Abstract
Orthodox Type IIP restriction endonucleases, which are commonly used in molecular biological work, recognize a single palindromic DNA recognition sequence and cleave within or near this sequence. Several new studies have reported on structural and biochemical peculiarities of restriction endonucleases that differ from the orthodox in that they require two copies of a particular DNA recognition sequence to cleave the DNA. These two sites requiring restriction endonucleases belong to different subtypes of Type II restriction endonucleases, namely Types IIE, IIF and IIS. We compare enzymes of these three types with regard to their DNA recognition and cleavage properties. The simultaneous recognition of two identical DNA sites by these restriction endonucleases ensures that single unmethylated recognition sites do not lead to chromosomal DNA cleavage, and might reflect evolutionary connections to other DNA processing proteins that specifically function with two sites.
INTRODUCTION
Specific DNA recognition by proteins is central to many biological processes like transcription, DNA repair, DNA recombination and DNA restriction by restriction endonucleases (REases). In bacterial cells, REases are generally accompanied by a cognate methyltransferase (MTase) (1). Both enzymes working together form a restriction-modification system (R-M system). R-M systems are thought to protect bacterial cells against invading DNA, because the REases cleave foreign DNA, which is not specifically methylated. In contrast, the host DNA will not be cleaved, because it is specifically methylated by the cognate MTase (1). The protective function of R-M systems is not questioned, because bacteriophages have developed an impressive variety of anti-restriction mechanisms to counteract the host’s R-M systems (2). Another hypothesis about the function, however, has accounted R-M systems functioning as selfish mobile genetic elements because REases can kill host cells that have eliminated the corresponding R-M system genes (3). Furthermore, it has been postulated that R-M enzymes are products of ‘evolution genes’ and function together with DNA repair enzymes to modulate the frequency of genetic variation to benefit bacteria (1,4).
REases typically recognize 4–8 bp long, palindromic DNA sequences (recognition sites) and cleave both DNA strands within or near these sequences (5). They require only divalent metal ions, preferably Mg2+, as co-factor and are homodimeric proteins that symmetrically contact their palindromic recognition sequences. In this way, the catalytic center of each monomer approaches the phosphodiester bond of one strand of the double-stranded DNA recognition sequence, which will be symmetrically cleaved. REases that match these criteria are named orthodox Type IIP REases. In recent years, however, it has been found that many Type II REases do not fulfil the criteria for Type IIP and several other subtypes of Type II REases have been defined (6). Enzymes of certain subtypes—Types IIE, IIF and IIS—exhibit the particular property that they have to simultaneously bind two copies of their DNA recognition sequence to cleave the DNA.
It has recently been reported that the orthodox Type IIP REases SsoII and StyD4I, the Type IIE endonuclease EcoRII, and the Type IIF endonuclease NgoMIV that all recognize a DNA sequence containing 5′-CCGG in a slightly different sequence context, possess a similar DNA binding motif and catalytic center (7,8). Therefore, Types IIE, IIF and the orthodox Type IIP REases seem to be evolutionary related to each other despite their different tertiary and quaternary structures and despite different substrate requirements for DNA cleavage (7,8).
A comparison of crystal structures of proteins that interact specifically with DNA suggests that REases appear to be related to other DNA processing proteins (9–12). Similar to DNA recombinases and transcription factors, it has been shown that Type IIE and Type IIF REases induce DNA loops in DNA molecules that contain at least two copies of their recognition sequences (13–15). The Type IIE restriction endonuclease EcoRII and the integrase family of site-specific recombinases share homologous amino acid motifs and NaeI, another Type IIE restriction endonuclease, shows topoisomerase activity when a single amino acid is replaced (16–19). The monomeric REase Sau3AI forms DNA loops similar to the Type IIE and IIF enzymes and shows sequence homology to the mismatch repair protein MutH (20). Furthermore, a protein scaffold like that of the Type IIS restriction endonuclease FokI has been reported for the TnsA protein of the Tn7 transposase (10), and DNA excision by the Type IIF restriction endonuclease SfiI is reminiscent of a recombinase that simultaneously catalyzes the cleavage of four DNA strands (14). Therefore, it seems that REases that have acquired the ability to function simultaneously with two DNA sites reflect the variety of evolutionary routes to other protein functions that include specific interaction with two DNA sites. However, so far two DNA site-recognizing REases have not been proved to have additional functions beyond phosphodiester bond cleavage either in vivo or in vitro.
In this paper, we review REases of Types IIE, IIF and IIS that all, in contrast to orthodox Type IIP REases, have the capacity for interacting with two DNA recognition sites. We propose a mechanistic model for Type IIE REases based on current biochemical and on structural data. Furthermore, we compare this model with those proposed for Type IIF and IIS REases.
Mechanism of Type IIE REases
Type IIE REases are homodimeric proteins that simultaneously bind two recognition sites to cleave the DNA within or near their recognition sites (Table 1). The best understood Type IIE REases are EcoRII and NaeI (21,22). Both enzymes form loops with DNA molecules that contain at least two of their recognition sites (in cis) as shown by electron microscopy (13,15). Furthermore, Type IIE REases can also interact with two recognition sites on two different DNA molecules (in trans). The cleavage efficiency in trans depends on DNA site concentration and is inversely correlated to the length of the DNA (21–26). However, Type IIE REases react preferably with two DNA sites in cis like all proteins that interact with two sites (27). The interaction in trans has been used to overcome the low cleavage efficiency of Type IIE REases on DNA substrates with only a single or a few DNA recognition sites separated by >1000 bp (21). By adding oligonucleotide duplexes with a recognition site for the respective enzyme, the low cleavage efficiency can be increased (22–26,28).
Table 1. Characteristics of orthodox Type IIP REases and of the subtypes IIE, IIF and IIS.
| Orthodox Type IIP | Type IIE | Type IIF | Type IIS | |
|---|---|---|---|---|
| DNA substrate requirements | Recognize palindromic DNA recognition sequences | Two copies of their usually palindromic recognition sequence | Two copies of their usually palindromic recognition sequence | Two copies of their asymmetric recognition sequence |
| Oligomeric status in solution | Dimer | Dimer | Tetramer | Monomer, but active as dimer |
| Domains per monomer | One | Two, an endonuclease-like catalytic domain and a DNA binding domain | One | Two, an endonuclease-like catalytic domain and a DNA binding domain |
| Cleavage | Within a single recognition site | Within one of the two recognition sites | Concertedly within or near both recognition sites | At a defined distance downstream of one recognition site |
| Prototypes | BamHI, EcoRI | EcoRII, NaeI | Cfr10I, SfiI | FokIa |
aIt recently has been reported that Type IIS REases follow different modes of action probably caused by different protein structures (54).
Reaction kinetics of the Type IIE REases NaeI and EcoRII using plasmid DNA containing a single recognition site or with two recognition sites for the respective enzyme further point out the requirement for a second DNA recognition site for efficient DNA cleavage. NaeI and EcoRII cleave supercoiled plasmids with two recognition sites faster than plasmids with a single site (29) (G.Tamulaitis, M.Mucke, M.Reuter and V.Siksnys, manuscript in preparation). Furthermore, NaeI also cleaves catenanes that consist of two plasmid rings with a single recognition site in each ring faster than plasmids with a single site (29). The kinetics using these different plasmids showed that Type IIE and IIF REases react differently, because they release different reaction intermediates and products. Type IIF REases convert a two-site plasmid directly to the products that result from concerted cleavage at both sites (30–32), whereas Type IIE REases predominantly release the full-length linear DNA that results from cleavage at one of the two sites (29); (G.Tamulaitis, M.Mucke, M.Reuter and V.Siksnys, manuscript in preparation). Thus, in contrast to Type IIF REases that cleave two of their DNA recognition sites in a concerted reaction, Type IIE enzymes, cleave only one of the two bound recognition sites. In the case of EcoRII, both strands of this recognition site are cut in one catalytic event (33). Cleavage of only one site and the lower cleavage efficiency of Type IIE compared to that of orthodox Type IIP REases can be explained by the domain structure of Type IIE REases.
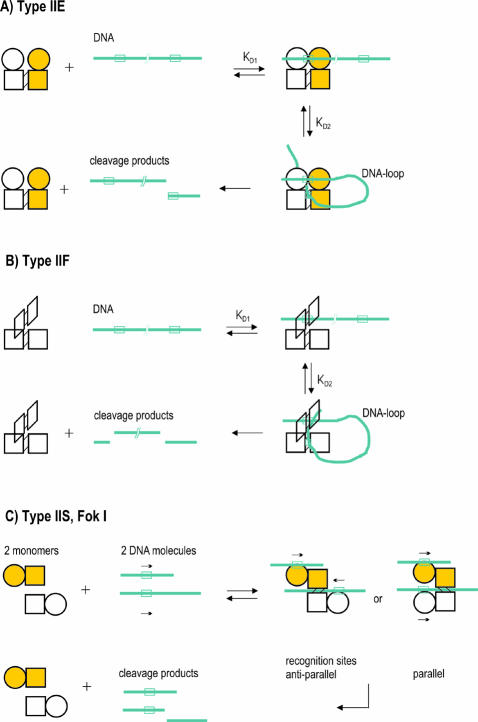
Type IIE REases contain two domains as shown for NaeI and EcoRII by biochemical and crystal structure analyses (34–37) (X.E.Zhou, Y.Wang, M.Reuter, M.Mucke, D.H.Kruger, E.J.Meehan and L.Chen, manuscript submitted). These two domains are an endonuclease-like catalytic domain and a DNA binding domain that both specifically interact with DNA (35,37,38). The DNA binding domain binds one of the recognition sites, the activator site. This binding activates the endonuclease-like domain to bind and to cleave the second DNA recognition site, the substrate site (38). The positive cooperativity observed for DNA cleavage by EcoRII and NaeI reflects this two-site DNA binding process (24,39,40). In the case of NaeI, the crystal structure revealed the presence of a structural motif characteristic for the DNA binding site of the catabolite activator protein in the DNA binding domain (35,38). In the case of EcoRII, the amino acid sequence of the DNA binding domain does not show any resemblance to any other known DNA binding motifs (X.E.Zhou, Y.Wang, M.Reuter, M.Mucke, D.H.Kruger, E.J.Meehan and L.Chen, manuscript submitted). Because the DNA binding dissociation constants (Kd) for the separately purified DNA binding domains of NaeI and of EcoRII differ only marginally from that for the respective complete enzyme, and because the isolated catalytic domain does not bind at all (NaeI) or only weakly (EcoRII) to DNA (34,37), we propose that in the complete enzymes the activating DNA binding domain initially binds to specific DNA (Fig. 1A). The binding of the activator site changes the protein conformation and promotes the binding of the substrate site by the endonuclease-like domain (35,37,38). Thus, the complex of a Type IIE REase dimer with two DNA recognition sites is the active form of the enzyme (38,41,42). In the active complex, the DNA recognition site, which is bound to the endonuclease-like domains of the dimer is the substrate site that will be cleaved.
Figure 1.
(A) Type IIE REase interaction with a DNA molecule containing two sites in cis. The two different domains of a monomer are symbolized by a circle and a rectangle, respectively. White, one monomer; yellow, the other monomer of the homodimer. Circle, DNA-binding domain; rectangle, endonuclease-like domain. The lines that link both rectangles symbolize dimerization. Blue stretches with boxes, DNA with respective recognition sites. The enzyme forms intermediate DNA loops with DNA containing two recognition sites. The DNA will be cleaved at one recognition site, which is bound to the endonuclease domain. (B) Type IIF REase interaction with a DNA molecule containing two sites in cis. Each of the four endonuclease subunits of the tetrameric enzyme is symbolized by a rectangle. Two monomeric subunits form a pseudo-dimer and two of these pseudo-dimers are packed back-to-back forming two identical DNA binding clefts. With DNA containing two recognition sites, the enzyme forms intermediate DNA loops. The DNA is concertedly cleaved at both DNA recognition sites. (C) One of the Type IIS REase interactions with DNA (named mode C, see text) as exemplified by the best understood enzyme FokI (53,54). Circle, DNA recognition domain; rectangle, cleavage domain of an enzyme monomer. The two enzyme monomers are yellow and white, respectively. The enzyme dimerizes probably after each monomer (via its DNA recognition domain) has specifically bound to a DNA recognition site, because specific DNA binding results in opening of the sequestered FokI monomer (53). This opening is most likely essential for the dimerization, which is mediated only by the cleavage domains. Two different complexes of a FokI dimer and two DNA recognition sites have been modeled depending on the relative orientation of the asymmetric recognition sites. The parallel orientation of the recognition sites indicated by the arrows could better stabilize the complex by possible protein–protein interactions than the anti-parallel orientation. Whether a preferred orientation exists remains to be determined as well as whether the enzyme forms DNA loops on a DNA molecule containing two recognition sites. The cleavage domains of two FokI monomers cooperate to cleave at either one of the two coordinated recognition sites that is in contact with both catalytic domains (9/13) bp downstream of this site (the first numeral in the parentheses indicates the position of cleavage in the strand written and the second numeral the cleavage position on the complementary strand).
In the crystals without DNA, only the endonuclease-like catalytic domain of NaeI contributes to dimerization (35,38). However, once the enzyme has bound the specific activator DNA, the additional DNA binding domain of NaeI also contributes to dimerization (38). Therefore, binding of the activator DNA by NaeI not only changes the conformation of the protein to promote binding of the substrate DNA to the endonuclease domain, but also leads to a more compact protein dimer.
For EcoRII, the C-terminal endonuclease-like domain can be separately purified as a dimer (37). This truncated REase specifically will now cleave DNA containing a single DNA recognition site and much more rapidly than the full-length EcoRII. One could consider this difference as a change in the enzyme’s site specificity from two copies of the recognition sequence [the complete sequence can be taken as 5′-CCWGG-(N)i-CCWGG] to a new specificity for a single recognition site (5′-CCWGG). Whereas the endonuclease domain alone is able to cleave DNA like an orthodox Type IIP restriction endonuclease, the second DNA binding domain at the N-terminus modulates the endonucleolytic activity. For EcoRII, we can only speculate about an additional function of the enzyme due to the second DNA binding capacity, but it is known that NaeI is only a single amino acid substitution away from topoisomerase activity (17). Additionally, there are conserved amino acid motifs between EcoRII and the integrase family of site-specific recombinases (16,19). The relationship of topoisomerases/recombinases and of Type IIE REases could reflect functions beyond endonucleolytic DNA cleavage.
Type IIF
Type IIF REases are homotetrameric enzymes and usually recognize palindromic DNA sequences. The crystal structures of the Type IIF REases Cfr10I, NgoMIV and Bse643I (31,43,44) show that two monomeric subunits together form one of the two equivalent DNA binding sites (Fig. 1B). Two of these virtual dimers are packed back-to-back so that the enzyme binds both DNA recognition sites on opposite sides of the tetramer (31,43,44). As both DNA binding sites are identical it is irrelevant which of the two virtual dimers binds first to the DNA. In this respect, the two equivalent DNA binding sites contrast with the non-equivalent activator and substrate binding sites of Type IIE REases.
The kinetic investigation of Type IIF enzymes on supercoiled plasmids with one or two recognition sites, first carried out for SfiI (30), demonstrated that these enzymes concertedly cleave at both recognition sites upon binding of two of their DNA recognition sites (31,32,44). Thus, the active complex is composed of a tetrameric enzyme and two recognition sites. This is in contrast to the active complex of Type IIE enzymes that consist of an enzyme dimer and of two recognition sites, where one of these recognition sites is the DNA substrate and the other functions as an allosteric activator.
Like Type IIE enzymes, Type IIF REases interact with two recognition sites in cis by forming DNA loops (14,32). The DNA loops formed by SfiI, NgoMIV and Cfr10I, respectively, vary in stability and the DNA loop stability seems to depend strongly on the reaction conditions (45). DNA substrates with a single recognition site can only be cleaved very slowly by interactions in trans. As with Type IIE REases, Type IIF REases prefer to interact with two DNA recognition sites in cis rather than with sites in trans (27).
Although the excision of the DNA in-between two recognition sites in a concerted reaction is similar to the reactions of some transposases (30), there is, at least for SfiI heterologously expressed in Escherichia coli, no evidence for other enzymatic activities than REase activity in vivo or in vitro (46).
Disruption of the tetramer interface of the Type IIF REase Cfr10I by mutating a single amino acid important for dimer–dimer contacts, did not result in a re-activated, orthodox Type IIP REase-like enzyme (32). On the contrary, the cleavage activity of the dimeric mutant W220A of Cfr10I is only 0.1% of that of the wild type (32). This again illustrates that characteristic Type IIF enzymes are normally active as tetramers.
Type IIS
Type IIS REases recognize asymmetric DNA sequences and cleave at a defined distance downstream (47). The best understood Type IIS REase is FokI. This enzyme has a two-domain structure (48,49), similar to that reported for Type IIE REases. Both domains of FokI can be purified following limited proteolysis (49). FokI consists of a sequence-specific N-terminal DNA binding domain and of a sequence-unspecific catalytic domain (49). The purified catalytic domain of FokI cleaves DNA unspecifically and has successfully been used to engineer chimeric endonucleases with new DNA specificities (50).
FokI and other Type IIS REases are monomers in solution. Because a single catalytic domain of a monomer contains only one catalytic center, FokI has to dimerize to catalyze the hydrolysis of two phosphodiester bonds in double-stranded DNA. Dimerization has first been suggested from FokI crystal structures, from cooperative DNA cleavage and from different FokI variants relevant to dimerization of the enzyme (51,52). Recently, it has been demonstrated that FokI dimerizes only in the presence of divalent metal ions and that the active dimeric FokI–DNA complex binds two recognition sites (53). Each asymmetric recognition site is thought to bind to one N-terminal DNA recognition domain, whereas both C-terminal catalytic domains dimerize to cleave together one DNA double strand at a distance of 9 and 13 bp, respectively from the recognition site.
The catalytic domain mediates the dimerization of FokI (52). This is similar to the dimerization mode of the Type IIE restriction endonuclease NaeI in the absence of DNA (35). Moreover, the DNA binding domains of FokI and of NaeI seem to be similar to each other as both contain a DNA binding motif that is related to the helix–turn–helix motif of the catabolite gene activator protein (35,48). However, the DNA binding domain of FokI does not contribute to enzyme dimerization, either in the absence or presence of DNA. In contrast, the DNA binding domain of NaeI contributes to dimerization after DNA binding.
Concerning the reaction pathway, it has recently been reported that Type IIS REases appear to follow different reaction schemes (54). Using supercoiled plasmid DNA with one or two recognition sites for estimating the cleavage kinetics of several Type IIS REases, four modes of action (called modes A–D) have been differentiated based on reaction intermediates, products and cleavage rates (54).
The REases that follow mode A, such as BsmI, cleave close to their asymmetric recognition sequence. Furthermore, they cleave plasmids containing a single site or two sites with a similar rate. These REases do not require two copies of their recognition sequence and seem to cleave the DNA like orthodox Type IIP REases (54). Because the oligomeric state and domain structure of the mode A Type IIS REases remain to be determined, their reaction mechanism is not yet clear.
The REases that follow mode B, such as BpmI, require two copies of their recognition sequence and cleave subsequently at both cleavage sites (54). They differ from Type IIF and IIE REases, which cleave concertedly at both and at only one cleavage site, respectively. Cleavage of the second site by mode B Type IIS enzymes is possible because they cleave the DNA outside of the recognition site, which remains intact and which can activate cleavage at another recognition site in trans.
FokI is a Type IIS REase following mode C (Fig. 1C). It requires two copies of its recognition sequence and cleaves at only one cleavage site similarly to Type IIE REases (53,54). In contrast to the Type IIE REases, which bind one DNA site by the activator binding site and the second by the substrate binding site, FokI binds each of the two DNA recognition sites to one of the two N-terminal DNA recognition domains. Thus, in the case of FokI, the active complex consists of an enzyme dimer and two DNA recognition sites. The cleavage domain of FokI is not involved in specific DNA recognition, but contacts in its dimeric form both DNA strands at one cleavage position (9/13) bp apart from one of the two coordinated DNA recognition sites (the first numeral in the parentheses indicates the position of cleavage in the strand written and the second numeral the cleavage position on the complementary strand).
The Type IIS REases following mode D, such as BspMI, are similar to Type IIF REases because they are also tetrameric enzymes, require two copies of their DNA recognition sequence and cleave concertedly at both cleavage sites (54). However, in contrast to Type IIF endonucleases, the mode D Type IIS enzymes recognize an asymmetric DNA sequence. Therefore, the domain organization of tetrameric Type IIS enzymes will probably differ from that of Type IIF enzymes.
Kinetic studies showed that the Type IIS REases follow different reaction mechanisms (54). Most probably, these different modes of action refer to different structures of the enzymes with respect to domain organization and to oligomerization state. Moreover, it became clear that many but not all of the Type IIS REases seem to require two copies of their asymmetric recognition sequence before they can cleave the DNA. Thus, despite obvious differences between the three subtypes IIE, IIF and IIS, some of the apparently monomeric Type IIS REases presumably do not remain monomeric when bound to DNA and probably interact with two sites like Type IIE and IIF endonucleases. On the other hand, it has been shown that the unusual REases SgrAI and Sau3AI are monomers in solution like Type IIS REases, but recognize palindromic recognition sequences and interact with DNA as tetramers or dimers similarly to Type IIF or Type IIE REases (20,55). Therefore, it appears that the transition between the three subtypes IIE, IIF and IIS is smooth.
CONCLUSIONS
We can only speculate about why some REases require binding to a second DNA recognition site, either using DNA binding domains that structurally differ from endonuclease-like domains (Type IIE and IIS) or by two additional endonuclease subunits (Type IIF). The dependence of cleavage on recognizing two identical and non-modified DNA sites reduces the probability that an REase will accidentally cleave the DNA of its host cell if a single DNA recognition site is unmethylated. Such rare unmethylated recognition sites can be due to incomplete DNA methylation or to DNA repair events.
It should be noted that other REases interact with two DNA sites in a different way. These are Type I (e.g. EcoKI), Type III (e.g. EcoP15I) and at least one of the methylation dependent restriction enzymes, McrBC. All of these enzymes depend on nucleotide triphosphate hydrolysis for activity (56) and communicate with two DNA sites by DNA translocation and not by DNA looping. For the Type III REases, the two non-palindromic DNA sites have to be oriented in the DNA molecule in opposite directions. Thus, the enzyme surveys the substrate not only for the number of DNA sites but also for their respective orientation (57,58). Altogether, the fascinating variety of REases that involve two DNA sites in their reaction mechanism reflects the diversity of their evolutionary origins.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Thomas A. Bickle and Alfred Pingoud for valuable discussions and critical reading of the manuscript. Work in the authors’ laboratory has been supported by Deutsche Forschungsgemeinschaft (grants RE879/2, KR1293/1), Universitäre Forschungsförderung of Humboldt University, Sonnenfeld-Stiftung and Fonds der Chemischen Industrie.
REFERENCES
- 1.Bickle T.A. and Kruger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruger D.H. and Bickle,T.A. (1983) Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol. Rev., 47, 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi I. (2001) Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res., 29, 3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber W. (2000) Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev., 24, 1–7. [DOI] [PubMed] [Google Scholar]
- 5.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts R.J., Belfort,M., Bestor,T., Bhagwat,A.S., Bickle,T.A., Bitinaite,J., Blumenthal,R.M., Degtyarev,S.K., Dryden,D.T., Dybvig,K. et al. (2003) A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res., 31, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pingoud V., Kubareva,E., Stengel,G., Friedhoff,P., Bujnicki,J.M., Urbanke,C., Sudina,A. and Pingoud,A. (2002) Evolutionary relationship between different subgroups of restriction endonucleases. J. Biol. Chem., 277, 14306–14314. [DOI] [PubMed] [Google Scholar]
- 8.Tamulaitis G., Solonin,A.S. and Siksnys,V. (2002) Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett., 518, 17–22. [DOI] [PubMed] [Google Scholar]
- 9.Kovall R.A. and Matthews,B.W. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol., 3, 578–583. [DOI] [PubMed] [Google Scholar]
- 10.Hickman A.B., Li,Y., Mathew,S.V., May,E.W., Craig,N.L. and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell, 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- 11.Tsutakawa S.E., Muto,T., Kawate,T., Jingami,H., Kunishima,N., Ariyoshi,M., Kohda,D., Nakagawa,M. and Morikawa,K. (1999) Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell, 3, 621–628. [DOI] [PubMed] [Google Scholar]
- 12.Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topal M.D., Thresher,R.J., Conrad,M. and Griffith,J. (1991) NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry, 30, 2006–2010. [DOI] [PubMed] [Google Scholar]
- 14.Wentzell L.M. and Halford,S.E. (1998) DNA looping by the SfiI restriction endonuclease. J. Mol. Biol., 281, 433–444. [DOI] [PubMed] [Google Scholar]
- 15.Mucke M., Lurz,R., Mackeldanz,P., Behlke,J., Kruger,D.H. and Reuter,M. (2000) Imaging DNA loops induced by restriction endonuclease EcoRII. A single amino acid substitution uncouples target recognition from cooperative DNA interaction and cleavage. J. Biol. Chem., 275, 30631–30637. [DOI] [PubMed] [Google Scholar]
- 16.Nunes-Duby S.E., Kwon,H.J., Tirumalai,R.S., Ellenberger,T. and Landy,A. (1998) Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res., 26, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jo K. and Topal,M.D. (1995) DNA topoisomerase and recombinase activities in NaeI restriction endonuclease. Science, 267, 1817–1820. [DOI] [PubMed] [Google Scholar]
- 18.Carrick K.L. and Topal,M.D. (2003) Amino acid substitutions at position 43 of NaeI endonuclease. Evidence for changes in NaeI structure. J. Biol. Chem., 278, 9733–9739. [DOI] [PubMed] [Google Scholar]
- 19.Topal M.D. and Conrad,M. (1993) Changing endonuclease EcoRII Tyr308 to Phe abolishes cleavage but not recognition: possible homology with the Int-family of recombinases. Nucleic Acids Res., 21, 2599–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedhoff P., Lurz,R., Luder,G. and Pingoud,A. (2001) Sau3AI, a monomeric type II restriction endonuclease that dimerizes on the DNA and thereby induces DNA loops. J. Biol. Chem., 276, 23581–23588. [DOI] [PubMed] [Google Scholar]
- 21.Kruger D.H., Barcak,G.J., Reuter,M. and Smith,H.O. (1988) EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res., 16, 3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad M. and Topal,M.D. (1989) DNA and spermidine provide a switch mechanism to regulate the activity of restriction enzyme NaeI. Proc. Natl Acad. Sci. USA, 86, 9707–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pein C.D., Reuter,M., Meisel,A., Cech,D. and Kruger,D.H. (1991) Activation of restriction endonuclease EcoRII does not depend on the cleavage of stimulator DNA. Nucleic Acids Res., 19, 5139–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabbara S. and Bhagwat,A.S. (1992) Interaction of EcoRII endonuclease with DNA substrates containing single recognition sites. J. Biol. Chem., 267, 18623–18630. [PubMed] [Google Scholar]
- 25.Pein C.D., Reuter,M., Cech,D. and Kruger,D.H. (1989) Oligonucleotide duplexes containing CC(A/T)GG stimulate cleavage of refractory DNA by restriction endonuclease EcoRII. FEBS Lett., 245, 141–144. [DOI] [PubMed] [Google Scholar]
- 26.Piatrauskene O.V., Tashlitskii,V.N., Brevnov,M.G., Bakman,I. and Gromova,E.S. (1996) Kinetic modeling of the mechanism of allosteric interactions of restriction endonuclease EcoRII with two DNA segments. Biokhimiia, 61, 1257–1269. [PubMed] [Google Scholar]
- 27.Schleif R. (1992) DNA looping. Annu. Rev. Biochem., 61, 199–223. [DOI] [PubMed] [Google Scholar]
- 28.Conrad M. and Topal,M.D. (1992) Modified DNA fragments activate NaeI cleavage of refractory DNA sites. Nucleic Acids Res., 20, 5127–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Embleton M.L., Siksnys,V. and Halford,S.E. (2001) DNA cleavage reactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol., 311, 503–514. [DOI] [PubMed] [Google Scholar]
- 30.Wentzell L.M., Nobbs,T.J. and Halford,S.E. (1995) The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol., 248, 581–595. [DOI] [PubMed] [Google Scholar]
- 31.Deibert M., Grazulis,S., Sasnauskas,G., Siksnys,V. and Huber,R. (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol., 7, 792–799. [DOI] [PubMed] [Google Scholar]
- 32.Siksnys V., Skirgaila,R., Sasnauskas,G., Urbanke,C., Cherny,D., Grazulis,S. and Huber,R. (1999) The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol., 291, 1105–1118. [DOI] [PubMed] [Google Scholar]
- 33.Petrauskene O.V., Babkina,O.V., Tashlitsky,V.N., Kazankov,G.M. and Gromova,E.S. (1998) EcoRII endonuclease has two identical DNA-binding sites and cleaves one of two co-ordinated recognition sites in one catalytic event. FEBS Lett., 425, 29–34. [DOI] [PubMed] [Google Scholar]
- 34.Colandene J.D. and Topal,M.D. (1998) The domain organization of NaeI endonuclease: separation of binding and catalysis. Proc. Natl Acad. Sci. USA, 95, 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huai Q., Colandene,J.D., Chen,Y., Luo,F., Zhao,Y., Topal,M.D. and Ke,H. (2000) Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase. EMBO J., 19, 3110–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuter M., Schneider-Mergener,J., Kupper,D., Meisel,A., Mackeldanz,P., Kruger,D.H. and Schroeder,C. (1999) Regions of endonuclease EcoRII involved in DNA target recognition identified by membrane-bound peptide repertoires. J. Biol. Chem., 274, 5213–5221. [DOI] [PubMed] [Google Scholar]
- 37.Mucke M., Grelle,G., Behlke,J., Kraft,R., Kruger,D.H. and Reuter,M. (2002) EcoRII: a restriction enzyme evolving recombination functions? EMBO J., 21, 5262–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huai Q., Colandene,J.D., Topal,M.D. and Ke,H. (2001) Structure of NaeI-DNA complex reveals dual-mode DNA recognition and complete dimer rearrangement. Nature Struct. Biol., 8, 665–669. [DOI] [PubMed] [Google Scholar]
- 39.Yang C.C. and Topal,M.D. (1992) Nonidentical DNA-binding sites of endonuclease NaeI recognize different families of sequences flanking the recognition site. Biochemistry, 31, 9657–9664. [DOI] [PubMed] [Google Scholar]
- 40.Kupper D., Reuter,M., Mackeldanz,P., Meisel,A., Alves,J., Schroeder,C. and Kruger,D.H. (1995) Hyperexpressed EcoRII renatured from inclusion bodies and native enzyme both exhibit essential cooperativity with two DNA sites. Protein Expr. Purif., 6, 1–9. [DOI] [PubMed] [Google Scholar]
- 41.Petrauskene O.V., Karpova,E.A., Gromova,E.S. and Guschlbauer,W. (1994) Two subunits of EcoRII restriction endonuclease interact with two DNA recognition sites. Biochem. Biophys. Res. Commun., 198, 885–890. [DOI] [PubMed] [Google Scholar]
- 42.Reuter M., Kupper,D., Meisel,A., Schroeder,C. and Kruger,D.H. (1998) Cooperative binding properties of restriction endonuclease EcoRII with DNA recognition sites. J. Biol. Chem., 273, 8294–8300. [DOI] [PubMed] [Google Scholar]
- 43.Bozic D., Grazulis,S., Siksnys,V. and Huber,R. (1996) Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 Å resolution. J. Mol. Biol., 255, 176–186. [DOI] [PubMed] [Google Scholar]
- 44.Grazulis S., Deibert,M., Rimseliene,R., Skirgaila,R., Sasnauskas,G., Lagunavicius,A., Repin,V., Urbanke,C., Huber,R. and Siksnys,V. (2002) Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res., 30, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milsom S.E., Halford,S.E., Embleton,M.L. and Szczelkun,M.D. (2001) Analysis of DNA looping interactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol., 311, 515–527. [DOI] [PubMed] [Google Scholar]
- 46.Bilcock D.T. and Halford,S.E. (1999) DNA restriction dependent on two recognition sites: activities of the SfiI restriction-modification system in Escherichia coli. Mol. Microbiol., 31, 1243–1254. [DOI] [PubMed] [Google Scholar]
- 47.Szybalski W., Kim,S.C., Hasan,N. and Podhajska,A.J. (1991) Class-IIS restriction enzymes—a review. Gene, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- 48.Wah D.A., Hirsch,J.A., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1997) Structure of the multimodular endonuclease FokI bound to DNA. Nature, 388, 97–100. [DOI] [PubMed] [Google Scholar]
- 49.Li L., Wu,L.P. and Chandrasegaran,S. (1992) Functional domains in FokI restriction endonuclease. Proc. Natl Acad. Sci. USA, 89, 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandrasegaran S. and Smith,J. (1999) Chimeric restriction enzymes: what is next? Biol. Chem., 380, 841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bitinaite J., Wah,D.A., Aggarwal,A.K. and Schildkraut,I. (1998) FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wah D.A., Bitinaite,J., Schildkraut,I. and Aggarwal,A.K. (1998) Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanamee E.S., Santagata,S. and Aggarwal,A.K. (2001) FokI requires two specific DNA sites for cleavage. J. Mol. Biol., 309, 69–78. [DOI] [PubMed] [Google Scholar]
- 54.Bath A.J., Milsom,S.E., Gormley,N.A. and Halford,S.E. (2002) Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem., 277, 4024–4033. [DOI] [PubMed] [Google Scholar]
- 55.Bitinaite J. and Schildkraut,I. (2002) Self-generated DNA termini relax the specificity of SgrAI restriction endonuclease. Proc. Natl Acad. Sci. USA, 99, 1164–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dryden D.T., Murray,N.E. and Rao,D.N. (2001) Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res., 29, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mucke M., Reich,S., Moncke-Buchner,E., Reuter,M. and Kruger,D.H. (2001) DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites. J. Mol. Biol., 312, 687–698. [DOI] [PubMed] [Google Scholar]
- 58.Janscak P., Sandmeier,U., Szczelkun,M.D. and Bickle,T.A. (2001) Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol., 306, 417–431. [DOI] [PubMed] [Google Scholar]