Abstract
Cre recombinase is extensively used to engineer the genome of experimental animals. However, its usefulness is still limited by the lack of an efficient temporal control over its activity. To overcome this, we have developed DiCre, a regulatable fragment complementation system for Cre. The enzyme was split into two moieties that were fused to FKBP12 (FK506-binding protein) and FRB (binding domain of the FKBP12–rapamycin-associated protein), respectively. These can be efficiently heterodimerized by rapamycin. Several variants, based on splitting Cre at different sites and using different linker peptides, were tested in an indicator cell line. The fusion proteins, taken separately, had no recombinase activity. Stable transformants, co-expressing complementing fragments based on splitting Cre between Asn59 and Asn60, displayed low background activity affecting 0.05–0.4% of the cells. Rapamycin induced a rapid recombination, reaching 100% by 48–72 h, with an EC50 of 0.02 nM. Thus, ligand-induced dimerization can efficiently regulate Cre, and should be useful to achieve a tight temporal control of its activity, such as in the case of the creation of conditional knock-out animals.
INTRODUCTION
Cre recombinase is a site-specific recombinase that catalyzes the recombination between two 34 bp sequences called LoxP to excise or invert an intervening sequence or create intermolecular recombinations, such as integration or translocation. Because of its simplicity and efficiency, Cre has been increasingly used to engineer the genome of experimental animals (1,2). However, a major limitation for its use is the lack of temporal control over its activity. Thus, when expressed early in development as is the case when a ‘deleter’ (Cre-expressing) mouse is crossed with a mouse that contains a floxed gene in its genome, the inactivation of the target gene may induce early embryonic death, precluding the study of the role of the gene at later stages. Moreover, even in the absence of embryonic lethality, the appearance of compensatory mechanisms during development can mask the ulterior role of the gene. Finally, when Cre is expressed for a prolonged period or at a high level, its interactions with LoxP-like sequences existing in the mammalian genome (3) can lead to chromosomal rearrangements and disturbances of cell physiology (4–6). Two basic approaches have been explored to circumvent these problems and be able to achieve recombination at any chosen stage during the lifetime of the animal. The first is based on the regulation of the transcription of Cre through the use of an inducible promoter (7–9), while the second uses a fusion Cre protein and post-translational regulation by synthetic steroids (10–13). However, these systems are often not tight enough, and a non-negligible level of leakiness is frequently observed. Given its potential importance, the development of an efficient modulatable Cre system thus remains an important task. In the present work, we have explored a new approach for the regulation of Cre, the controlled complementation of inactive Cre fragments, that we have called DiCre (for dimerizable Cre).
Reconstitution of enzymatic activity through the non-covalent association of complementing fragments had been developed to monitor protein–protein interactions. For that, the two candidate proteins are fused to the inactive fragments of an ‘indicator’ enzyme (14–16). Interaction of the proteins leads to the association of the enzyme fragments that, in turn, leads to the appearance of enzymatic activity. In the converse manner, making the interaction of a given protein partner pair regulatable may allow the control of the activity of the enzyme. The ligand-induced dimerization system (17) based on the heterodimerization by rapamycin of the immunophilin FK506-binding protein (FKBP12) (18) and FKBP12– rapamycin-associated protein (FRAP) (19) offers the possibility of achieving that. Indeed, FKBP12 and the FKBP12–rapamycin-binding (FRB) domain of FRAP (20) can be fused to the inactive fragments of the enzyme. The association of the enzyme moieties, and potentially enzymatic activity, can then be regulated by the addition of rapamycin that will bring about the association of the fused FKBP12 and FRB domain. This approach has been shown to work with some simple monomeric enzymes (14,15,21). The mechanism of action of Cre involves the formation of a highly ordered protein–DNA complex between the two LoxP sequences and four enzyme molecules interacting with each other, followed by a succession of cleavage–religation steps (22). We show that despite this complexity, ligand-induced dimerization can efficiently regulate Cre and represents a promising alternative for the regulation of this enzyme.
MATERIALS AND METHODS
In silico engineering
The crystallographic structures of the Holliday junction (23) (PDB_ID 3crx) and of the FKBP12–rapamycin–FRB complex (24) (PDB_ID 2fap) were used to make structural models of the proposed constructs. A 2-fold axis symmetry was applied to the Cre dimer complex for generating the complete Holliday junction. Analyses of the crystallographic structures were achieved by using the INSIGHT II package (Accelrys, San Diego, CA). Intra- as well as intermolecular close contacts between Cre monomers and LoxP in the Holliday junction were analyzed and computed by using the CSU online server (http://bioinfo.weizmann.ac.il:8500/oca-bin/csu?PDB_ID=3crx) (25). Building of the linkers and merging the Cre domains with linkers and FKBP12 or FRB were performed with the BIOPOLYMER module of INSIGHT II. Energy minimization of the obtained models was performed by using DISCOVER with the DNA structure and the four Cre backbones fixed. The FRB–FKBP–rapamycin complex was tethered on its initial position, while the artificial linkers were completely free.
Vector construction
Sequences coding for the fusion proteins have been constructed by PCR cloning (26) starting from pNLS-iCre (27), a humanized version of Cre recombinase, pCF1E for human FKBP12 and pCGNN-FRB(H1) for FRB [both from Ariad Pharmaceuticals, Cambridge, MA (www.ariad.com/regulationkits)] and cloned into pcDNA3.1 expression vector (Invitrogen, Cergy Pontoise, France). FRB corresponds to the rapamycin-binding domain of human FRAP (amino acids 2021–2113) with a T2098L point mutation. Retroviral vectors for the Cre constructs were obtained by inserting the cassette coding for the constructs into the pQCXIP or pQCXIH bicistronic retroviral backbones, upstream of the IRES sequence (BD Clontech, Palo Alto, CA). The bicistronic MuMLV retroviral backbone p1704 was used to construct the retroviral vector expressing CrePR1 (12) and hygromycin-B-phosphotransferase. The CALNLZ cassette (28) was inserted into a self-inactivating retroviral backbone (pSIR; BD Clontech). Constructs were controlled by sequencing.
Cell culture techniques
Cell culture media and reagents were obtained from Invitrogen. The Rat2 fibroblast line was obtained from ATCC (Rockville, MD) and was grown in standard tissue culture conditions. Cells were transfected using polyethylenimine (29) (25 kDa, Sigma-Aldrich, Saint Quentin Fallavier, France). Retroviral supernatants were produced by transient transfection of the Phoenix-Eco packaging cells and used for infection in the presence of 8 µg/ml polybrene (Sigma-Aldrich). Infected cells were selected using the appropriate selection marker and cloned using limiting dilution. The antibiotics were maintained during expansion and propagation of the clones, and were removed before the experiments. Growth curves were obtained by plating cells in 24-well plates (2.0 × 104 cells/well) and counting the number of cells after trypsinization between 4 and 72 h post plating.
Test of Cre activity
Rat2/CALNLZ cells were plated in 24-well plates and transfected 6 h later with pcDNA3.1 expression vector containing the constructs to be tested (1 µg DNA/well). The relative quantities of the constructs were set to have an equal copy number of the plasmids coding for the complementing Cre moieties. The medium was changed the next day and rapamycin was added. β-Galactosidase staining was performed 48 h after transfection and the number of blue (X-gal-positive) cells/well was counted. To analyze Cre activity in stable transfectants, cells were plated in 24-well plates and grown for 48–72 h in the presence of the dimerizer. After fixation and β-galactosidase staining, the percentage of blue cells was evaluated from five randomly taken micrographs per well. Experiments were repeated at least three times. Data were analyzed using the GraphPad Prism software package (GraphPad Software, San Diego, CA).
Northern and Southern blotting
Nucleic acids were prepared from 60 mm semi-confluent plates (26). Genomic DNA was digested to completion with the chosen restriction enzymes before electrophoresis. Following the transfer onto Hybond-N+ membranes (Amersham Biosciences, Little Chalfont, UK), hybridization was performed using digoxigenin-labeled probes prepared according the manufacturer’s (Roche Diagnostics, Meylan, France) instructions. Bound probes were revealed by chemiluminescence using CSPD as substrate (Roche Diagnostics). Signals were acquired on a ChemiGeniusII station (Syngene, Cambridge, UK) and analyzed with Genetools (Syngene).
RESULTS
General principles of the construction of DiCre
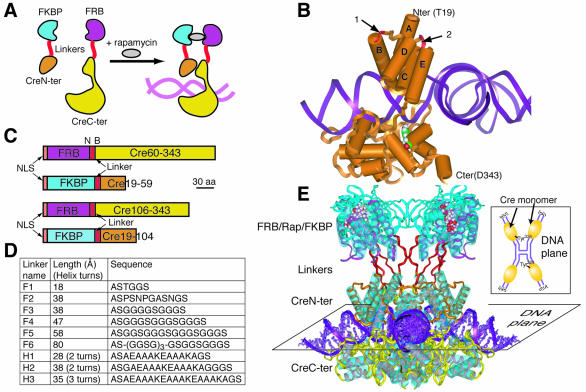
DiCre is based on splitting Cre recombinase into two fragments, devoid of enzymatic activity, that can be reassociated by dimerization to reconstitute activity (Fig. 1A). Cre is composed of two major helical domains joined by a flexible hinge region (22) (Fig. 1B) that form a C-shaped clamp around the DNA. To avoid disrupting the active site, located in the larger C-terminal domain, it was decided to leave this domain intact. Thus, based on the analysis of the published structure of Cre (23), two alternative sites were defined in the N-terminal domain for cutting (Fig. 1B). The first is located between Asn59 and Asn60, producing the fragments designated Cre(59) and Cre(60) for the N- and C-terminal moieties, respectively. The second site is located between Leu104 and Arg106, resulting in the Cre(104) and Cre(106) moieties (note that in this case Pro105 has been deleted to avoid the imposition of a directionality on the linker fused to the N-terminus of the fragment). Both sites are within interhelical loops fully exposed to the solvent and not involved in extensive intra- or intermolecular interactions. Splitting at these sites preserves subdomains of secondary structures (α-helices) that may remain structured when isolated in the fragment.
Figure 1.
Principles of the DiCre system. (A) Scheme of the DiCre system. The enzyme is split into two fragments without enzymatic activity, that are fused to proteins (here FKBP12 and FRB) that can be dimerized by a small molecule ligand. Dimerization leads to the association of the complementing Cre moieties and the reconstitution of enzymatic activity. (B) Ribbon/cylinder model of Cre recombinase, based on the published structure (23). Arrows point to the sites of splitting within the interhelical loops B/C (1) or D/E (2). The Tyr324 residue in the active site within the C-terminal domain has been colored. (C) Schematic representation of the constructs used for DiCre. The translation initiation codon has been placed into the context of Kozak’s consensus sequence (36). NLS corresponds to a nuclear localization signal peptide (37). The unique NheI (N) and BamHI (B) sites flanking the linker sequence allow its simple replacement with variants. (D) List of the different linkers tested. Sequences were defined to obtain flexible (38) (linkers F1–F6) or α-helical [AAAKE motif (38); linkers H1–H3] peptides of variable length. The lengths correspond to that of the maximally extended configuration of the linker peptide. (E) Conceptual model of DiCre within the Holliday structure. The insert shows the scheme of the planar Holliday junction, with the four Cre monomers bound to the half LoxP sites. The different portions of the fusion molecules are assembled in distinct planes that are parallel to that of the DNA.
The regulation of DiCre relies on the non-covalent association of the two complementing enzyme moieties through the heterodimerization of FKBP12 and FRB fused to them. To avoid a major steric hindrance that could be induced by the fusion of both the linker + FKBP12 and the linker + FRB peptides with the new termini created by the cutting, and that would impede the reconstitution of an active structure, FKBP12 and FRB were both fused to the N-terminus of the Cre moieties (Fig. 1C). Moreover, the inspection of the structure of Cre indicated that Thr19 lies relatively close to the B/C and D/E loops that are cut (23) and preliminary experiments have indicated that Cre(19–343) possesses the same recombinase activity as full-length Cre (results not shown). Thus, to position FKBP12 in the vicinity of FRB, the first 18 amino acids of Cre were deleted and FKBP12 was fused directly to Thr19. The resulting constructs are designated as 59.X, 104.X, 60.Y and 106.Y (where X and Y stand for the name of the linker, see below).
To evaluate whether the projected constructs were compatible with the spatial constraints of the recombination synapse, they were conceptually modeled using the published structures for Cre within the Holliday junction (23) and for the FKBP12–rapamycin–FRB complex (24). From these models it appears that the peptides merged with Cre can extend orthogonally relative to the plane of the Holliday junction, with the FKBP–rapamycin–FRB complexes assembled in a plane parallel to that plane (see Fig. 1E).
It should be mentioned that prior to the present study, a first DiCre version was constructed and tested. It involved splitting between Arg130 and Ala131, i.e. in the hinge region between the two major domains of Cre, and fusion of the linker–FKBP and linker–FRB to the C- and N-terminal amino acids of the N- and C-terminal fragments. However, no reconstitution of activity was obtained with these constructs (results not shown).
In vivo characterization by transient expression
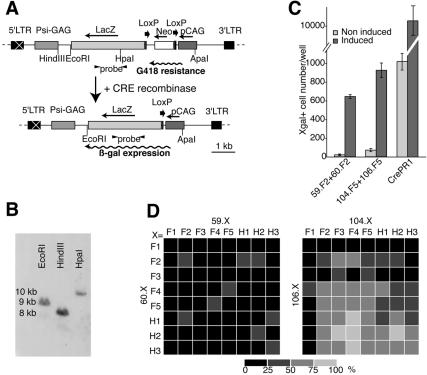
The activity of the different constructs has been evaluated using an indicator cell line corresponding to a rat fibroblast line (Rat2 cells) expressing the construct CALNLZ (28) (Rat2/CALNLZ line; Fig. 2A). To test DiCre in a situation mimicking conditions of potential in vivo applications where single alleles will have to be excised, CALNLZ was introduced in Rat2 cells by a retroviral expression vector to obtain clones with single insertions. One of these clones, for which the existence of a single insertion has been ascertained (Fig. 2B), was used in this study. Note that we have never observed spontaneous excision, i.e. not involving Cre, in these cells.
Figure 2.
Analysis of DiCre by transient transfection. (A) Scheme of the vector used to establish the Rat2/CALNLZ indicator cell line. The transcription of the CALNLZ unit (28) is driven by the CAG promoter. LacZ is transcribed only if a floxed fragment comprising the aminoglycosyd phosphotransferase (neo) gene with a translational stop codon and polyadenylation signal is excised. ‘Probe’ corresponds to the 890 bp fragment used for Southern blots. Neo allows the selection of infected cells. The restriction sites indicated correspond to those used to examine viral integration (B) or recombination (EcoRI–ApaI; Fig. 3D). (B) Southern blot analysis of viral integration of the Rat2/CALNLZ clone 1 used throughout this study. Genomic DNA was digested with EcoRI, HindIII or HpaI. The existence of a single band in each case indicates a single insertion. (C) Example of the recombination in Rat2/CALNLZ cells following transient transfection with different Cre constructs. The example shown corresponds to the results obtained with pairs that were used thereafter to characterize the DiCre system, i.e. 59.F2–60.F2 and 104.F5–106.F5. CrePR1 corresponds to a previously described Cre construct regulated by RU-486 (12). Cre activity has been induced by 50 nM rapamycin (DiCre constructs) or 1 µM RU-486 (CrePR1). (D) Comparison of the level of rapamycin-induced recombination following the transient transfection of the different pairs of complementing Cre fragments. The number of X-gal-positive cells/well was expressed as the percentage of the maximal value obtained (with the 104.F4–106.H2 combination). The percentages were coded by levels of gray as indicated on the right. The results correspond to the mean of four experiments.
For each fusion protein, several variants, differing in the linker peptide inserted between the Cre moiety and FKBP12 or FRB, were tested (Fig. 1D). We tested semi-flexible or semi-rigid linkers (linkers F2 and H1) and also completely flexible or rigid α-helical linkers of variable length (F1 and F3–F5, H2–H3). Each construct was expressed transiently in Rat2/CALNLZ cells either separately or paired, in all pair-wise combinations, with a variant of the complementing Cre fragment, and the number of X-gal-positive cells was evaluated 48 h after the transfection (Fig. 2C and D).
These tests demonstrate that recombination can be regulated by dimerization of complementing Cre constructs. Indeed, when expressed alone, neither of the constructs induced any recombination. When Cre(104) and Cre(106) or Cre(59) and Cre(60) (the Cre moieties without FKBP12 or FRB fused to them) were co-expressed, some recombinase activity, as evidenced by the appearance of a few rare blue cells, could be observed, but their level was independent of the presence of rapamycin. The basal activity was at the same low level after co-transfection of the different 59.X–60.Y or 104.X–106.Y pairs. However, in these cases, the presence of 50 nM rapamycin led to a marked increase in the number of blue cells (Fig. 2C). Note that, independent of the actual combination, the level of recombination obtained with rapamycin was always lower than that obtained using an equimolar amount of Cre(19–343). Similarly, comparison with a previously described regulatable version of Cre, CrePR1 (12), indicated that the activation of CrePR1 by 1 µM RU-486 led to a number of blue cells that was 10–20 times higher than that observed following rapamycin-induced activation of the different DiCre variants (Fig. 2C). However, basal activity of CrePR1 was also considerably higher, and thus, its final performance, in terms of activation ratio (10×), was clearly worse than that of the DiCre constructs (up to 200×).
The level of recombination obtained for the 59.X–60.Y pairs was generally lower than that obtained for the 104.X–106.Y pairs, and this was true for both the non-activated, basal level of excision and the rapamycin-activated excision. The use of different linker arms induced no significant variations of basal activity. There was, conversely, a significant variability of rapamycin-induced activity, with the ratio of the highest to lowest level of excision observed being about 1:5. No systematic trend was evident for the 59.X–60.Y constructs (Fig. 2D) and the variations could not be related to specific features of the linkers. On the other hand, for the 104.X–106.Y combinations, there seems to exist some trend for a lower activated level of excision when the length of the linkers is decreased (especially in the 106.Y constructs). In fact, the use of the 106.F1 construct results in a complete absence of recombination. To control this hypothesis, we designed a long linker (F6, Fig. 1D), to test whether its use would further increase the efficiency of recombination for the 104.X–106.Y variants. We were indeed able to observe that 104.F6–106.F6 had a somewhat increased activity relative to the maximal level observed with the previous constructs, but this increase was limited (∼1.25-fold).
Stable clones
Based on the previous results, three combinations were chosen to obtain stable subclones of Rat2/CALNLZ cells, co-expressing complementing Cre constructs: one mediating a high (use of linker F6) and one a moderate (use of linker F5) level of activated recombination for the 104.X–106.Y version, and one mediating the highest level of recombination for the 59.X–60.Y version (linker F2). Cells were sequentially infected and selected before cloning, and the resulting clones (designated 104.F6–106.F6, 104.F5–106.F5 and 59.F2–60.F2, respectively) were screened for both basal and rapamycin-induced recombination.
There was no difference between the three variants in terms of activated recombination, and exposure to 50 nM rapamycin led to recombination in 100% of the cells for most clones obtained with the DiCre constructs (Table 1). This lack of difference, contrasting with the differences observed following transient expression, might be related to a higher quantity of proteins accumulated within the stable transformants that masked the differences in intrinsic activity of the different constructs. Conversely, there was a significant difference for the basal, unstimulated activity that was low for 59.F2–60.F2 cells, intermediate for 104.F5–106.F5 clones and high for 104.F6–106.F6 clones. Interestingly, clones expressing the Cre moieties without FKBP and FRB fused to them [59/60 and 104/106 clones harboring Cre(59) + Cre(60) and Cre(104) + Cre(106), respectively] also displayed spontaneous recombination, that was, moreover, significantly higher than for cells with the DiCre constructs. In fact, in the case of 104/106 cells, basal activity reached 100% in a number of clones. Finally, for Rat2/CALNLZ cells stably infected with a retroviral vector expressing CrePR1 (CrePR1 cells), background activity was also very high (>60%).
Table 1. Basal and rapamycin-induced recombination in Rat2/CALNLZ clones with stable expression of DiCre.
| Clones | 59.F2–60.F2 (n = 43) | Cre(59)–Cre(60) (n = 33) | 104.F5–106.F5 (n = 36) | 104.F6–106.F6 (n = 23) | Cre(104)–Cre(106) (n = 21) | CrePR1 (n = 4) |
|---|---|---|---|---|---|---|
| Control | 0.20 ± 0.03 | 0.58 ± 0.08* | 4.87 ± 0.83 | 20.14 ± 3.87 | 81.05 ± 4.05* | 66.87 ± 3.64 |
| Activated | 95.1 ± 1.5 | ND | 97.4 ± 2.5 | 98.3 ± 1.2 | ND | 91.4 ± 1.4 |
| Activation ratio | 1038.0 ± 108.5 | – | 40.7 ± 9.4 | 8.4 ± 1.2 | – | 1.4 ± 0.1 |
Results correspond to the percentage of X-gal+ cells present in the absence (‘Control’) or presence (‘Activated’) of dimerizer or RU-486, and are expressed as mean ± SEM. *P < 0.001 relative to corresponding DiCre constructs. ND, not determined. The activation ratio corresponds to the means of the activation ratios of the individual clones.
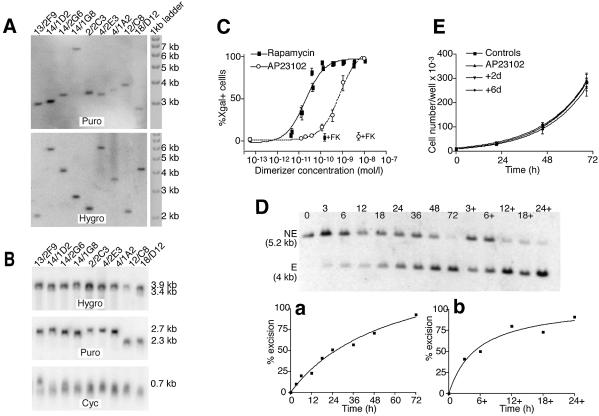
The differences in the background activities seem to be an inherent property of each construct, rather than being related to differences in insertion number or expression levels. Indeed, the analysis of a few representative clones selected from each kind of transformant indicated that they all have a single insertion of each construct (Fig. 3A). Similarly, the mRNA expression for the different constructs was comparable for the different clones and did not correlate with the observed background levels (Fig. 3B).
Figure 3.
Analysis of DiCre expressing stable Rat2/CALNLZ transformants. (A) Southern blot analysis of the integration for the Cre constructs in clones displaying different levels of background activity. The background values (percentage of X-gal+ cells in the absence of dimerizer) for the clones are 59(F2)/60(F5) clones: 13/2F9, 0.1%; 14/1D2, 0.1%; 14/2G6, 0.4%; 14/1G8, 0.05%; 104.F6–106.F6 clone 2/2C3, 25.1; 104.F5–106.F5 clones, 4/2E3, 3.1%; 4/1A2, 36%; 59/60 clone 12/C8, 0.7 %; 104/106 clone 18/D12, 100%. To analyze the 59(F2), 104(F5), 104(F6), Cre(59) and Cre(104) constructs, hybridization was performed with a probe homologous to a 760 bp sequence of the puromycin (Puro) resistance gene of the pQCXIP bicistronic vector, while for the 60(F2), 106(F5), 106(F6), Cre(60) and Cre(106) constructs, an RNA probe of 1030 nucleotides, homologous to the hygromycin (Hygro) resistance gene of the pQCXIH bicistronic vector, was used. For each clone, a single band, indicative of a single insertion, was obtained for both constructs. (B) Northern blot analysis of the level of transcription of the transduced Cre construct in the clones analyzed in (A). The same probes were used as in (A) with an additional 630 bp probe homologous to cyclophilin A (Cyc). The differences in mRNA levels found between the different clones are relatively weak and do not correlate with the level of background activity. (C) Dose–response curves for rapamycin and AP23102. Cells were exposed to the indicated concentrations of the dimerizer for 72 h and the percentage of recombined (X-gal-positive) cells was evaluated by direct counting. +FK corresponds to the level of excision observed when FK-506 is added with the dimerizer (200 nM against rapamycin and 500 nM against AP23109). The results shown correspond to the means of three experiments performed each time on three different 59.F2–60.F2 clones. (D) Kinetics of recombination in 59.F2–60.F2 cells (clone 14/1G8) following exposure to 10 nM AP23102. The dimerizer was added at t = 0 and removed at the indicated times. Genomic DNA was prepared either immediately after the removal of the dimerizer or at t = 72 h (lanes 3+, 6+, 12+, 18+ and 24+). Hybridization was performed with a probe homologous to an internal segment of the provirus (Fig. 2A.); NE corresponds to the fragment obtained when no recombination has occurred; E is the fragment obtained after recombination. (a) Kinetics of recombination when the dimerizer is present continuously for the indicated length of time. (b) Level of recombination observed at t = 72 h with the dimerizer present only for the indicated length of time, beginning at t = 0. (E) Cell proliferation following the transient activation of DiCre by AP23102. Cells were exposed to 10 nM of the dimerizer for 24 h. The cells were cultured for a further 2 (group +2d) or 6 (group +6d) days before the evaluation of proliferation over a subsequent 3-day period. Controls correspond to the cells not exposed to the dimerizer; AP23102 correspond to cells exposed to 10 nM AP23102 only during the period of evaluation of cell proliferation. The results shown are the means obtained with four different 59.F2–60.F2 clones.
Despite the fact that it possesses a lower intrinsic activity, as shown following transient expression, the 59.F2–60.F2 variant appears, among those tested, to be the optimal one to obtain a well regulated system in stable transformants, due to its lower background activity. Thus, four 59.F2–60.F2 clones (13/2F9, 14/1G8, 14/2G6 and 14/1D2) were chosen to characterize the DiCre system further.
The dose–response analysis revealed a high potency for rapamycin, with an EC50 value of 0.02 nM and a maximal activation reached between 0.3 and 1 nM (Fig. 3C). We also examined the potency of the rapamycin analog AP23102 that has fewer side effects than rapamycin, in particular no influence on cell proliferation (T.Clackson and V.Rivera, personal communication; Fig. 3E). Indeed, rapamycin induces numerous side effects, through its interference with endogenous FRAP (30), such as inhibition of cell proliferation, immunosuppression, etc. AP23102, which does not bind wild-type FRAP, is devoid of such side effects, but remains a potent inducer of dimerization due to its high affinity binding to the mutated FRB that was used here. For this reason, AP23102, or other similar analogs of rapamycin, are to be preferred to activate DiCre, both in cell cultures and in whole animals. In the present system, AP23102 has an EC50 value of 0.6 nM and maximal activation is reached at ∼3 nM (Fig. 3C). The effect of the dimerizers was blocked by FK-506, a compound that binds FKBP12 but not FRB and thus acts as a competitive inhibitor of heterodimerization (31) (Fig. 3C). On the other hand, FK-506 had no effect on the excision brought about by infection of Rat2/CALNLZ cells with a lentiviral vector expressing constitutively active Cre (results not shown), indicating that the antagonization of the rapamycin- and AP23102-induced excision is specific.
The kinetics of recombination were examined using Southern blots following exposure of the cells to 10 nM AP23102. Some excision could be observed as soon as 3 h after the beginning of the exposure to the dimerizer, and it increased progressively thereafter, to reach completion by 72 h (Fig. 3D). Moreover, the dimerizer did not have to be present in the incubation medium for all that time, and exposure for only 12–24 h is sufficient to bring about a complete excision 72 h later. Similar results were obtained when the kinetics of excision were assessed by direct counting of the proportion of X-gal+ cells (results not shown).
It has been described that prolonged and/or high level expression of Cre can lead to growth inhibition in mammalian cells, presumably due to its interactions with endogenous LoxP-like sequences (5,6). To test whether Cre expression induced by dimerization for the time needed to achieve full recombination would lead to a similar adverse effect, we exposed cells for 24 h to 10 nM AP23102, and evaluated the cell division rate between 3 and 6 days or between 8 and 10 days following the beginning of the treatment. β-Galactosidase staining confirmed that 100% of the cells had undergone recombination following exposure to the dimerizer. However, no modification of cell proliferation was found (Fig. 3E).
DISCUSSION
The major conclusion of our study is that the controlled complementation of the Cre fragments FKBP-Cre(59) and FRB-Cre(60) can be used to obtain a tight regulation of recombinase activity within the cell. Indeed, background activity of the system is low and induction is both rapid, related to the constitutive expression of the components of the system within the nucleus, and efficient, leading to full recombination in the cells within 48–72 h. In contrast, and somewhat surprisingly, a previously described regulatable form of Cre, CrePR1 (12), displayed a high background activity, affecting >60% of the cells. This background activity is much higher than that originally described for CrePR1, a discrepancy that could be related to the use of different promoters (retroviral versus SV40 promoter) resulting in different levels of expression, of different cell lines (Rat2 versus CV1 cells) that might express different levels of heat shock proteins necessary for the sequestration of CrePR1 in the cytoplasm, and/or the use, in the present study, of cells with single copies for both the indicator and the recombinase constructs. Irrespective of the origin of this discrepancy, when tested in parallel in the same conditions, the dimerizable system appears to work better than CrePR1 as it possesses a much lower background activity and a much higher induction ratio. Although direct comparison has not been performed with other constructs, inspection of the data published for another variant of the RU-486-regulated system (13) or for the tamoxifen-regulated system (10,11) suggest that DiCre possesses a higher induction ratio than these systems, too. It should also be noted that DiCre has the additional advantage of being applicable for modulation of recombinase activity in prokaryotic cells.
The DiCre system displays very high sensitivity to the dimerizers and there is an apparent leftward shift of the dose–response curves relative to published data (15,17,32). However, recombination can occur at a relatively low level of active Cre in the nucleus (6). Thus, with a strong promoter inducing a high level of expression, i.e. when the Cre moieties are present in the nucleus at a high concentration, even a low concentration of the dimerizer, well below its Kd value for the association of FKBP12 and FRB, might be sufficient to bring about a level of activated Cre complex sufficient for recombination, even if the proportion of this complex relative to the total concentration of Cre moieties, as described by the classical binding kinetics, is low.
We found a high level of recombinase activity following co-expression of Cre(104) and Cre(106), i.e. the naked complementary Cre moieties. It is likely that Cre(106), similarly to Cre(119–343) (33), binds to LoxP. However, to achieve recombination, the correct positioning and/or stabilization of the right conformation of Cre(106) by the N-terminal fragment Cre(104) seems to be required. In the native Cre molecule, helices B and D contact the LoxP site, while the loop A/B is engaged in interactions with helix E of the same or adjacent Cre protein (23), and it can be supposed that these interactions also occur for the isolated Cre(104) moieties. Moreover, given the interactions of helices A and B with helices C and D, it is likely that the isolated Cre(104) fragment possesses a structure close to the one it adopts within the intact Cre molecule, and that would also favor its ability to stabilize the complex. The appearance of recombinase activity would then be the consequence of the stepwise assembly, on the LoxP site, of a native-like conformation of Cre by the isolated moieties, involving, first, the binding of a Cre(106) molecule to the LoxP half site, followed by the binding of Cre(104) and the subsequent stabilization of the ensemble through interactions of Cre(104) with Cre(106), and also with Cre(104) moieties fixed to the adjacent LoxP half site. According to this model, the influence of FKBP12 and FRB fused to the Cre moieties is dual. On one hand, due to steric hindrance, they weaken the direct interactions between the two Cre moieties. This is reflected by the decrease of the basal activity observed in 104.F5–106.F5 or 104.F6–106.F6 cells relative to 104–106 cells. On the other hand, their strong association induced by the dimerizer compensates for the first effect and in fact potentiates the interactions between the Cre moieties, helping the emergence of the correct conformation of the assembly and, through that, of enzymatic activity.
The mechanism may be similar for the Cre(59)–Cre(60) pair. However, the interactions of the Cre(59) portion of Cre with the rest of the molecule and with LoxP are less numerous and weaker than those of Cre(104). Moreover, the structure of the isolated Cre(59) fragment reproduces probably less well the structure it adopts within the intact Cre protein. In fact, one can hypothesize that the two α-helices do not form their V-like native structure in the isolated fragment, and that it is the concurrent binding of Cre(59) to LoxP and the Cre(60) fragment bound to the same site that will lead it to adopt a native-like conformation that, in turn, will stabilize the whole complex. This mechanism of action, i.e. interaction-induced folding of proteins from complementary peptide fragments, has been invoked to explain the protein fragment complementation assay (34) that presents similarities with our approach. Note that this mechanism was postulated to work only for monomeric enzymes, while the present example suggests that it might also work for more complex, multimeric, enzymes. These aspects, i.e. weaker binding to LoxP, fewer interactions with Cre(60) and the possible need to restore first a native-like conformation, explain that the stabilization brought about by Cre(59) is much less efficient than that of Cre(104) and, therefore, the background activity in this case is much lower. The fused FRAP and FRB peptides further decrease these interactions, similar to what has been invoked for the Cre(104)–Cre(106) pair. However, in this case, ligand-induced dimerization increases the probability of interactions of the Cre(59) and Cre(60) moieties, well above what the weak DNA binding of Cre(59) can achieve, leading to an efficient restoration of enzymatic activity. Note that if the above model is valid, decreasing the initial binding of the Cre(59) moiety should lower the activity of the Cre(59)–Cre(60) pair and, consequently, the background activity of the FKBP-Cre(59)–FRB-Cre(60) pair. However, if this effect on the stabilization of the complex can be compensated by dimerization, similar to what has been described for other systems (35), one should obtain a globally more efficient system. This hypothesis is currently examined by mutating the residues involved in the interactions of Cre(59).
Our experiments based on transient expression suggest that the intrinsic activity of the DiCre variants based on 104.X–106.Y constructs is higher than that of the variants based on the 59.X–60.Y constructs. This can be related to the more efficient stabilization brought about by the N-terminal Cre-moiety. However, in a stable expression context, the limiting factor seems not to be the intrinsic activity, presumably because of the accumulation of enough protein in the cell to compensate for the differences in the intrinsic activities. The same conclusion can be drawn from the global comparison of CrePR1 and the different DiCre variants. The prime criterion should rather be the background activity of the system, and from this point of view the DiCre system based on Cre(59) and Cre(60) clearly performs better, as not only does it achieve full recombination but it also possesses a low background activity. Thus, it should represent a good solution when a tight temporal control of recombinase activity is required, as in the case of the creation of conditional knock-out animals, and we are presently examining the applicability of this system for that purpose.
Finally, and at a more general level, our results demonstrate that the regulated dimerization of inactive fragments may represent an efficient approach for the regulation, within the cell, of the activity of a given enzyme, even in the case of enzymes possessing a relatively complex mechanism of action. It may thus complement existing approaches, such as the regulation of expression through modulatable promoters or the use of drugs interacting specifically with this enzyme.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr J. Majors (Washington University, St Louis, MO) for the p1704 retroviral backbone, Dr G. Nolan (Stanford University, Stanford, CA) for the ecotropic Phoenix packaging cell line, Dr I. Saito (University of Tokyo, Tokyo, Japan) for pCALNLZ, Dr F. Stewart (Technische Universität, Dresden, Germany) for pNLS-iCre, Dr F. Tronche (Collège de France, Paris, France) for the pZCrePR1(3) construct, Ariad Pharmaceutical (Cambridge, MA; www.ariad.com/regulationkits) for the plasmids pCF1E and pCGNN-FRB(H1) as well as the rapamycin analog AP-23102, and Drs H. Cremer and J.-L. Franc for critical reading of the manuscript. This work was supported by institutional funding from the CNRS and Université de la Méditerranée and grants from the Association Française contre les Myopathies (J.-P.H.)
REFERENCES
- 1.Sauer B. (1998) Inducible gene targeting in mice using the Cre/lox system. Methods, 14, 381–392. [DOI] [PubMed] [Google Scholar]
- 2.Nagy A. (2000) Cre recombinase: the universal reagent for genome tailoring. Genesis, 26, 99–109. [PubMed] [Google Scholar]
- 3.Thyagarajan B., Guimaraes,M.J., Groth,A.C. and Calos,M.P. (2000) Mammalian genomes contain active recombinase recognition sites. Gene, 244, 47–54. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt E.E., Taylor,D.S., Prigge,J.R., Barnett,S. and Capecchi,M.R. (2000) Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl Acad. Sci. USA, 97, 13702–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loonstra A., Vooijs,M., Beverloo,H.B., Allak,B.A., van Drunen,E., Kanaar,R., Berns,A. and Jonkers,J. (2001) Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl Acad. Sci. USA, 98, 9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silver D.P. and Livingston,D.M. (2001) Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell, 8, 233–243. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn R., Schwenk,F., Aguet,M. and Rajewsky,K. (1995) Inducible gene targeting in mice. Science, 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- 8.St-Onge L., Furth,P.A. and Gruss,P. (1996) Temporal control of the Cre recombinase in transgenic mice by a tetracycline responsive promoter. Nucleic Acids Res., 24, 3875–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utomo A.R., Nikitin,A.Y. and Lee,W.H. (1999) Temporal, spatial and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat. Biotechnol., 17, 1091–1096. [DOI] [PubMed] [Google Scholar]
- 10.Feil R., Brocard,J., Mascrez,B., LeMeur,M., Metzger,D. and Chambon,P. (1996) Ligand-activated site-specific recombination in mice. Proc. Natl Acad. Sci. USA, 93, 10887–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Riesterer,C., Ayrall,A.M., Sablitzky,F., Littlewood,T.D. and Reth,M. (1996) Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res., 24, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellendonk C., Tronche,F., Monaghan,A.P., Angrand,P.O., Stewart,F. and Schutz,G. (1996) Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res., 24, 1404–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wunderlich F.T., Wildner,H., Rajewsky,K. and Edenhofer,F. (2001) New variants of inducible Cre recombinase: a novel mutant of Cre–PR fusion protein exhibits enhanced sensitivity and an expanded range of inducibility. Nucleic Acids Res., 29, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelletier J.N., Campbell-Valois,F.X. and Michnick,S.W. (1998) Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl Acad. Sci. USA, 95, 12141–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galarneau A., Primeau,M., Trudeau,L.E. and Michnick,S.W. (2002) β-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein–protein interactions. Nat. Biotechnol., 20, 619–622. [DOI] [PubMed] [Google Scholar]
- 16.Paulmurugan R. and Gambhir,S.S. (2003) Monitoring protein–protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal. Chem., 75, 1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera V.M., Clackson,T., Natesan,S., Pollock,R., Amara,J.F., Keenan,T., Magari,S.R., Phillips,T., Courage,N.L., Cerasoli,F.,Jr et al. (1996) A humanized system for pharmacologic control of gene expression. Nature Med., 2, 1028–1032. [DOI] [PubMed] [Google Scholar]
- 18.Siekierka J.J., Hung,S.H., Poe,M., Lin,C.S. and Sigal,N.H. (1989) A cytosolic binding protein for the immunosuppressant FK506 has peptidyl–prolyl isomerase activity but is distinct from cyclophilin. Nature, 341, 755–757. [DOI] [PubMed] [Google Scholar]
- 19.Chiu M.I., Katz,H. and Berlin,V. (1994) RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl Acad. Sci. USA, 91, 12574–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Zheng,X.F., Brown,E.J. and Schreiber,S.L. (1995) Identification of an 11-kDa FKBP12–rapamycin-binding domain within the 289-kDa FKBP12–rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl Acad. Sci. USA, 92, 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remy I. and Michnick,S.W. (1999) Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl Acad. Sci. USA, 96, 5394–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Duyne G.D. (2001) A structural view of Cre–loxP site-specific recombination. Annu. Rev. Biophys Biomol. Struct., 30, 87–104. [DOI] [PubMed] [Google Scholar]
- 23.Guo F., Gopaul,D.N. and van Duyne,G.D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature, 389, 40–46. [DOI] [PubMed] [Google Scholar]
- 24.Liang J., Choi,J. and Clardy,J. (1999) Refined structure of the FKBP12–rapamycin–FRB ternary complex at 2.2 Å resolution. Acta Crystallogr. D, 55, 736–744. [DOI] [PubMed] [Google Scholar]
- 25.Sobolev V., Sorokine,A., Prilusky,J., Abola,E.E. and Edelman,M. (1999) Automated analysis of interatomic contacts in proteins. Bioinformatics, 15, 327–332. [DOI] [PubMed] [Google Scholar]
- 26.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (2003) Current Protocols in Molecular Biology. John Wiley, New York. [Google Scholar]
- 27.Shimshek D.R., Kim,J., Hubner,M.R., Spergel,D.J., Buchholz,F., Casanova,E., Stewart,A.F., Seeburg,P.H. and Sprengel,R. (2002) Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis, 32, 19–26. [DOI] [PubMed] [Google Scholar]
- 28.Kanegae Y., Lee,G., Sato,Y., Tanaka,M., Nakai,M., Sakaki,T., Sugano,S. and Saito,I. (1995) Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res., 23, 3816–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boussif O., Lezoualc’h,F., Zanta,M.A., Mergny,M.D., Scherman,D., Demeneix,B. and Behr,J.P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham R.T. (1998) Mammalian target of rapamycin: immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr. Opin. Immunol., 10, 330–336. [DOI] [PubMed] [Google Scholar]
- 31.Abraham R.T. and Wiederrecht,G.J. (1996) Immunopharmacology of rapamycin. Annu. Rev. Immunol., 14, 483–510. [DOI] [PubMed] [Google Scholar]
- 32.Pollock R., Issner,R., Zoller,K., Natesan,S., Rivera,V.M. and Clackson,T. (2000) Delivery of a stringent dimerizer-regulated gene expression system in a single retroviral vector. Proc. Natl Acad. Sci. USA, 97, 13221–13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoess R., Abremski,K., Irwin,S., Kendall,M. and Mack,A. (1990) DNA specificity of the Cre recombinase resides in the 25 kDa carboxyl domain of the protein. J. Mol. Biol., 216, 873–882. [DOI] [PubMed] [Google Scholar]
- 34.Michnick S.W. (2001) Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr. Opin. Struct. Biol., 11, 472–477. [DOI] [PubMed] [Google Scholar]
- 35.Johnsson N. and Varshavsky,A. (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA, 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 37.Kalderon D., Roberts,B.L., Richardson,W.D. and Smith,A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- 38.Arai R., Ueda,H., Kitayama,A., Kamiya,N. and Nagamune,T. (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng., 14, 529–532. [DOI] [PubMed] [Google Scholar]