efficiency at 20 ms) (see graphics (b)). (c) Cross-peak intensity build-ups in [U-13C,15N]-N-f-MLF-OH as a function of 15N-15N PAR mixing time. The PAR mixing consisted of ~ 4 kHz 15N and ~ 53 kHz 1H C.W. irradiations for both (a) and (c).
At 20 ms mixing time, the spectrum displays two sequential contacts in the tripeptide N-f-MLF-OH corresponding to the 15N-15N distances of 2.7 Å and 3.6 Å respectively.72 Although the involved 15N-[1H]-15N TSAR recoupling mechanism does not rely on the 15N-15N couplings and thus does not directly rely on the 15N-15N distances,29 the strongest cross-peak corresponds to the shortest 15N-15N distance. This is illustrated in Fig. 4c that shows the polarization transfer (under the TSAR settings mentioned above) as a function of the mixing time. In this case, the sequential transfer appears “indirectly” sensitive to the 15N-15N distances since the corresponding PAR couplings are proportional to the sequential 15N-1H distances.
N-f-MLF-OH is a well-suited model system for testing typical 15N-15N sequential spin topologies present in proteins. The LN-FN topology is similar to that encountered for neighboring residues in α-helices (~ 2.8 Å 15N-15N, ~ 2.4 Å and ~2.8 Å 1H-15N distances). On the other hand, the MN-LN arrangement corresponds to neighboring residues in β-sheets (~ 3.5 Å 15N-15N, ~ 3.4 Å and ~3.9 Å 1H-15N distances). In Fig. 4 we can clearly distinguish between these two different topologies simply on the basis of the cross-peak intensity.
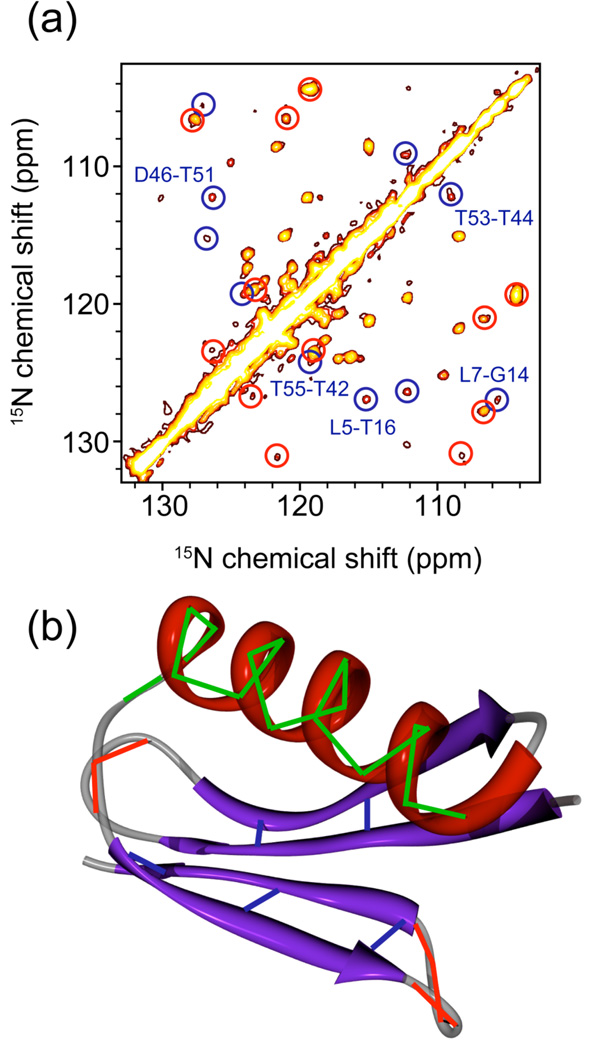
4.2 15N-15N PAR on microcrystalline protein GB1
Figure 5 shows a 2D 15N-15N PAR correlation spectrum on [U-15N,1,3-13C] protein GB1 obtained at ω0H/2π= 900 MHz and ωr/2π= 20 kHz using 18 ms mixing with ω1H/2π ~ 49 kHz and ω1N/2π ~ 52 kHz (see Fig. SI1 for the spectrum obtained using 22 ms mixing with ω1N/2π ~ 4 kHz 15N and ω1H/2π ~ 55 kHz ω0H/2π= 900 MHz and ωr/2π= 20 kHz and Fig. SI3 for spectrum obtained using 20 ms mixing with ω1N/2π ~ 71 kHz and ω1H/2π ~ 69 kHz at ω0H/2π= 500 MHz and ωr/2π= 11 kHz). With this mixing time the spectrum contains two important categories of cross peaks corresponding to the strongest PAR couplings that are well above the noise level (see the cross-peak list in Table SI4). The first contains short (≤ 3.2 Å) sequential 15N-15N contacts (see Table I), which are primarily observed in α-helical regions and occasionally in loops and turns. The second category consists of 15N-15N contacts between residues participating in β-bridges involving antiparallel β-sheets (see Table I). Note that for these particular settings the sequential cross-peaks in the β-sheets are generally weak or below the noise level since the corresponding PAR couplings are not favorable. Indeed the sequential 1H to 15N distances in β-sheets correspond to ~3.8 – 4.1 Å whereas the inter-strand 1H to 15N distances in antiparallel β-sheets are generally smaller (~3.3 – 3.7 Å). Observation of sequential cross peaks in β-sheets require longer PAR mixing times and increased signal averaging (see Fig. 4).
Figure 5.
(a) 2D 15N-15N PAR correlation spectrum on [1,3-13C,U-15N]-GB1. The spectrum was obtained using 18 ms PAR mixing with ω1N/2π ~ 52 kHz and ω1H/2π ~ 49 kHz at ωr/2π = 20 kHz and ω0H/2π = 900 MHz. The cross-peaks circled in red correspond to sequential contacts in loop regions that are also indicated with red lines in (b)). The cross-peaks circled in blue correspond to contacts between the strands in antiparallel β-sheets (nitrogens for the residues participating in a β-bridge) that are also indicated with blue lines in (b). The unmarked cross-peaks correspond primarily to the sequential contacts in the α-helix that are marked with green lines in (b).
Table I.
Average N-N and H-N distances in typical elements of secondary structure in proteins. The values were extracted based on 100 randomly chosen protein structures in the program STARS.82.
| Type of contact | N1-N2 (Å) | N1-H2 (Å) | N2-H1 (Å) |
|---|---|---|---|
| Sequential Ni-Ni+1 in β-sheet | 3.5±0.2 | 3.9±0.3 | 3.7±0.7 |
| Sequential - Ni-Ni+1 in α-helix | 2.8±0.1 | 3.3±0.1 | 2.5±0.1 |
| Sequential - Ni-Ni+2 in α-helix | 4.3±0.1 | 3.6±0.2 | 5.0±0.1 |
| Sequential - Ni-Ni+3 in α-helix | 4.8±0.2 | 6.8±2.4 | 7.6±1.8 |
| β-bridge partners in antiparallel β-sheet | 4.5±0.4 | 3.8±0.7 | 3.8±0.7 |
| β -bridge partners in parallel β-sheet | 4.8±0.2 | 4.0±0.4 | 5.7±0.5 |
These experimental observations can be fully supported by numerical simulations. In the next section we study the relationship between PAR buildups, 15N-15N distances and the type of contacts involved.
5. 15N-[1H]-15N PAR experiments applied to structure determination
The relationship between the TSAR buildups and the inter-nuclear distances is discussed in detail for the case of the 13C-[1H]-13C TSAR mechanism by De Paëpe et al.29 If only three spins are considered, i.e. two carbons/nitrogens and a single proton, it was shown that the TSAR coupling was proportional to the product of 13C-1H/15N-1H couplings, independent from the 13C-13C/15N-15N distance and strongly dependent on the angle between the heteronuclear interactions involved.29
In the case where multiple protons are involved, e.g. fully protonated systems, the TSAR buildup analysis is more complicated, at least analytically. Indeed, the TSAR polarization transfer in this case is the result of the superposition of multiple contributions involving nearby protons (typically protons which are closer than 2.5 Å for the 13C-[1H]-13C case). However, it was found experimentally that the 13C-[1H]-13C buildups recorded on fully protonated [U-13C, 15N]-Crh can, to a large extent, be classified in different distance classes and used to perform a 3D structure calculation.29
As we have already mentioned above, the spatial distribution of backbone 15N’s and amide 1H’s is intimately linked to the secondary, tertiary and often quaternary structure of proteins and nucleic acids through the pattern of hydrogen bonds. Table I lists 15N-15N and important 1H-15N distances in some typical motifs encountered in proteins. Because PAR polarization transfer is proportional to the product of the 1H-15N couplings, it ideally suited for probing geometries imposed by hydrogen bonding patterns. We illustrate this in the next sections where we consider 15N-15N PAR polarization transfer in three different typical secondary and tertiary structural motifs encountered in proteins: α-helix, antiparallel β-sheet and parallel β-sheet.
5.1 Sequential 15N-15N contacts in an α-helix
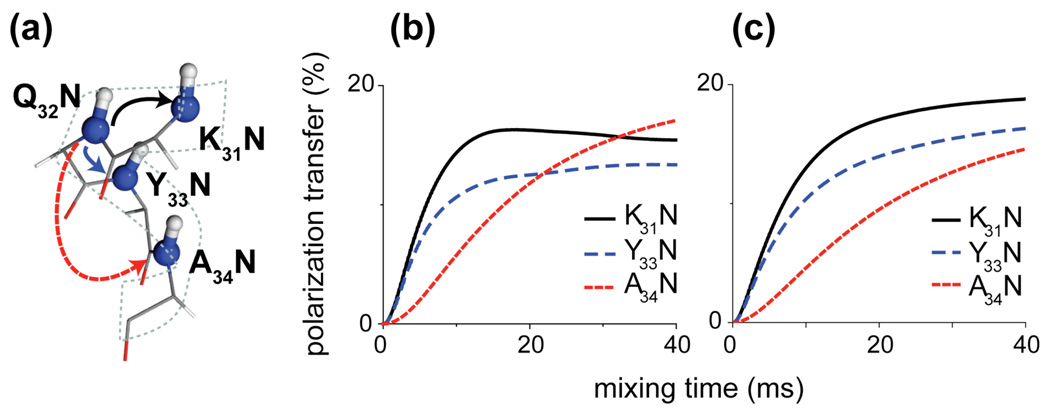
Figure 6 shows numerical simulations of the 15N-15N polarization transfer in a typical α-helical spin system taken from the x-ray structure of protein GB1 (PDB 2GI9). The spin system is depicted in Fig. 6a and consists of four backbone 15N’s and amide 1H’s from four consecutive residues in an α-helix. The initial magnetization is placed on Q32N and the polarization transfer to the other 15N’s is monitored as a function of time. Note that the distances between amide protons (1Hn) to sequential nitrogens (15Nn±1) in α-helices are the shortest 1H-15N distances (excluding directly bonded spins) of all the spin topologies presented in Table I. Consequently the corresponding 15N-[1H]-15N polarization transfer, simulated in Fig. 6b, displays the most rapid (10–20 ms) buildup time and is consistent with the experimental data.
Figure 6.
Numerical simulations of 15N-15N PAR polarization transfer in an α-helix. The spin system (a) consists of 4 backbone 15N’s and amide 1H’s only for simulation in (b) and amide protons plus 3 Hα’s for simulation in (c). The coordinates were taken from residues 31 to 34 in the x-ray structure of GB1 (PDB ID 2GI9)46 – see Table SI2). Simulations include nitrogen and proton chemical shifts (see Table SI2). The initial magnetization is placed on Q32N. Simulations were performed at ωr/2π=20 kHz MAS and ω0H/2π=750 MHz with pN=2.7 and pH=2.5.
The spin system used in the simulations in Fig. 6b includes only the amide protons, so strictly speaking it corresponds to a perdeuterated sample with back-exchanged amide protons. We have shown that in the case of 13C-[1H]-13C TSAR usually multiple protons participate and influence polarization transfer between any two given 13C sites. In order to evaluate the influence of protons other than amide protons we have performed a series of multispin simulations on the α-helix spin system. Figure 6c shows simulations for an α-helix with amide protons and alpha protons (which are, besides the amide 1H’s, consistently the most strongly coupled to the backbone 15N’s). The addition of Hα’s only slightly affects the overall polarization transfer with the change more pronounced for Ni-Ni+2 polarization transfer. This suggest that in order to predict the general trends of 15N-15N PAR polarization in proteins we can restrict our analysis to nitrogens and the amide protons (though for a precise analysis requires complex multiple spin simulations).
The simulations in Fig. 6 suggest that for mixing times longer than the one we employed in the experiment in Fig. 5, we should also observe cross-peaks to Nn±2. In fact many Ni – Ni±2 contacts in the helix are also detectable in the data presented in Fig. 5 but are much weaker and closer to the noise level.
5.2 15N-15N contacts in β-sheets
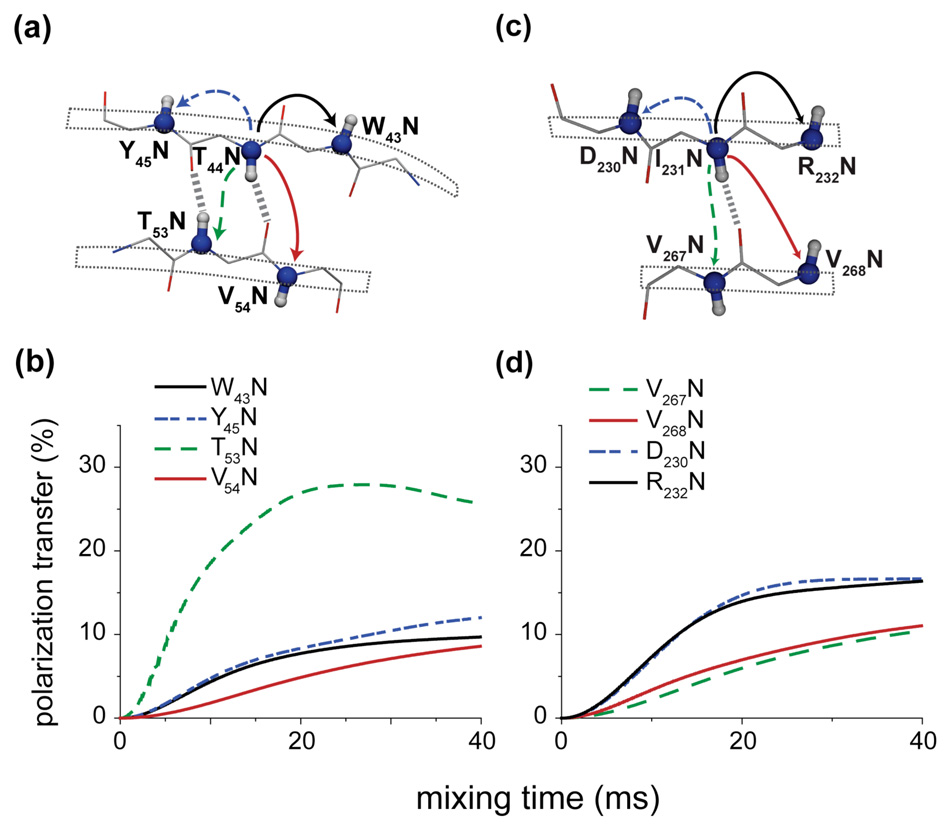
Figure 7 illustrates numerical simulations of 15N-15N polarization transfer in two typical β-sheets geometries: the antiparallel β-sheet arrangement shown in Fig. 7a (coordinates from PDB 2GI9)46) and the parallel β-sheet arrangement shown in Fig. 7c (with coordinates taken from the SSNMR structure of the Het-s prion2).
Figure 7.
Numerical simulations of 15N-15N PAR polarization transfer in an antiparallel β-(a–b) and parallel β-sheet (c–d). In (a) the spin system consists of 5 backbone nitrogens with directly bonded protons from two strands in an antiparallel β-sheet (coordinates for residues 43–45 and 53–55 from x-ray structure of GB1, PDB ID 2GI946 – see Table SI3). The spin system consists of 5 backbone nitrogens with directly bonded protons from two strands in an parallel β-sheet (coordinates from SSNMR structure of the HET-s(218–289) prion, PDB ID 2RNM2 – see Table SI4). Simulations include nitrogen and proton chemical shifts (see Table SI3 and SI4). The initial magnetization is placed on the T44N in (b) and I231N in (d). Simulations were performed at ωr/2π=20 kHz MAS and ω0H/2π=750 MHz with pN=2.7, pH=2.5.
The spin system (Fig 7a) used in the simulation in Fig. 7b consists of five backbone 15N’s in an antiparallel β-sheet with their amide 1H’s. In the case of the antiparallel β-sheet arrangement, the inter-strand polarization transfer between the β-bridge partners (T44N and T53N) is clearly preferred over the transfer to the sequential nitrogens within the strands. Such a situation is a direct consequence of the topology imposed by the hydrogen bonding pattern: the amide protons from the β-bridge partners are pointing towards the nitrogens in the other strand, leading to strong PAR couplings. Moreover, the N1-H2 and N2-H1 couplings are identical (see Table I) or very close to each other which results in ideal or close to ideal compensation of the heteronuclear autocross term and consequently no effective tilting of the PAR recoupling axis.
The spin system of Fig. 7c consists of five backbone 15N’s in a parallel β-sheet and their amide 1H’s. The geometry imposed by the hydrogen bonding pattern is not as favorable as in the case of an antiparallel β-sheet for observing inter-strand contacts (see also Table I). In this case the sequential polarization transfer between the neighboring 15N’s is preferred over the polarization transfer between the 15N’s in the neighboring strands (which is also consistent with the distribution of NH dipolar couplings in Table I).
Naturally, the Ni-Ni±1 polarization transfer in both parallel and antiparallel β-sheets have similar characteristics (since the NH couplings for sequential sites are similar – see Table I), even though overall efficiency of such transfers in the antiparallel β-sheet are lower due to the presence of more favorable transfer between strands.
The simulations suggest τmix ≥ 30 ms for PAR is required for optimal polarization transfer between sequential contacts in β-sheets. This is consistent with our observation of only a few of such cross peaks in the data presented in Fig. 5, which uses τmix = 18 ms.
6. 15N-15N PAR in the context of other methods
To complete our discussion, we briefly compare the 15N-15N PAR to PDSD and NHHN experiments – two other popular alternatives for 15N-15N polarization transfer.
As we have already mentioned above, the 15N-15N PAR experiment accelerates the polarization transfer between nitrogens by two to three orders of magnitude compared to PDSD (milliseconds in PAR versus seconds in PDSD17,46,74,80,81). Optimal PDSD mixing times increase with the spinning frequency, rendering it practical for 15N-15N correlation experiments employing spinning frequencies ωr/2π ≤ 12–14 kHz.83 For instance, the LN-FN polarization transfer efficiency in the tripetide [U-13C,15N]-f-MLF-OH at ω0H/2π = 750 MHz drops from 15% to 4% when going from 10 kHz to 20 kHz spinning frequency in an experiment with 2 s of PDSD mixing time. The required increasing PDSD mixing times at higher spinning frequencies also becomes a limiting factor both in terms of loss associated with relaxation and the experimental time per scan. In contrast, according to simulations, 15N-15N PAR should be applicable at all spinning frequencies presently accessible in solid-state magic angle spinning NMR (up to 70 kHz) requiring reasonable mixing times (on the order of tens milliseconds).
Moreover, as was shown by Castellani et al.45 due to the decreasing overlap integral the increase of the magnetic field strength adversely affects the polarization transfer in 15N-15N PDSD experiment. As a consequence, even though at fields < 14 T the correlation between the 15N-15N distances and the polarization transfer buildups can be established quite straightforwardly, recovering any such correlation at fields > 14 T requires prior knowledge of the undecoupled nitrogen linewidths and correction for the chemical shift differences between the recoupled sites. 45,80
However, it is important to note that 15N-15N PAR and 15N-15N PDSD experiments run under optimal conditions provide spectra with quite different information contents and can thus be used jointly.
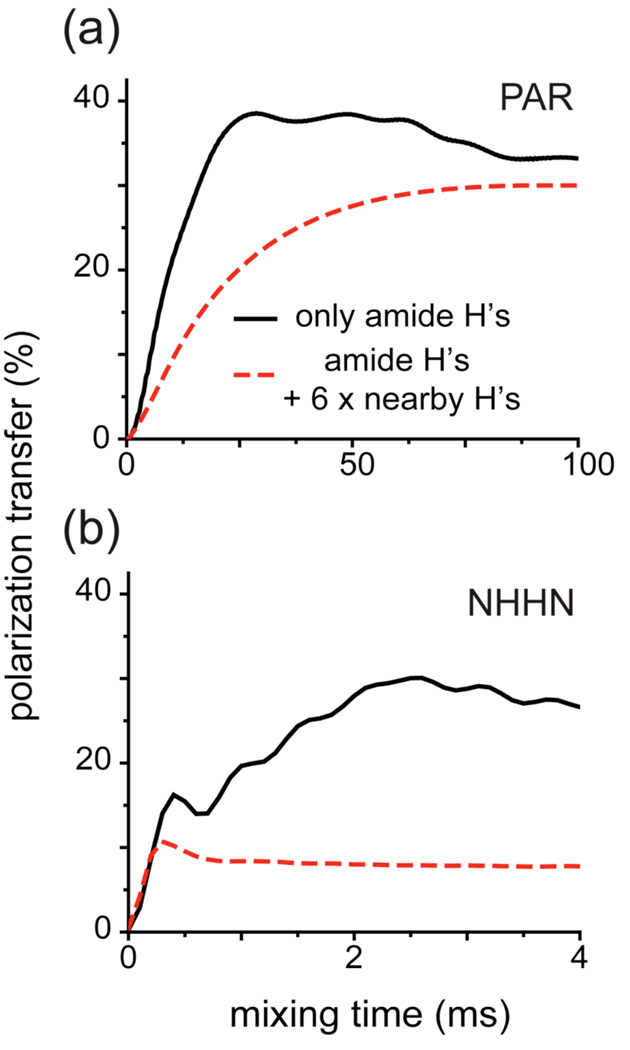
The NHHN experiment was demonstrated to provide valuable structural information on perdeuterated back-exchanged samples.84 For example, similarly to the PAR experiment presented here, NHHN yields contacts between strands in antiparallel β-sheets (though the crowding should be reduced in PAR spectra with sequential cross-peaks in β-sheets significantly attenuated at mixing times favoring the inter-strand polarization transfer). However, it was also noted that the performance of the NHHN experiment deteriorates significantly in fully protonated samples, where mostly sequential cross-peaks are retained.84 It transpires that 15N-15N PAR experiment should be more sensitive than the NHHN experiments for probing 15N-15N contacts in a fully protonated sample and yield comparable structural information. This is illustrated in Fig. 8, which shows a comparison of the polarization transfer between the β-bridge nitrogen partners in an antiparallel β-sheet in NHHN and 15N-15N PAR experiments. It turns out that the addition of 6 closest protons (see Table SI5 for the details on the spin system) leads to substantial reduction of polarization transfer efficiency in the NHHN experiment, but only a few percent reduction of polarization transfer efficiency in the PAR experiment. Note that in general the number of neighboring protons is much larger than the number of protons that we have included in these simulations, which means that the experimental performance of NHHN may actually deteriorate even further. For example, on the [U-13C,15N]-f-MLF-OH sample at ω0H/2π = 750 MHz and ωr/2π = 20 kHz the LN-FN polarization transfer in a 15N-15N PAR experiment is almost 7 times more efficient than in NHHN experiment run under the same set of conditions (see Figure SI 6).
Figure 8.
Numerical simulation of PAR (a) and NHHN (b) polarization transfer between nitrogens from a β-bridge partner residues in an antiparallel β-sheet. The black solid line represents simulations with only amide protons included, and the red dashed line represents simulation with amide protons plus 6 other closest protons. The simulations were performed at ωr/2π= 20 kHz and ω0H/2π= 750 MHz and include all chemical shifts (see Table SI5). The 1H-15N CP steps in NHHN are simulated explicitly using 0.15 ms contact time with ω1H/2π= 100 kHz and ω1N/2π= 80 kHz. The PAR mixing settings are: pN = 2.7 and pH = 2.5.
7. Conclusion
We have described a new experiment for performing 15N-15N MAS correlation spectroscopy that provides direct access to secondary and tertiary structural information of proteins. 15N-15N PAR accelerates the 15N-15N polarization transfer up to three orders of magnitude compared to spin diffusion experiments. Moreover, in fully protonated samples, 15N-15N PAR yields interstrand cross-peaks in antiparallel β-sheets as well as the sequential contacts in helices. Most transmembrane proteins consist of either β-barrel or α-helical structural motifs. Provided that sufficient sensitivity is available, our results suggests that the 15N-15N PAR method should allow straightforward identification of α-helical segments and should permit one to establish connectivities between β-strands in β-barrels, which typically consist of antiparallel β-sheets, and thus provide valuable structural information about membrane proteins. Moreover, the fact that the interstrand 15N-15N contacts for the β-bridge partners in antiparallel β-sheets are substantially larger than sequential 15N-15N contacts within the strands should lead to significant simplification of the spectra without need for deuteration or other specific labeling – a feature that should be greatly appreciated in larger systems with significant spectral overlap.
15N-15N PAR is applicable over almost the entire range of MAS frequencies currently available (10–70 kHz) and could be used as a building block for more sophisticated SSNMR experiments. More importantly, 15N-15N spectroscopy should benefit strongly from the development of sensitivity enhanced techniques like DNP, and become an integral part of the SSNMR toolkit for structural characterization of proteins.
8. Material and methods
8.1 Sample preparation
Preparation of N-[U-13C,15N]-f-MLF-OH
N-f-MLF-OH peptide was obtained by solid phase peptide synthesis from CS Bio Inc. (Menlo Park, CA). The peptide was prepared with uniformly 13C and 15N labeled amino acids from Cambridge Isotope Laboratories (Andover, MA). The peptide was crystallized from isopropanol and packed in a 2.5mm Bruker rotor.
Preparation of GB1 Samples
Two labeled samples were prepared for 15N-15N TSAR studies: one [1,3 13C, U-15N] and one [12C, U-15N]. Samples were prepared according to previously published protocol.85 E. coli BL21 (DE3) cells (Invitrogen) were transformed with the T2Q mutant of GB1. The [1,3 13C, U-15N] sample was grown in M9 minimal media containing 2.0 grams of [1,3-13C] glycerol and 2.0 grams 12C NaHCO3 as the sole carbon sources and 1.0 gram 15N ammonium chloride as the sole nitrogen source; the U-15N sample was prepared in M9 minimal media containing 1.0 gram 15N ammonium chloride and 8.0 g 12C glucose. Protein expression, extractions, and purification were done according to previous studies. Microcrystalline samples were prepared according to ref. 85 by dialysis in 50 mM phosphate buffer (pH 5.7) and precipitated with 3 aliquots of 2:1 MPD:IPA at a protein concentration of 25 mg/mL. One sample containing ~9–10 mg of [1,3 13C, U-15N] labeled protein was centrifuged into a 2.5 mm Bruker rotor, while ~ 20 mg of [12C, U-15N] protein was centrifuged into a 4.0 mm Varian rotor. Both rotors were sealed with epoxy to maintain sample hydration levels throughout the studies.
8.2 NMR Spectroscopy
The experiments were carried out using a commercial Bruker spectrometer operating at 900.1 MHz 1H Larmor frequency using a Bruker triple resonance (HCN) probe equipped with a 2.5 mm spinner module. Spinning frequencies of 20 kHz were used in all experiments and regulated to ±2 Hz with a Bruker spinning frequency controller (Bruker BioSpin, Billerica MA). The PAR experiment was optimized by matching the interference pattern with the simulated PAR optimum (a comparison of the polarization transfer map and the interference map can be found in the Fig. SI2). With an optimization of this kind we take advantage of the fact that the conditions leading to destructive interference of nitrogen polarization (i.e. rotary resonance and 1H-15N Hartmann-Hahn conditions) are also outlined as features in the PAR optimization map. The 15N power was set to ~52 kHz or ~4 kHz (i.e. pN=2.6 or 0.2 – the value that leads to appreciable TSAR mechanism in simulations) and 1H rf was scanned through to identify Hartmann-Hahn conditions. 1H rf power leading to minimal interference just under the n=0 condition was used for the first case and just under the n=3 Hartmann-Hahn condition for the second case.
The 1H decoupling during t1 evolution and acquisition was implemented through optimized 100 kHz TPPM24. The recycle delay was 3 s. For the 2D 15N-15N PAR correlation spectrum on [U-13C,15N]-f-MLF-OH, acquisition times were 20 ms in t2 and 12.8 ms in t1 (64 × 200 µs; spectral width 54.8 ppm) with 4–16 scans per t1 point. One of the 2D 15N-15N PAR correlation spectrum on [1,3-13C,15N]-GB1 was obtained with 18 ms mixing time using ca. 52 kHz 15N and 49 kHz 1H irradiation; acquisition times were 25.6 ms in t2 and 16 ms in t1 (80 × 200 µs spectral width 54.8 ppm) with 224 scans per t1 point. Second of the 2D 15N-15N PAR correlation spectrum on [1,3-13C,15N]-GB1 was obtained with 22 ms mixing time using ~4 kHz 15N and ~55 kHz 1H irradiation; acquisition times were 25.6 ms in t2 and 16 ms in t1 (64 × 250 µs spectral width 43.8 ppm) with 96 scans per t1 point. The temperature was regulated using Bruker BCU-X (target temperature −18°C, flow 1400L/h, resulting in a sample temperature between 0 to 5 °C as indicated by the water 1H chemical shift referenced to PEG (3.74 ppm, referenced externally to DSS).86
8.3 Numerical simulations and data analysis
Numerical simulations were performed using SPINEVOLUTION 3.3. The NH bonds were set to 1.04 Å for the simulations. For viewing and processing PDB files we used Chimera87 and DS Visualizer 2.0 (Accelrys). Chimera was also used for producing some of the graphics used in figures. Data was processed using NMRPipe88 and analyzed in Sparky (T. D. Goddard and D. G. Kneller, University of California).
Supplementary Material
Acknowledgement
We would like thank Dr. Patrick van der Wel for insightful discussions. We are very grateful to Dr. Mikhail Veshtort for providing the SPINEVOLUTION software that has been used throughout the course of this work. This work was supported by the National Institute of Health Grants EB-001960, EB-003151, and EB-002026.
Footnotes
Supporting Information Available: Details of the spin systems used in numerical simulations in the text. Supplemental figures. Low power 15N-15N PAR spectrum on GB1. List of observed cross-peaks. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Andrew ER, Bradbury A, Eades RG. Nature. 1958;182:1659–1659. [Google Scholar]
- 2.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 3.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. J. Am. Chem. Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 4.Franks WT, Wylie BJ, Schmidt HLF, Nieuwkoop AJ, Mayrhofer RM, Shah GJ, Graesser DT, Rienstra CM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frericks HL, Zhou DH, Yap LL, Gennis RB, Rienstra CM. J. Biomol. NMR. 2006;36:55–71. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- 6.Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M. Nature. 2006;440:959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 8.Petkova AT, Ishii Y, Tycko R. Biophys. J. 2002;82:320A–320A. [Google Scholar]
- 9.Thompson LK, Mcdermott AE, Raap J, Vanderwielen CM, Lugtenburg J, Herzfeld J, Griffin RG. Biochemistry. 1992;31:7931–7938. doi: 10.1021/bi00149a026. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Paramasivan S, Marulanda D, Cataidi M, Tasayco ML, Polenova T. Magn. Reson. Chem. 2007;45:S73–S83. doi: 10.1002/mrc.2092. [DOI] [PubMed] [Google Scholar]
- 11.Loquet A, Bardiaux B, Gardiennet C, Blanchet C, Baldus M, Nilges M, Malliavin T, Boeckmann A. J. Am. Chem. Soc. 2008;130:3579–3589. doi: 10.1021/ja078014t. [DOI] [PubMed] [Google Scholar]
- 12.Creuzet F, Mcdermott A, Gebhard R, Vanderhoef K, Spijkerassink MB, Herzfeld J, Lugtenburg J, Levitt MH, Griffin RG. Science. 1991;251:783–786. doi: 10.1126/science.1990439. [DOI] [PubMed] [Google Scholar]
- 13.Hong M. J. Am. Chem. Soc. 2000;122:3762–3770. [Google Scholar]
- 14.Martin RW, Zilm KW. J. Magn. Reson. 2003;165:162–174. doi: 10.1016/s1090-7807(03)00253-2. [DOI] [PubMed] [Google Scholar]
- 15.Lemaster DM. Prog. Nucl. Magn. Reson. Spectrosc. 1994;26:371–419. [Google Scholar]
- 16.LeMaster DM, Kushlan DM. J. Am. Chem. Soc. 1996;118:9255–9264. [Google Scholar]
- 17.Goldbourt A, Day LA, McDermott AE. J. Magn. Reson. 2007;189:157–165. doi: 10.1016/j.jmr.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Hong M, Jakes K. J. Biomol. NMR. 1999;14:71–74. doi: 10.1023/a:1008334930603. [DOI] [PubMed] [Google Scholar]
- 19.Linge JP, Habeck M, Rieping W, Nilges M. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 20.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 21.Manolikas T, Herrmann T, Meier BH. J. Am. Chem. Soc. 2008;130:3959–3966. doi: 10.1021/ja078039s. [DOI] [PubMed] [Google Scholar]
- 22.Zech SG, Wand AJ, McDermott AE. J. Am. Chem. Soc. 2005;127:8618–8626. doi: 10.1021/ja0503128. [DOI] [PubMed] [Google Scholar]
- 23.Bennett AE, Ok JH, Griffin RG, Vega S. J. Chem. Phys. 1992;96:8624–8627. [Google Scholar]
- 24.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 25.Caravatti P, Braunschweiler L, Ernst RR. Chem. Phys. Lett. 1983;100:305–310. [Google Scholar]
- 26.De Paepe G, Bayro MJ, Lewandowski J, Griffin RG. J. Am. Chem. Soc. 2006;128:1776–1777. doi: 10.1021/ja0550430. [DOI] [PubMed] [Google Scholar]
- 27.De Paepe G, Elena B, Emsley L. J. Chem. Phys. 2004;121:3165–3180. doi: 10.1063/1.1773155. [DOI] [PubMed] [Google Scholar]
- 28.De Paepe G, Hodgkinson P, Emsley L. Chem. Phys. Lett. 2003;376:259–267. [Google Scholar]
- 29.De Paepe G, Lewandowski J, Locquet A, Bockmann A, Griffin RG. J. Chem. Phys. 2008;129:245101, 1–21. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Paepe G, Lewandowski JR, Griffin RG. J. Chem. Phys. 2008;128:124503–124526. doi: 10.1063/1.2834732. [DOI] [PubMed] [Google Scholar]
- 31.Jaroniec CP, Filip C, Griffin RG. J. Am. Chem. Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- 32.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. J. Am. Chem. Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 33.Maricq MM, Waugh JS. J. Chem. Phys. 1979;70:3300–3316. [Google Scholar]
- 34.Nielsen NC, Bildsoe H, Jakobsen HJ, Levitt MH. J. Chem. Phys. 1994;101:1805–1812. [Google Scholar]
- 35.Oas TG, Griffin RG, Levitt MH. J. Chem. Phys. 1988;89:692–695. [Google Scholar]
- 36.Raleigh DP, Levitt MH, Griffin RG. Chem. Phys. Lett. 1988;146:71–76. [Google Scholar]
- 37.Schaefer J, Mckay RA, Stejskal EO. J. Magn. Reson. 1979;34:443–447. [Google Scholar]
- 38.Stejskal EO, Schaefer J, Waugh JS. J. Magn. Reson. 1977;28:105–112. [Google Scholar]
- 39.Takegoshi K, Nakamura S, Terao T. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 40.Bayro MJ, Ramachandran R, Caporini MA, Eddy MT, Griffin RG. J. Chem. Phys. 2008;128 doi: 10.1063/1.2834736. [DOI] [PubMed] [Google Scholar]
- 41.Lewandowski JR, De Paepe G, Griffin RG. J. Am. Chem. Soc. 2007;129:728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran R, Lewandowski JR, van der Wel PCA, Griffin RG. J. Chem. Phys. 2006;124 doi: 10.1063/1.2194905. [DOI] [PubMed] [Google Scholar]
- 43.Lewandowski JR, De Paepe G, van der Wel PCA, Birkett NR, Belenky M, Maly T, Bayro MJ, Sivertsen AC, Dobson CM, Herzfeld J, Griffin RG. submitted. 2008 [Google Scholar]
- 44.Reif B, Hohwy M, Jaroniec CP, Rienstra CM, Griffin RG. J. Magn. Reson. 2000;145:132–141. doi: 10.1006/jmre.2000.2067. [DOI] [PubMed] [Google Scholar]
- 45.Castellani F. Freien Universität Berlin. 2003 [Google Scholar]
- 46.Franks WT, Wylie BJ, Stellfox SA, Rienstra CM. J. Am. Chem. Soc. 2006;128:3154–3155. doi: 10.1021/ja058292x. [DOI] [PubMed] [Google Scholar]
- 47.Szeverenyi NM, Sullivan MJ, Maciel GE. J. Magn. Reson. 1982;47:462–475. [Google Scholar]
- 48.Baldus M. Prog. Nucl. Magn. Reson. Spectrosc. 2002;41:1–47. [Google Scholar]
- 49.Baldus M, Geurts DG, Hediger S, Meier BH. J. Magn. Reson. Ser. A. 1996;118:140–144. [Google Scholar]
- 50.Castellani F, van Rossum BJ, Diehl A, Rehbein K, Oschkinat H. Biochemistry. 2003;42:11476–11483. doi: 10.1021/bi034903r. [DOI] [PubMed] [Google Scholar]
- 51.Detken A, Hardy EH, Ernst M, Kainosho M, Kawakami T, Aimoto S, Meier BH. J. Biomol. NMR. 2001;20:203–221. doi: 10.1023/a:1011212100630. [DOI] [PubMed] [Google Scholar]
- 52.Heise H, Seidel K, Etzkorn M, Becker S, Baldus M. J. Magn. Reson. 2005;173:64–74. doi: 10.1016/j.jmr.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Marulanda D, Tasayco ML, Cataldi M, Arriaran V, Polenova T. J. Magn. Reson. 2005;109:18135–18145. doi: 10.1021/jp052774d. [DOI] [PubMed] [Google Scholar]
- 54.Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Chembiochem. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 55.Rienstra CM, Hohwy M, Hong M, Griffin RG. J. Am. Chem. Soc. 2000;122:10979–10990. [Google Scholar]
- 56.Sun BQ, Rienstra CM, Costa PR, Williamson JR, Griffin RG. J. Am. Chem. Soc. 1997;119:8540–8546. [Google Scholar]
- 57.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 58.Caravatti P, Bodenhausen G, Ernst RR. Chem. Phys. Lett. 1982;89:363–367. [Google Scholar]
- 59.Egorova-Zachernyuk TA, Hollander J, Fraser N, Gast P, Hoff AJ, Cogdell R, de Groot HJM, Baldus M. J. Biomol. NMR. 2001;19:243–253. doi: 10.1023/a:1011235417465. [DOI] [PubMed] [Google Scholar]
- 60.Hong M, Griffin RG. J. Am. Chem. Soc. 1998;120:7113–7114. [Google Scholar]
- 61.van der Wel PCA, Lewandowski JR, Griffin RG. J. Am. Chem. Soc. 2007;129:5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 62.Hong M, Gross JD, Griffin RG. J. Magn. Reson. 1997;101:5869–5874. [Google Scholar]
- 63.Hong M, Gross JD, Hu W, Griffin RG. J. Magn. Reson. 1998;135:169–177. doi: 10.1006/jmre.1998.1573. [DOI] [PubMed] [Google Scholar]
- 64.Ladizhansky V, Jaroniec CP, Diehl A, Oschkinat H, Griffin RG. J. Am. Chem. Soc. 2003;125:6827–6833. doi: 10.1021/ja029082c. [DOI] [PubMed] [Google Scholar]
- 65.Ladizhansky V, Veshtort M, Griffin RG. J. Magn. Reson. 2002;154:317–324. doi: 10.1006/jmre.2001.2488. [DOI] [PubMed] [Google Scholar]
- 66.Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaroniec CP, MacPhee CE, Bajaj VS, Dobson CM, Griffin RG. Biophys. J. 2003;84:154A–154A. [Google Scholar]
- 68.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. Proc. Natl. Acad. Sci. U. S. A. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Biophys. J. 2001;80:368A–368A. [Google Scholar]
- 70.Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. J. Am. Chem. Soc. 1999;121:10237–10238. [Google Scholar]
- 71.Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. J. Magn. Reson. 2000;146:132–139. doi: 10.1006/jmre.2000.2128. [DOI] [PubMed] [Google Scholar]
- 72.Rienstra CM, Tucker-Kellogg L, Jaroniec CP, Hohwy M, Reif B, McMahon MT, Tidor B, Lozano-Perez T, Griffin RG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10260–10265. doi: 10.1073/pnas.152346599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sein J, Giraud N, Blackledge M, Emsley L. J. Magn. Reson. 2007;186:26–33. doi: 10.1016/j.jmr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Giraud N, Blackledge M, Bockmann A, Emsley L. J. Magn. Reson. 2007;184:51–61. doi: 10.1016/j.jmr.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 75.Giraud N, Blackledge M, Goldman M, Bockmann A, Lesage A, Penin F, Emsley L. J. Am. Chem. Soc. 2005;127:18190–18201. doi: 10.1021/ja055182h. [DOI] [PubMed] [Google Scholar]
- 76.Chevelkov V, Zhuravleva AV, Xue Y, Reif B, Skrynnikov NR. J. Am. Chem. Soc. 2007;129:12594–12595. doi: 10.1021/ja073234s. [DOI] [PubMed] [Google Scholar]
- 77.Barnes AB, De Paepe G, van der Wel PCA, Hu KN, Joo CG, Bajaj VS, Mak-Jurkauskas ML, Sirigiri JR, Herzfeld J, Temkin RJ, Griffin RG. Applied Magnetic Resonance. 2008;34:237–263. doi: 10.1007/s00723-008-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maly T, Debelouchina GT, Bajaj VS, Hu KN, Joo CG, Mak-Jurkauskas ML, Sirigiri JR, van der Wel PCA, Herzfeld J, Temkin RJ, Griffin RG. J. Chem. Phys. 2008;128:052211. doi: 10.1063/1.2833582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou DH, Shea JJ, Nieuwkoop AJ, Franks WT, Wylie BJ, Mullen C, Sandoz D, Rienstra CM. Angew. Chem. Int. Edit. 2007;46:8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marulanda D, Tasayco ML, McDermott A, Cataldi M, Arriaran V, Polenova T. J. Am. Chem. Soc. 2004;126:16608–16620. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- 81.Seidel K, Etzkorn M, Heise H, Becker S, Baldus M. Chembiochem. 2005;6:1638–1647. doi: 10.1002/cbic.200500085. [DOI] [PubMed] [Google Scholar]
- 82.Zheng Y, Yang DW. Bioinformatics. 2005;21:2925–2926. doi: 10.1093/bioinformatics/bti437. [DOI] [PubMed] [Google Scholar]
- 83.Krushelnitsky A, Bräuniger T, Reichert D. J. Magn. Reson. 2006;182:339–342. doi: 10.1016/j.jmr.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 84.Reif B, van Rossum BJ, Castellani F, Rehbein K, Diehl A, Oschkinat H. J. Am. Chem. Soc. 2003;125:1488–1489. doi: 10.1021/ja0283697. [DOI] [PubMed] [Google Scholar]
- 85.Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. J. Am. Chem. Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 86.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ, Rance M. Protein NMR Spectroscopy: Principles and Practice. Academic Press; 2006. [Google Scholar]
- 87.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 88.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.