Abstract
Udu has been shown to play an essential role during blood cell development; however, its roles in other cellular processes remain largely unexplored. In addition, ugly duckling (udu) mutants exhibited somite and myotome boundary defects. Our fluorescence-activated cell sorting analysis also showed that the loss of udu function resulted in defective cell cycle progression and comet assay indicated the presence of increased DNA damage in udutu24 mutants. We further showed that the extensive p53-dependent apoptosis in udutu24 mutants is a consequence of activation in the Atm–Chk2 pathway. Udu seems not to be required for DNA repair, because both wild-type and udu embryos similarly respond to and recover from UV treatment. Yeast two-hybrid and coimmunoprecipitation data demonstrated that PAH-L repeats and SANT-L domain of Udu interacts with MCM3 and MCM4. Furthermore, Udu is colocalized with 5-bromo-2′-deoxyuridine and heterochromatin during DNA replication, suggesting a role in maintaining genome integrity.
INTRODUCTION
ugly duckling (udutu24) mutant was first isolated from the 1996 Tübingen screen and has been shown to exhibit a short body-axis with massive cell death (Hammerschmidt et al., 1996). Another udu allele, udusq1, was isolated in a genetic screen aiming at mutants with defects in hematopoiesis (Liu et al., 2007). Positional cloning revealed that Udu protein encodes a novel nuclear factor consisting of three conserved regions (CR-1, CR-2, and CR-3) that do not share similarity with any known domains, two paired amphipathic α-helix like (PAH-L) repeats, and one putative SW13, ADA2, N-Cor and TFIIIB like (SANT-L) domain. The C-terminal PAH-L and SANT-L domains have been shown to be essential in primitive erythroid cell development (Liu et al., 2007). The PAH domain, first identified in yeast SIN3 (Wang et al., 1990), has been demonstrated to mediate protein–protein interactions (Spronk et al., 2000). SIN3 has no intrinsic DNA binding ability and has to be targeted to gene promoters by interacting with DNA binding proteins, thereafter positively and negatively regulating genes involved in diverse cellular functions (Silverstein and Ekwall, 2005). The SANT domain is a highly conserved motif with similarity to Myb DNA binding domain (Aasland et al., 1996), which has been shown to be critical in regulating chromatin accessibility (Boyer et al., 2002, 2004).
The DNA damage response pathway is a cellular surveillance system that senses the presence of damaged DNA and elicits checkpoint activation and subsequent lesion repair in preventing amplification or loss of genes or chromosomes (Zhou and Elledge, 2000). The critical components of DNA damage checkpoint are two phosphatidyl inositol 3-kinase-like kinase family proteins: Ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) (Abraham, 2003; Shiloh, 2003). ATR functions as a sensor and transducer in response to UV light and presumably to genotoxic agents that give rise to stalled replication forks, and subsequently activates Checkpoint kinase (Chk) 1 (Melo et al., 2001), whereas ATM is a central signaling protein that functions in response to DNA double-stranded breaks (DSBs), leading to activation of Chk2 (Shiloh, 2003), which ultimately activates the tumor suppressor p53 (Helton and Chen, 2007). On p53 phosphorylation through ATM, its interaction with Mdm2 is inhibited, resulting in p53 stabilization (Shieh et al., 1997). p53 accumulates in the nucleus and regulates transcription of genes involved in DNA damage response by eliciting cell cycle arrest and/or apoptosis and thereby constraining tumor progression (Lane, 1992; Ljungman, 2000).
Most of the genes targeted by p53 are associated with the regulation of cell cycle arrest, apoptosis and/or DNA repair processes, which function to prevent proliferation of damaged cells. The central components of the cell cycle system are cyclins and cyclin-dependent kinases (CDKs). A major player in the p53-mediated G1 arrest is p21WAF/CIP1 that inhibits cyclin E-CDK2. Thus, an accumulation of p21WAF/CIP1 prevents G1-to-S transition and aberrant replication of damaged DNA (Waldman et al., 1995). p53 also plays a critical role in G2 arrest that allows cells to avoid segregation of defective chromosomes through the regulation of many target genes during G2/M arrest (Bunz et al., 1998). For example, growth-arrest and DNA damage-inducible 45 (GADD45) proteins can associate with p21WAF/CIP1, which inhibits G1-to-S phase transition and also promotes dissociation of the cell division control (Cdc)2/cyclin B1 complex, inducing a G2/M arrest (Fornace et al., 1992; Vairapandi et al., 1996; Wang et al., 1999; Mak and Kultz, 2004).
Minichromosome maintenance (MCM) proteins are conserved in eukaryotes and have essential roles in initiation and elongation during DNA replication (Tye, 1999; Labib et al., 2000; Forsburg, 2004; Pacek and Walter, 2004; Shechter et al., 2004). MCM4, -6, and -7 have been shown to unwind DNA helices in a ATP-dependent manner, whereas the heterohexameric MCM2-7 does not, suggesting that MCM4, -6, and -7 may function as the catalytic core and the replicative helicase during DNA replication (Fujita et al., 1997; Kubota et al., 1997; Labib et al., 2000). Association of the MCM complex with the chromatin is cell cycle regulated: MCM2-7 binds to chromatin only during the G1/S phase and dissociates from as replication proceeds (Kearsey and Labib, 1998). Moreover, loss of MCM function has been demonstrated to cause DNA damage and genome instability (Bailis and Forsburg, 2004).
Here, we investigated what causes p53 up-regulation and developmental defects that are associated with the loss of udu function. We demonstrated that the DNA damage response pathway, containing Atm-Chk2-p53, is activated in udutu24 mutants. The PAH-L and SANT-L domains of Udu were found to be involved in protein–protein interactions with MCM3 and MCM4. Our data indicate that Udu deficiency during zebrafish development causes genomic instability, leading to an activation of the DNA damage checkpoint, which activates p53 and subsequently results in the cell cycle arrest and apoptosis. To our knowledge, this work provides the first biochemical link between PAH-L and SANT-L domains and MCM proteins and a possible functional link between Udu and DNA replication.
MATERIALS AND METHODS
Embryos
Wild-type and udutu24 (Hammerschmidt et al., 1996) embryos were staged as described previously (Kimmel et al., 1995). Dechorinated embryos were placed ∼65 cm from a UV bulb (SYLVANIA Germicidal G30T8/30 W), irradiated for 15 min, and subsequently rinsed several times in E3 medium and then incubated at 28.5°C for 1 h before any subsequent experiments were carried out. Embryos were soaked in KU55933 (ATM kinase inhibitor) at 15 μM and CGK733 (ATM/ATR kinase inhibitor) at 200 μM for 20 h. All experiments on zebrafish embryos were approved by the Biological Research Centre, Agency for Science, Technology and Research (Singapore).
Injection Experiments
Morpholinos (MOs) (Gene Tools, Philomath, OR) were dissolved in 1× Danieau buffer and stored at −20°C. We injected 1.77 pmol of p53-MO (5′-GCGCCATTGCTTTGCAAGAATTG-3′; Langheinrich et al., 2002), 0.78 pmol of atm-MO (5′-GAAAACGGCACCACCTGGTAAA-3′; Yamaguchi et al., 2008), and 2.08 pmol of chk2-MO (5′-CAGACATGATGCTTTTATTCTGGAC-3′; Yamaguchi et al., 2008) into the yolks of one- to four-cell stage embryos.
Genotyping of udutu24 Embryos
Homozygous udutu24 embryos injected with p53-MO were selected based on somite phenotype described and shown in Supplemental Figure S1E. Homozygous udutu24 embryos injected with chk2-MO have tail-tips that were not round-shaped but tapered (see Figure 4N). Selected p53-MO– and chk2-MO–injected udutu24 embryos were subjected to sequencing experiments to confirm their genotype. Homozygous udutu24 embryos were identified via polymerase chain reaction (PCR) amplification of genomic DNA by using forward (5′-TTGGCTCAACCAGTGTAAA-3′) and reverse (5′-TGTGAATGTTACCTAATAGC-3′) primers. Sequencing of these PCR fragments revealed a T-to-A transition in exon 12 of the mutant embryos, shown in Supplemental Figure S1F.
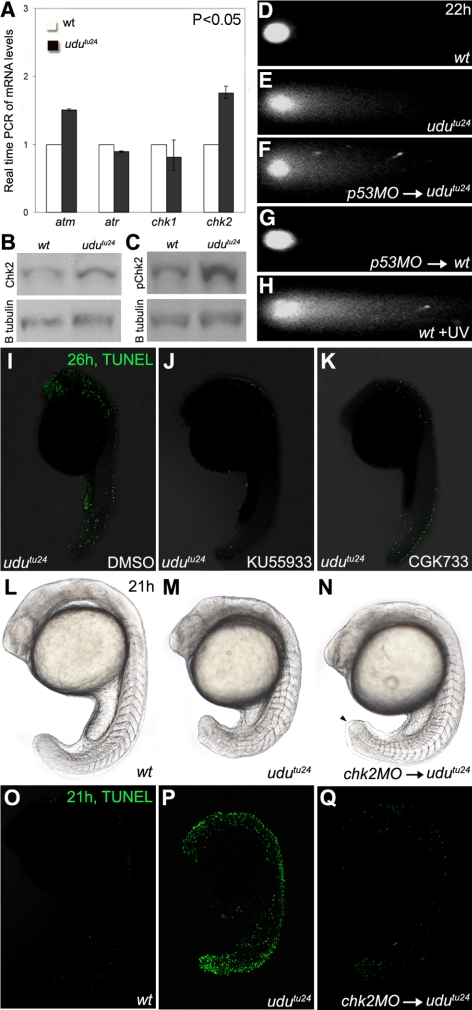
Figure 4.
Activation of Atm-Chk2 pathway in udutu24 mutants. (A) Real-time PCR of atm, atr, chk1 and chk2 mRNA levels extracted from 22 hpf embryos. The histogram is depicted as average ± SD format from three independent experiments (p < 0.05, Student's t test). The transcript level of atm and chk2 are significantly up-regulated in udutu24 mutants. (B and C) Western analyses showed increased levels of Chk2 and phospho-Chk2 (Ser33) in udutu24 embryos compared with wild-type embryos at 22 hpf. (D–H) Neutral single-cell electrophoresis or comet assay of untreated wild-type cells (D), untreated udutu24 cells (E), cells from p53-MO–injected udutu24 embryos (F), cells from p53-MO–injected wild-type embryos (G), and wild-type cells (H) isolated from embryos irradiated with UV for 1 h. The head is composed of intact DNA, whereas the tail consists of DNA with DSB. (I–K) TUNEL staining of udutu24 embryos treated with DMSO (I), 15 μM KU55933 (J), and 200 μM CGK733 (K) at 26 hpf. The number of TUNEL-positive cells is significantly reduced in embryos treated with KU55933 (62.5%; n =5/8) and CGK733 (80%; n = 8/10). (L–N) Phenotypes of chk2 morphants at 21 hpf. (N) Homozygous udutu24 embryos injected with chk2-MO have tail-tips that were not round-shaped but tapered (arrowhead). Somite structures are restored in 2.08 pmol chk2-MO–injected udutu24 embryos, compared with (M) noninjected udutu24 embryos. (O–Q) TUNEL staining of embryos at 21 hpf. (Q) The number of TUNEL-positive cells is significantly reduced in chk2-MO–injected udutu24 embryos, compared with (P) noninjected udutu24 embryos. (L and O) Wild-type embryos as the positive control for phenotypic observations and TUNEL assay, respectively.
RNA Isolation and Quantitative Real-Time PCR
RNA was isolated from wild-type and udutu24 embryos at the respective stage by using TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA was reversely transcribed using Superscript II reverse transcriptase (Invitrogen) and cDNA was amplified with the respective primers designed using Primer Express software, version 3.0 (Applied Biosystems, Foster City, CA). The SYBR Green-based quantitative real-time PCR was carried out on an ABI 7300 real-time PCR system. Reactions were performed using SYBR Green PCR master mix (Applied Biosystems) for one cycle of 95°C for 10 min, followed by 44 cycles of 95°C for 15 s and 60°C for 1 min. elf1a was used as an endogenous control. Relative quantification was performed using Prism Sequence Detection software (Applied Biosystems). Gene accession numbers and primer sequences used for the quantitative real-time PCR are presented in Supplemental Table 1.
Plasmids
The construct of PAH-L and SANT-L domains was made by subcloning the PCR-amplified fragments into pcDNA3.1(+) vector. We used forward (5′-CCTCCACAGATCAGCGAGGGTACCATGGATTACAAGGACGACGATGACAAGGGAGGCCGT-3′) and reverse (5′-CTGCTCTGCAGCCTCGAGCTCTGAACTGGCCTG-3′) primers to amplify the PAH-L and SANT-L domains. HP1β, MCM3, and MCM4 PCR-amplified fragments were made by subcloning the PCR-amplified fragments into pCS2+MT vector. We used forward (5′-AAAAGATCTGGGAATTCCATGGCTGCTGAAGTTGTG-3′) and reverse (5′-ACTGATATTGTCCCGCTCGAGCGGTTAAATCAGGAA-3′) to amplify mcm3; forward (5′-CGCACGTTTTGGAATTCCATGTCTTCACCATCA-3′) and reverse (5′-TCCTGGGATCAGTTGCTCTAGAGCTCAGGGCTTCTCGAT-3′) to amplify mcm4; forward (5′-AGAGCCTCGGGATCCATGAGCCAACCTACA-3′) and reverse (5′-TGACTTGCCATCGATCTAGTTCTTGTCATC-3′) to amplify hp1β. Total RNA was reversely transcribed using SuperScript II reverse transcriptase (Invitrogen), and all PCR amplification experiments were done using Expand High Fidelity PCR System (Roche, Basel, Switzerland). All PCR amplified sequences were verified using SeqMan II, expert sequence analysis (DNASTAR, Madison, WI).
Whole-Mount mRNA In Situ Hybridization (WISH) and Immunohistochemical Staining
Digoxigenin-labeled antisense RNA probes were generated with a Stratagene RNA transcription kit. Single WISH was done as described previously (Qiu et al., 2004). For immunohistochemical staining, embryos were anesthetized, dechorinated and fixed in 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) at 4°C, unless otherwise stated. The following antibodies were used: mouse anti-β-catenin (Abcam, Cambridge, United Kingdom) and polyclonal rabbit anti-phosphorylated histone H3 (pH3) (Cell Signaling Technology, Danvers, MA). The appropriate AlexaFluor-conjugated secondary antibodies (Invitrogen) were used for signal detection. Embryos were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO) to visualize cell nuclei. Embryos labeled with pH3 antibody and DAPI were scanned using confocal microscope, and the numbers of dividing cells and total cells were counted. Immunohistochemical staining of γ-H2AX antibody (Novus Biologicals, Littleton, CO) was carried out on embryos fixed in acetone:methanol (50:50).
5-Bromo-2′-deoxyuridine (BrdU) Incorporation
For labeling of S phase cells, BrdU was incorporated by incubating dechorinated live zebrafish embryos in BrdU solution (10 mM BrdU; 15% dimethyl sulfoxide [DMSO] in E3 medium) for 20 min on ice, followed by washes in E3 medium and further incubation for 1 h at 28.5°C. Embryos were then fixed in 4% PFA/PBS overnight at 4°C. After several washes in 0.1% Tween in PBS (PBST), embryos were incubated in 100% methanol overnight at −20°C. Embryos were permeabilized with proteinase K, refixed in 4% PFA for 20 min, and then rinsed in PBST and 2 N HCl, and followed by incubation in 2 N HCl for 1 h. Subsequently, embryos were blocked for 1 h in blocking solution and incubated with primary anti-BrdU antibody (Sigma-Aldrich) and secondary AlexaFluor488-conjugated anti-mouse antibody (Invitrogen).
Fluorescence-Activated Cell Sorting (FACS) and Cell Cycle Analysis
Whole embryos or tail region posterior to yolk extension at 22 h postfertilization (hpf) were used. Single-cell suspension was obtained as described previously (Ryu et al., 2005). Cells were resuspended in 4 mM citrate buffer, pH 6.5, containing 0.1 mg/ml propidium iodide, 200 μg/ml RNase, and 0.1% Triton X-100. Cell cycle progression was analyzed using FACScan machine (BD Biosciences, San Jose, CA).
Detection of Apoptotic Cells in Whole-Mount and Cryostat Section
The fragmented DNA of apoptotic cells was identified by the terminal deoxynucleotidyl transferase-mediated biotinylated dUTP nick end labeling (TUNEL) method, by using the AP, In Situ Cell Death Detection kit (Roche) and the DeadEnd TUNEL system (Promega, Madison, WI), according to the manufacturer's instructions. For cryostat sections, embryos were embedded in OCT medium as described in the zebrafish book (http://www.zfin.org).
Comet Assays
Neutral comet assay was performed using the CometAssay kit (4250-040-K; Trevigen, Gaithersburg, MD) according to the manufacturer's instruction. DNA was stained with SYBR Green as provided (Trevigen). Comet images were taken by fluorescence microscopy.
Nuclear Protein Extraction
Yolks were removed from zebrafish embryos (22 hpf) by using ice-cold PBS with protease inhibitor cocktail (Roche) by pipetting with a 200-μl tip. Nuclear protein was obtained from the yolk-extirpated embryos according to the protocol described in Davuluri et al. (2008). The extracted proteins were electrophoresed on an SDS-polyacrylamide gel followed by Western blotting with rabbit anti-Chk2 (Cell Signaling Technology) and rabbit anti-phospho-Chk2 (Ser33) (Cell Signaling Technology). We used 5% bovine serum albumin for blocking. β-Tubulin (Abcam) was used as a loading control.
Cell Culture, Transfection, Immunoprecipitation, and Synchronization
COS7 cells were cultured in DMEM supplemented with 10% (vol/vol) fetal bovine serum and 1% penicillin-streptomycin. Cells were transiently transfected using Lipofectamine LTX transfection reagent (Invitrogen). In immunoprecipitation assay, cells were harvested 24 h after transfection in IONIC buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% NP-40, 0.5% deoxycholic acid, and 0.05% SDS, containing protease inhibitor cocktail [Roche]). Cell lysate from 100-mm dish was clarified by centrifugation and incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 4 h at 4°C. The eluted proteins were electrophoresed on an SDS-polyacrylamide gel followed by Western blotting with rabbit anti-Myc (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-FLAG antibodies (Santa Cruz Biotechnology). For cell synchronization, COS7 cells were incubated in 1 μg/ml aphidicolin (Calbiochem, San Diego, CA) for 20 h and then released into aphidicolin-free medium. Cells were confirmed in G1/S border by FACS analysis. Synchronized cells were then incubated in 10 μM BrdU for 10 min at 37°C.
Immunofluorescence
COS7 cells were grown on coverslips under the conditions as described in Cell Culture, Transfection, Immunoprecipitation, and Synchronization. After 24-h transfection, cells were fixed in 4% PFA/PBS for 10 min at room temperature. Fixed cells were permeabilized in 0.1% Triton X-100 in PBS for 10 min and then blocked in blocking solution (10% goat serum in PBS) for 1 h. The cells were then sequentially treated with primary and secondary antibodies diluted in blocking solution for 1 h each at room temperature. The primary antibodies used were rabbit anti-FLAG (Santa Cruz Biotechnology), mouse anti-BrdU (Sigma-Aldrich), and mouse anti-Myc (Santa Cruz Biotechnology) and polyclonal anti-Udu (Liu et al., 2007). The secondary antibodies used were Alexa488 goat anti-rabbit and Alexa568 goat anti-mouse antibodies (Invitrogen).
Microscopy
Embryos were observed on an Axioskop microscope (Carl Zeiss, Jena, Germany) equipped with a camera (Nikon, Tokyo, Japan) for digital image capture. Confocal images of TUNEL and BrdU assays were taken on an inverted 510 LSM laser scanning confocal microscope (Carl Zeiss), and images were processed using LSM image browser (Carl Zeiss). pH3 and γ-H2AX immunohistochemical staining and immunofluorescence assays were taken on a FluoView upright confocal microscope (Olympus, Tokyo, Japan) equipped with FV10-ASW 1.6 viewer. All images were assembled using Photoshop (Adobe Systems, Mountain View, CA).
RESULTS
udutu24 Mutants Exhibit Defects in Somites and Myotome Boundaries
The earliest morphological udutu24 phenotype could be observed at 15.5 hpf (13 somite stage [ss]) to 16.5 hpf (15 ss): the regular somite morphology was lost and tail failed to extend properly (data not shown). At 20 hpf, the short body-axis was more evident; the boundaries of the posterior somites were not chevron-shaped and lacked clear boundaries. Overall, udutu24 mutants displayed a retarded growth, with reduced number of formed somites (Figure 1, A and B). At day 2, udutu24 mutants showed a significant reduction in body length, yolk sac extension as well as smaller head and eyes (Figure 1, C and D). Maternal udu mRNA transcript was detected from one-cell stage and diminished during gastrula; zygotic transcript reappeared in a ubiquitous manner and was enriched in specific tissues later (Liu et al., 2007). The maternal udu contribution may attribute to the late onset of observable phenotypes in udutu24 mutants.
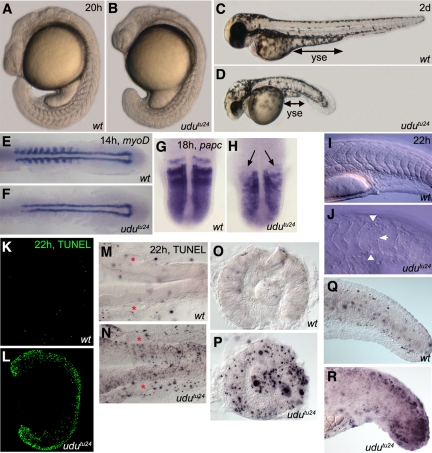
Figure 1.
Developmental and apoptotic defects of udutu24 mutants. Morphology of wild-type and udutu24 mutants at 20 hpf (A and B) and 2 dpf (C and D). (E and F) WISH for myoD at 14 hpf. myoD is only expressed in the adaxial cells and is not detected in the posterior half of formed somites in udutu24 embryos. (G and H) WISH for papc at 18 hpf in dorsal view, with anterior upward. papc is not expressed in the nascent somites of udutu24 embryos; indicated by arrows. (I and J) Normaski images of wild-type and udutu24 embryos' somites (approximately the 12th to 24th somite region). Somites of wild-type embryos were chevron- shaped with clear boundaries; however, mutant somites were U-shaped and their boundaries were less distinct, indicated by arrowheads. All embryos are represented in lateral view with anterior to the left and dorsal upwards, unless otherwise stated. wt, wild type; yse, yolk sac extension. (K and L) TUNEL assay of whole-mount wild-type and udutu24 embryos. Massive amount of apoptotic cells are found throughout udutu24 embryos at 22 hpf. (M–R) Examination of sections of TUNEL staining in lateral view and anterior to the left, unless otherwise stated. Midbrain-hindbrain region, dorsal view (M and N), lens and retina (O and P), and tail region (Q and R) of udutu24 mutants consisted of many apoptotic cells compared with wild-type embryos. Areas marked by red asterisks denote the otic placode; eyes are removed and mounted in lateral view.
Next, we investigated the somite defects in udutu24 mutants with various somite markers. At 14 hpf (10 ss), myoD was expressed in adaxial cells and in the posterior half of the formed somites of wild-type embryos (Weinberg et al., 1996); however, myoD-expressing cells were only found in the adaxial cells of udutu24 mutants, an indication of developmental retardation (Figure 1, E and F). In 18 hpf (18 ss) wild-type embryos, papc is expressed in the anterior parts of nascent somites, segmental stripes in the anterior presomitic mesoderm (PSM) and at lower level in the posterior PSM (Yamamoto et al., 1998). The absence of papc expression in the anterior parts of nascent somites in udutu24 mutants indicated the defects in anterior somite specification (Figure 1, G and H). Normaski images have demonstrated that the somite boundaries of udutu24 embryos were not formed properly (Figure 1, I and J), which were further highlighted by membrane-localized β-catenin antibody (Supplemental Figures S2, A and B).
To examine whether the oscillator mechanism regulating zebrafish somite segmentation is affected, we analyzed deltaC expression, which is a readout of the cycling segmentation clock and prefigures the reiterated somites 60–90 min, time for generating two to three somites, before they physically form a boundary (Jiang et al., 2000). In the mutants, cycling deltaC expression in the PSM was maintained at 12 and 15 ss, suggesting that the clock functions normally, at least at the onset of udu somite phenotype and is not responsible for the somite boundary defects (Supplemental Figure S3).
Dramatically Increased Apoptotic Cells Are Found in Specific Tissues of udutu24 Mutants
Apoptotic cells have been found in udu mutants previously (Hammerschmidt et al., 1996; Liu et al., 2007). We further analyzed the developing embryos for TUNEL-positive cells, to investigate whether the severe morphological abnormality of udutu24 mutants results from a spatial activation of the apoptotic pathway. The udutu24 embryos had an extensive amount of TUNEL-positive cells in head, neural tube, and tail compared with wild-type embryos at 22 hpf (Figure 1, K and L). Examination of the TUNEL staining by sectioning revealed massive apoptotic cells in the midbrain–hindbrain region, the lens, and retina of udutu24 mutants, whereas the otic placode remained unaffected (Figure 1, M–P). The mutant tail region was particularly concentrated with apoptotic cells, corresponding to the observed somite defects and short body length (Figure 1, Q and R). This analysis demonstrated that the increased activation of cellular apoptotic pathway correlates with regions of developmental abnormalities found in udutu24 mutants.
Elevated Level of p53 Causes Apoptosis in udutu24 Embryos
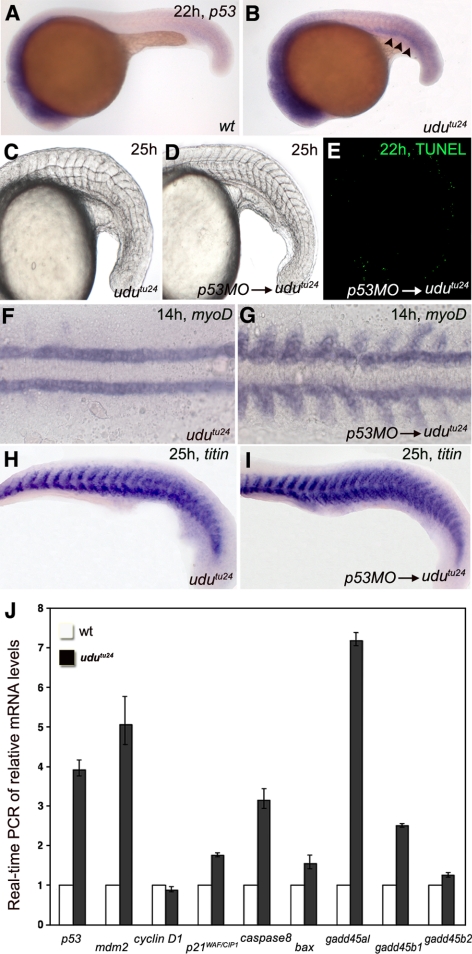
To examine whether the p53 pathway was activated in the apoptotic udutu24 mutants, we examined p53 transcript levels. Up-regulation of p53 mRNA was obvious in the developing brain, pronephron, posterior trunk, and tail bud regions of udutu24 embryos (Figure 2, A and B), which was correlated with the massive amount of TUNEL-positive cells found in corresponding regions (Figure 1, M–R). Next, p53-morpholino (MO) (Langheinrich et al., 2002) injection was performed to examine whether elevated level of p53 in udutu24 mutants causes apoptosis. The knockdown of p53 could significantly reduce the amount of TUNEL-positive cells and rescue somite defects in 65.9% (n = 29/44) of the injected mutants (Figure 2, C–E, compared with Figure 1L). High-resolution images of somite structures are shown in Supplemental Figure S1. Consistently, p53 MO-injected udutu24 mutants displayed a normal expression pattern of myoD and titin (Figure 2, F–I). Further evidence for the rescue of somite boundaries by p53-MO was presented as high-resolution confocal images of embryos labeled with β-catenin antibody (Supplemental Figure S2, C and D). Thus, inhibition of p53 translation can suppress apoptosis and rescue developmental defects in udutu24 mutants.
Figure 2.
p53 and its downstream target genes induce apoptosis in udutu24 embryos. (A and B) WISH of p53 in the zebrafish embryos at 22 hpf displays a higher level of expression in many parts of udutu24 mutants. Arrowheads indicate pronephron. (C–E) Developmental and apoptotic defects in udutu24 embryos can be rescued by inactivating p53 function with MO. (E) TUNEL staining of udutu24 embryos injected with 1.77 pmol of p53-MO. (F and G) WISH of myoD at 14 hpf. Note the myoD-striped pattern in the somites of injected embryos, whereas myoD-expressing cells were only found in the adaxial cells of udutu24 embryos. (H and I) Myotome boundaries of udutu24 embryos can be rescued with p53-MO injection as shown by titin at 25 hpf. (J) Real-time PCR of relative mRNA levels of 22 hpf wild-type and udutu24 embryos. elf1a is the endogenous control for the experiment. The histogram is depicted as average ± SD format from two independent duplicated experiments. Homozygous udutu24 embryos have elevated levels of p53, mdm2, p21WAF/CIP1, caspase 8, bax, gadd45αl, gadd45β1, and gadd45β2 and barely reduced level of cyclin D1 compared with wild-type embryos. All embryos are showed in anterior to the left and dorsal upward.
To investigate whether genes involved in the p53 pathway were up- or down-regulated in udutu24 embryos, real-time PCR was performed to determine the expression levels of p53, mdm2, cyclin D1, p21WAF/CIP1, caspase 8, and bax and genes of the gadd45 family. p53 mRNA was obviously up-regulated in the mutants (Figure 2J), which corresponded to elevated level of p53 shown previously with WISH. The level of cyclin D1 was almost unaltered in udutu24 embryos, whereas downstream targets of the p53 pathway, including mdm2, p21WAF/CIP1, caspase 8, and particularly gadd45αl, were up-regulated. Similarly, WISH confirmed that gadd45αl was indeed up-regulated in the trunk and tail regions of udutu24 embryos (data not shown). These data suggest that the apoptotic udutu24 phenotype is attributed to the activation of p53 and its downstream target genes.
Loss of udu Function Results in Cell Cycle Defects
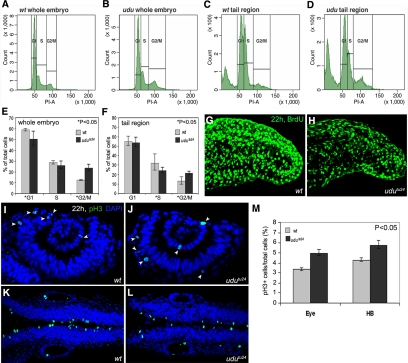
With the up-regulation of p53 and its response genes that are known to be involved in cell cycle and apoptosis, we performed FACS analysis of dissociated cells from whole body and tail region posterior to yolk extension of wild-type and udutu24 embryos stained with propidium-iodide (PI) to examine cell cycle progression. At 22 hpf, udutu24 mutants had a lower fraction of cells in the G1 and S phases and an accumulation of cells in the G2/M phase (Figure 3, A–F). To further verify the reduction of cell number in the S phase, incorporation of BrdU was carried out. The number of BrdU-positive cells was indeed much more reduced in udutu24 tail region posterior to yolk extension (Figure 3, G and H). Anti-pH3 antibody was used to label proliferating cells in the mitotic phase of 22 hpf embryos. Quantification of pH3-positive cells showed that the ratio of pH3-positive cells to the total number of cells at 22 hpf was higher in udutu24 mutants compared with wild-type embryos in the eye and hindbrain regions (Figure 3, I–M). In summary, these results showed that udu mutation indeed affects cell cycle progression.
Figure 3.
Loss of udu function results in aberrant cell cycle. (A–D) Representative traces from FACS analyses after PI staining of dissociated cells at 22 hpf from wild-type and udutu24 embryos. (A and B) Dissociated cells from whole wild-type and udutu24 embryos. (C and D) Dissociated cells from wild-type and udutu24 embryos' tail region posterior to yolk extension. (E and F) Histograms summarize experimental data from FACS analyses after PI staining of dissociated cells from (E) whole embryos and (F) tail region posterior to yolk extension. The histograms are depicted as average ± SD format from triplicate experiments. udutu24 embryos contain fewer cells in the G1 and S phases and have an accumulation of cells in the G2/M phase (asterisks marked those p <0.05, Student's t test; p value for whole embryo: *G1, p = 0.041; S, p = 0.151; *G2/M, p = 0.007; for tail region: G1, p = 0.05; *S, p = 0.035; *G2/M, p = 0.016). (G and H) Projection of the lateral stacks of S phase cells labeled with BrdU at 22 hpf. Images are tail region posterior to yolk extension shown in lateral view with anterior to the left. (I–L) Wild-type and udutu24 embryos stained with pH3 antibody (green, arrowheads); nuclei are counterstained with DAPI (blue): eye region (I and J) and hindbrain region (K and L). (M) Percentage of pH3-positive cells in eye and hindbrain regions are obtained by counting pH3-positive cells versus total cells labeled with DAPI and are summarized in histogram, expressing in average ± SD format from four wild-type and four udutu24 embryos (p < 0.05, Student's t test).
The Atm–Chk2 Pathway Is Activated in udutu24 Mutants Due to DNA Damage
It has been shown that a p53 downstream target gene p21WAF/CIP1, encoding a cyclin-dependent kinase inhibitor, mediates DNA damage-induced cell cycle arrest during G1/S and G2/M transition (Niculescu et al., 1998). In combination with the above-described observations, we suspected that the DNA damage pathway, which functions upstream of p53, may be responsible for udutu24 phenotypes. Real-time PCR was performed to determine the expression levels of atm, atr, chk1, and chk2. It was found that atm and chk2 levels are significantly up-regulated in udutu24 mutants (Figure 4A), suggesting an activation of the Atm–Chk2 pathway. Western analyses further demonstrated increased levels of Chk2 and phospho-Chk2 in udutu24 mutants (Figure 4, B and C), confirming Chk2 activation. Because Atm is activated in response to DSBs (Shiloh, 2003), we then used the neutral comet assay to analyze DSBs in dissociated cells from 22 hpf wild-type and udutu24 embryos. The resulting images resemble a “comet” with a distinct head and tail. The head is composed of intact DNA, whereas the tail consists of DSB DNA. In wild-type embryos, the fluorescence was confined to the comet head, indicating intact and undamaged DNA, whereas in udutu24 mutants, the presence of DSB DNA was depicted by the comet tail (Figure 4, D and E). Comet tail was detected in p53-MO-injected udutu24 mutants, in which apoptosis is inhibited, excluding the possibility that DNA defect originates from apoptosis-induced DNA fragmentation (Figure 4, F and G). Wild-type embryos subjected to UV treatment, which also causes DSB via propagation of clustered single-stranded breaks (SSBs) that occur at closely spaced DNA lesions (Jenner et al., 2001; Rapp and Greulich, 2004; Garinis et al., 2005), were used as a positive control (Figure 4H).
To elucidate whether the cellular phenotype of p53-dependent apoptosis in udutu24 mutants relies on the DNA damage checkpoint, we injected atm-MO and chk2-MO into udutu24 mutants. atm-MO–injected embryos showed severe developmental arrest (data not shown) as reported previously (Yamaguchi et al., 2008). We further tried chemical inhibitors, KU55933 and CGK733, which are kinase inhibitors of ATM and ATM/ATR, respectively (Hickson et al., 2004; Won et al., 2006). Although the somite phenotype could not be rescued, the TUNEL assay showed that treatment with KU55933 (62.5%; n = 5/8) and CGK733 (80%; n = 8/10) could significantly reduce the number of apoptotic cells (Figure 4, I–K). Moreover, posterior segmented somites were restored in 57.4% (n = 27/47) of udutu24 mutants injected with chk2-MO compared with uninjected udutu24 mutants at 21 hpf (Figure 4, L–N). The total number of apoptotic cells was also significantly reduced in the somite-rescued chk2-MO–injected udutu24 mutants (Figure 4, O–Q). These data demonstrate an activation of the Atm–Chk2–p53 signaling pathway in response to DNA damage in udutu24 mutants.
DNA Repair for DSB Seems Functional in udutu24 Mutants
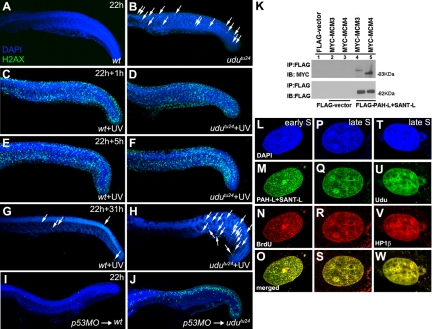
To dissect the mechanism that could potentially induce DNA DSB and activate the Atm–Chk2–p53 pathway, we investigated the role of Udu in cellular processes involving DNA repair and replication. Phosphorylated H2AX, referred to as γ-H2AX, can be detected within minutes after the induction of DSB and is required for the recruitment of several DNA repair proteins to the DNA damage sites (Paull et al., 2000). Each γ-H2AX focus is assumed to label one DSB site (Rothkamm and Löbrich, 2003). The slightly increased number of γ-H2AX foci found in non–UV-treated udutu24 mutants indicated the presence of DNA DSB, which is consistent with the previously-shown comet assay (Figure 5, A and B). After 1-h irradiation, the number of γ-H2AX foci was significantly increased, and they dispersed throughout the trunk and tail regions of both wild-type and udutu24 embryos (Figure 5, C and D). Thus, udutu24 mutants were able to response to DNA damage by recruiting DNA repair proteins via γ-H2AX. The density and intensity of γ-H2AX signals 5 h post-UV irradiation was similarly maintained in both wild-type and udutu24 embryos (Figure 5, E and F). It has been reported that repair of DNA DSBs takes a longer time compared with DNA SSBs (Rapp and Greulich, 2004). Indeed, our experimental results showed that cells with γ-H2AX signals were significantly reduced in the trunk and tail regions of both wild-type and udutu24 embryos after 24 h (data not shown) and further reduced after 31 h (Figure 5, G and H) UV irradiation. To further interrogate the effects of inhibition of apoptosis, wild-type and udutu24 embryos were injected with p53-MO. Increased γ-H2AX staining due to damaged DNA can be observed in p53-injected udutu24 embryos, because damaged cells were not cleared via apoptosis and therefore accumulated (Figure 5, I and J). Together, these results further link the loss of udu function with DNA damage and suggest that Udu is not essential for DSB DNA repair.
Figure 5.
Udu dynamics and localization during DNA repair and replication. (A and B) Immunohistochemical staining of γ-H2AX in nonirradiated embryos at 22 hpf embryos: γ-H2AX foci in wild-type embryos (A) and udutu24 embryos (B). (C–H) Wild-type and udutu24 embryos were irradiated with 15 min of UV and stained with γ-H2AX at different time after irradiation. (C and D) γ-H2AX staining after 1 h post-UV treatment. Both UV-treated wild-type and udutu24 embryos showed an increased in γ-H2AX foci. Hence, in the presence of extrinsic DNA damage agents, udutu24 mutants were able to response accordingly by recruiting DNA repair proteins via the activation of γ-H2AX as indicated by the amplified fluorescence staining. (E and F) γ-H2AX staining of embryos 5 h post-UV irradiation. The level of DNA damage after 5 h of repair were similar in both wild-type and udutu24 embryos. (G and H) The number of cells with γ-H2AX signals was significantly reduced in the trunk and tail regions of wild-type and udutu24 embryos after 31 h post-UV irradiation. (I and J) γ-H2AX staining of wild-type and udutu24 embryos injected with p53-MO. White arrows indicate the γ-H2AX foci. (K) Coimmunoprecipitation of MYC-tagged MCM proteins and FLAG-tagged PAH-L+SANT-L domains. FLAG-tagged PAH-L+SANT-L is immunoprecipitated from the cell lysates and detected with anti-FLAG antibody as a control and with anti-MYC antibody to detect the interacting proteins, MCM3 and MCM4, shown in lanes 4 and 5. (L–O) PAH-L and SANT-L domains are associated with replication foci during S phase. COS7 cells were synchronized at the G1/S border with aphidicolin, released into S phase, labeled with BrdU and fixed at different time points and submitted to laser scanning confocal microscopy for detection: DAPI (blue) (L and P), PAH-L+SANT-L (green) (M and Q), BrdU (red) (N and R), and merged images (yellow) (O and S) show the pattern of replication in early S phase (L–O) and late S phase (P–S). (T–W) Localization of Udu in pericentromeric heterochromatin: DAPI (blue) (T), Udu (green) (U), HP1β (red) (V), and merged image (yellow) (W) shows that Udu is colocalized with HP1β, a protein marker for pericentromeric heterochromatin. IB, immunoblotting; IP, immunoprecipitation.
Udu Binds MCM Proteins and Localizes in Chromatin during DNA Replication
To further explore the function of Udu, we engaged in the yeast two-hybrid (Y2H) screen service of Hybrigenics (Paris, France) to discover Udu-interacting partners in a cDNA library made from 18 to 20 hpf zebrafish embryos. Using PAH-L and SANT-L domains together as a bait, MCM3 and MCM4 were found to be interesting among the proteins identified from the Y2H screen. We next performed coimmunoprecipitation to examine whether both MCM3 and MCM4 really bind to PAH-L and SANT-L domains. FLAG-tagged proteins (PAH-L+SANT-L) and MYC-tagged interacting proteins (MCM3 and MCM4) were used and results demonstrated that MCM3 and MCM4 indeed bind to PAH-L and SANT-L domains of Udu (Figure 5K).
Udu protein, containing both PAH-L and SANT-L domains, has been shown to reside in the nucleus (Liu et al., 2007). However, it was done in unsynchronized cells. The available Udu antibody (Liu et al., 2007) failed to detect Udu in zebrafish embryos via immunohistochemical method and to gain better understanding of its function during DNA replication, colocalization experiments were performed. COS7 cells were synchronized at the G1-S border with DNA polymerase inhibitor, aphidicolin (data not shown). After released from the aphidicolin block, BrdU was incorporated into the synchronized cells, fixed at different time points and coimmunostained with antibodies against BrdU and FLAG-tagged PAH-L and SANT-L. During the early S phase, BrdU incorporation was seen in granular pattern throughout the nucleus (Figure 5, L–O). Five hours after being released into S phase, cells exhibited large BrdU foci that are colocalized with PAH-L and SANT-L domains, indicating the involvement of PAH-L and SANT-L domains during DNA replication (Figure 5, P–S).
Replication of euchromatin occurs in early S phase, whereas pericentromeric heterochromatin is replicated in late S phase (Fox et al., 1991; O'Keefe et al., 1992). We further proceeded to investigate whether Udu is localized in the replicating pericentromeric heterochromatin by staining the cells for Udu or PAH-L and SANT-L domains and HP1β, a marker for pericentromeric heterochromatin (Wreggett et al., 1994). Indeed, Udu (and PAH-L+SANT-L) foci were colocalized with HP1β foci (Figure 5, T–W; data not shown). Together, these data suggest a possible role of Udu in protecting the genome integrity. The loss of Udu function, particularly lacking of PAH-L and SANT-L domains, can cause DNA damage, resulting in the activation of DNA damage pathway and, subsequently, cell cycle arrest and apoptosis.
DISCUSSION
Udu Deficiency Activates the Atm–Chk2–p53 Signaling Pathway
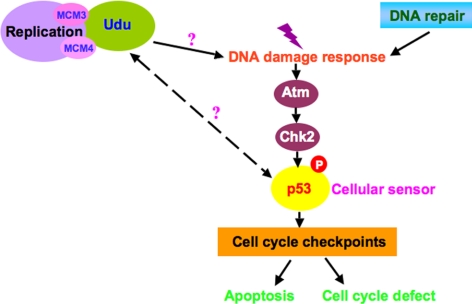
Previous studies have shown that Udu protein is critical in regulating cell cycle progression and differentiation of the primitive erythroid lineage in a p53-dependent manner in udusq1 mutants. Both udutu24 and udusq1 alleles displayed similar phenotypes and sequence analysis revealed that mutations are respectively located in exon 12 (T1461 to A) and exon 21 (T2976 to A), resulting in a premature stop codon (Liu et al., 2007). Although apoptosis involving in the p53 pathway has been described in udusq1 mutants, cellular events leading to an up-regulation of p53 have not been investigated. Here, we demonstrated that udu apoptosis is p53-dependent and established a major role of the Atm-Chk2 signaling pathway in response to DNA damage as summarized in Figure 6.
Figure 6.
Model of Udu function in DNA damage response. Udu may be required for DNA replication by interacting with MCM3 and MCM4. Without Udu, particularly its PAH-L and SANT-L domains, replication cannot proceed properly and DNA damage response is activated via Atm-Chk2-p53 pathway. However, the molecular mechanism leading to DNA damage response is not clear. Furthermore, it is unknown whether Udu has a more direct role in regulating p53 transcriptionally. The cell cycle defects found in udu mutants could be replication-linked, in addition to the effect of p53.
The Mdm2 protein is a key regulator for both p53 nuclear localization and stability by actively participating in the nuclear export and degradation of p53 (Lane and Hall, 1997; Boyd et al., 2000; Geyer et al., 2000). Unexpectedly, the up-regulated mdm2 mRNA shown in our real-time PCR is not sufficient to reverse the effects of increased p53 activation, presumably due to the increased p53 transcript, in udutu24 mutants. Our results indicated that the Atm–Chk2 pathway is activated in response to DNA damage, which thereafter causes p53-dependent apoptosis in udutu24 mutants. It has been shown that Atm phosphorylates p53 on serine-15 and Mdm2 on serine-395; Chk2 phosphorylates p53 on serine-20 (Chehab et al., 1999; Maya et al., 2001; Shiloh, 2001). These phosphorylation modifications prevent p53–Mdm2 association that targets p53 for proteolysis and hence may contribute to DNA damage-induced p53 stabilization (Khosravi et al., 1999).
Is Udu Required for DNA Replication?
Our findings have raised the interesting question of what actually causes DNA damage in udutu24 mutants. Based on the following reasons, it is possible that DNA damage found in udutu24 mutants is caused by the dysfunction in DNA replication. First, the ability to form γ-H2AX foci after UV treatment in udutu24 mutants suggests that the DNA repair mechanism and, presumably, most of its corresponding proteins are largely unaffected. Second, Y2H and coimmunoprecipitation show that Udu binds to MCM3 and MCM4. Third, Udu localizes in chromatin during the S phase in synchronized cells. Fourth, mcm2, mcm4, and mcm5 expression level is down-regulated in udusq1 mutants (Liu et al., 2007), probably due to the elevated p53 expression (Scian et al., 2008) and/or cross-regulation between MCM proteins (Fitch et al., 2003). And last, Udu contains PAH-L and SANT-L domains, which have been shown to be involved in transcription and/or chromatin remodeling.
DNA replication occurs at specialized sites, known as the replication origins, during the S phase of the cell cycle. Before DNA replication starts, the DNA helix must be unwound by MCM proteins that act as DNA helicase to allow the access of numerous accessory proteins, including DNA polymerases, proliferating cell nuclear antigen, primase, topoisomerase, and DNA ligase (Bell and Dutta, 2002). Hence, DNA replication involves dynamic changes of chromatin structure. Studies have shown that the PAH domain is a flexible domain that could function with numerous sequence specific transcription factors and regulate gene transcription (Silverstein and Ekwall, 2005). This implies that Udu protein may form complexes with interacting partners via PAH-L repeats, together with SANT-L domain functioning as a chromatin remodeling complexes (Boyer et al., 2002, 2004). Our data demonstrated that PAH-L and SANT-L domains of Udu binds to MCM3 and MCM4. Hence, Udu in udutu24 mutants, which lacks both PAH-L and SANT-L domains, may be unable to interact with MCM proteins, probably leading to increased DNA damage, indicated by our comet assay. The reduction of BrdU-positive cells in udutu24 mutants further supports our notion that progression though the S phase is delayed or prevented.
MCMs proteins have been shown to be direct targets of ATM/ATR kinases (Cortez et al., 2004; Ishimi et al., 2003; Shi et al., 2007; Yoo et al., 2004), suggesting that the MCMs may be targets or effectors of the replication checkpoint. It has been shown that the phosphorylation of MCM4 in the checkpoint control inhibits DNA replication, which includes blockage of DNA fork progression through inactivation of the MCM complex (Ishimi et al., 2003). Thus, the DNA sequence fidelity, together with associated chromatin structure must be accurately replicated to maintain genetic and epigenetic information through cell generations. To achieve this, it is plausible that chromatin remodeling may play important roles to facilitate the many steps of replication process.
In conclusion, we provide evidence that PAH-L and SANT-L domains of Udu associates with MCM proteins and localizes in chromatin during the S phase, suggesting that these domains may be required for DNA replication. We speculate that the association of Udu with MCM proteins may provide additional factors and/or activation steps on structural modulation of MCM proteins in DNA unwinding at replication origins. Our findings warrant further study of the Udu function in DNA replication.
Is Udu Coupled with Cell Cycle?
To maintain the genome stability, the cell cycle control system must ensure that replication is completed before chromosome segregation can occur. Blast searches in database revealed that the Udu protein had the highest homology to the human and mouse GON4L in both protein sequence and gene structure (Kuryshev et al., 2006; Liu et al., 2007). Caenorhabditis elegans gon-4, from which the name of the mammalian orthologue was derived, was identified as cell lineage regulator in gonadogenesis of Caenorhabditis elegans. It was also suggested that gon-4 may control expression of genes that drive the cell cycle (Friedman et al., 2000). Further evidence for the involvement of GON4L in cell cycle control comes from the association of the Drosophila GON4L homolog, Cdp1, with cyclin D demonstrated in a Y2H screen (Zhong et al., 2003). Hence, these data suggest that the function of GON4L may be linked to cell cycle control. Actually, we also identified cell cycle defects in udutu24 mutants, although mainly through p53. It will be intriguing to revisit the phenotypes of C. elegans gon-4 and Drosophila cdp1 mutants and examine whether the Atm-Chk2-p53 pathway is activated and whether Udu proteins regulate cell cycle coupling with DNA replication.
In zebrafish, mutants of several proteins required for DNA replication have been identified and characterized: mcm5 (Ryu et al., 2005), DNA polymerase delta1/fla (Plaster et al., 2006) and primase/piy (Yamaguchi et al., 2008). Similar to udu mutants, these mutants all show apoptosis, two of which are also p53-dependent and show cell cycle defects (Ryu et al., 2005; Plaster et al., 2006). Although not yet examined, retinal apoptosis in mcm5 and fla mutants may depend on Atm and/or Chk2. It is unclear why piy mutants activate the DNA damage response via Atm-Chk2-p53 without any or little defects in DNA replication. Nevertheless, the characterization of these mutants suggests that cell cycle and DNA replication are tightly coregulated.
In summary, we have demonstrated the involvement of the Atm–Chk2–p53 pathway in causing the apoptosis in udutu24 mutants and the association of Udu with MCM proteins as well as a possible link between PAH-L and SANT-L domains and DNA replication. These findings lead us to hypothesize a possible role of Udu in maintaining or protecting the genome integrity, as the loss of udu function leads to an activation of the DNA damage response pathway, which potentially eliminates damaged cells via apoptosis. Overall, this could lead us to have a better understanding of Udu and its related members in the regulation of replication and the mechanism underlying p53-dependent apoptosis in maintaining stability and integrity of genome during development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Ichiro Masai for providing chk2 MO and ATM/ATR inhibitors and members of the Jiang lab for helpful discussions. This work was supported by the Biomedical Research Council of Agency for Science, Technology and Research, Singapore.
Abbreviations used:
- ATM
Ataxia telangiectasia mutated
- ATR
ATM- and Rad3-related
- Chk
checkpoint kinase
- GADD45
growth-arrest and DNA damage-inducible 45
- hpf
hours postfertilization
- MCM
minichromosome maintenance
- MO
morpholino
- PAH-L
paired amphipathic α-helix like
- pH3
phosphorylated histone H3
- SANT-L
SW13, ADA2, N-Cor and TFIIIB like
- ss
somite stage
- udu
ugly duckling
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0109) on August 5, 2009.
REFERENCES
- Aasland R., Stewart A. F., Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Abraham R. T. Checkpoint signaling: epigenetic events sound the DNA strand-breaks alarm to the ATM protein kinase. Bioessays. 2003;25:627–630. doi: 10.1002/bies.10310. [DOI] [PubMed] [Google Scholar]
- Bailis J. M., Forsburg S. L. MCM proteins: DNA damage, mutagenesis and repair. Curr. Opin. Genet. Dev. 2004;14:17–21. doi: 10.1016/j.gde.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Boyd S. D., Tsai K. Y., Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Langer M. R., Crowley K. A., Tan S., Denu J. M., Peterson C. L. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Latek R. R., Peterson C. L. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Chehab N. H., Malikzay A., Stavridi E. S., Halazonetis T. D. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Glick G., Elledge S. J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davuluri G., Gong W., Yusuff S., Lorent K., Muthumani M., Dolan A. C., Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000240. e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch M. J., Donato J. J., Tye B. K. Mcm7, a subunit of the presumptive MCM helicase, modulates its own expression in conjunction with Mcm1. J. Biol. Chem. 2003;278:25408–25416. doi: 10.1074/jbc.M300699200. [DOI] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Jackman J., Hollander M. C., Hoffman-Liebermann B., Liebermann D. A. Genotoxic-stress-response genes and growth-arrest genes. gadd, MyD, and other genes induced by treatments eliciting growth arrest. Ann. N.Y. Acad. Sci. 1992;663:139–153. doi: 10.1111/j.1749-6632.1992.tb38657.x. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M. H., Arndt-Jovin D. J., Jovin T. M., Baumann P. H., Robert-Nicoud M. Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J. Cell Sci. 1991;99:247–253. doi: 10.1242/jcs.99.2.247. [DOI] [PubMed] [Google Scholar]
- Friedman L., Santa Anna-Arriola S., Hodgkin J., Kimble J. gon-4, a cell lineage regulator required for gonadogenesis in Caenorhabditis elegans. Dev. Biol. 2000;228:350–362. doi: 10.1006/dbio.2000.9944. [DOI] [PubMed] [Google Scholar]
- Fujita M., Kiyono T., Hayashi Y., Ishibashi M. In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J. Biol. Chem. 1997;272:10928–10935. doi: 10.1074/jbc.272.16.10928. [DOI] [PubMed] [Google Scholar]
- Garinis G. A., et al. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 2005;24:3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R. K., Yu Z. K., Maki C. G. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
- Helton E. S., Chen X. p53 modulation of the DNA damage response. J. Cell. Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Komamura-Kohno Y., Kwon H. J., Yamada K., Nakanishi M. Identification of MCM4 as a target of the DNA replication block checkpoint system. J. Biol. Chem. 2003;278:24644–24650. doi: 10.1074/jbc.M213252200. [DOI] [PubMed] [Google Scholar]
- Jenner T. J., Fulford J., O'Neill P. Contribution of base lesions to radiation-induced clustered DNA damage: implication for models of radiation response. Radiat. Res. 2001;156:590–593. doi: 10.1667/0033-7587(2001)156[0590:cobltr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jiang Y.-J., Aerne B. L., Smithers L., Haddon C., Ish-Horowicz D., Lewis J. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- Kearsey S. E., Labib K. MCM proteins: evolution, properties, and role in DNA replication. Biochim. Biophys. Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- Khosravi R., Maya R., Gottlieb T., Oren M., Shiloh Y., Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc. Natl. Acad. Sci. USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Mimura S., Nishimoto S., Masuda T., Nojima H., Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryshev V. Y., et al. An anthropoid-specific segmental duplication on human chromosome 1q22. Genomics. 2006;88:143–151. doi: 10.1016/j.ygeno.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Labib K., Tercero J. A., Diffley J. F. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lane D. P. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Lane D. P., Hall P. A. MDM2–arbiter of p53's destruction. Trends Biochem. Sci. 1997;22:372–374. doi: 10.1016/s0968-0004(97)01119-5. [DOI] [PubMed] [Google Scholar]
- Langheinrich U., Hennen E., Stott G., Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr. Biol. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- Liu Y., et al. The zebrafish udu gene encodes a novel nuclear factor and is essential for primitive erythroid cell development. Blood. 2007;110:99–106. doi: 10.1182/blood-2006-11-059204. [DOI] [PubMed] [Google Scholar]
- Ljungman M. Dial 9-1-1 for p 53, mechanisms of p53 activation by cellular stress. Neoplasia. 2000;2:208–225. doi: 10.1038/sj.neo.7900073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak S. K., Kultz D. Gadd45 proteins induce G2/M arrest and modulate apoptosis in kidney cells exposed to hyperosmotic stress. J. Biol. Chem. 2004;279:39075–39084. doi: 10.1074/jbc.M406643200. [DOI] [PubMed] [Google Scholar]
- Maya R., et al. ATM-dependent phosphorylation of Mdm2 on serine 395, role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A., Cohen J., Toczyski D. P. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu A. B., 3rd, Chen X., Smeets M., Hengst L., Prives C., Reed S. I. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol. Cell. Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe R. T., Henderson S. C., Spector D. L. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M., Walter J. C. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Plaster N., Sonntag C., Busse C. E., Hammerschmidt M. p53 deficiency rescues apoptosis and differentiation of multiple cell types in zebrafish flathead mutants deficient for zygotic DNA polymerase delta1. Cell Death Differ. 2006;13:223–235. doi: 10.1038/sj.cdd.4401747. [DOI] [PubMed] [Google Scholar]
- Qiu X., Xu H., Haddon C., Lewis J., Jiang Y.-J. Sequence and embryonic expression of three zebrafish fringe genes, lunatic fringe, radical fringe, and manic fringe. Dev. Dyn. 2004;231:621–630. doi: 10.1002/dvdy.20155. [DOI] [PubMed] [Google Scholar]
- Rapp A., Greulich K. O. After double-strand break induction by UV-A, homologous recombination and nonhomologous end joining cooperate at the same DSB if both systems are available. J. Cell Sci. 2004;117:4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]
- Rothkamm K., Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl. Acad. Sci. USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S., Holzschuh J., Erhardt S., Ettl A. K., Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc. Natl. Acad. Sci. USA. 2005;102:18467–18472. doi: 10.1073/pnas.0506187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scian M. J., et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27:2583–2593. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- Shechter D., Ying C. Y., Gautier J. DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J. Biol. Chem. 2004;279:45586–45593. doi: 10.1074/jbc.M407772200. [DOI] [PubMed] [Google Scholar]
- Shi Y., Dodson G. E., Mukhopadhyay P. S., Shanware N. P., Trinh A. T., Tibbetts R. S. Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies. J. Biol. Chem. 2007;282:9236–9243. doi: 10.1074/jbc.M609256200. [DOI] [PubMed] [Google Scholar]
- Shieh S. Y., Ikeda M., Taya Y., Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Silverstein R. A., Ekwall K. Sin 3, a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Spronk C. A., Tessari M., Kaan A. M., Jansen J. F., Vermeulen M., Stunnenberg H. G., Vuister G. W. The Mad1-Sin3B interaction involves a novel helical fold. Nat. Struct. Biol. 2000;7:1100–1104. doi: 10.1038/81944. [DOI] [PubMed] [Google Scholar]
- Tye B. K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Vairapandi M., Balliet A. G., Fornace A. J., Jr., Hoffman B., Liebermann D. A. The differentiation primary response gene MyD118, related to GADD45, encodes for a nuclear protein which interacts with PCNA and p21WAF1/CIP1. Oncogene. 1996;12:2579–2594. [PubMed] [Google Scholar]
- Waldman T., Kinzler K. W., Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- Wang H., Clark I., Nicholson P. R., Herskowitz I., Stillman D. J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol. Cell. Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. W., Zhan Q., Coursen J. D., Khan M. A., Kontny H. U., Yu L., Hollander M. C., O'Connor P. M., Fornace A. J., Jr., Harris C. C. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl. Acad. Sci. USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E. S., Allende M. L., Kelly C. S., Abdelhamid A., Murakami T., Andermann P., Doerre O. G., Grunwald D. J., Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Won J., Kim M., Kim N., Ahn J. H., Lee W. G., Kim S. S., Chang K. Y., Yi Y. W., Kim T. K. Small molecule-based reversible reprogramming of cellular lifespan. Nat. Chem. Biol. 2006;2:369–374. doi: 10.1038/nchembio800. [DOI] [PubMed] [Google Scholar]
- Wreggett K. A., Hill F., James P. S., Hutchings A., Butcher G. W., Singh P. B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell. Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Fujimori-Tonou N., Yoshimura Y., Kishi T., Okamoto H., Masai I. Mutation of DNA primase causes extensive apoptosis of retinal neurons through the activation of DNA damage checkpoint and tumor suppressor p53. Development. 2008;135:1247–1257. doi: 10.1242/dev.011015. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Amacher S. L., Kim S.-H., Geissert D., Kimmel C. B., De Robertis E. M. Zebrafish paraxial protocadherin is a downstream target of spadetail involved in morphogenesis of gastrula mesoderm. Development. 1998;125:3389–3397. doi: 10.1242/dev.125.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Y., Shevchenko A., Shevchenko A., Dunphy W. G. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- Zhong J., Zhang H., Stanyon C. A., Tromp G., Finley R. L., Jr. A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res. 2003;13:2691–2699. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. B., Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.