SUMMARY
We investigated whether a continuous transcript is necessary for co-transcriptional pre-mRNA processing. Cutting an intron with the fast-cleaving hepatitis δ ribozyme, but not the slower hammerhead, inhibited splicing. Exon tethering to RNA pol II therefore cannot rescue splicing of a transcript severed by a ribozyme that cleaves rapidly relative to splicing. Ribozyme cutting also released cap-binding complex (CBC) from the gene, suggesting that exon 1 is not tethered. Unexpectedly, cutting within exons inhibits splicing of distal introns where exon definition is not affected, probably due to disruption of interactions with CBC and the pol II CTD that facilitate splicing. Ribozyme cutting within the mRNA also inhibited 3’ processing and transcription termination. We propose that damaging the nascent transcript aborts pre-mRNA processing and this mechanism may help prevent association of processing factors with pol II that is not productively engaged in transcription.
Keywords: splicing, cleavage/polyadenylation, nascent transcript tethering, cap binding complex, termination, ribozyme
INTRODUCTION
Efficient and accurate mRNA biogenesis is achieved by coupling synthesis of the transcript by RNA polymerase II (pol II) with RNA processing by capping, splicing and cleavage/polyadenylation. The coupling of pre-mRNA processing with transcription is thought to occur by the association of processing factors with the transcription elongation complex (TEC) largely via protein:protein interactions with the C-terminal heptad repeat domain (CTD) of the pol II large subunit. The CTD enhances splicing and 3’ end processing and is thought to act as a landing pad for processing factors during transcription 1–3. The CTD also helps coordinate splicing with RNA editing 4 in a process that requires intra-molecular base-pairing often between exon and intron sequences. These various co-transcriptional functions of the CTD presumably require that the nascent transcript remains in close proximity to the RNA polymerase.
The nature of the interface between the nascent transcript and the TEC is poorly understood. A key question is whether proximity of the pre-mRNA to the CTD is maintained by protein:RNA tethering contacts between exons and pol II in addition to its anchor at the RNA-DNA hybrid in the active site of the enzyme. Such additional tethering contacts could be made by direct binding of RNA to the CTD 5. Alternatively tethering could be indirect through RNA binding proteins that contact the TEC including SR proteins, snRNP’s 6,7, U2AF 8, HIV Tat 9, and cleavage stimulation factor CstF 10. It has also been suggested that CBC acts as a bridge between the nascent transcript and pol II 11,12. CBC localizes with pol II 13,14 and enhances splicing and 3’ processing 15–18.
Consistent with RNA tethering, transcripts cleaved at the poly A site are not always liberated from the template 19,20 in coupled transcription and 3’ processing reactions in vitro. In vivo, tethering of exons to pol II was proposed to account for the unexpected observation that cleavage of nascent pre-mRNA by the hammerhead ribozyme within an intron failed to inhibit splicing of β-globin transcripts 21. Similarly, splicing in yeast was resistant to cutting within the intron by the same ribozyme 22. Exon tethering is also consistent with observations that co-transcriptional excision of miRNA’s from introns by Drosha cleavage does not inhibit splicing 23,24. Tethering of exons to pol II is not necessarily required for splicing across a discontinuous intron however, as the spliceosome is capable of bimolecular exon ligation 25. Exon tethering to pol II was also proposed to account for the finding that in nuclear runon assays treated with urea, pol II is released from the template in association with a transcript that has not been processed at the poly (A) site 26. It remains possible however that under in vitro conditions, RNA binding to pol II is a consequence of incomplete packaging into hnRNPs. Although exon tethering could enhance co-transcriptional mRNA maturation, such interactions might also produce non-productive interfering complexes if they formed with pol II that was not engaged in productive transcription.
In many circumstances, splicing requires exon definition through communication between proteins across the exon 27. Exon 1 is defined by interaction between CBC and U1 or U6 snRNPs 16,28 and internal exons are defined by interactions between U1 and U2snRNP’s and SR proteins bound to splicing enhancer elements 29,30. The last exon is defined by interactions of U2 snRNP and U2AF with cleavage/polyadenylation factors and pol II 31,32 33. Ribozyme cleavage of β-globin exon 2 disrupted splicing of intron 1 but unexpectedly splicing of intron 2 was unaffected 21. The inhibition of splicing by exon cutting suggests that exon definition operates in vivo in mammalian cells. On the other hand, it is not known whether exon cutting affects splicing under conditions when exon definition does not occur. Nor is it known how ribozyme cleavage within the first and last exons affects splicing.
Other experiments employing similar RNA cutting strategies are not readily reconciled with exon tethering. We and others reported that poly (A) site cleavage and splicing of the last intron are disrupted by severing the nascent RNA downstream of the poly (A) site either by a ribozyme in vivo 34 or by RNAse H digestion in vitro 20,33. This effect is most easily explained if cutting the RNA disrupts its association with pol II and CTD-associated processing factors. Surprisingly, although ribozyme cutting close to the poly (A) site interferes with processing, it was reported not to inhibit transcription termination 26, in an apparent exception to tight coupling of termination with 3’ end processing 35.
We re-examined the question of transcript tethering to the TEC in vivo by severing the nascent RNA within exons or introns using self-cleaving hepatitis δ or hammerhead ribozymes. Our results suggest that splicing is facilitated by a link with pol II that can be disrupted by a ribozyme situated either in an exon or an intron provided that RNA cleavage is fast enough to compete with splicing. These results therefore argue against exon tethering and suggest that in response to endonucleolytic damage inflicted on a nascent transcript, splicing and 3’ end processing can be aborted.
RESULTS
Decapitation by ribozyme cleavage in exon 1 inhibits splicing
To investigate the effects of severing the RNA connection between CBC and the rest of the transcription complex, we incorporated the hepatitis δ variant ribozyme or an inactive point mutant 34 into exon 1 of a CMV β-globin reporter gene 63 bases from the cap. A histone stem loop sequence was included upstream of the ribozyme cut site to stabilize the 5’ product 36. Transcripts made in transiently transfected cells were analyzed by RNAse protection assay (RPA) that permits quantitation of processed:unprocessed transcripts. RPA with a probe spanning the ribozyme cut site showed complete cleavage by the WT but not the mutant ribozyme (Fig. 1a). 3’ products of ribozyme cleavage that lack a cap were relatively stable (data not shown) presumably because the 5’OH and secondary structure of the ribozyme are poor substrates for 5’ exonucleases.
Figure 1. Ribozyme cleavage in globin exon 1 inhibits splicing of introns 1 and 2 and 3’ end processing.

a. RPA of ribozyme cutting in exon 1. Wild-type (WT) and mutant (Mut) hepatitis δ (δ) ribozymes were inserted in exon 1 of pCMVHA β-glob (Supplementary Table 1, plasmids 2, 3).
b. RPA of intron 1 and intron 2 splicing in exon 1 ribozyme constructs (Supplementary Table 1 plasmids 1–3). VA is a loading control. Ratios of spliced:unspliced transcripts are calculated from Phosphorimager analysis after compensating for 32P-U content of the protection products. Multiple unspliced intron 2 bands are due to RNAse nibbling. Quantitation of RPA results for all experiments is shown in Supplementary Table 2.
c. RPA of intron 1 and intron 2 splicing in exon 2 ribozyme constructs with a mutant poly (A) site (A2GA3) and fibronectin splicing enhancer (FN) in exon 3 (Supplementary Table 1 plasmids 6, 7).
The maps depict antisense RPA probes (black arrows), the WT or mutant ribozymes (downward gray arrow), the fibronectin splicing enhancer (FN), the poly (A) site (downward black arrow) and the poly (A) site mutation (AAGAAA).
As expected, removal of the cap by WT ribozyme in exon 1 markedly reduced the steady state ratio of spliced:unspliced intron 1 transcripts consistent with inhibition of splicing relative to the mutant ribozyme control (Fig. 1b lanes 1–3). More surprisingly, severing exon 1 also inhibited intron 2 splicing and poly (A) site cleavage (Fig. 1b lanes 4–6, Supplementary Fig. 1a). The magnitude of the splicing defects caused by ribozyme cutting in our experiments would be underestimated if unspliced precursors are less stable than spliced RNAs. This possibility is suggested by the fact that ribozyme cleavage in exon 1 (Fig. 1b) and elsewhere (see below) reduced the accumulation of spliced RNA without an equivalent increase of unspliced precursors (Fig. 1b). This apparent instability of unspliced transcripts was reversed by mutation of the poly (A) site (Fig. 1c) as discussed below. Inhibition of NMD or the exosome by dominant negative Upf1 or PM-Scl-75 shRNAs did not affect the ratio of spliced:unspliced transcripts (data not shown) arguing against the unlikely possibility that spliced transcripts are preferentially degraded following ribozyme cleavage. In summary the experiments in Figure 1 suggest that a continuous RNA connection between the cap and the rest of the transcript enhances all steps in processing of the β-globin pre-mRNA, not just splicing of the first intron.
Severing exon 2 can inhibit processing upstream and downstream of the cut site
We next examined the effects of cutting the β-globin transcript in exon 2. The hepatitis δ ribozyme inserted in exon 2 119 bases upstream of the 5’ splice site, strongly reduced the ratio of spliced:unspliced transcripts of intron 1 (Supplementary Fig. 1b, lanes 1–3) as previously reported for the hammerhead 21. Unexpectedly, δ ribozyme cutting in exon 2 also impaired splicing of intron 2 and poly (A) site cleavage (Supplementary Fig. 1b, lanes 4–9) consistent with the idea that an intact 5’ end is important for these downstream processing events. We note however that in the context of a stably integrated chromosomal β-globin reporter gene, ribozyme cutting in exon 2 inhibited intron 1 splicing (Supplementary Fig. 2a), but had little effect on 3’ end processing (data not shown). This discrepancy may reflect a change in the kinetics of co-transcriptional processing, transcription elongation, or pausing in a chromosomal context relative to a transiently transfected plasmid.
It is possible that inhibition of intron 1 splicing by severing exon 2 is caused by disruption of interactions with the poly (A) site which stimulates splicing of both introns 10. We therefore asked how cutting exon 2 affects splicing of transcripts with a mutant poly (A) site (A2GA3) and a fibronectin splicing enhancer (FN) inserted in exon 3 (Fig. 1c). The results in Fig. 1c show that splicing of introns 1 and 2 was inhibited by the WT but not the mutant ribozyme in exon 2. The shutdown of splicing caused by severing exon 2 is therefore independent of a functional poly (A) site.
In constructs with the A2GA3 mutant poly (A) site, ribozyme cleavage always elevated the level of unspliced precursors relative to the VA loading control (Fig. 1c, see also Fig. 5a lanes 4, 5). This apparent stabilization of unspliced transcripts may be due to their retention at the site of transcription 34. In summary, the experiments in Figure 1 and Supplementary Figure 1 show ribozyme cutting in exons 1 and 2 causes a general shutdown of splicing and 3’ end processing of β-globin transcripts.
Figure 5. Ribozyme cleavage in exon 3 inhibits splicing independently of effects on exon definition.

a. RPA of intron 2 splicing in RNA from cells transfected with pCMV β-globin control (lane 1) and constructs with WT and mutant δ ribozyme in exon 3 with WT poly (A) site (lanes 2, 3) or FN A2GA3 (lanes 4, 5) (Supplementary Table 1 plasmids 25–33). The FN splicing enhancer (lanes 6, 7) does not rescue splicing that is inhibited by the WT ribozyme. Ribozyme cutting upstream of the mutant poly (A) site (lanes 4, 5) has a greater effect on splicing than cutting downstream (lanes 8, 9). The map depicts antisense RPA probes, the positions of ribozymes either within exon 3 or 115 bp downstream, the FN splicing enhancer, the poly (A) site, and A2GA3 mutation as in Fig. 2.
b. Inhibition of intron 1 splicing by ribozyme cutting in exon 3 with and without the FN splicing enhancer. VA is a loading control (Supplementary Table 1 plasmids 33, 25, 26, 29, 30).
Transcript release by ribozyme cutting
To test whether the transcript is tethered at the gene following ribozyme cleavage in exon 2, we monitored CBC, which binds the 5’ ends of nascent transcripts, by ChIP with anti-CBP80 antibody. If exon1 is tethered, then CBC ChIP signal should be unaffected by the ribozyme. On the other hand, if the RNA is released by the ribozyme, then the CBC ChIP signal should diminish at positions downstream of the cut. We analyzed pol II and CBP80 by ChIP at 12 amplicons along the β-globin gene with the WT or mutant hepatitis δ ribozyme in exon 2. Because high resolution ChIP was not possible with plasmids, we integrated a single copy of the CMV driven, tet-inducible, β-globin genes by site-specific recombination. As expected, intron 1 splicing was inhibited by the WT but not the mutant ribozyme in these cell lines (Supplementary Fig. 2a). The results in Figure 2a show that the distribution of pol II on β-globin was not detectably affected by ribozyme cutting in exon 2. In contrast, CBP80 cross-linking was specifically reduced in the vicinity of the WT δ ribozyme and at positions downstream compared to the mutant control (Fig. 2b). The effect of the ribozyme on CBP80 ChIP signals is only about 2-fold at most positions possibly because not all transcripts are co-transcriptionally cleaved before polymerase reaches the end of the gene. The fact that a WT ribozyme reproducibly reduces the CBP80 ChIP signal relative to the mutant, strongly suggests that nascent RNA with CBC bound to the 5’ cap is released from the gene. In summary these results argue that rather than being tethered to the template, exon 1 is released by RNA cleavage in exon 2.
Figure 2. Ribozyme cleavage in exon 2 releases CBC from the gene.
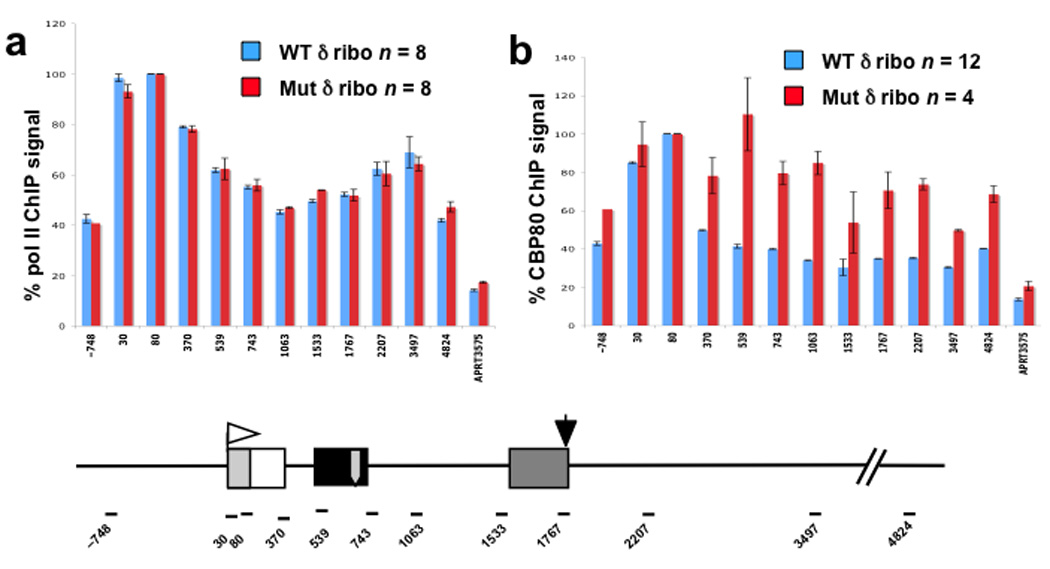
a. b. ChIP of pol II and CBP80 on integrated CMV β-globin in CHO Flp-in T-REx cells (Invitrogen) with WT (blue bars) or mutant hepatitis δ ribozyme (red bars) in exon 2. Relative ChIP signals are shown for 12 amplicons in CMV-globin gene and one in APRT (primers are in Supplementary Table 3). Mean values normalized to the maximal signal at β-globin amplicon +80 and standard errors of the mean (s.e.m.) are shown. Results are from two independent cells lines for the WT and mutant ribozyme constructs. Note that ribozyme cutting has no effect on pol II distribution (a) but specifically reduces CBP80 levels within the gene (amplicons +370 to +4824) relative to the 5’ end (amplicons +30, +80). The map shows PCR amplicon positions relative to the CMV start site, Tet operators (gray box), and δ ribozyme (gray arrow).
Severing an intron with hepatitis δ, but not hammerhead ribozyme, inhibits splicing
We next addressed whether splicing can occur across a cut within an intron by introducing the hepatitis δ ribozyme into intron 2, positioning it 200 bases from the 5’ splice site. The WT ribozyme consistently inhibited splicing of intron 2 whereas mutant ribozyme had no effect (Fig. 3a lanes 1, 4, 5). A similar inhibition of splicing was caused by the WT δ ribozyme in intron 2 of a β-globin gene with a poly (A) site mutation and fibronectin splicing enhancer (FNA2GA3) (Fig. 3a lanes 11, 12). To control for the possibility that the spliced:unspliced ratio is biased by selective degradation in the cytoplasm, we analyzed fractionated nuclear and cytoplasmic RNA. The experiment in Figure 3a lanes 13, 15 shows the reduction of spliced:unspliced transcripts caused by intronic δ ribozyme, is reproduced in nuclear RNA.
Figure 3. Hepatitis δ but not hammerhead ribozyme cleavage in intron 2 inhibits splicing.
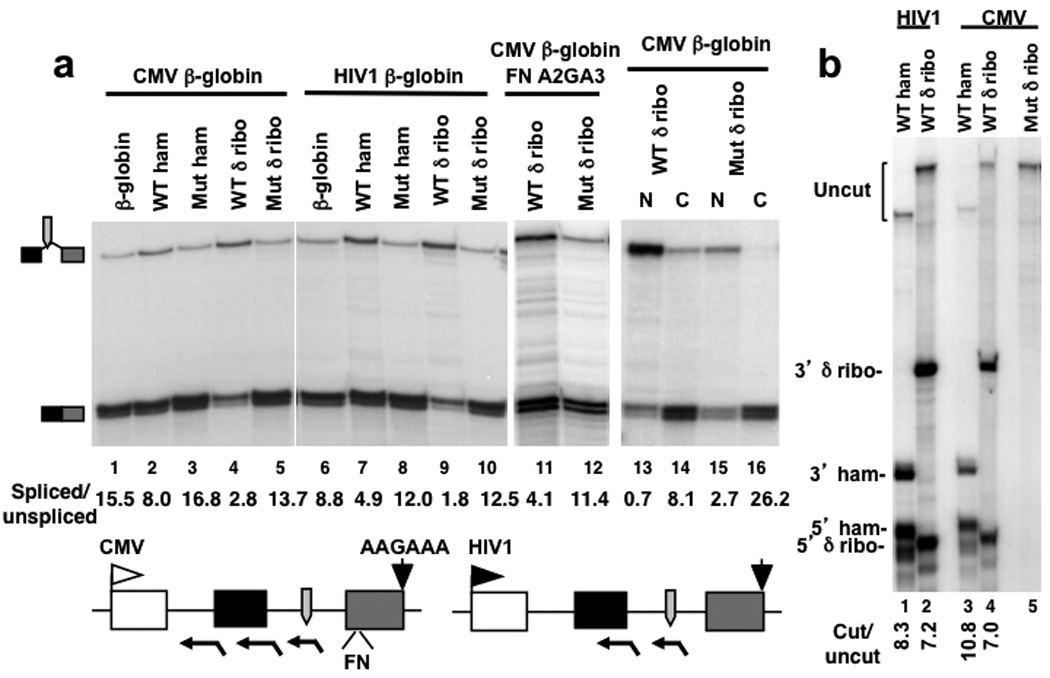
a. RPA of intron 2 splicing in RNA from cells transfected with pCMV β-globin and pHIV1HA β-globin plasmids (lanes 1, 6) and derivatives with WT or Mut hepatitis δ (δ̣) or hammerheaḍ (ham) ribozymes inserted in intron 2 (Supplementary Table 1 plasmids 33, 8–16). The HIV1 promoter (lanes 6–10) was activated by co-transfecting pSVTat. Lanes 11, 12: RPA of FN A2GA3 constructs with WT and Mut δ ribozymes in intron 2 (Supplementary Table 1 plasmids 17,18). Lanes 13–16: RPA of nuclear (N) and cytoplasmic (C) RNA from equal numbers of cells transfected with pCMV β-globin intron 2 δ ribozyme constructs as in lanes 4, 5. Note the inhibition of splicing by WT δ ribozyme relative to mutant in the nuclear fraction.
The map depicts RPA probes, ribozymes, the FN splicing enhancer, the poly (A) site and A2GA3 mutation as in Fig. 2.
b. Hammerhead and hepatitis δ ribozymes in intron 2 self-cleave to approximately equal extents and produce 5’ and 3’ ends that remain intact. RPA with antisense probes spanning the cut sites. Protection products corresponding to uncut RNA and 5’ and 3’ self-cleavage products are indicated.
The inhibition of splicing by the δ ribozyme in intron 2 contrasts with previous observations using the minimal hammerhead ribozyme 21 so we compared splicing with the two ribozymes at the same position in intron 2. The experiment in Figure 3a (lanes 1–3) shows that compared to the δ ribozyme, the WT hammerhead causes a much smaller inhibition of splicing. The hammerhead and hepatitis δ ribozymes in intron 2 cut with approximately equivalent efficiencies as monitored by RPA with probes spanning the ribozyme cut sites (Fig. 3b). The different effects of these two ribozymes on intron 2 splicing therefore can not be explained by differential cutting. Furthermore the 5’ and 3’ ends made by the hammerhead and hepatitis δ ribozymes were discrete, and accumulated to approximately equivalent levels indicating that they are not rapidly degraded. Because both ribozymes generate RNA fragments with identical end structures, the specific reduction in spliced:unspliced transcripts caused by the δ ribozyme can not be explained by differential degradation of spliced versus unspliced RNA in response to ribozyme-generated 5’ and 3’ ends. To account for the different effects of the two intronic ribozymes we propose that because the hepatitis δ ribozyme cuts far more rapidly than the hammerhead 37 38, it is able to compete effectively with spliceosome assembly. We also tested whether precursors for miR21 or miR30 39 affected splicing when inserted in intron 2 of constructs with the A2GA3 mutant poly (A) site to enhance their processing 40. Consistent with previous findings 23 24, the miRNA precursors had little or no effect on intron2 splicing (Supplementary Fig. 2b) suggesting that like the hammerhead, cleavage by Drosha is relatively slow, and does not compete with splicing.
Previous investigations of ribozyme cleavage in intron 2 (ref. 21) were performed with a reporter gene activated by HIV1 Tat which could facilitate tethering because it binds RNA and pol II. We therefore asked how the δ ribozyme in intron 2 affected splicing of transcripts driven by the Tat activated HIV1 LTR compared to the CMV promoter. The results in Fig. 3a show that intron 2 splicing was inhibited by the WT δ ribozyme relative to the mutant regardless of the promoter (compare lanes 4, 5 with 9, 10). The hammerhead had a small inhibitory effect on splicing with both promoters (lanes 2, 3, 7, 8). We conclude that inhibition of splicing by cutting an intron is not promoter-specific. Although ribozyme cleavage in intron 2 is not expected to affect definition of exon 2, we consistently observed that splicing of intron 1 was inhibited (Fig. 4a lanes 1–3) but to a smaller extent than intron 2 (Fig. 3a). Cutting intron 2 with the δ ribozyme, but not the hammerhead, also inhibited 3’ processing of CMV and HIV1 driven transcripts (Fig. 4a lanes 5, 7, Supplementary Fig. 3, lanes 2, 4).
Figure 4. Hepatitis δ ribozyme cleavage in introns inhibits mRNA processing, protein expression and RNA editing.

a. δ ribozyme cutting in intron 2 inhibits intron1 splicing and poly (A) site cleavage. RPA of pCMVβ-globin with hepatitis δ or hammerhead (ham) ribozymes in intron 2 as in Fig. 3a.
b. δ ribozyme cutting in β-globin intron 2 blocks protein expression. Western blot of HA tagged β-globin and GFP loading control in cells transfected with pCMVHAβ-glob (lane 1) and derivatives with WT or Mut hepatitis δ or hammerhead (ham) ribozymes in intron 2 (Supplementary Table 1 plasmids 1, 19–22).
c. δ ribozyme cleavage in intron 11 of the GluR-B minigene inhibits splicing and editing at the Q/R site. Left panel: Sequencing runs of the GluR-B Q/R site in RT-PCR products from cells transfected with pcDNA Flag QR/SS WT and Mut δ ribo plasmids (Supplementary Table 1 plasmids 23, 24). Note reduced editing of A to G with the WT ribozyme. Right panel: δ ribozyme cleavage in GluR-B intron 11 inhibits splicing. RPA with a 5’ splice site probe. Map depicts the RPA probe (black arrow), Q/R editing site (downward arrowhead) and ribozyme (gray arrow) within the sequence complementary to the guide.
To ask how ribozyme cleavage of intron 2 affected protein expression, we monitored accumulation of the HA-tagged globin protein. The intronic δ ribozyme abolished protein expression relative to the co-transfected GFP control (Fig. 4b lane 2), whereas the hammerhead had little or no effect (Fig. 4b lane 4). The protein expression data are therefore consistent with the RNA analysis that demonstrates cutting intron 2 (Fig. 3, Fig. 4a) shuts down splicing and 3’ end processing.
Severing the nascent transcript in an intron inhibits ADAR-mediated RNA editing
We also examined the effects of ribozyme cleavage in an intron of the glutamate receptor subunit B (GluR-B) gene 4 at a site in between the Q/R editing site in exon 11 and the complementary sequence in intron 11 that directs A-I editing by ADARs. If ribozyme cutting results in physical separation of the upstream and downstream RNA fragments, it should disrupt editing. On the other hand, if the exons are tethered to the TEC, then ribozyme cutting should not impede the base pairing that enables editing. cDNA sequencing showed that editing of the Q/R site from A to G was inhibited by the WT ribozyme in intron 11 relative to the mutant (Fig. 4c left panel). We suggest that severing the intron impairs editing by preventing base pairing between the Q/R site upstream of the cut and the complementary sequence downstream of the cut. Ribozyme cutting in the GluR-B intron 11 also strongly reduced the ratio of spliced to unspliced transcripts (Fig. 4c right panel) consistent with inhibition of splicing as we observed for the ribozyme in β-globin intron 2.
Inhibition of splicing by exon cleavage without disrupting exon definition
Communication across the last exon between the splicing and 3’ end processing proteins normally enhances removal of the last intron and cleavage at the poly (A) site. We addressed whether RNA continuity of the terminal exon is required for splicing of the last intron by incorporating the WT and mutant δ ribozymes at the EcoRI site in exon 3 50 bases downstream of the 3’ splice site and 190 bases upstream of the poly (A) site. The experiment in Figure 5a (lanes 1–3) shows that severing exon 3 markedly inhibited intron 2 splicing assayed by RPA with a probe spanning the 5’ splice site. A similar effect on intron 2 splicing was observed when nuclear RNA was analyzed thereby eliminating the possibility of a bias introduced by selective RNA degradation in the cytoplasm (Supplementary Fig. 4 lanes 1, 3). The WT δ ribozyme in exon 3 also inhibited splicing to a cryptic 3’ splice site within intron 2 when the natural 3’ splice site was deleted (data not shown). Splicing of intron 2 was also strongly inhibited by the WT ribozyme in exon 3 when the FN splicing enhancer was inserted 10 bases upstream of the ribozyme (Fig. 5a lanes 6, 7). We conclude that the inhibition of intron 2 splicing by ribozyme cutting in exon 3, is not rescued by a splicing enhancer.
If the ribozyme in exon 3 disrupts splicing by preventing exon definition, then it should have no effect when exon definition is prevented. To test this prediction, we blocked exon definition by inactivating the poly (A) site in constructs with the WT and mutant δ ribozyme in exon 3 (Fig. 5). In these experiments intron 2 splicing was stimulated by the FN splicing enhancer upstream of the ribozyme. The experiment in Figure 5a (lanes 4, 5) shows that the WT ribozyme inhibited splicing about 5 fold relative to the mutant even when exon definition is prevented by mutating the poly (A) site. A ribozyme situated downstream of exon 3 however has little effect on splicing presumably because in this case, self-cleavage mostly occurs after commitment to splicing (Fig. 5a lanes 8, 9).
Unexpectedly, splicing of intron 1 in total and nuclear RNA fractions monitored by RPA with a probe spanning the 3’ splice site was inhibited by ribozyme cutting in exon 3 to about the same extent as splicing of intron 2 (Fig 5, Supplementary Fig. 4). Similar results were obtained with exon 3 ribozyme constructs with and without the FN splicing enhancer (Fig. 5b). In summary ribozyme cleavage within exon 3 inhibited intron 1 splicing and intron 2 splicing even after inactivation of the poly (A) site. Neither of these effects is easily explained by disruption of exon definition. These results therefore suggest that another mechanism is responsible for aborting pre-mRNA processing when the nascent RNA is damaged by an endonuclease.
Inhibition of poly (A) site cleavage and transcription termination by ribozyme cutting
Ribozyme cutting of exon 3 specifically reduced the ratio of transcripts cleaved at the poly (A) site relative to uncleaved precursors consistent with inhibition of 3’ end processing (Fig. 6a lanes 1–3). These results therefore suggest that cross-exon communication that enhances cleavage/polyadenylation is sensitive to disruption of RNA continuity within the last exon. If 3’ end processing is really aborted in response to ribozyme cleavage, then we predict there should be a defect in termination, which is normally tightly coupled to processing. On the other hand if 3’ end processing were not directly affected by ribozyme cleavage upstream of the poly (A) site, then termination should occur normally. We investigated whether ribozyme cleavage affects transcription termination by using RPA to quantify correctly initiated 5’ ends and readaround transcripts that run all the way around the plasmid because they failed to terminate 41. To validate this assay we compared WT with the A2GA3 poly (A) site mutant and confirmed that the mutation greatly enhanced readaround transcription with and without the FN splicing enhancer in exon 3 (Fig. 6b lanes 1–3). We note that while termination in these experiments is dependent on the poly (A) site, it is independent of the CoTC termination element 26,42 that is absent from our constructs. Ribozyme cutting in exon 3 strongly inhibited termination relative to the mutant ribozyme control (Fig. 6b lanes 4, 5). Similarly hepatitis δ, but not hammerhead, ribozyme cutting in intron 2 inhibited termination as determined by the readaround transcription assay (Supplementary Fig. 5a). Overall we found a strong correlation between the extent of cleavage at a WT poly (A) site and termination downstream, as predicted by the torpedo model for coupling of termination with 3’ end processing 41. These results contrast with a previous report that hammerhead ribozyme cutting in exon 3 disrupted 3’ processing without affecting termination 26 however 3’ end processing in this case was not assayed by quantifying cleaved and uncleaved transcripts. This apparent discrepancy may be due to slower cutting by the hammerhead. The fact that ribozyme cleavage upstream of the poly (A) site prevents transcription termination shows that ribozyme cutting occurs before the transcript is released from the template. More importantly, the inhibition of termination confirms that processing of a wild-type poly (A) site, which is coupled to termination, is disrupted by ribozyme cutting presumably by preventing assembly of a functional 3’ end processing complex.
Figure 6. Inhibition of poly (A) site cleavage and transcription termination by ribozyme cleavage in exon 3.
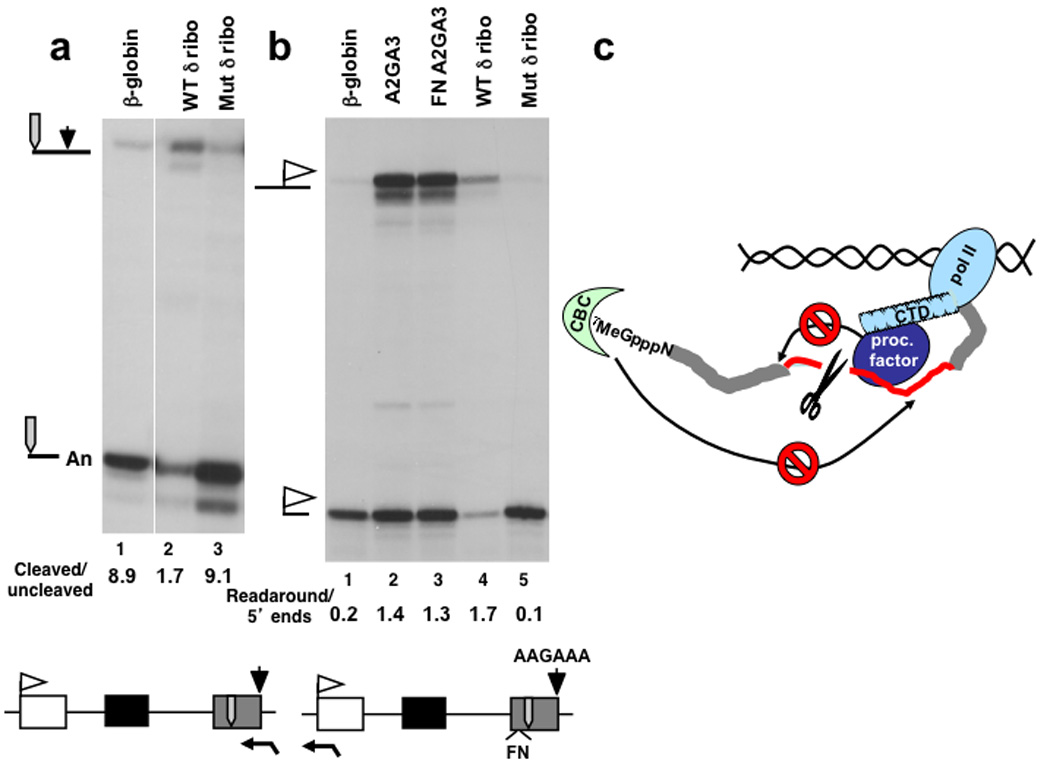
a. RPA of processing at the poly (A) site in constructs with WT and mutant hepatitis δ ribozyme in exon 3 (Supplementary Table 1 plasmids 33, 25, 26).
b. RPA of 5’ ends and transcripts that read around the plasmid. Readaround transcription indicates failure to terminate that correlates with poor cleavage at the poly (A) site in A2GA3 mutants and with the WT δ ribozyme in exon 3 (lanes 2–4)(Supplementary Table 1 plasmids 33–34, 25, 26).
c. Model for inhibition of co-transcriptional pre-mRNA processing at positions upstream and downstream of a cut in the transcript (scissors) through disruption of interactions with CBC bound to the cap (7MeGpppN) or CTD-bound processing factors (proc. factor).
DISCUSSION
Disruption of splicing by an intronic ribozyme: implications for exon tethering
We report that a contiguous nascent RNA chain is necessary for processing by splicing and cleavage polyadenylation. Both these co-transcriptional processing events are vulnerable to disruption by endonucleolyic damage to exons or introns. During transcription, exon definition and coupling between capping, splicing, and 3’ end processing are facilitated by interactions between proximal and distal segments of the pre-mRNA. It was previously proposed that tethering of exons to pol II enhances co-transcriptional processing because of the surprising finding that a hammerhead ribozyme in β-globin intron 2 does not disrupt splicing 21 43. We found that in contrast to the hammerhead, intron cleavage by the hepatitis δ ribozyme does impair splicing as well as 3’ end processing and protein expression (Fig. 3, Fig. 4, Supplementary Fig. 3). δ ribozyme cleavage between the self-complementary sequences in exon 11 and intron 11 of GluR-B also preventing RNA editing by ADAR (Fig. 4c). Therefore exon-intron base-pairing appears to be prevented if the complementary sequences in the nascent RNA are separated by a ribozyme cut. These observations suggest that exons upstream of a ribozyme cut site dissociate from the TEC rather than being tethered to it. Consistent with this model, ribozyme cutting in β-globin exon 2 released the RNA 5’ end from the gene as determined by ChIP of the cap binding complex (Fig. 2b).
The hepatitis δ ribozyme cuts about an order of magnitude faster than the minimal hammerhead 37,38 and we suggest that splicing is only disrupted if an intronic ribozyme cuts fast enough to interfere with co-transcriptional spliceosome assembly. Intron cleavage by Drosha during miRNA excision probably does not disrupt splicing (Supplementary Fig. 2b)23,24 because like the hammerhead, it is slow relative to spliceosome assembly. Kinetic competition between splicing and ribozyme cutting is also suggested by the observation that the hammerhead inserted in a yeast intron only inhibited splicing if transcription was slowed by mutation of elongation factors 22.
Inhibition of processing upstream and downstream of ribozyme cut sites
Processing is inhibited at positions upstream and downstream of a ribozyme cut site in the nascent pre-mRNA. Cutting within exons is predicted to inhibit splicing by disrupting exon definition 27 21 but our experiments show such an effect can not completely account for the effects of exonic ribozymes. For example intron 1 splicing was diminished by cutting exon 3 and intron 2 splicing was impaired by cutting exon 1 (Fig. 5b, Fig. 1b). Furthermore, severing exon 3 disrupted splicing of intron 2 even when exon 3 definition was prevented by mutating the poly (A) site (Fig. 5a lanes 4, 5).
We suggest that in addition to effects on exon definition, two factors which contribute to disruption of processing upstream and downstream of a ribozyme cut site are: 1) separation of the TEC with its CTD-associated processing factors from sequences upstream of the cut (Fig. 6c) and 2) separation of CBC bound to the 5’ cap from sequences downstream of the cut (Fig. 2b). For example, the inhibition of intron 1 splicing by cleavage in intron 2 or exon 3 (Fig. 4a, Fig. 5b, Supplementary Fig. 4 lanes 5, 7) is most easily explained by release of the 5’ segment of the transcript from the TEC (Fig. 2b) so that it is less accessible to processing factors associated with the CTD (Fig. 6c). Conversely inhibition of intron 2 splicing by cutting in exons 1 and 2 (Fig. 1b, c, Supplementary Fig. 1) is most easily explained by release of the RNA 5’ end (Fig. 2b) because CBC stimulates splicing and cleavage/polyadenylation 15–18 (Fig. 6c). Similarly 3’ end processing and coupled transcription termination were both inhibited by ribozyme cutting upstream in intron 2 or exon 3 (Fig. 6a, b and Supplementary Fig. 5a).
In summary these results show that damage inflicted in exons or introns causes processing events upstream and downstream of the cut site to be aborted. This conclusion is consistent with the model that the nascent transcript has an important function in stabilizing complexes between processing factors and the pol II TEC 20,34 and that damage to the RNA would therefore destabilize such complexes. Such a “bail-out” mechanism would have the potential advantage of avoiding formation of non-productive complexes between processing factors and pol II that is not engaged in synthesis of a functional transcript.
METHODS
Transient Transfections and RNA analysis
293 cells (150 mm plates) were transiently transfected with 10 µg of reporter plasmid and 1 µg of pSPVA using linear polyethyleneimine (M.Wt. 25,000). VA RNA transcribed by pol III is a control for transfection efficiency and recovery. It was quantified by RPA in all experiments and is shown in selected figures. HIV1 transcription was activated by co-transfection with 2 µg of pSVTat. GFP loading control was expressed using pEGFP2.2 (Clontech) (Fig. 4b). Cells were harvested after 40 hours. The results shown in the figures are representative of those obtained in multiple independent transfections and mean values for multiple experiments are in Supplementary Table 2. Nuclear and cytoplasmic RNA was prepared as described 34.
Total RNA was digested with DNAse I to remove all contaminating DNA (Supplementary Fig. 5b) and analyzed by RNAse protection as described 10 In all cases undigested probe (not shown) was clearly resolved from RNAse protection products. Results were quantified by Molecular Dynamics phosphor-imager. Ratios of processed:unprocessed RNA were calculated after compensating for 32P-U content and are summarized in Supplemental Table 2.
Cell lines
Human HA-β-globin genes with WT and mutant hepatitis δ ribozyme in the BamHI site in exon 2 were cloned into the pcDNA5/FRT/TO vector (Invitrogen) and integrated by Flp recombinase-mediated site-specific recombination into CHO Flp-in T-REx cells (Invitrogen). Two independent lines of each integrant were selected by hygromycin B resistance.
ChIP was as described previously with anti pan pol II CTD 44 and anti-CBP80, a generous gift of Dr. E. Izaurralde. Real-time PCR was carried out in a Roche LC-480 with Sybr Green as described 44. Results are from at least two PCR’s with IP’s from each of two independent cell lines. Transcription was induced with doxycycline (2 µg ml−1) overnight before cross-linking. Amplicons in Figure 2 are labeled by the position of the middle of the amplicon relative to the transcription start site. Primers are described in Supplementary Table 3.
Plasmids
A list of plasmids used in this study is in Supplementary Table 1. pCMV β-globin reporter gene and pBS CMV globin used to make the 5’ end RPA probe have been described 45 as have plasmids for VA transfection control and those for globin splice sites and poly (A) site 10.
Ribozymes were inserted into exon 1 (ex1) at a XhoI site introduced at position −30 relative to the ATG of the N-terminal HA tag. Ribozymes were inserted in exon 2 (ex2) at the BamHI site, and in exon 3 (ex3) at the EcoRI site. The fibronectin (FN) splicing enhancer (EDA) was inserted in exon 3 at the BstX1 site. Ribozymes and miRNA precursors were inserted in intron 2 (int 2) at the MunI site.
The HIV1 LTR +TAR (-453 - +83) was PCR amplified and inserted into the BglII-XhoI sites replacing the CMV promoter of pCMVHA β-glob.
Ribozyme cleavage occurs 1 base after the histone stem loop sequence GGCCCT TATC AGGGCC (stem underlined). The variant hepatitis δ ribozyme sequence that immediately follows the stem-loop is: A*GGGCGGCATGGTCCCAGCCTCCTCGCTGGCGCCGCCTGGGCAACATGCTTCGGCATGGCGAATGGGACCAAAT.
C76 that is substituted with T in the mutant is underlined and the cut site is marked by * 37.
The hammerhead sequence that immediately follows the histone stem loop is: ACCTGTC*ACCGGATGTGTTTTCCGGTCTGATGAGTCCGTGAGGACGAAACAGG
The G that is substituted with C in the mutant is underlined and the cut site is marked by * 46. Note the minimal hammerhead sequence used here and in a previous study 21 lacks the sequences in stem-loops I and II of full-length hammerhead that greatly enhance its activity 47.
Pri-miR-21 and –miR-30 sequences were PCR amplified from pCMV-miR-21 and pCMV-miR-30 39 and inserted in intron 2 of pCMVβ-globFNA2GA3 at MunI.
The GluR-B minigene pcDNA3Flag/GQRSS has been described 4. WT and mutant δ ribozyme was inserted at an EcoRI site 65 bases from the 5’ splice site in intron 11 sequence. The RPA probe (Fig. 4c) was made from pBSKS- QRSS that contains a fragment spanning the 5’ splice site.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NIH grant GM58613 to D.B. We thank T. Blumenthal, R. Davis, R. Ferguson (U. Colorado), J. Underwood (UC Santa Cruz), J. Manley (Columbia U.), K. Ryman (U. Stockholm), B. Cullen (Duke U.), S. Chavez (U. Sevilla) for helpful suggestions, E. Izaurralde (U. Tubingen) for anti CBP80, B. Cullen for miR21 and miR30 plasmids, J. Lykke-Andersen (U. Colorado) for Upf1 and PM-Scl75 plasmids, and the UCHSC Cancer Center sequencing facility.
Contributor Information
Nova Fong, Dept. Biochemistry and Molecular Genetics, University of Colorado School of Medicine, UCDHSC, MS8101, P.O. Box 6511, Aurora CO. 80045..
Marie Ohman, Department of Molecular Biology and Functional Genomics, Stockholm University, S-106 91 Stockholm, Sweden..
David L. Bentley, Dept. Biochemistry and Molecular Genetics, University of Colorado School of Medicine, UCDHSC, MS8101, P.O. Box 6511, Aurora CO. 80045..
REFERENCES
- 1.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 2.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 4.Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko S, Manley JL. The Mammalian RNA Polymerase II C-Terminal Domain Interacts with RNA to Suppress Transcription-Coupled 3' End Formation. Mol Cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 6.Mortillaro MJ, et al. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Ujvari A, Luse DS. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J Biol Chem. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- 9.Keen NJ, Churcher MJ, Karn J. Transfer of Tat and release of TAR RNA during the activation of the human immunodeficiency virus type-1 transcription elongation complex. EMBO J. 1997;16:5260–5272. doi: 10.1093/emboj/16.17.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong N, Bentley D. Capping, Splicing and 3’ Processing are Independently Stimulated by RNA Polymerase II: Different Functions for Different Segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:236–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol Cell Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 15.Izaurralde E, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 16.Lewis JD, Izaurralde E, Jarmolowski A, Mcguigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of u1-snrnp with the cap-proximal 5' splice-site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 17.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3' processing. Proc Natl Acad Sci U S A. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamson TE, Shutt DC, Price DH. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J Biol Chem. 2005;280:32262–32271. doi: 10.1074/jbc.M505532200. [DOI] [PubMed] [Google Scholar]
- 20.Rigo F, Kazerouninia A, Nag A, Martinson HG. The RNA tether from the poly(a) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 22.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson K, Moore MJ. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 26.West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 28.O'Mullane L, Eperon IC. The pre-mRNA 5' cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5' splice sites. Mol Cell Biol. 1998;18:7510–7520. doi: 10.1128/mcb.18.12.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 30.Lam BJ, Hertel KJ. A general role for splicing enhancers in exon definition. RNA. 2002;8:1233–1241. doi: 10.1017/s1355838202028030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyburz A, Friedlein A, Langen H, Keller W. Direct Interactions between Subunits of CPSF and the U2 snRNP Contribute to the Coupling of Pre-mRNA 3' End Processing and Splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 32.Millevoi S, et al. An interaction between U2aF 65 and CF I(m) links the splicing and 3' end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3'-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008;28:849–862. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. Ribozyme Cleavage Reveals Connections between mRNA Release from the Site of Transcription and Pre-mRNA Processing. Mol Cell. 2005;20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, Carmichael GC. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 38.Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 42.Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- 43.de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3'-end formation. Mol Cell Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samarsky DA, et al. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc Natl Acad Sci U S A. 1999;96:6609–6614. doi: 10.1073/pnas.96.12.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


