Abstract
Eukaryotic cells ensure accurate chromosome segregation in mitosis by assembling a microtubule-binding site on each chromosome called the kinetochore that attaches to the mitotic spindle. The kinetochore is assembled specifically during mitosis on a specialized region of each chromosome called the centromere, which is constitutively bound by >15 centromere-specific proteins. These proteins, including centromere proteins A and C (CENP-A and -C), are essential for kinetochore assembly and proper chromosome segregation. How the centromere is assembled and how the centromere promotes mitotic kinetochore formation are poorly understood. We have used Xenopus egg extracts as an in vitro system to study the role of CENP-C in centromere and kinetochore assembly. We show that, unlike the histone variant CENP-A, CENP-C is not maintained at centromeres through spermatogenesis but is assembled at the sperm centromere from the egg cytoplasm. Immunodepletion of CENP-C from metaphase egg extract prevents kinetochore formation on sperm chromatin, and depleted extracts can be complemented with in vitro–translated CENP-C. Using this complementation assay, we have identified CENP-C mutants that localized to centromeres but failed to support kinetochore assembly. We find that the amino terminus of CENP-C promotes kinetochore assembly by ensuring proper targeting of the Mis12/MIND complex and CENP-K.
INTRODUCTION
Cell proliferation requires the equal segregation of the genome between daughter cells during division. Eukaryotic chromosome segregation is accomplished by attaching each replicated chromosome to opposite poles of the mitotic spindle so that chromosomes are equally distributed in anaphase. The interaction site between chromosomes and the mitotic spindle is the kinetochore, a multiprotein complex that assembles in mitosis to bind spindle microtubules. Kinetochores also monitor improper attachment to the spindle through the mitotic checkpoint and directly couple the chromosomes to spindle forces during anaphase segregation (Inoue and Salmon, 1995; Nicklas, 1997; Rieder and Salmon, 1998; Cleveland et al., 2003).
Although kinetochores only assemble in mitosis, the site of kinetochore assembly, the centromere, persists throughout the cell cycle. The first identification of centromere-specific binding proteins came from analysis of scleroderma patient sera and led to the identification of three proteins that constitutively localize to centromeres (Moroi et al., 1980; Earnshaw and Rothfield, 1985). Two of these proteins, CENP-A and -C, are essential for centromere formation in all eukaryotes. CENP-A is a histone H3 variant that replaces histone H3 in centromeric nucleosomes and is required for kinetochore formation (Palmer et al., 1991; Cleveland et al., 2003; Carroll and Straight, 2006). CENP-A chromatin directs the assembly of at least 19 additional centromere proteins (CENP-C, -H, -I, K–U, and -W and the Mis12/MIND proteins: Mis12, Nsl1, Nnf1, and Dsn1) that are required for formation of the mitotic kinetochore (Cheeseman and Desai, 2008; Hori et al., 2008).
Inhibition of CENP-C, another essential centromere protein, either by antibody microinjection or depletion, mimics the phenotype of CENP-A disruption, resulting in a general failure in centromere and kinetochore assembly (Saitoh et al., 1992; Brown et al., 1993; Tomkiel et al., 1994; Fukagawa and Brown, 1997; Fukagawa et al., 1999; Kwon et al., 2007). The mechanism for specifically targeting CENP-C to centromeres is not known. In human cells depletion of the CENP-K member of the CENP-H/I/K complex results in a 50% depletion of CENP-C at centromeres (Cheeseman et al., 2008), but CENP-C is able to localize to centromeres in the absence of the CENP-H or -I proteins (Goshima et al., 2003; Liu et al., 2006). Studies in chicken cells have shown that targeting of CENP-C to the interphase centromere requires the CENP-H/I/K proteins but that, in mitosis, CENP-C can associate with centromeres in the absence of CENP-H/I/K (Fukagawa et al., 2001a; Okada et al., 2006; Kwon et al., 2007; Cheeseman et al., 2008). CENP-C binds to DNA and immunoprecipitates with histone H3-containing nucleosomes but has not been shown to interact directly with CENP-A nucleosomes (Yang et al., 1996; Ando et al., 2002; Hori et al., 2008); thus, how CENP-C is specifically targeted to centromeres remains to be determined.
Several domain mapping and mutagenesis studies have defined regions of CENP-C that are required for stability, DNA binding, dimerization, and centromere localization. The amino terminus of CENP-C confers instability on CENP-C, and fusion of the amino terminus of human CENP-C with heterologous proteins is sufficient to destabilize the fusion protein (Lanini and McKeon, 1995). Both CENP-C's central region and carboxy terminus have been implicated in targeting CENP-C to the centromere and binding alpha-satellite DNA (Lanini and McKeon, 1995; Yang et al., 1996; Song et al., 2002; Trazzi et al., 2002). All known CENP-C homologues contain a highly conserved Mif2 homology/CENP-C signature motif near their carboxy terminus (Brown, 1995; Talbert et al., 2004), and mutants in this conserved domain prevent centromere localization (Meluh and Koshland, 1995; Fukagawa et al., 2001b; Heeger et al., 2005). Another conserved region in the carboxy terminus of CENP-C has been shown to dimerize the protein (Sugimoto et al., 1997; Cohen et al., 2008), implying that the interpretation of previous cellular localization studies of CENP-C mutants may have been confounded by heterodimerization of mutant CENP-C with endogenous wild type CENP-C.
To understand how CENP-C assembles at centromeres and how centromeric CENP-C directs centromere and kinetochore formation, we used an established in vitro system for kinetochore assembly onto sperm chromatin in Xenopus laevis egg extracts. We chose this system for several reasons. First, mitotic Xenopus egg extracts assemble functional kinetochores after sperm addition (Desai et al., 1997; Wood et al., 1997). Second, because egg extracts can be depleted of endogenous proteins and complemented with mutants, they provide an excellent system for dissecting the centromere and kinetochore assembly process. Third, most chromatin proteins are removed from sperm chromatin during spermatogenesis and do not reassemble until fertilization (Philpott and Leno, 1992), making sperm chromatin a relatively naive template for analyzing centromere and kinetochore assembly.
Here we demonstrate that CENP-C is not packaged into sperm chromatin but assembles at centromeres from the egg cytoplasm upon sperm addition. Depletion of CENP-C from egg extract prevents centromere and kinetochore assembly. By complementing the depletion phenotype with CENP-C mutant proteins, we show that the localization of CENP-C to centromeres is primarily controlled through the CENP-C motif and the central DNA-binding domain. We further show that the amino terminus of CENP-C directs centromere and kinetochore assembly through the localization of the Mis12/MIND and CENP-H/I/K complexes.
MATERIALS AND METHODS
Cloning and Antibody Generation
X. laevis CENP-C (GenBank accession number FJ791250) was cloned by screening a lambda phage library made from X. laevis ovary RNA with a PCR fragment of X. tropicalis CENP-C. CENP-K (GenBank accession number NP_001088353) and Nsl1 (GenBank accession number FJ791251) were identified through BLAST analysis (Altschul et al., 1990) and were amplified from a Xenopus ovary cDNA library by PCR. CENP-C sequences from different organisms were aligned using MAFFT (Katoh et al., 2002).
For CENP-C antibody production, a fragment of XlCENP-C (amino acids 207–296) was cloned into pGEX-6P-1 (GE Healthcare, Waukesha, WI) to yield plasmid ASP382, which was expressed in Escherichia coli and purified using glutathione agarose (Sigma, St. Louis, MO) according to the manufacturer's instructions. To generate an affinity column for antibody purification, a six-histidine fusion to the same CENP-C fragment was purified on Ni-NTA agarose (Qiagen, Chatsworth, CA) and coupled to Affigel-10–activated NHS agarose (Bio-Rad, Richmond, CA). Rabbit polyclonal antibodies were affinity-purified against the antigen and eluted in 100 mM glycine, pH 2.5, with 100 mM NaCl. XlCENP-A antibodies were generated as previously described (Maddox et al., 2003). XlCENP-E antibodies were generated as previously described (Wood et al., 1997) using the XlCENP-E rod construct provided by Ken Wood (Cytokinetics Inc., South San Francisco, CA) and Don Cleveland (Ludwig Institute for Cancer Research, San Diego, CA). Peptide antibodies were generated as previously described (Field et al., 1998) against peptides for CENP-K (acetyl-CRHPEDPKRIRLE-amide) and Nsl1 (acetyl-CRPVETTPRETEAKVK-amide) synthesized by Bio-Synthesis (Lewisville, TX). MYC antibody was purified from the supernatant of 9E10 mouse hybridoma cells by protein G affinity chromatography. The Mad2 antibody was provided by Andrew Murray (Harvard University, Cambridge, MA) and Rey-Huei Chen (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan), the Zw10 and XRod antibodies were provided by Geert Kops (University Medical Center Utrecht, Utrecht, The Netherlands), the dynein antibody was provided by Suzanne Pfeffer (Stanford University School of Medicine, Stanford, CA), and P. Todd Stukenberg (University of Virginia, Charlottesville, VA) provided the Nnf1 and Dsn1 antibodies.
For expression of CENP-C using rabbit reticulocyte lysate, the XlCENP-C sequence was codon-optimized for E. coli and rabbit reticulocyte expression by gene synthesis (DNA 2.0, Menlo Park, CA). The resulting sequence and truncation fragments were cloned into modified pCS2+ vectors to generate the MYC-tagged constructs listed in Supplemental Table S1. All constructs except 1533 and 1540 were cloned into a modified pCS2+ backbone with AscI and PacI sites inserted after six copies of the MYC tag (polylinker: CCATGGAGCAAAAGCTCATTTCTGAAGAGGACTTGAATTCGAGGCGCGCCAAATTAATTAACTCGAGCCTCTAGA). For 1020, 1147, 1148, 1150, and 1151, ACC was inserted between the AscI site and the codon for the first indicated amino acid.
During the course of our complementation experiments, we found that N-terminally tagged full-length CENP-C did not complement CENP-K localization as well as untagged CENP-C, even though the N-terminally tagged protein localized normally and complemented CENP-E assembly. We constructed C-terminally tagged versions of CENP-C, ASP1533 and ASP1540, and verified that their loading to centromeres was equivalent to N-terminal tagged versions (Supplemental Figure S3). These C-terminal truncations were used in the experiments shown in Figures 5 and 6 and Supplemental Figure S3. C-terminal MYC-tagged fusions to CENP-C were constructed by cloning the codon-optimized XlCENP-C sequence cloned into a modified pCS2+ backbone with AscI and PacI sites inserted (polylinker: GCAGGATCCCATCGATTCGAATTCGAGGCGCGCCAAATTAATTAACTCGAGCCTCTAGA) after PCR to remove the stop codon and add three copies of the MYC tag at the carboxy terminus followed by a stop codon.
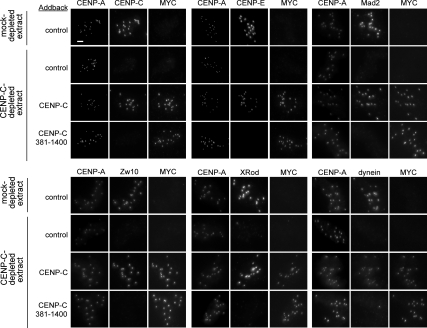
Figure 5.
Kinetochore assembly is rescued by full-length CENP-C but not CENP-C 381-1400. The localization of CENP-A and MYC-CENP-C or MYC-CENP-C 381–1400 with CENP-C, CENP-E, Mad2, Zw10, XRod, and dynein at kinetochores of sperm nuclei incubated in either mock- or CENP-C-depleted extracts after complementation with buffer control, tagged full-length CENP-C, or tagged CENP-C 381–1400 addback. CENP-C is not detected in the CENP-C 381–1400 addback because the antibody was made to the N-terminus of CENP-C. Bar, 5 μm.
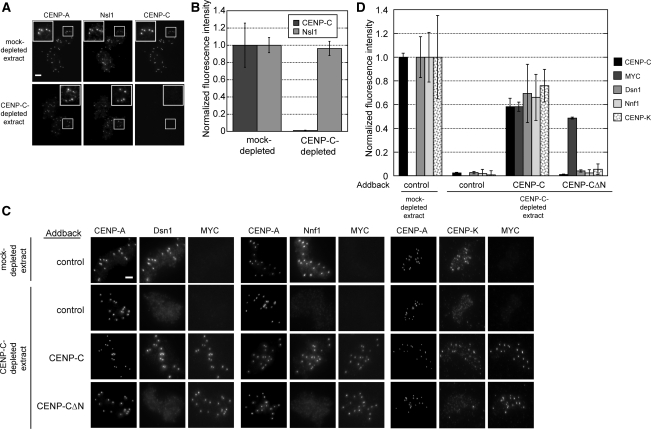
Figure 6.
CENP-C is necessary for the assembly of the Mis12/MIND complex and CENP-K. (A) Nsl1 localization is independent of CENP-C. Representative immunofluorescence image of Nsl1 and CENP-C localization in mock- or CENP-C-depleted extracts are shown. Bar, 5 μm. (B) Quantification of Nsl1 and CENP-C levels (normalized to the levels in mock-depleted extracts) after CENP-C depletion. Error bars, SEM; n = 3. (C) Dsn1, Nnf1, and CENP-K localization in mock- and CENP-C–depleted extracts after complementation with buffer control, full-length CENP-C, or tagged CENP-C 381–1400. Bar, 5 μm. (D) Quantification of Dsn1, Nnf1, and CENP-K levels at sperm centromeres in CENP-C–depleted extracts after buffer control, CENP-C, or CENP-C 381–1400 addback. Error bars, SEM; n = 3.
Xenopus Extracts and Tissue Culture
X. laevis CSF extracts and demembranated sperm were prepared as previously described (Murray, 1991). Nocodazole was added to extracts to 100 μg/ml, and recombinant nondegradable cyclin B was added to extracts to 45 μg/ml.
Xenopus tissue culture (XTC) cells were grown in 70% Leibovitz L-15 media (Invitrogen, Carlsbad, CA) supplemented with 15% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (0.1 mg/ml).
Immunofluorescence
XTC cells grown on coverslips were fixed for 5 min in 2% formaldehyde and then postfixed for 3 min in ice-cold methanol. Coverslips were then rehydrated, blocked, and treated with CENP-A and -C antibodies at 1 μg/ml followed by DNA staining with Hoechst33342 at 1 μg/ml. The localization of CENP-C and -A on demembranated sperm nuclei was assayed by adhering sperm to poly-l-lysine–coated acid-washed coverslips without fixative and then treating sperm with 5 μM recombinant Xenopus Nap-1 (Shintomi et al., 2005) in sperm dilution buffer (10 mM K+HEPES, pH 7.7, 1 mM MgCl2, 100 mM KCl, and 150 mM sucrose) for 30 min. Sperm nuclei from this and egg extract experiments were fixed for 5 min using 2% formaldehyde and processed for immunofluorescence as previously described (Desai et al., 1999) with minor modifications (McClelland et al., 2007). Alexa-conjugated secondary antibodies were used according to the manufacturer's specification (Molecular Probes, Eugene, OR), as was a MYC antibody directly conjugated to Alexa fluor (Cell Signaling Technology, Danvers, MA). Where required, coverslips were blocked using rabbit whole IgG (Jackson ImmunoResearch, West Grove, PA). For three-wavelength detection of centromere proteins, CENP-A and -C antibodies were directly coupled to Alexa fluors (Molecular Probes).
Immunodepletion
Depletion experiments were performed using affinity-purified antibodies bound to Dynabeads protein A beads (Invitrogen). For 100 μl of extract, 0.6 μg of CENP-C antibody or whole rabbit IgG was bound to 33 μl of beads in 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Triton X-100 for 1 h at 4°C. The beads were then washed and resuspended in extract for 1 h at 4°C. Beads were removed from the extract by two 5-min rounds of exposure to a magnet.
In Vitro Translation of Recombinant XlCENP-C
The TnT Sp6-coupled rabbit reticulocyte system (Promega, Madison, WI) was used for in vitro transcription/translation (IVT) of plasmid DNA according to the manufacturer's protocol, except twice the recommended DNA was added for maximum extract expression. For a 40 μl egg extract reaction, 2 μl of IVT reaction were added and mixed before addition of sperm chromatin.
Immunoblotting
Samples were separated by SDS-PAGE and transferred onto nitrocellulose in transfer buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11.3, 0.1% SDS, and 20% methanol] for 2 h and 15 min for detection of CENP-A and -C. CENP-A and -C primary antibodies were both used at 1.5 μg/ml, and either horseradish-peroxidase–conjugated anti-rabbit antibody (1:5000; GE Healthcare) or Alexa488-conjugated anti-rabbit antibody (1:500; Molecular Probes) was used for detection by chemiluminescence or fluorescence, respectively. Fluorescence of detected bands was background-corrected and quantified using ImageJ (http://rsb.info.nih.gov/ij/; Abramoff et al., 2004); background correction was performed by measuring the integrated intensity of a region encompassing the CENP-C band and then subtracting from that the intensity as measured in an identical region above the band in the same lane. CENP-C was quantified relative to the glutathione S-transferase (GST)-tagged portion of CENP-C used as a blotting standard. For egg extract experiments, 1.5 μl of extract was loaded onto each lane. Frog tissue culture nuclei were prepared for immunoblot by swelling XTC cells in 10 mM K+HEPES, pH 8.0, 1.5 mM MgCl2, 10 mM KCl, and 1 mM DTT for 15 min on ice and then shearing by passage through a 23-gauge needle.
Microscopy and Analysis
Immunofluorescence microscopy images were obtained with a 60× 1.4 NA PlanApo objective and a CoolSnap-HQ CCD camera (Photometrics, Tucson, AZ) mounted on an Olympus IX70 microscope (Melville, NY) using a motorized stage and a DeltaVision Spectris system (Applied Precision, Seattle, WA) and deconvolved with Applied Precision software. Axial images were collected at 0.2-μm steps through the entire cell or nucleus and used to construct maximum intensity projections. The fluorescence intensity of centromeres was quantified using MetaMorph software (Molecular Devices, Sunnyvale, CA) by constructing a mask using the CENP-A channel and then applying that mask to images and reading signal intensity at each centromere. More than 100 centromeres were analyzed per condition in each experimental trial. To control for variations in noncentromeric background fluorescence, eight 121-pixel boxes were positioned in noncentromeric regions over each nucleus. The background fluorescence per pixel was calculated for each channel and subtracted from centromere fluorescence measurements in that nucleus.
RESULTS
Characterization of X. laevis CENP-C
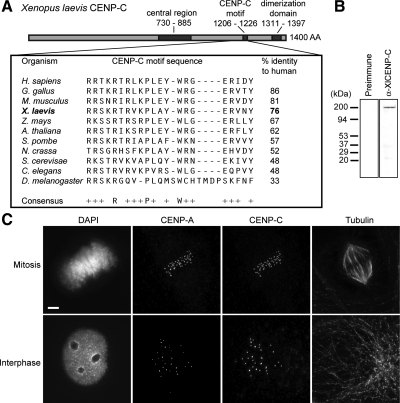
To examine centromere assembly using the frog egg extract system, we cloned the X. laevis CENP-C homologue (XlCENP-C; Figure 1A and Supplemental Figure S1). The 1400-amino acid XlCENP-C is highly homologous to all known CENP-C proteins within the conserved CENP-C signature motif (residues 1206–1226). XlCENP-C also contains the C-terminal cupin domain (residues 1311–1397) that has been shown to dimerize CENP-C (Sugimoto et al., 1997; Cohen et al., 2008) and a central region (residues 730–885) previously shown to be sufficient for DNA binding and centromere localization in human cells (Yang et al., 1996; Sugimoto et al., 1997; Trazzi et al., 2002). We generated a polyclonal antibody to an 89-amino acid fragment in the amino terminus of XlCENP-C (Figure 1B). We stained cultured X. laevis S3 cells with this antibody and observed that XlCENP-C localizes to centromeres in both interphase and mitosis (Figure 1C), consistent with the constitutive cell cycle localization of CENP-C in other eukaryotes (Saitoh et al., 1992; Knehr et al., 1996; Kalitsis et al., 1998b; Dawe et al., 1999; Ogura et al., 2004; Heeger et al., 2005).
Figure 1.
X. laevis CENP-C contains the universal CENP-C motif and localizes to centromeres in tissue culture cells. (A) Domain organization (top) of X. laevis CENP-C and its homology in the CENP-C signature motif (boxed) to organisms ranging from yeast to humans. (B) Western blot of Xenopus tissue culture cell lysate with preimmune sera (Preimmune) or affinity-purified Rabbit antibody raised against Xenopus CENP-C (α-XlCENP-C). (C) Colocalization of CENP-C with the centromeric histone CENP-A by immunofluorescence in interphase and mitotic Xenopus tissue culture cells. Bar, 5 μm.
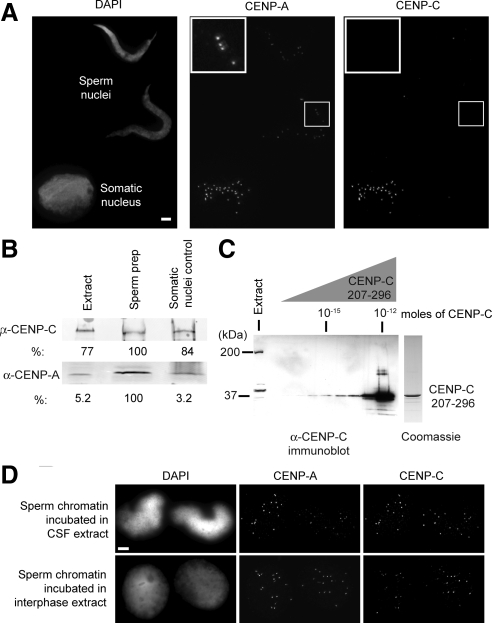
During spermatogenesis, sperm chromatin is extensively remodeled through the replacement of histones H2A and H2B with protamines and the removal of histone H1 (Philpott and Leno, 1992). CENP-A is retained at sperm centromeres during spermatogenesis (Haaf et al., 1990; Palmer et al., 1990; Zeitlin et al., 2005), and CENP-A chromatin serves as the centromere and kinetochore assembly site for each paternal chromosome after fertilization. We characterized CENP-C's assembly in the Xenopus egg extract system in order to determine whether CENP-C is also preserved in sperm chromatin or whether it is assembled at fertilization. We treated demembranated sperm with the histone chaperone nucleosome assembly protein 1 (Nap1) to artificially decondense the sperm chromatin (Mosammaparast et al., 2005; Shintomi et al., 2005) and performed immunofluorescence using CENP-A and -C antibodies. CENP-A was retained on sperm chromatin, but we did not detect CENP-C staining on sperm centromeres (Figure 2A). The lack of detectable CENP-C on sperm chromatin was not likely due to removal by Nap1, as CENP-C is localized at centromeres on contaminating somatic nuclei in the sperm preparation (Figure 2A, Somatic nucleus).
Figure 2.
CENP-C assembly in the Xenopus egg extract system. (A) CENP-C is not retained in Xenopus sperm chromatin. After decondensation with Xenopus Nap-1, Xenopus sperm nuclei show staining for CENP-A but not CENP-C, whereas Xenopus somatic cell nuclei stain for both CENP-A and -C. Bar, 5 μm. (B) Immunoblot of Xenopus sperm preparation and XTC cell nuclei, indicating the levels of CENP-A and -C in the sperm preparation and in somatic nuclei. Crude Xenopus egg extract (Extract), nuclei from the sperm preparation (Sperm prep, 5 × 106 total nuclei with 3.95 × 105 contaminating somatic nuclei), and XTC cell nuclei corresponding to the number of contaminating somatic nuclei in the sperm preparation (Somatic nuclei control, 3.95 × 105 nuclei from XTC cells). (C) CENP-C is ∼10 nM in CSF egg extract by immunoblot. The level of CENP-C in crude Xenopus egg extract (Extract) was compared by Western blotting to serial dilutions of a purified GST fusion of CENP-C (AA207–296) as a standard. A Coomassie-stained gel of the purified GST fusion is shown on the right. (D) CENP-C assembles onto sperm chromatin centromeres after incubation of sperm in either metaphase arrested CSF (top) or interphase (bottom) egg extract. Bar, 5 μm.
Our immunofluorescence data suggested that CENP-A is retained in sperm chromatin but CENP-C is not. By Western blotting we detected both CENP-A and -C in our sperm preparation (Figure 2B); however, ∼8% of the nuclei in our sperm preparation are from contaminating somatic cells. We immunoblotted somatic cell nuclei isolated from cultured Xenopus cells in equivalent number to those contaminating our sperm preparation (Figure 2B). We found that the levels of CENP-C were similar between the sperm preparation and the purified somatic cell nuclei, indicating that the levels of CENP-C in our sperm preparation can be largely accounted for by the contribution from contaminating somatic cells. This is in contrast to the levels of CENP-A in our sperm preparation that are considerably higher than can be accounted for by contaminating somatic cells and thus are consistent with CENP-A being retained in sperm chromatin (Figure 2B).
CENP-C is not present on sperm chromatin, but it is readily detectable in Xenopus egg extracts (Figure 2B). We determined the CENP-C concentration in Xenopus egg extract to be ∼10 nM by comparing the levels in egg extract to a standard of the CENP-C fragment used to generate our antibody (Figure 2C). To assay whether CENP-C could assemble from the egg cytoplasm onto sperm chromatin, we incubated demembranated sperm in either metaphase or interphase egg extract followed by immunofluorescent localization of CENP-C and -A. CENP-C assembles from the egg cytoplasm and colocalizes with CENP-A at centromeres within an hour after sperm addition to the egg extract (Figure 2D). Together, these data demonstrate that CENP-C is absent from sperm chromatin and assembles from the Xenopus egg cytoplasm upon introduction of sperm. Thus, CENP-C assembly onto sperm chromatin is likely to be an early step in centromere assembly after fertilization of the egg.
CENP-C Is Required for Kinetochore Assembly in Xenopus Egg Extract
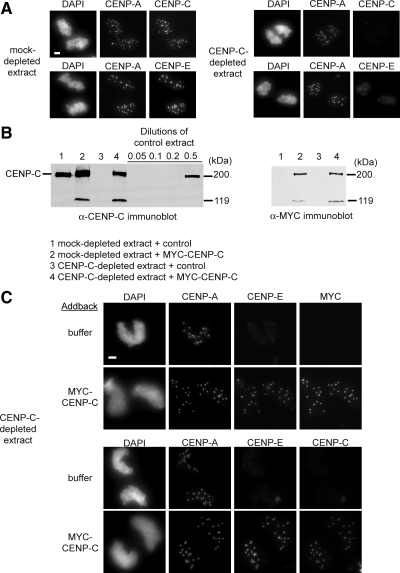
The kinetochore kinesin CENP-E is essential for chromosome alignment, mitotic checkpoint signaling, and kinetochore function in Xenopus egg extract (Wood et al., 1997; Abrieu et al., 2000). In human cells, CENP-E localization to kinetochores depends upon CENP-C (Liu et al., 2006). We used the localization of CENP-E to kinetochores as an assay for kinetochore assembly in mitotic egg extracts. CENP-A, -C, and -E localized normally in mock-depleted egg extracts, whereas extracts depleted of CENP-C showed normal CENP-A localization but lacked CENP-C and -E at centromeres (Figure 3A). To test whether we could rescue the kinetochore assembly defect in CENP-C-depleted extracts, we added in vitro–translated CENP-C, tagged at the amino terminus with six copies of the MYC tag, back into the depleted extract at approximately the same levels as the endogenous protein (Figure 3B). MYC-CENP-C addback rescued both the CENP-C and -E localization defect (Figure 3C). MYC-CENP-C efficiently complemented the depleted extracts as assayed by the percentage sperm nuclei that exhibited CENP-A and -C colocalization at all 18 haploid centromeres (mock depleted, 97 ± 0.57% CENP-C-depleted, 97 ± 0.57% mean ± SEM; n = 3). Our data indicate that CENP-C is required for CENP-E localization in Xenopus egg extracts and establishes an in vitro assay for studying the role of CENP-C in centromere and kinetochore assembly.
Figure 3.
CENP-C is required for CENP-E assembly in Xenopus egg extract. (A) CENP-C–depleted CSF egg extracts fail to assemble CENP-C and -E at centromeres. Xenopus sperm chromatin incubated in mock-depleted extracts (left) and CENP-C–depleted extracts (right) was stained for CENP-A and -C or -E. Bar, 5 μm. (B) Addition of MYC-tagged CENP-C to CENP-C–depleted extracts restores CENP-C to endogenous levels (left). An additional ∼119-kDa results from the in vitro translation reaction used to complement the extract, as seen in the MYC immunoblot (right). (C) Addback of MYC-CENP-C rescues CENP-E kinetochore localization in extracts depleted of endogenous CENP-C. Bar, 5 μm.
Mutants of CENP-C Separate Its Targeting to the Centromere from Its Role in Kinetochore Assembly
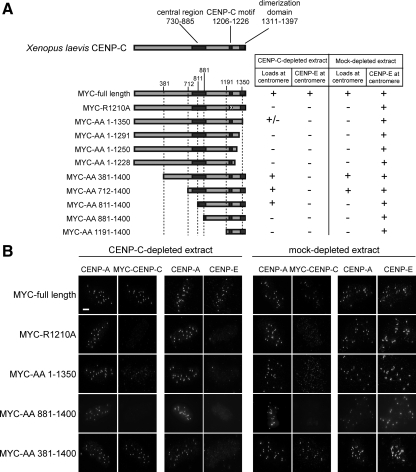
To analyze the determinants of CENP-C assembly at the centromere, we generated a CENP-C truncation series spanning the known functional domains of CENP-C, as well as an arginine to alanine point mutant in a critical residue of the CENP-C motif (R1210A; Fukagawa et al., 2001b; Heeger et al., 2005) and analyzed the ability of each mutant to localize and to support CENP-E assembly in the presence or absence of the endogenous CENP-C (Figure 4A, Supplemental Figure S2). The full-length CENP-C and the truncation mutants (381–1400 and 712–1400) that retained the intact cupin homology domain, the CENP-C signature motif, and the central region were able to localize to centromeres in mock-depleted extracts. The expression of the mutants in the presence of endogenous CENP-C did not have dominant effects, because CENP-E localized normally in all cases. In CENP-C–depleted extracts, the full-length protein and the truncations retaining all three CENP-C domains (381–1400 and 712–1400) localized to centromeres. A partial truncation of the central region (811–1400) and a truncation of the dimerization domain (1–1350) were able to target to centromeres only when CENP-C was depleted, although the truncation of the dimerization domain was present at reduced levels. This result suggests that these truncations contain sufficient targeting information for centromere localization but that the endogenous CENP-C prevents the localization of these mutants. Our complementation experiments show that all three conserved regions of CENP-C are essential for proper localization of CENP-C to the centromere.
Figure 4.
Domain analysis of CENP-C. (A) The CENP-9193C truncations used in the experiment are diagrammed on the left. The regions spanning the central domain, CENP-C motif and cupin domain (from left to right) are shaded dark gray. The results of the extract complementation results for MYC-tagged CENP-C localization and CENP-E localization are indicated on the right. (B) Representative immunofluorescence images of complementation experiments showing CENP-A and MYC-CENP-C or CENP-E. Bar, 5 μm. See also Supplemental Figure S2.
The N-terminal truncation mutants that localize normally to the centromere (381–1400 and 712–1400) are unable to support mitotic kinetochore assembly as assayed by CENP-E localization (Figure 4, A and B). This indicates that the amino terminus of CENP-C is essential for kinetochore assembly. Our mutant analysis demonstrates that the localization of CENP-C to centromeres can be separated from the ability of CENP-C to support kinetochore formation.
The Amino Terminus of CENP-C Is Necessary for Centromere and Kinetochore Assembly
We determined whether the amino terminus of CENP-C was generally required for kinetochore assembly, in addition to being required for CENP-E localization, by analyzing the localization of other mitotic kinetochore proteins in our complementation assay. In Xenopus egg extracts, the Rod/Zw10/Zwilch (RZZ) complex controls the kinetochore localization of the checkpoint protein Mad2 and the motor protein dynein but not CENP-E (Kops et al., 2005). In extracts depleted of CENP-C, we did not detect Mad2, Zw10, Rod, or dynein localization at kinetochores (Figure 5). Our data are consistent with previous experiments in human and chicken cells demonstrating the dependence of these other kinetochore proteins on CENP-C for kinetochore localization (Liu et al., 2006; Kwon et al., 2007).
Complementation of CENP-C depleted extracts with full-length CENP-C rescued the localization defect of all of these kinetochore proteins, indicating that CENP-C depletion did not also deplete other factors required for kinetochore assembly. Complementation of CENP-C–depleted extracts with the CENP-C mutant that lacks the first 381 amino acids (CENP-C 381–1400) failed to rescue the assembly of the RZZ complex, Mad2, dynein, and CENP-E (Figure 5). Our results demonstrate that the assembly of multiple distinct kinetochore proteins depends on the amino terminus of CENP-C.
CENP-C is a member of a group of ∼15 proteins, called the constitutive centromere-associated network (CCAN), that is bound to the centromere throughout the cell cycle and are required for kinetochore assembly. As cells enter mitosis, the CCAN directs the assembly of the KNL-1 protein and Mis12/MIND complex (Mis12, Dsn1, Nnf1, and Nsl1), which in turn recruit the Ndc80 complex (Spc24, Spc25, Ndc80, and Nuf2) during kinetochore assembly (reviewed in Cheeseman and Desai, 2008). In vertebrates, the CENP-H/I/K complex of CCAN proteins is required at the centromere for the assembly of the Ndc80 complex but not the Mis12 complex (Goshima et al., 2003; Liu et al., 2006; McClelland et al., 2007). To determine whether CENP-C directs the centromere localization of the Mis12 or the CENP-H/I/K complexes in Xenopus extracts, we depleted CENP-C from egg extract, incubated sperm in the depleted extract, and assayed the resulting sperm centromeres for the localization of three Mis12 complex proteins (Nsl1, Dsn1, and Nnf1) and the CENP-K protein. In contrast to results in other metazoan systems (Kwon et al., 2007; Przewloka et al., 2007), Nsl1 localization at centromeres was independent of CENP-C (Figure 6, A and B). In Xenopus extracts, the other Mis12 complex proteins, Dsn1 and Nnf1, required CENP-C for their targeting to centromeres (Figure 6C). In human cells, the centromeric localization of Mis12, Dsn1, Nnf1, and Nsl1 is interdependent, and depletion of Nsl1, Nnf1, or Mis12 causes loss of cellular Dsn1 protein (Kline et al., 2006). Our data indicate that Xenopus Nsl1 can be localized to centromeres independently of the other Mis12 complex members. The defect in Mis12 complex localization in CENP-C–depleted extracts is not likely due to codepletion with CENP-C or protein instability because CENP-C depletion does not affect the levels of Mis12, Dsn1, or Nsl1 in the extract (Supplemental Figure S4). The CENP-K protein also required CENP-C for its localization to centromeres, suggesting that the CENP-H/I/K complex is dependent on CENP-C in Xenopus (Figure 6C). Complementation of the depleted extracts with full-length CENP-C rescued Dsn1, Nnf1, and CENP-K targeting. However, addback of CENP-C 381-1400 failed to complement the targeting of Dsn1, Nnf1, and CENP-K, indicating that the amino terminus of CENP-C also directs their localization (Figure 6, C and D).
DISCUSSION
CENP-C is an essential component of the centromere and kinetochore in all eukaryotes. Here we use an in vitro system in Xenopus egg extracts to study the role of CENP-C in centromere and kinetochore assembly. We demonstrate that, before fertilization, sperm chromatin lacks CENP-C and that CENP-C is assembled from the egg cytoplasm onto sperm centromeres. Removal of CENP-C from egg extracts prevents centromere and kinetochore formation, and readdition of CENP-C to the extract rescues centromere and kinetochore assembly. Using this complementation assay, we demonstrate that CENP-C targeting to the centromere requires the central region of CENP-C thought to be involved in DNA binding, the CENP-C signature motif, and (to a lesser extent) the CENP-C cupin domain that may mediate CENP-C dimerization. We generated mutants that separate the targeting of CENP-C to the centromere from its role in centromere and kinetochore assembly. We show that the amino terminus of CENP-C is required for proper kinetochore targeting of proteins involved in spindle checkpoint function and microtubule binding. We also find that CENP-C directs centromere and kinetochore assembly by facilitating the assembly of the Mis12 complex at the centromere, as well as the localization of CENP-K.
Domain Organization of Xenopus CENP-C
Our domain mapping studies have shown that the overall structure of Xenopus CENP-C is conserved with other vertebrate CENP-C proteins. Xenopus CENP-C contains the CENP-C signature motif, and mutation of a conserved arginine (R1210A) in this region prevents CENP-C localization, consistent with mutational studies of the CENP-C motif in other organisms (Fukagawa et al., 2001b; Heeger et al., 2005). The central region of vertebrate CENP-C is thought to mediate DNA binding and has been shown to be important for centromere localization in human and chicken cells (Yang et al., 1996; Sugimoto et al., 1997; Song et al., 2002; Trazzi et al., 2002). We find that this region is required for CENP-C localization to Xenopus centromeres. Yeast CENP-C (Mif2) lacks this conserved region, but it has recently been shown that Mif2 binds specifically to yeast centromere DNA (Cohen et al., 2008); thus, DNA binding appears to be an important feature of CENP-C proteins. The third conserved region between CENP-C proteins is the C-terminal cupin domain, which has been shown to be required for dimerization of the CENP-C protein (Sugimoto et al., 1997; Cohen et al., 2008). Deletion of the cupin domain severely compromises the ability of CENP-C to assemble at sperm centromeres. The amino terminus of CENP-C has previously been shown to confer instability on the protein and has been proposed to mediate oligomerization of CENP-C in vivo (Lanini and McKeon, 1995; Sugimoto et al., 1997). We find that the amino terminus of CENP-C is dispensable for proper centromere localization but essential for kinetochore assembly.
An important feature of our in vitro complementation system in Xenopus egg extracts is that it allows us to test the functions of CENP-C mutants in the absence of significant levels of endogenous protein. In addition, monitoring kinetochore assembly in egg extracts does not require that a specific mutant maintain the viability of the organism or cell. In the presence of the endogenous CENP-C, mutants of CENP-C that lack part of the DNA binding domain or lack the cupin domain (811–1400 and 1–1350, respectively) are unable to localize to centromeres. However, these mutants can bind to the centromere when the endogenous protein is removed, indicating that they possess sufficient information to target properly but cannot compete with wild type CENP-C.
The Amino Terminus of CENP-C Directs Centromere and Kinetochore Assembly
The importance of CENP-C in the process of kinetochore formation is well established from conditional mutant, antibody injection, gene knockout, and siRNA depletion studies (Meeks-Wagner et al., 1986; Brown et al., 1993; Tomkiel et al., 1994; Fukagawa and Brown, 1997; Kalitsis et al., 1998a; Fukagawa et al., 1999; Moore and Roth, 2001; Oegema et al., 2001; Kwon et al., 2007). Using our complementation assay, we have been able to separate the functional domains of CENP-C that direct kinetochore formation and centromere localization. A CENP-C truncation that localizes normally to the centromere but lacks the first 381 amino acids of the amino terminus cannot rescue kinetochore formation in extracts depleted of the endogenous CENP-C. We assayed the assembly of several different protein complexes, including the CENP-E motor protein important for chromosome congression and mitotic checkpoint signaling (Wood et al., 1997; Abrieu et al., 2000), the Mad2 protein required for the mitotic checkpoint (Li and Murray, 1991), and the dynein motor and the RZZ complex necessary for lateral chromosome microtubule interaction and for proper spindle checkpoint signaling (Starr et al., 1998; Howell et al., 2001; Wojcik et al., 2001; Gassmann et al., 2008). Previous work in Xenopus extracts showed that CENP-E assembly at the kinetochore is independent of the RZZ complex, whereas both Mad2 and dynein require the RZZ complex for their localization (Kops et al., 2005). All of these factors were dependent on the amino terminus of CENP-C for their localization to the kinetochore. This is consistent with depletion studies that place other kinetochore proteins downstream of CENP-C (Liu et al., 2006; Kwon et al., 2007) and demonstrates the importance of the amino terminus of CENP-C in directing the assembly of these complexes.
Kinetochore microtubule binding is regulated by the functions of the KMN (KNL1/Mis12/Ndc80) network. The recruitment of the Mis12/MIND complex (Mis12, Nsl1, Dsn1, and Nnf1) to the kinetochore is a key step in bringing together the microtubule binding activities of the KNL1 protein and the Ndc80 complex (Cheeseman et al., 2006; Kline et al., 2006). Previous studies in vertebrate cells had shown that the Mis12/MIND complex proteins required CENP-C for their centromeric localization (Kwon et al., 2007). Surprisingly, we found that the Nsl1 component of the Mis12/MIND complex could be independently associated with centromeres in the absence of CENP-C, but that Dsn1 and Nnf1 required the amino terminus of CENP-C for their localization to centromeres. This suggests the interesting possibility that CENP-C may facilitate the assembly of the Mis12 complex at centromeres by recruiting Dsn1, Nnf1 and Mis12 to centromeric Nsl1.
CENP-C is a component of the CCAN that includes CENP-C, -H, -I, -K–U, and -W (Cheeseman and Desai, 2008; Hori et al., 2008). Previous studies in chicken cells have implicated the CCAN proteins CENP-H, -I, -K, and -L in controlling both the incorporation of CENP-A into centromeric chromatin and the association of CENP-C with centromeres (Okada et al., 2006). In human cells, CENP-C and the Mis12 complex appear to localize to centromeres after CENP-H or -I depletion (Goshima et al., 2003; Liu et al., 2006). However, CENP-K depletion reduces the level of CENP-C at centromeres by one-half (Cheeseman et al., 2008). The depletion of CENP-C from Xenopus egg extract prevented CENP-K assembly, and this defect could be rescued by addback of CENP-C but not the amino terminal CENP-C truncation. Although we have been unable to localize CENP-H or -I in the Xenopus egg extract system, the failure of CENP-K to localize after CENP-C depletion suggests that CENP-C directs the assembly of the CENP-H/-I/-K complex to centromeres. This is consistent with our observation that Xenopus CENP-C depletion prevents CENP-K and Mad2 assembly; analogous to the failure of Mad2 localization in human or Schizosaccharomyces pombe cells lacking CENP-I (Liu et al., 2003; Saitoh et al., 2005).
Functional Roles for CENP-C in Xenopus
We have found that CENP-C is excluded from sperm chromatin and must be assembled upon fertilization in the early embryo. Our observations suggest that the site of the centromere is predetermined by the location of CENP-A chromatin and that, after fertilization, the recruitment of CENP-C to centromeric sites is a key step in centromere and kinetochore assembly. CENP-C appears to play a bridging role by both recognizing CENP-A chromatin and directing the assembly of the centromere and kinetochore. A major unresolved question is what molecular mechanisms direct the centromere-specific binding of CENP-C. The Xenopus in vitro system we have described should allow us to directly test specificity determinants for CENP-C association with centromeric chromatin. Identification of factors that associate with the amino terminus of CENP-C is a promising future direction for understanding how CENP-C facilitates the assembly of the centromere and kinetochore.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Straight lab, especially Christopher Carroll, and the Biochemistry Department for discussions on this project and manuscript. We are grateful to Rey-Huei Chen, Jennifer DeLuca, Geert Kops, Andrew Murray, Suzanne Pfeffer, and P. Todd Stukenberg for antibodies. K.J.M. was supported by the Department of Defense's National Defense Science and Engineering Grant Fellowship, the National Science Foundation's Graduate Research Fellowship, and the National Institutes of Health (NIH) Grant T32GM007276. B.M. was supported by NIH Grant T32GM007276. A.F.S. is the Gordon Family Scholar of the Damon Runyon Cancer Research Foundation, and this work was supported by NIH Grant R01GM074728 to A.F.S.
Abbreviations used:
- C-
carboxy
- CENP
centromere protein
- CSF
cytostatic factor
- IVT
in vitro transcription/translation
- N-
amino.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0378) on July 29, 2009.
REFERENCES
- Abramoff M. D., Magelhaes P. J., Ram S. J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Abrieu A., Kahana J. A., Wood K. W., Cleveland D. W. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. T., Goetsch L., Hartwell L. H. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. T. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Carroll C. W., Straight A. F. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Hori T., Fukagawa T., Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Cohen R. L., Espelin C. W., De Wulf, P., Sorger P. K., Harrison S. C., Simons K. T. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe R. K., Reed L. M., Yu H. G., Muszynski M. G., Hiatt E. N. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell. 1999;11:1227–1238. doi: 10.1105/tpc.11.7.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Deacon H. W., Walczak C. E., Mitchison T. J. A method that allows the assembly of kinetochore components onto chromosomes condensed in clarified Xenopus egg extracts. Proc. Natl. Acad. Sci. USA. 1997;94:12378–12383. doi: 10.1073/pnas.94.23.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T. J., Walczak C. E. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Field C. M., Oegema K., Zheng Y., Mitchison T. J., Walczak C. E. Purification of cytoskeletal proteins using peptide antibodies. Methods Enzymol. 1998;298:525–541. doi: 10.1016/s0076-6879(98)98043-0. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Brown W. R. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 1997;6:2301–2308. doi: 10.1093/hmg/6.13.2301. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001a;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Pendon C., Morris J., Brown W. CENP-C is necessary but not sufficient to induce formation of a functional centromere. EMBO J. 1999;18:4196–4209. doi: 10.1093/emboj/18.15.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Regnier V., Ikemura T. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 2001b;29:3796–3803. doi: 10.1093/nar/29.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Grunenberg H., Schmid M. Paired arrangement of nonhomologous centromeres during vertebrate spermiogenesis. Exp. Cell Res. 1990;187:157–161. doi: 10.1016/0014-4827(90)90130-3. [DOI] [PubMed] [Google Scholar]
- Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann, S., Lehner C. F. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 2005;19:2041–2053. doi: 10.1101/gad.347805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Salmon E. D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Fowler K. J., Earle E., Hill J., Choo K. H. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl. Acad. Sci. USA. 1998a;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., MacDonald A. C., Newson A. J., Hudson D. F., Choo K. H. Gene structure and sequence analysis of mouse centromere proteins A and C. Genomics. 1998b;47:108–114. doi: 10.1006/geno.1997.5109. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S. L., Cheeseman I. M., Hori T., Fukagawa T., Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knehr M., Poppe M., Schroeter D., Eickelbaum W., Finze E. M., Kiesewetter U. L., Enulescu M., Arand M., Paweletz N. Cellular expression of human centromere protein C demonstrates a cyclic behavior with highest abundance in the G1 phase. Proc. Natl. Acad. Sci. USA. 1996;93:10234–10239. doi: 10.1073/pnas.93.19.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G. J., Kim Y., Weaver B. A., Mao Y., McLeod I., Yates J. R., 3rd, Tagaya M., Cleveland D. W. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M. S., Hori T., Okada M., Fukagawa T. CENP-C Is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini L., McKeon F. Domains required for CENP-C assembly at the kinetochore. Mol. Biol. Cell. 1995;6:1049–1059. doi: 10.1091/mbc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Liu S. T., Hittle J. C., Jablonski S. A., Campbell M. S., Yoda K., Yen T. J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003;5:341–345. doi: 10.1038/ncb953. [DOI] [PubMed] [Google Scholar]
- Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P., Straight A., Coughlin P., Mitchison T. J., Salmon E. D. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S. E., Borusu S., Amaro A. C., Winter J. R., Belwal M., McAinsh A. D., Meraldi P. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Wood J. S., Garvik B., Hartwell L. H. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell. 1986;44:53–63. doi: 10.1016/0092-8674(86)90484-8. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. L., Roth M. B. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi Y., Peebles C., Fritzler M. J., Steigerwald J., Tan E. M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Del Rosario, B. C., Pemberton L. F. Modulation of histone deposition by the karyopherin Kap114. Mol. Cell. Biol. 2005;25:1764–1778. doi: 10.1128/MCB.25.5.1764-1778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nicklas R. B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Shibata F., Sato H., Murata M. Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet. Syst. 2004;79:139–144. doi: 10.1266/ggs.79.139. [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Palmer D. K., O'Day K., Margolis R. L. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma. 1990;100:32–36. doi: 10.1007/BF00337600. [DOI] [PubMed] [Google Scholar]
- Palmer D. K., O'Day K., Trong H. L., Charbonneau H., Margolis R. L. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A., Leno G. H. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- Przewloka M. R., Zhang W., Costa P., Archambault V., D'Avino P. P., Lilley K. S., Laue E. D., McAinsh A. D., Glover D. M. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L., Salmon E. D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C. A., Ratrie H. 3rd, Maurer M., Rothfield N. F., Earnshaw W. C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Ishii K., Kobayashi Y., Takahashi K. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol. Biol. Cell. 2005;16:3666–3677. doi: 10.1091/mbc.E05-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Iwabuchi M., Saeki H., Ura K., Kishimoto T., Ohsumi K. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc. Natl. Acad. Sci. USA. 2005;102:8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Gronemeyer B., Lu W., Eugster E., Tomkiel J. E. Mutational analysis of the central centromere targeting domain of human centromere protein C (CENP-C) Exp. Cell Res. 2002;275:81–91. doi: 10.1006/excr.2002.5495. [DOI] [PubMed] [Google Scholar]
- Starr D. A., Williams B. C., Hays T. S., Goldberg M. L. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Kuriyama K., Shibata A., Himeno M. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 1997;5:132–141. doi: 10.1023/a:1018422325569. [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Bryson T. D., Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel J., Cooke C. A., Saitoh H., Bernat R. L., Earnshaw W. C. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Bernardoni R., Diolaiti D., Politi V., Earnshaw W. C., Perini G., Della Valle, G. In vivo functional dissection of human inner kinetochore protein CENP-C. J. Struct. Biol. 2002;140:39–48. doi: 10.1016/s1047-8477(02)00506-3. [DOI] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaerou F., Karess R., Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- Wood K. W., Sakowicz R., Goldstein L. S., Cleveland D. W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yang C. H., Tomkiel J., Saitoh H., Johnson D. H., Earnshaw W. C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S. G., Patel S., Kavli B., Slupphaug G. Xenopus CENP-A assembly into chromatin requires base excision repair proteins. DNA Repair. 2005;4:760–772.. doi: 10.1016/j.dnarep.2005.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.