Abstract
Specific p53 binding-induced DNA bending and its underlying driving forces are crucial for the understanding of selective transcription activation. Diverse p53-response elements exist in the genome. However, it is not known what determines the DNA bending and to what extent. In order to gain knowledge of the forces that govern the DNA bending, molecular dynamics simulations were performed on a series of p53 core domain tetramer-DNA complexes in which each p53 core domain was bound to a DNA quarter site specifically. By varying the sequence of the central 4-base pairs of each half site, different DNA bending extents were observed. Our analysis shows that the interactions between p53 dimer-dimer were similar in all complexes; on the other hand, specific interactions between the p53 and DNA, including the interactions of Arg280, Lys120 and Arg248 with the DNA varied more significantly. In particular, the Arg280 interactions were better maintained in the complex with the CATG-containing DNA sequence, and were mostly lost in the complex with the CTAG-containing DNA sequence. Structural analysis shows that the base pairings for the CATG sequence were stable throughout the simulation trajectory while those for the CTAG sequence was partially dissociated in part of the trajectory, which affected the stability of nearby Arg280-Gua base interactions. Thus, DNA bending depends on the balance between the p53 dimer-dimer interactions and p53-DNA interactions, which in turn are related to the DNA sequence and DNA flexibility.
Keywords: p53-DNA binding, sequence-dependent DNA bending, p53 response elements
Introduction
Tumor suppressor p53 is responsible for human genome stability through its function to transactivate many genes upon cellular stress such as DNA damage, preventing uncontrolled cell development 1–4. The crucial biochemical event for the p53 transactivation activity is its sequence-specific binding to DNA 5. p53 binding sites are usually composed of two 10-base pair (bp) repeats, or two half sites, with the consensus of 5’-PuPuPuC(A/T)(A/T)GPyPyPy-3’, where Pu and Py stand for purine and pyrimidine bases, respectively 6–8; the two half sites can be separated by as many as 13 base pairs 7,9. More than 100 different specific p53 response elements have been characterized in the human genome 7. The variance of the DNA sequence and the consequent difference in p53 binding affinity was suggested to be important in the selective activation of target genes.
The p53 protein is a tetramer of four homologous peptide chains, with each chain composed of several domains including the N-terminal domain which regulates the p53 transactivation activity 10, the specific DNA-binding core domain 11, the tetramerization domain 12, and the C-terminal domain. When p53 is bound to DNA, DNA experiences conformational changes, notably axial bending 13–15, and p53 adjusts its conformation accordingly to adapt to the new environment 16. The proposed DNA bending in the p53-DNA complex in the middle of the half site and at the center of the full site points to the major role of the DNA-binding domain in DNA bending. Further experimental evidence indicates that p53 binding affinity is correlated with the DNA sequence 17. In particular, the p53-binding sites that contain CATG at the centers of the half sites usually have a higher binding affinity while those containing CTAG show low binding strength 17. The flexibility (bendability under axial stress) of a DNA segment with CATG at the center of the half sites is also shown to be higher than with other sequences at the center 6. However, there are no direct experimental structural data to show how the DNA bends and whether the binding affinity is truly correlated with the bending extent. Hence, important questions which might be the key to the differential transactivation, such as how the p53 interacts with different DNA sequences and how those interactions will affect the extent of DNA bending, can not be fully addressed.
Several structures involving p53 core domain-DNA specific binding became available during the last decade 11,18–20. Biophysical and biochemical data 11,21–24 were obtained and a similar p53 core domain tetramer-DNA complex model was proposed 11,21–24. The specific ways of association between p53 and DNA and between p53 monomers are supported by experiments 18,25–27. In addition, biophysical studies show that the p53 core domain alone can bend the DNA by only 30–35 degrees while the full length p53 can induce bending of 50–70 degrees, depending on the techniques used in the estimation 14. Molecular dynamics simulation also demonstrates a DNA bending of 35 degrees by p53 core domain alone 28 . This information provided the structural basis for computational investigation of the underlying forces that trigger the DNA bending. Here, we performed molecular dynamics simulations on the p53 core domain tetramer-DNA complexes and show that the DNA bending extent was correlated with the concurrent interactions between p53 dimer-dimer and p53-DNA.
Materials and Methods
Model construction
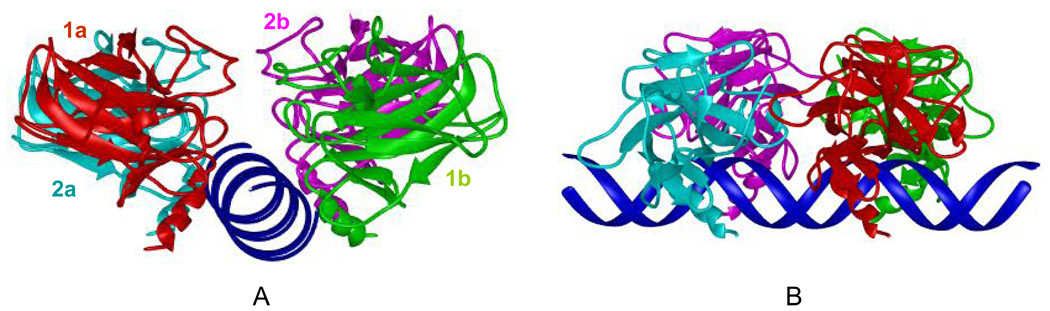
The method for building the p53 tetramer-DNA complex model was described previously 28. Briefly, the DNA segment and the B chain of p53 in the crystal structure that was bound to DNA specifically were extracted from the PDB file 11. We superimposed the base pairs from two copies of the extracted p53-DNA complex in reverse order so that the two copies of p53 were bound to two consecutive quarter sites of the DNA. The resulting DNA-p53 dimer complex structure ensures specific DNA-p53 binding and that the two copies of p53 have a C2 symmetry, with formation of the two salt bridges between Arg180 and Glu181 from the H1 helices of the p53 core domains. The p53 tetramer-DNA model was then constructed by connecting two copies of p53 dimer-DNA complex such that the resulting DNA segment has a continuous p53 full site binding with no insertion of base pairs and that the two dimers of p53 were side by side (Figure 1). The DNA sequence was 5’ATAATTGAGCA|TGCTC||GAGCA|TGCTCAGGAA3’, with the binding site (in bold) taken from the crystal structure solved by Ho et al 19 and is referred to below as the Ho complex. In order to study the relationship between DNA bending and the composition of the four base pairs at the centers of the half sites, the central four base pairs for each half site were mutated to CAAG, CTAG and CTTG. These complexes are named Ho_CAAG, Ho_CTAG and Ho_CTTG complexes, respectively. To ensure the reliability of the simulations results, a second set of simulations for the Ho, Ho_CAAG, Ho_CTAG and Ho_CAAG complexes were performed with slightly different starting configurations by varying the number of minimization steps during the preparation.
Figure 1.
The starting structure conformation for the simulations. The four p53 core domains were bound to the DNA specifically. The helical DNA is in linear conformation. For clarity, only the backbones of p53 and DNA are shown.
MD simulation protocol
Each starting structure was first solvated with a TIP3P water box 29 with a margin of at least 10 Å from any edge of the water box to any protein or DNA atom. Solvent molecules within 1.6 Å of the DNA or within 2.5 Å of the protein were removed. The systems were then neutralized by adding sodium ions. The resulting systems were subjected to a series of minimizations and equilibrations using the CHARMm program 30 and the CHARMm 22 and 27 force field for the protein and nucleic acid, respectively 31. Prior to the MD simulations, each system was minimized for 500 steps with the steepest decent algorithm, and 500 steps with the ABNR algorithm. For the second set of the simulations, additional 500 steps of ABNR minimization were applied to generate the slightly different configuration for the starting structures. The production MD simulations were performed at 300 degrees Kelvin using the NAMD program 32 and the CHARMm force field. Periodic boundary conditions were applied and the non-bonded lists were updated every 20 steps. NPT ensemble was applied and the pressure kept at 1 atom using Langevin-Nose-Hoover coupling. SHAKE constraints on all hydrogen atoms and a time step of 2 fs and a nonbonded cutoff of 12 Å were used in the trajectory production. The sizes of the systems were about 110,000 atoms and the duration for each simulation was 60 ns except for the second set of the simulations each of which lasted for 30 ns.
Results
DNA bending of the four complexes
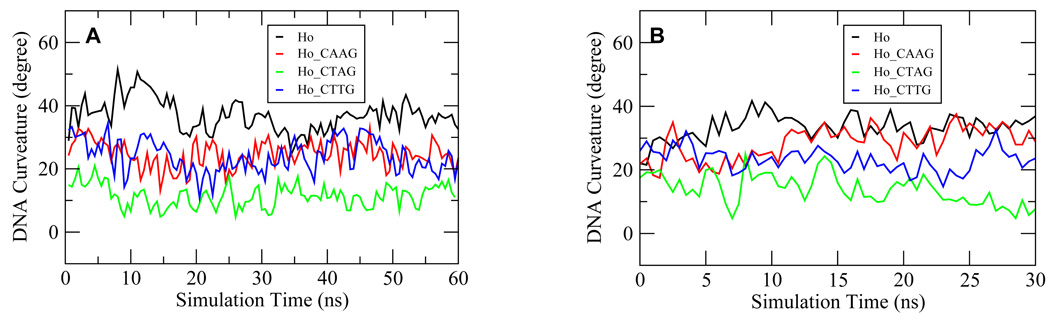
It has been shown that CATG is the most frequent sequence at the center of the half sites for the p53-response elements 33. However, other sequences such as CAAG, CTAG and CTTG also exist in many p53-response elements. In order to delineate the relationship between the composition of the central 4 base pairs and the p53-induced DNA bending, we mutated the CATG in the Ho complex into CAAG, CTAG, and CTTG to obtain the starting structures for the corresponding Ho_CAAG, Ho_CTAG and Ho_CTTG complexes and subjected them to MD simulations. Figure 2A shows the time series of DNA bending over the 60 ns trajectory for each of the complexes. The Ho complex, with the CATG sequence at the middle of the DNA sequence, induced the largest DNA bending extent with an average of 36.8 degrees for the last 20 ns (Table 1). The complexes Ho_CAAG and Ho_CTTG were expected to yield similar results because of the symmetric nature of the DNA sequences and their overall DNA bending extents were indeed similar to each other, with the average DNA bending for the last 20 ns at 25.1 degrees in both cases (Figure 2A, Table 1). In contrast, throughout the 60-ns trajectory the DNA bending in the Ho_CTAG complex was not significant, with an average DNA bending of about 11.5 degrees for the last 20 ns (Figure 2A, Table 1). The DNA essentially stayed in a straight conformation with visually undetectable bending. These results show that the DNA bending ability is in decreasing order for CATG, CAAG/CTTG, and CTAG.
Figure 2.
The extent of DNA bending for p53 core domain tetramer-DNA complexes Ho, Ho_CAAG, Ho_CTAG and Ho_CTTG from the 60 ns (A) and the 30 ns (B) trajectories. The DNA curvature was calculated with the CURVES program 35,50. The starting structure difference between (A) and (B) was the one following the minimization steps in the preparation stage.
Table 1.
Starting structure information for the MD simulations
| Complex Name | Binding site DNA sequence | Average Curvature (degrees, last 20 ns)a |
|---|---|---|
| Ho | GAGCA|TGCTC GAGCA|TGCTC | 36.8±4.2/33.7±4.5 |
| Ho_CAAG | GAGCA|AGCTC GAGCA|AGCTC | 25.1±6.2/31.4.0±4.4 |
| Ho_CTAG | GAGCT|AGCTC GAGCT|AGCTC | 11.5±4.4/10.6±5.1 |
| Ho_CTTG | GAGCT|TGCTC GAGCT|TGCTC | 25.1±7.2/22.2±7.1 |
The two averages were extracted from the two trajectories of 60 ns and 30 ns long respectively. The difference between the two simulations for each complex was the number of minimization steps in the preparation stage (see methods).
In order to further confirm the DNA bending extent for the different sequences, the complexes Ho, Ho_CAAG, Ho_CTAG and Ho_CTTG were simulated again starting from slightly different configurations, each with duration of 30 ns (see Methods). Overall, the new simulation results revealed a similar DNA bending trend as in the first simulation (Figure 2B, Table 1). For the same DNA sequence, the two sets of simulations also yielded consistent DNA bending, except that in the new simulations the Ho_CAAG complex displayed a bending extent that was comparable to the Ho complex (Figure 2, Table 1). However, the overall trend among the complexes did not change.
Difference in base pair parameters
In order to understand the structural basis of the DNA bending difference between the complexes, representative global inter-base pair parameters were measured for the last 20 ns (Figure S1). Calculations of the base pair parameters have been pioneered by Calladine 34, Lavery 35, and others. Conformational changes such as DNA bending might be reflected in the details of the base-step parameters, particularly the base-pair rolls and twists. Interestingly, although calculations clearly showed that the major groove narrowed and became deeper at the centers of the half sites (Figures S1–A, B), the inter-base pairs roll and twist parameters for the Ho complex, which involved the largest DNA bending, did not show significant variation among the 20 base pairs of the p53-response elements (Figures S1–C, D). There was no apparent difference between the central 4 bases and the rest of the bases within the DNA for the same Ho complex either (Figures S1, C, D). In addition, other parameters including shift (E), slide (F), rise (G) and tilt (H) did not show much difference among the 20 base pairs either. In general, comparison of these parameters among the Ho, Ho_CAAG and Ho_CTAG complexes revealed little difference, although for the Ho_CTAG complex these parameters fluctuated dramatically near the centers of the half sites where the CTAG motifs reside. Structural analysis revealed that this fluctuation was caused by the structural deviation from the WC base pairing for the Ho_CTAG complex: during the simulation, the AT base pairing for this complex was not well maintained in the Ho_CTAG complex unlike the Ho and Ho_CAAG complexes. Figure S2 shows that while the AT base pairs at the center of the half sites were well associated in the Ho complex, the base pairs at the same positions were unraveled to some extent in the Ho_CTAG complex. It seems that when the p53 attempted to bend the DNA, the base pairing at the center of the half sites were perturbed and the hydrogen bonding for the AT base pairs became vulnerable to conformational changes. Previous experimental and simulation results showed that in the CATG motif, the G or A base cross-strand stacking helped maintain the stability of the duplex while in the CTAG motif such G or A base stacking is not present 36,37. It seems that large base stacking may also help in stabilizing the bent DNA conformation upon p53 binding.
Conformational changes of the p53 tetramer
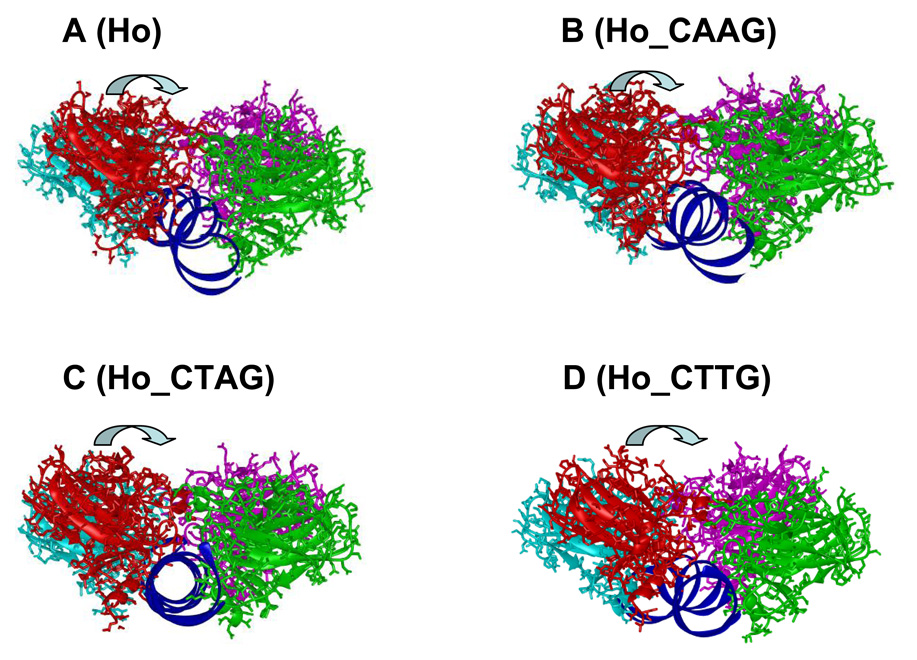
Structural fluctuations for individual domains of p53 were minimal, with an RMSD of 1.5 Å or less for the backbones compared with the starting structure. A simulation work on both free p53 and DNA-bound p53 core domains obtained similar results 11,38 and the small fluctuation in the p53 tetramer-DNA complex context is expected. The salt bridges within each of the two p53 core domain dimers were also largely intact throughout the trajectories for each complex. The most interesting conformational changes compared with the starting structures were in the p53 dimer-dimer organization. Figure 3 shows the averaged structures of four complexes from the respective trajectories. In each of the conformations for the Ho, Ho_CAAG, Ho_CTAG, and Ho_CTTG complexes, the dimer in the front (red and green) rotated clockwise with respect to the p53 dimer in the back (cyan and magenta), as indicated by the arrows (Figure 3). To quantify the extent of the rotation, a dihedral angle was defined as D1a1b2b2a where 1a, 1b, 2a and 2b were the four Cα atoms of residues Met160 from each of the four p53 core domains defined in Figure 1A. Met160 is located near the center of the core domain (not shown). The average dihedral angles for the last 10 ns of the trajectories were −15, −10, −13 and −14 degrees for the Ho, Ho_CAAG, Ho_CTAG and Ho_CTTG complexes, respectively. Although the magnitude was small, it did show that the two dimers did not favor the totally eclipsed conformation. In addition, the dimer in the front also slightly shifted toward the right, with the shift being slightly less significant in the Ho_CTAG complex than in the others. The similarity of the relative movement of the two p53 dimers with respect to each other among the complexes suggests that the p53 core domain tetramer preferred a similar organization regardless of the DNA sequence, implying that the DNA bending difference may be caused by the p53-DNA interactions.
Figure 3.
The average structures over the 25–30 ns period for the Ho, Ho_CAAG, Ho_CTAG, and Ho_CTTG complex simulations, showing the relative movement of the p53 dimers with respect to each other and their conformational changes from the starting conformation illustrated in Figure 1. The direction of the movement of the front p53 dimer is shown with the arrows.
Analysis of the p53-DNA and the p53-p53 interactions
Overall DNA-p53 interactions
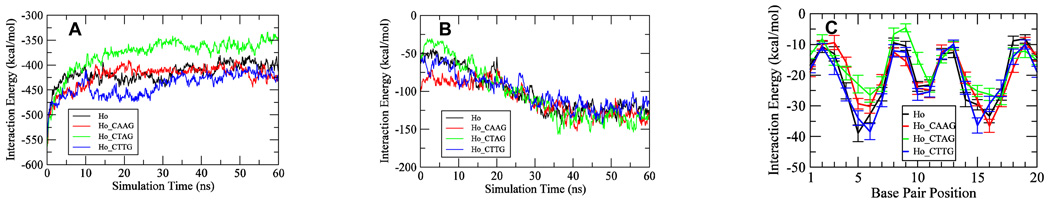
In the first 10 ns of the trajectories, the DNA-p53 interaction energy changed dramatically and became less favorable for each of the complexes (Figure 4A). This result is expected since the initial specific interactions were optimized for the p53 core domain monomer-DNA contact; that is, it was not in the tetramer complex environment. The interactions for each p53 core domain-DNA relaxed to some extent in the p53 tetramer-DNA complex environment, as observed previously 28. After 30 ns, each of the systems became equilibrated and the interaction energies were stabilized. The average interaction energies for the last 20 ns were −400±11, −413±13, −369±12 and −420±10 kcal/mol for the Ho, Ho_CAAG, Ho_CTAG, and Ho_CTTG complexes, respectively (Figure 4A). The large difference in interaction energy between the Ho_CTAG and other complexes suggests a more significant loss of interactions between the DNA and p53 for the Ho_CTAG complex than for the other three complexes. We subsequently observed that the p53-DNA interactions, specifically the hydrogen bonding, for the Ho_CTAG complex were indeed less populated in the simulations. On the other hand, interaction energies for the Ho, Ho_CAAG, and Ho_CTTG complexes were comparable with one another (Figure 4A) with similar p53-DNA interaction strength, as shown later.
Figure 4.
Time series for the p53-p53 and p53-DNA interactions for the Ho, Ho_CAAG, the Ho_CTAG, and Ho_CTTG complexes. (A) Total p53-DNA interaction energies. (B) p53 dimer-dimer interactions energies. (C) The interaction energies between p53 tetramer and each of the 20 base pairs of the response elements, averaged over the period of 20–30 ns of the trajectory. Interaction energies were calculated with the interaction module of the CHARMm program and distance-dependent dielectric constant of 4 was used for electrostatic energy calculations and a nonbonded cutoff of 12 Å.
Base-pair based p53-DNA interactions
Figure 4C shows that the interactions between the absolutely conserved GC base pair (the 4th position within the quarter-site Pu1Pu2Pu3C4(A/T)5) and p53 was the second most favorable among the five base pairs. The most favorable interaction energy was from the 5th position base pair, consistent with previous results 28, while the interaction of the second most conserved base pair at the 2nd position was much weaker. Therefore, 4th and 5th position base pairs dominate the DNA quarter site interactions with p53. Comparing these interactions among the Ho, Ho_CAAG, Ho_CTAG and Ho_CTTG complexes further shows that these interactions for the Ho_CTAG complex were the least favorable at both half sites among the four complexes (Figure 4C, the two peaks near base pairs 5 and 15); The same interactions for the other complexes were comparable with each other (Figure 4C). These results were consistent with the overall interaction energy trend shown in Figure 4A, suggesting that the associations between p53 and DNA was weaker for the Ho_CTAG complex after dynamic equilibration.
p53-p53 interactions
Opposite to the trend of the p53-DNA interactions, the p53 dimer-dimer interactions became increasingly favorable in the first 35-ns simulations (Figure 4B). The interaction energies became relatively stabilized in the last 20 ns for each of the trajectories. In the last two sections we show that the association between the protein and DNA in the Ho_CTAG complex was weaker than in the other three complexes (Figure 4A, C). However, after the systems reached equilibration, the p53 dimer-dimer interactions approached a similar level for all four complexes (Figure 4B). These data suggest that regardless of the DNA sequence differences, the p53 dimers tend to reach similar extent of favorable interactions. Such typical p53 dimer-dimer interface has been proposed previously 28 and the organizations of the complexes at the dimer-dimer interface were similar to each other as well (Figure 3). Comparison of the interfaces for the four complexes further shows that important interactions between the p53 monomers at the dimer-dimer interface and the residues involved were similar among the four complexes. Ser96, VAl97, Pro98, Gly99, Lys101, Ser166, Gln167, Met169, Thr170 and Glu171 were among the common residues at the interface within 4.5 Å of one p53 core domain, although the interacting partners were slightly shifted.
Specific p53-DNA interactions
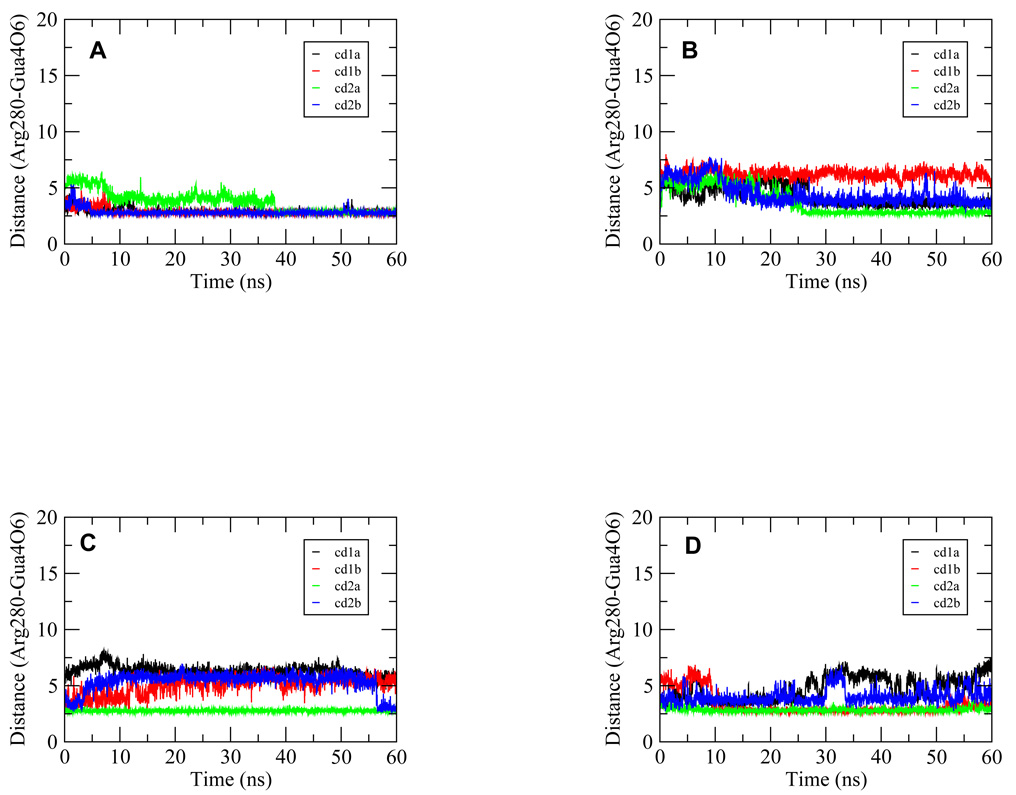
p53 interacts with DNA mainly through the hydrogen bonding of Arg280 and Lys120 with DNA bases at the major groove and through van der Waals contact of Arg248 with the DNA minor groove. These interactions were therefore monitored in order to compare the interaction extents among the complexes. Arg280 can form two hydrogen bonds with a G base. Here, we presented only one distance (Figure 5) since the behavior of the two were very similar and fluctuated concurrently. Figure 5A shows that for each of the p53 core domains the distance was largely maintained within the hydrogen bonding range throughout the trajectory for the Ho complex. Only the distance for one of the core domains was not within the hydrogen bonding distance in the first 38 ns, but the hydrogen bond reformed in the remaining trajectory (Figure 5A). For the Ho_CAAG complex, all the four hydrogen bonds were initially disrupted and were not present until about 25 ns where one hydrogen bond was formed and the other two distances were very close to the hydrogen bonding distance for most of the trajectory (Figure 5B). Only one distance was never within the hydrogen bonding range for the length of the trajectory (Figure 5B). In the case of the Ho_CTAG complex, only one core domain was able to maintain the hydrogen bonding, although another one formed at the end of the trajectory (Figures 5C). For the Ho_CTTG complex, two of the hydrogen bonding distances were maintained for most of the trajectory and a third one was very close to the hydrogen bonding distance for most of the trajectory (Figures 5D). The fourth distance was also fluctuating near the hydrogen bonding range for the first 25 ns although it did not come close again in the remaining simulation (Figure 5D).
Figure 5.
Time series from the 60-ns trajectories for the distances between the Arg280 guanidium nitrogen atom and the fourth Gua base of the p53 half site for complexes Ho (A), Ho_CAAG (B), Ho_CTAG (C) and Ho_CTTG (D), respectively. The atom specification shown on the Y-axis uses the CHARMm atom type convention. The pairs of atoms formed hydrogen bonds in the crystal structure and therefore were monitored for their dynamics behavior. These distances were shown for each of the four p53 monomers for every complex with the p53 core domains denoted as 1a, 1b, 2a and 2b in the inset legend as in Figure 1. The numerical notation for the base indicates its position in the quarter site G1A2G3C4A/T5.
Overall, these results show that in the initial phase of the simulation, the Arg280 hydrogen bonds were disrupted to different extents in different complexes. Clearly, the Ho complex was able to retain or quickly reform the hydrogen bonds. While for the Ho_CTAG complex, this hydrogen bond was almost completely abolished for most of the core domains. The hydrogen bonding populations are shown in Table 2.
Table 2.
Time percentage (%) for each of the distances that are shorter than the cutoffs a
| Arg280 | Lys120 | Arg248 | Arg280 | Lys120 | Arg248 | |
|---|---|---|---|---|---|---|
| From the 60 ns trajectory | From the 30 ns trajectory | |||||
| Ho | 79.3 | 33.8 | 88.1 | 60.4 | 44.2 | 88.5 |
| Ho_CAAG | 41.6 | 43.8 | 98.7 | 78.5 | 39.4 | 77.0 |
| Ho_CTAG | 17.8 | 40.8 | 85.3 | 18.3 | 24.0 | 72.2 |
| Ho_CTTG | 47.5 | 38.3 | 81.6 | 65.0 | 46.3 | 92.8 |
For each residue of Lys120, Arg248 and Arg280, averages were based on the data set collected at every 20 ps from the trajectory. Cutoff distance for the hydrogen bonds (Lys120 and Arg280) was 3.5 angstroms and that for the Arg248 interactions was 6.0 angstroms.
The interactions between Arg280 and the G base at the 4th position of a quarter site of the DNA have been shown previously to be the most critical to the p53-DNA binding. The large loss of such interactions for the Ho_CTAG complex suggests a weaker association in this complex and the results were consistent with the implications from Figures 4A and 4C. As discussed earlier, under the stress of DNA bending the association of AT base pairing at the center of the half sites was less stable in the Ho_CTAG complex than in the Ho complex possibly due to the less favorable inter-strand base-pair stacking. Such fluctuation of the AT base pairing, which is located right next to the 4th Gua base, may have directly affected the specific hydrogen bonding between Arg280 and the Gua base at the 4th position of the quarter site.
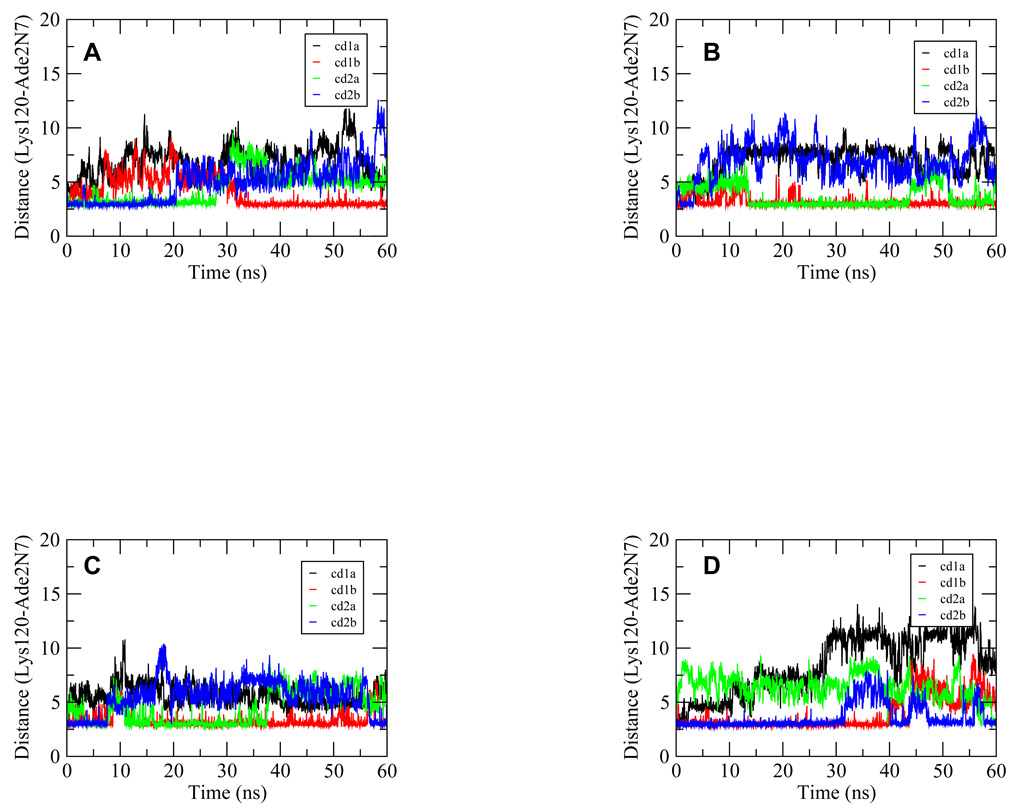
The second key residue involved in hydrogen bonding at the major groove is Lys120 with the less conserved Ade base at the 2nd position of the quarter site. Overall, the stability of these interactions among the four complexes did not show as significant a difference as displayed in the Arg280 hydrogen bonding (Figure 6). The overall population of the Lys120 hydrogen bonds for the Ho complex was lower than the Arg280 hydrogen bonds. Given that none of the complexes yielded high stability for this hydrogen bond, the result suggested that this hydrogen bond was flexible and may be not as important as the Arg280 interactions.
Figure 6.
Time series from the 60-ns trajectories for the distances between the Lys120 side-chain nitrogen atom and the second Ade base of the p53 half site for complexes Ho (A), Ho_CAAG (B), Ho_CTAG (C) and Ho_CTTG (D), respectively. The pair of atoms shown in the figure formed hydrogen bonds in the crystal structure and therefore their distances were monitored for their dynamics behavior. These distances were shown for each of the four p53 monomers for every complex with the p53 core domains denoted as 1a, 1b, 2a and 2b in the inset legend as in Figure 1. The numerical notation for the base indicates its position in quarter site G1A2G3C4A/T5.
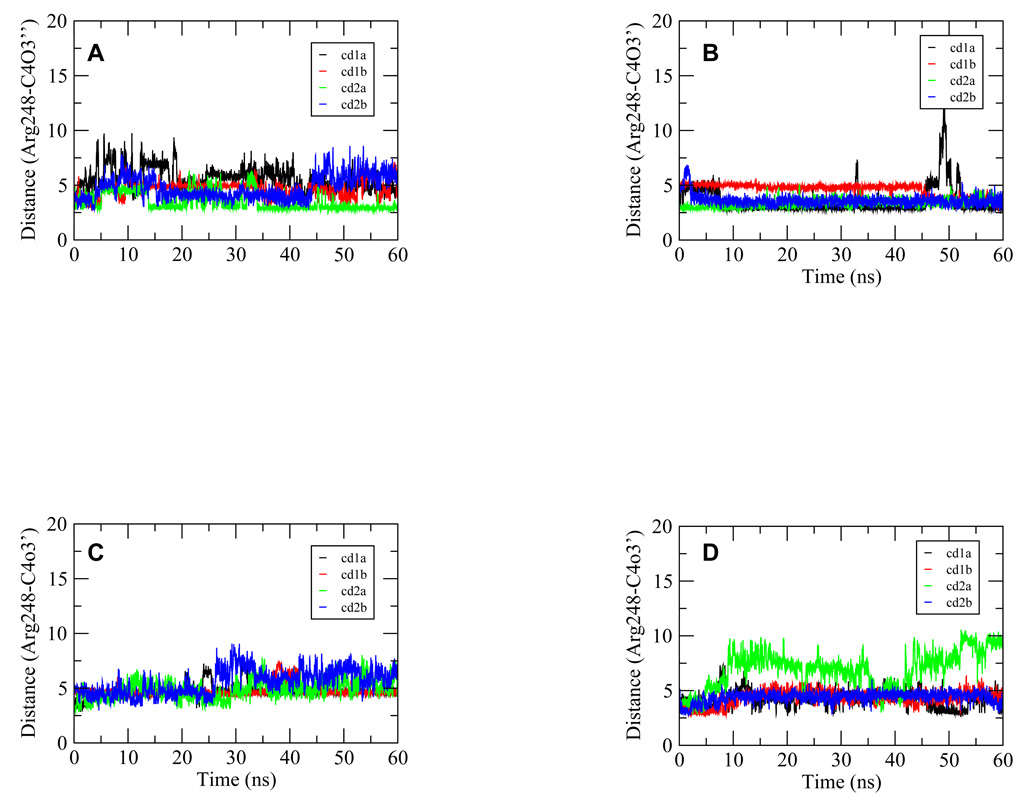
Arg248 is the key residue that interacts with the minor groove through electrostatic and van der Waals forces. The interactions were monitored through two distances between the guanidine carbon in the side chain of Arg248 and the nearest DNA backbone oxygen (3rd position Gua base and 4th position C base). Figure 7 presents only one distance because of their similarity. Overall, all four complexes maintained this interaction well, with Ho_CAAG having the highest stability (Figure 7B). Interestingly, the Ho complex displayed some fluctuation (Figure 7A) while the Ho_CTTG complex lost one of the contacts for most of the trajectory (Figure 7D). Regardless of the different behavior of the complexes, the overall populations of the distance within the cutoff were all high (Table 2), suggesting the relative stability of this interaction.
Figure 7.
Time series from the 60-ns trajectories for the distances between the atoms of Arg248 guanidium carbon and the backbone oxygens of the fourth Cyt base of the p53 half site for complexes Ho (A), Ho_CAAG (B), Ho_CTAG (C) and Ho_CTTG (D), respectively. The pair of atoms formed tight interactions in the crystal structure and therefore were monitored for their dynamics behavior. These distances were shown for each of the four p53 monomers for every complex with the p53 core domains denoted as 1a, 1b, 2a and 2b in the legend as in Figure 1. The numerical notation for the base indicates its position in quarter site G1A2G3C4A/T5.
In the second set of the simulations, similar results were observed for each of the interactions. Again, the Arg280 hydrogen bonds were better retained in the Ho, Ho_CAAG, and Ho_CTTG complexes (Figure S3–A, B, D), while it was largely disrupted in the Ho_CTAG complex (Figure S3–C). The difference for the Lys120 and Arg248 among the complexes was smaller compared with that for the Arg280 interactions (Figure S4, Figure S5). However, comparison of the percentage of the distances with the cutoff (Table 2) shows the relative lower stability (lower percentage) of these interactions for the Ho_CTAG complex.
Comparing all the interaction data among the complexes, it seems clear that the disruption of these specific interactions was responsible for the loss of total favorable interactions between p53 and DNA. The importance of Arg280 in the stabilization of p53-DNA interactions was highlighted in both Figure 4 and Figure 5–Figure 7. Results from Figure 5–Figure 7 further show that the stability of this interaction was important for the DNA conformational change during p53 binding. Indeed, Arg280 was anchored in the helix motif (the H2 helix of p53) of the core domain as in many other DNA binding proteins, rendering it as the major DNA binding motifs. The role of the specific interaction of Lys120 with DNA is unclear because of its variable presence in existing crystal structures. Lys120 was attached to the mobile loop L1 of the p53 protein and it is likely that because of this flexible structural feature, the Lys120-DNA interactions were well maintained in some crystal structures under one condition 11, while in others these interactions were not observed 19,20. Arg248, on the other hand, interacts with the DNA backbone and therefore was less specific in its interactions with the DNA, and these interactions were relatively better maintained in all of the complexes in our simulations. It should be noted that when the specific interactions were disrupted during the simulations, residues Arg280, Lys120 and Arg248 still interacted with the DNA, but non-specifically, with the DNA backbone. As a result, the p53 core domain only touched the surface of the DNA, instead of the previous clamp-like tight “grip” of the DNA which was necessary for the DNA bending upon p53 dimer-dimer interactions.
Discussion and Conclusions
Base pair composition is known to play crucial roles in structure and stability properties. A/T rich segments such as a TATA box are more flexible/bendable than others 39–41. How the change of sequence affects the DNA and the DNA-protein complex structures depends on the content of the complex. The sequence-dependent DNA deformation has been systematically studied with molecular modeling and compared with experimental data 42. Sarai and his colleagues have shown that the AT dimer has the highest stability while the PyPu dimers such as TA present large fluctuations, depending on the flanking sequences. In our simulations, the dissociation of A/T base pairs was also observed more frequently in the Ho_CTAG complex than in others. A recent study has shown that the flexibility and bending extent of the TATA box is dependent on the context of nearby sequences, classified into flexible and rigid groups 43. Even the concentration and type of ions present in the environment can have large impact on DNA conformations 44. From our simulations, with everything else equal, the CATG containing DNA segment seems to be more stable and could maintain its helical conformation even in the bent state. While the more rigid CTAG containing DNA was less tolerable to conformational change and easy to suffer from base pairing disruption. It should be noted that there were other A/T base pairs in the p53-response elements. However, only the segments at the middle of the half sites were not in extensive contact with the p53 protein and therefore can undergo conformational changes.
Among the p53 response elements, the majority of the 20-base pair segments contain CATG at the centers of the half sites; however, there are also combinations of CATG, CAAG, CTTG or CTAG at the same positions 33. Nature seems to have used the variations of the DNA sequence to modulate the response to the environmental stress. This was illustrated by previous experimental and simulation results which showed that CATG and CAAG containing p53 response elements can bind p53 efficiently and bend significantly 45,19,28. Many factors may contribute to the DNA bending; other parts of the protein or base pairs flanking the p53-response elements may also be involved in triggering additional conformational change and in stabilizing the bent DNA conformation. There is evidence indicating that efficient p53 binding requires both the core domain and the C-terminal domain of the p53 protein 46,47. In addition, base pairs other than the central 4 could also contribute to different extents. However, current experimental data suggest that the bending of the DNA induced by specific p53-binding is correlated with the presence of the CATG motif at the center of the half sites 45. Balagurumoorthy et al. further observed that the CATG region in the p53 response elements is kinked; the less flexible CAAG/CTTG containing DNA segments are less so 8; and the CTAG containing DNA segments were not observed to bend. It seems that in most cases nature has chosen more flexible sequences to allow the DNA bending required for efficient p53 binding, as was found in the majority of the p53-response elements 33. Although other sequences such as CTAG also exist, comparative study of the p53 response elements shows that most high-affinity segments contain CATG, while the CTAG containing response elements showed binding affinities at the lower end of the spectrum 17.
To further illustrate the correlation between DNA bending/efficient binding and the transactivational function, Inga et al. measured the capacity of various response elements for transactivation 48. They found that there is a good correlation between the binding affinity of p53 and its transactivation capacity. They attempted to elucidate the relationship between features of the DNA sequence and the transactivation capacity using several parameters, such as the number of non-consensus base pairs or the location of the non-consensus pairs or both. However, no obvious trend was detected. In the end, they proposed that the transactivation capacity is correlated with the conformational change of the DNA and p53 upon p53 tetramer-DNA association. Their hypothesis was strongly supported by the measurements of these parameters for sequences that are different only in the central 4 base pairs.
While DNA bending for the 20 contiguous base pair binding site appears to be the key for the differential binding affinity, the functions of the low affinity p53 response elements and those with base-pair insertions are largely unknown. Even those with only a couple of base-pair insertions will greatly alter the p53 core domain dimer-dimer interactions and are likely to be very different from those without base-pair insertions. Studies of the structural properties could present significant difficulties for structural biologists. However, with the accumulating experimental structural results 19,20,49 and with more advanced computational power, computational studies can play a more important role in revealing the relationship between the complex structure and transcriptional selection and activity.
In summary, we have performed MD simulations on p53-DNA complexes with different DNA sequences containing CATG, CAAG, CTAG and CTTG at the centers of the half sites. Our results show that the extent of DNA bending depends on the ability of DNA to maintain its specific interactions with p53 while the p53 dimers approach each other to maximize the p53-p53 interactions. Complexes with DNA containing the CATG motif can better retain the specific interactions than the complexes with DNA containing the CTAG motif, which is associated with the intrinsic bendability of the CATG motif or the rigidity of the CTAG segment of the DNA.
Supplementary Material
Acknowledgements
This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The research of R. Nussinov in Israel has been supported in part by the Center of Excellence in Geometric Computing and its Applications funded by the Israel Science Foundation. This study utilized the high-performance computational capabilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
References
- 1.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Cancer Res. 1991;51:6304. [PubMed] [Google Scholar]
- 2.Vousden KH. Biochim Biophys Acta. 2002;1602:47. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Nat Rev Cancer. 2002;2:594. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.el-Deiry WS. Semin Cancer Biol. 1998;8:345. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 5.Bargonetti J, Friedman PN, Kern SE, Vogelstein B, Prives C. Cell. 1991;65:1083. doi: 10.1016/0092-8674(91)90560-l. [DOI] [PubMed] [Google Scholar]
- 6.Balagurumoorthy P, Sakamoto H, Lewis MS, Zambrano N, Clore GM, Gronenborn AM, Appella E, Harrington RE. Proc Natl Acad Sci U S A. 1995;92:8591. doi: 10.1073/pnas.92.19.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat Genet. 1992;1:45. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 8.Balagurumoorthy P, Lindsay SM, Harrington RE. Biophys Chem. 2002;101–102:611. doi: 10.1016/s0301-4622(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 9.Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. Mol Cell Biol. 1992;12:2866. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields S, Jang SK. Science. 1990;249:1046. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 11.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Science. 1994;265:346. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 12.Clore GM, Omichinski JG, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn AM. Science. 1994;265:386. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- 13.Nagaich AK, Appella E, Harrington RE. J Biol Chem. 1997;272:14842. doi: 10.1074/jbc.272.23.14842. [DOI] [PubMed] [Google Scholar]
- 14.Nagaich AK, Zhurkin VB, Durell SR, Jernigan RL, Appella E, Harrington RE. Proc Natl Acad Sci U S A. 1999;96:1875. doi: 10.1073/pnas.96.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherny DI, Striker G, Subramaniam V, Jett SD, Palecek E, Jovin TM. J Mol Biol. 1999;294:1015. doi: 10.1006/jmbi.1999.3299. [DOI] [PubMed] [Google Scholar]
- 16.Ceskova P, Chichger H, Wallace M, Vojtesek B, Hupp TR. J Mol Biol. 2006;357:442. doi: 10.1016/j.jmb.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. J Mol Biol. 2005;348:589. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Rippin TM, Freund SM, Veprintsev DB, Fersht AR. J Mol Biol. 2002;319:351. doi: 10.1016/S0022-2836(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 19.Ho WC, Fitzgerald MX, Marmorstein R. J Biol Chem. 2006;281:20494. doi: 10.1074/jbc.M603634200. [DOI] [PubMed] [Google Scholar]
- 20.Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Mol Cell. 2006;22:741. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Waterman JL, Shenk JL, Halazonetis TD. Embo J. 1995;14:512. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Schwedes JF, Parks D, Mann K, Tegtmeyer P. Mol Cell Biol. 1995;15:2157. doi: 10.1128/mcb.15.4.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLure KG, Lee PW. Embo J. 1998;17:3342. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaich AK, Zhurkin VB, Sakamoto H, Gorin AA, Clore GM, Gronenborn AM, Appella E, Harrington RE. J Biol Chem. 1997;272:14830. doi: 10.1074/jbc.272.23.14830. [DOI] [PubMed] [Google Scholar]
- 25.Klein C, Planker E, Diercks T, Kessler H, Kunkele KP, Lang K, Hansen S, Schwaiger M. J Biol Chem. 2001;276:49020. doi: 10.1074/jbc.M107516200. [DOI] [PubMed] [Google Scholar]
- 26.Sun XZ, Vinci C, Makmura L, Han S, Tran D, Nguyen J, Hamann M, Grazziani S, Sheppard S, Gutova M, Zhou F, Thomas J, Momand J. Antioxid Redox Signal. 2003;5:655. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- 27.Dehner A, Klein C, Hansen S, Muller L, Buchner J, Schwaiger M, Kessler H. Angew Chem Int Ed Engl. 2005;44:5247. doi: 10.1002/anie.200501887. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Nussinov R. J Biol Chem. 2007;282:691. doi: 10.1074/jbc.M605908200. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- 30.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187. [Google Scholar]
- 31.MacKerell AD, Jr, Bashford D, Jr, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. Journal of Physical Chemistry B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 32.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. Cell. 2006;124:207. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 34.el Hassan MA, Calladine CR. J Mol Biol. 1995;251:648. doi: 10.1006/jmbi.1995.0462. [DOI] [PubMed] [Google Scholar]
- 35.Swaminathan S, Ravishanker G, Beveridge DL, Lavery R, Etchebest C, Sklenar H. Proteins. 1990;8:179. doi: 10.1002/prot.340080208. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, McDowell JA, Kierzek R, Krugh TR, Turner DH. Biochemistry. 2000;39:8970. [PubMed] [Google Scholar]
- 37.Pan Y, Priyakumar UD, MacKerell AD., Jr Biochemistry. 2005;44:1433. doi: 10.1021/bi047932q. [DOI] [PubMed] [Google Scholar]
- 38.Lu Q, Tan YH, Luo R. J Phys Chem B. 2007;111:11538. doi: 10.1021/jp0742261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Souza ON, Ornstein RL. Biopolymers. 1998;46:403. doi: 10.1002/(SICI)1097-0282(199811)46:6<403::AID-BIP5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 40.Strahs D, Schlick T. J Mol Biol. 2000;301:643. doi: 10.1006/jmbi.2000.3863. [DOI] [PubMed] [Google Scholar]
- 41.Davis NA, Majee SS, Kahn JD. J Mol Biol. 1999;291:249. doi: 10.1006/jmbi.1999.2947. [DOI] [PubMed] [Google Scholar]
- 42.Fujii S, Kono H, Takenaka S, Go N, Sarai A. Nucleic Acids Res. 2007;35:6063. doi: 10.1093/nar/gkm627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faiger H, Ivanchenko M, Haran TE. Nucleic Acids Res. 2007;35:4409. doi: 10.1093/nar/gkm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heddi B, Foloppe N, Hantz E, Hartmann B. J Mol Biol. 2007;368:1403. doi: 10.1016/j.jmb.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Zhou H, Zhang Y, Ou-Yang Z, Lindsay SM, Feng XZ, Balagurumoorthy P, Harrington RE. J Mol Biol. 2001;306:227. doi: 10.1006/jmbi.2000.4370. [DOI] [PubMed] [Google Scholar]
- 46.McKinney K, Mattia M, Gottifredi V, Prives C. Mol Cell. 2004;16:413. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 47.McKinney K, Prives C. Mol Cell Biol. 2002;22:6797. doi: 10.1128/MCB.22.19.6797-6808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inga A, Storici F, Darden TA, Resnick MA. Mol Cell Biol. 2002;22:8612. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, Orlova EV. Embo J. 2006;25:5191. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. J Biomol Struct Dyn. 1989;6:669. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.