Abstract
The efficient enzymatic detection of damaged bases concealed in the DNA double helix is an essential step during DNA repair in all cells. Emergent structural and mechanistic approaches have provided glimpses into this enigmatic molecular recognition event in several systems. A ubiquitous feature of these essential reactions is the binding of the damaged base in an extrahelical binding mode. The reaction pathway by which this remarkable extrahelical state is achieved is of great interest and even more debate.
Keywords: DNA damage, DNA glycosylase, extrahelical base recognition, base flipping, DNA base dynamics
Introduction
For small molecules in solution, simple bond breaking and bond making reactions occur when the reactants are positioned closely, and with the proper geometries, to allow the chemical transformation. To achieve close proximity and optimal reaction geometry, many enzyme catalyzed reactions involving large substrates such as proteins or nucleic acids require large conformational changes in the substrate to allow the enzyme to access and optimally position the site of reaction. One of the most dramatic substrate conformational transformations in site-specific nucleic acid recognition by enzymes is the complete rotation of a base and its attached sugar from the base stack through either the major or minor groove, a transition commonly called “base flipping” (Figure 1) [1, 2]. The ubiquitous occurrence of this mechanism for a variety of enzymes that break or form new bonds involving the base, sugar or phosphate moieties of RNA or DNA suggests strong convergent evolution for this mechanistic strategy (Table 1 and Scheme 1).
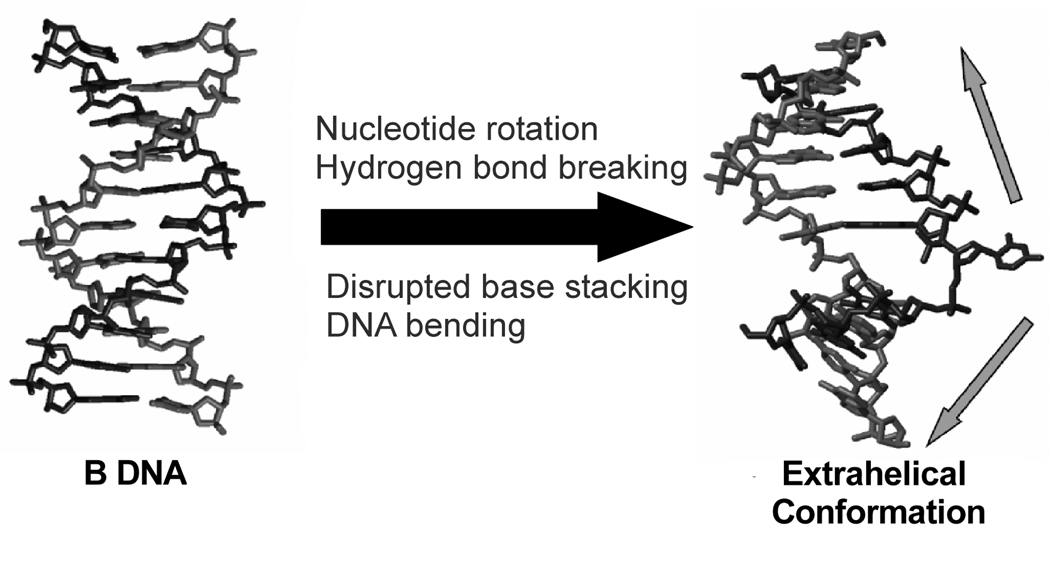
Figure 1.
DNA deformations occuring during the process of enzymatic base flipping. The unfavorable energetic events during the reaction are listed. The enzyme must pay for these costs through the use of favorable binding energy, which would be expected increase over the reaction pathway. The extrahelical conformation on the right was extracted from the complex of uracil DNA glycosylase bound to uracilated DNA (pdb 1EMH).
Table 1.
Enzymes that Flip Bases
| Enzyme [a] | Reaction | PDB [b] |
|---|---|---|
| Uracil DNA glycosyase (UNG, 1) | C1′-N bond cleavage of deoxyuridine in DNA | 1EMH, 2OXM |
| human 8-oxoguanine DNA glyocsylase (hOGG1, 2) | C1′-N bond cleavage of 8-oxoguanine in DNA | 1EBM, 1YQK |
| adenine, thymine, hypoxantihine, and various photodamaged, oxidized and alkylated base DNA glycosylases | C1′-N bond cleavage of cognate base in DNA | [3] |
| DNA methyltransferase (MTase, 3) | Methylation of the C5 and N4 of C, N6 of A and O6 of G | 2HMY |
| RNA methyltransferase (2’-OMTase, 4) | Methylation of nucleoside- 2′-O | 1EIZ, 1EJ0 |
| pseudouridine synthase (5) | Isomerization of uridine to pseudouridine in RNA | 2I82 |
| T4 5-hydroxymethyl cytosine glucosyltransferase (5-HMC GTase, 6) | Transfer of glucose from UDP-glucose to Cytosine 5-CH2OH | 1Y6F |
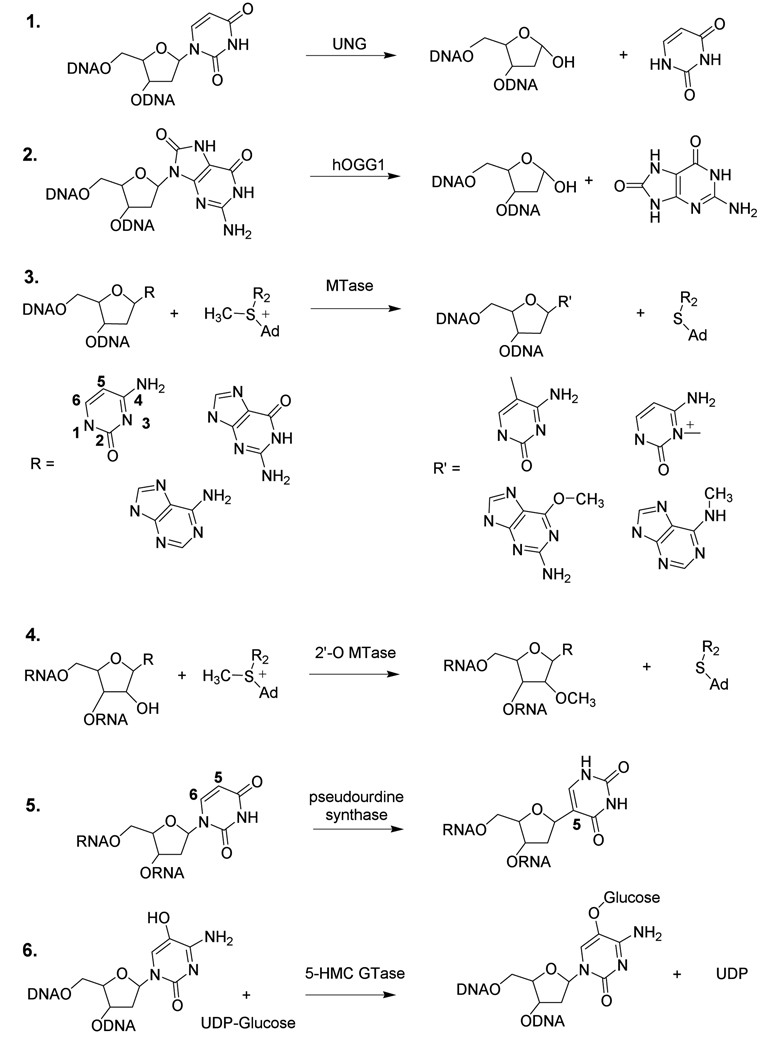
Representative enzymes that have been shown crystallographically to flip bases. The list is not comprehensive and the reader should refer elesewhere for additional examples [4]. The numbers in parentheses refer to the reactions shown in Scheme 1.
The pdb structural files listed are of a complex with a fully flipped cognate base or a trapped intermediate state along the flipping coor dinate.
Scheme 1.
Reactants and products of several enzymes that use a base flipping mechanism. R2 = (CH2)2CHNH2CO2H, Ad = 5′-adenosyl
Why would so many diverse enzymes share a mechanism that requires such an energetically unfavorable conformational transition in the substrate? Since biological processes seldom if ever occur by the unnecessary expenditure of energy, we must assume that there is a unifying reason that a base flipping mechanism is followed. A passing consideration of the base flipping process points to several obvious energetic barriers: the breaking of Watson-Crick hydrogen bonds, the disruption of base stacking interactions, and the introduction of unfavorable backbone conformations that are required to allow a rotation of the base from the base stack [1]. One rationale for the investment of binding energy in this process stems from simple steric considerations involving access of the enzyme to the reactive site in the substrate. In addition, catalytic requirements of the given chemical transformation must be considered (Table 1). For instance, DNA damage specific glycosylases perform nucleophilic attack at the C1′ position of damaged or mismatched bases in DNA resulting in cleavage of the C1′-N glycosidic bond (Scheme 1). Such enzymes comprise the initial step in a pathway for the ultimate excision and replacement of the entire damaged nucleotide in DNA. For these enzymes, that act on B-form DNA, the access of a water or enzyme nucleophile to C1′ is enhanced by rotation of the base sugar from the base stack. In addition, the difficult chemical problem of glycosidic bond cleavage also requires access to the hydrogen bond acceptor atoms on the base leaving group so that the enzyme can neutralize the negative charge that develops in the transition state [3]. In the absence of base flipping, these groups are buried in the DNA base stack and the enzyme is unable to employ this essential catalytic strategy. Steric access and catalytic considerations are also major problems in the mechanisms of both 5-methyl cytosine DNA methyltransferase (MTase) and pseudouridine synthase (Table 1 and reactions 3 and 5 in Scheme 1)[4, 5]. These enzymes both require attack of a cysteine nucleophile at C6 of a pyrimidine base (cytosine or uracil) to generate a carbanion at C5 to initiate the reaction (see Scheme 1 for ring numbering). To further the need for base flipping in the methyltransferase reaction, the second step involves transfer of a methyl group from S-adenosylmethionine to the nucleophilic C5 carbanion of cytosine-cysteine adduct. Thus cytosine methylation requires access to both C6 and C5, as well as protonation at N3 to provide an electron sink for stabilization of the carbanion[6]. Similar steric and/or chemical considerations apply to each enzyme system listed in Table 1 and provide the evolutionary driving force for base flipping.
Although base flipping was first observed almost fifteen years ago[7], a mechanistic dissection of the reaction pathway has only recently become possible through the use of new structural and biophysical tools. It is the purpose of this review to highlight the findings from two DNA repair glycosylases that have benefited most from these approaches. Since several reviews have previously dealt with the subject of enzymatic base flipping, the reader is referred to these for further information[1, 2, 4, 8]. To avoid redundancy, this article emphasizes recent findings that have provided important insights into the nature of the reaction pathway for enzymatic base flipping, and importantly, elucidation of the dynamic properties of DNA that impact the recognition of damaged DNA bases. These informative new discoveries have pushed the measurement envelope so as to provoke new questions for mechanistic investigation.
The Energetics of Flipping Bases
Computational Studies
What does the reaction coordinate for spontaneous opening of DNA base pairs look like, and what are the energetic barriers? For base flipping from duplex DNA, the reaction coordinate may be described by the angle of rotation of the flipped nucleotide from its most energetically stable conformation within the DNA duplex, with a 180 degree rotation somewhat arbitrarily defined as complete base flipping. Depending on the base (pyrimidine or purine) and the sequence context, flipping may occur from either the minor or major groove. The free energy profile for such a reaction cannot be directly measured experimentally, but can be calculated using computational methods[8–14]. A schematic profile mimicking that obtained from a potential of mean force calculation is shown in Figure 2. At small rotation angles the free energy shows a sharp increase reflecting the breaking of hydrogen bonds and optimal stacking interactions in B form DNA. At larger rotations (∼40 to 180 degrees), the base and attached sugar become solvent exposed, and the reaction coordinate becomes fairly flat. Rotation along the coordinate requires changes in the various dihedral angles for the two phosphodiester linkages that flank the flipping base. A key implication of the flat region in the trajectory is that opening of a base pair generates a population of conformationally heterogeneous extrahelical states.
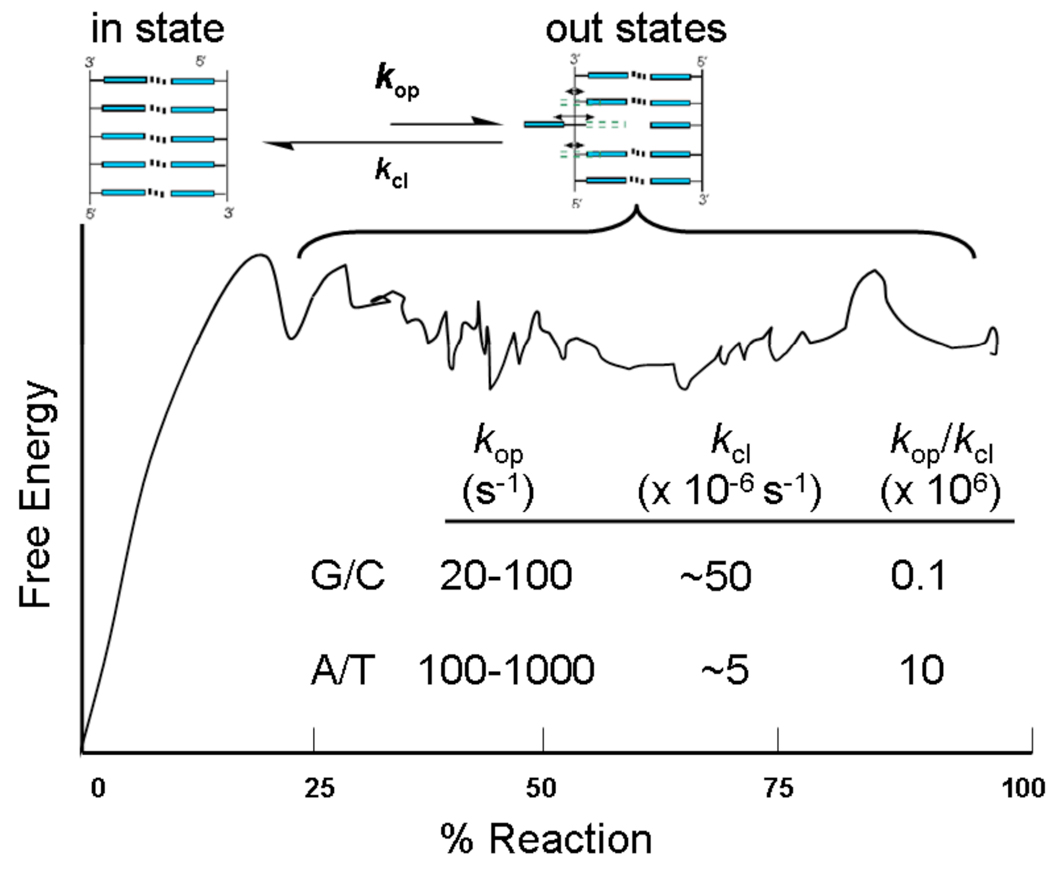
Figure 2.
Reaction coordinate (schematic) for spontaneous base pair opening in B DNA through a 180° rotation along a major groove pathway as determined in potential of mean force calculations [9–11]. Characteristic opening equilibria (kop/kcl), and opening and closing rates (kop and kcl) for G:C and A:T base pairs are noted (T = 15 °C) [15,17]. The bracket denotes a population of isoenergetic out conformations that are in rapid fluctation.
NMR Dynamic Measurements
NMR spectroscopy is the only experimental method capable of providing kinetic and thermodynamic information on spontaneous base pair opening[15–17]. This method indirectly measures the both the rates (kop, kcl) and equilibrium (Kop = kop/kcl) for base pair opening and closing by monitoring exchange of the imino protons of T or G with solvent protons that have been magnetically labeled by selective inversion (Figure 2). This approach reasonbly assumes that exchange occurs only from the open state. Thus, when imino exchange is fast relative to opening, the observed rate is equal to the rate limiting step, base pair opening. Although it is tempting to infer that the base pair opening rates measured by NMR reflect the flipping and exposure of the T and G bases that contain the observable imino proton, this may not be the case. It is possible that for T imino exchange, the opposite adenine flips leading to exposure of the T imino proton without requiring T to leave the base stack. Similarly, imino proton exchange in G/C pairs could occur by flpping the opposing cytosine base. In fact, computational studies suggest that exchange always reflects flipping of the purine base of the pair[18]. For G/C pairs, the bias is predicted to be greatest with G flipping dominating 1,000-fold over C, whereas flipping of A from an A/T pair is calculated to be only 6-fold more favorable than T. These findings, if confirmed experimentally, have major implications for the mechanism of enzyme mediated flipping (see below).
Linear Free Energy Relationships
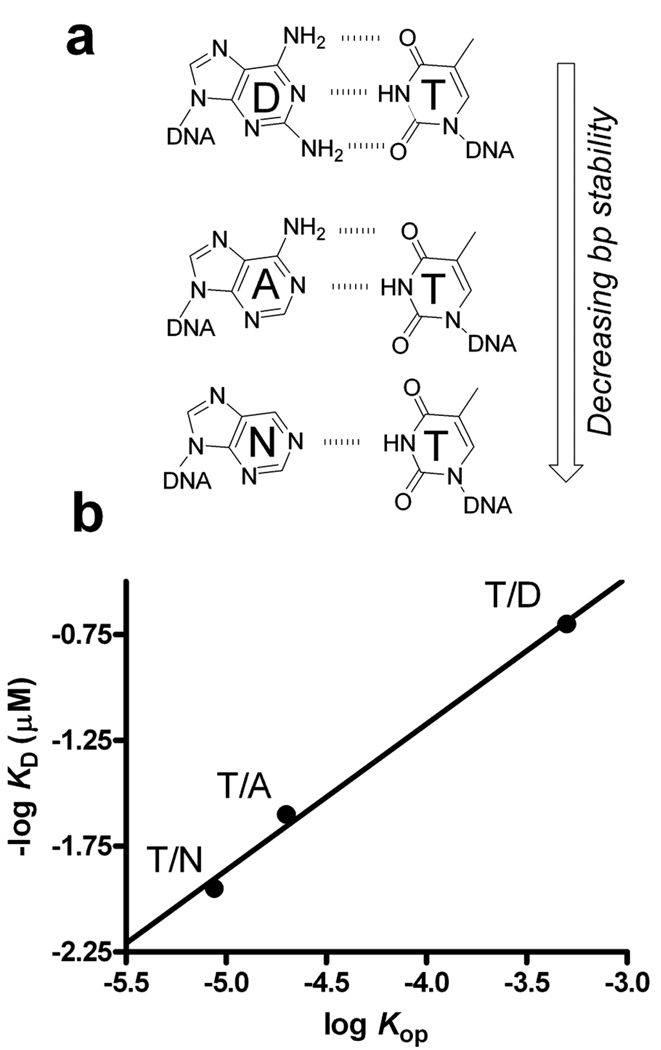
A reasonable person might ask if an energetic correlation exists between the rate and/or equilibrium for base pair opening and the preference for an enzyme to interact with the site. This question has great import because the increased flexibility or dynamics of a damaged base pair could guide an enzyme to the site. A systematic study of this question has been undertaken for UNG (Scheme 1, reaction 1). In these studies, DNA duplexes containing a single U/X or T/X base pair were used, where X was a adenine analogue capable of forming one, two or three hydrogen bonds with the opposing U or T (Figure 3a)[19–21]. [As further elaborated below, UNG can bind T or U in an extrahelical mode, but the shared binding site for T and U is not the active site pocket that only accomodates U.] Thus, progressive destabilization of the base pair by removal of hydrogen bonds would be expected to favor base pair opening and also enzyme binding, because less binding energy would be required to open a destabilized base pair. Two linear free energy correlations have been measured in this regard. In the first, ΔG for DNA melting was correlated with the free energy for UNG binding using the DNA series with U:X pairs[21]. This correlation was linear with a slope of 0.3, indicating that base pairs with stronger hydrogen bonds inhibit uracil flipping and enzyme binding. Second, the equilibria for T:X base pair opening and imino proton exchange were correlated with log KD for enzyme binding to T/X pairs (Figure 3b). Once again a linear correlation was observed (slope = 0.7), indicating that base pairs with larger equilibrium constants for opening lead to enhanced binding. These energetic correlations between the intrinsic stability and dynamics of the base pair and enzyme binding to both uracil and thymine suggest that initial damage recognition may rely on the intrinsic dynamic and physical properties of the base pair.
Figure 3.
UNG binds more tightly to T/X (or U/X) base pairs that have large opening equilibrium constants: X = D (diaminopurine), A. (adenine), and N (nebularine). (a) In this series of base pairs, the number of hydrogen bonds in the T/X pair is incrementally decreased from three to one while keeping the shape and electronic properties of the X partner constant. (b) The opening equilibrium constant was measured using NMR imino proton exchange,[19] and then compared with the dissociation constant for UNG binding to each construct (Krosky and Stivers, unpublished).
Piecing Together the Enzymatic Pathway
Experimental and computational descriptions of the energetics of spontaneous base pair opening reveal the lowest energy pathway for flipping bases in the absence of an enzyme. Although an enzyme could follow an entirely different pathway, this would seem highly unlikely given the constraints on base flipping imposed by B DNA structure. Thus it seems likely that enzymes would take advantage of the same lowest energy trajectory, and then use binding energy to overcome the thermodynamic and kinetic problems that prevent bases from remaining extrahelical in B DNA. A related issue is whether DNA glycosylases use the spontaneous breathing dynamics of base pairs to “inspect” bases while they are in a transient extrahelical state. The correlation between base pair opening and the binding affinity for UNG suggests that extrahelical inspection occurs, at least in this system (Figure 3).
What are the kinetic and thermodynamic problems that enzymes must overcome to rapidly and stably bind bases in their active sites? As revealed by parameters listed in Figure 2, base pair opening is fairly rapid even at 10 or 15 °C, where most of the NMR measurements are performed. If an extrapolation to 25 °C is made using estimates of the activation enthalpies for opening[22], then an opening rate of 1,400 s−1 may be calculated for the T/A DNA used in the UNG free energy correlations[23]. This opening rate is faster than the measured rates of uracil flipping by UNG at 25 °C (<700 s−1)[24, 25]. Thus UNG, as a catalyst, need not find a way to enhance the initial motions that lead to opening. In contrast, the open state(s) that are achieved from base pair opening exist only for about 0.1 microseconds or less at T = 25 °C (Figure 2). Thus, if enzymes are to utilize the spontaneous opening rate to initiate the flipping process, they must possess a means of rapidly and efficiently grabbing onto an extrahelical state that has only a fleeting lifetime. Then, this intermediate must be rapidly funnelled forward before it has an opportunity to fall back into the DNA base stack. Finally, the flat profile for flipping in the absence of an enzyme is not conducive to highly productive catalysis, and enzymes must find ways to energetically stabilize conformations along this trajectory and guide the base into the active site. Many insights into the above mechanistic questions have recently been elucidated using structural trapping methods and NMR dynamic studies.
Lessons from Crystal Structures
Novel methods have been recently employed to trap and characterize unstable and fleeting intermediates that occur along enzymatic base flipping pathways. The two most informative studies that have been performed are on human 8-oxoguanine (hOGG1) and uracil DNA glycosylases (UNG)(Table 1), enzymes originally discovered by Boiteux and Lindahl, respectively [26, 27]. Both studies were designed to ask the question of whether these enzymes extrahelically inspect normal bases such as guanine and thymine during the hunt for their structurally similar cognate damaged bases 8-oxoG and U (Scheme 1).
hOGG1
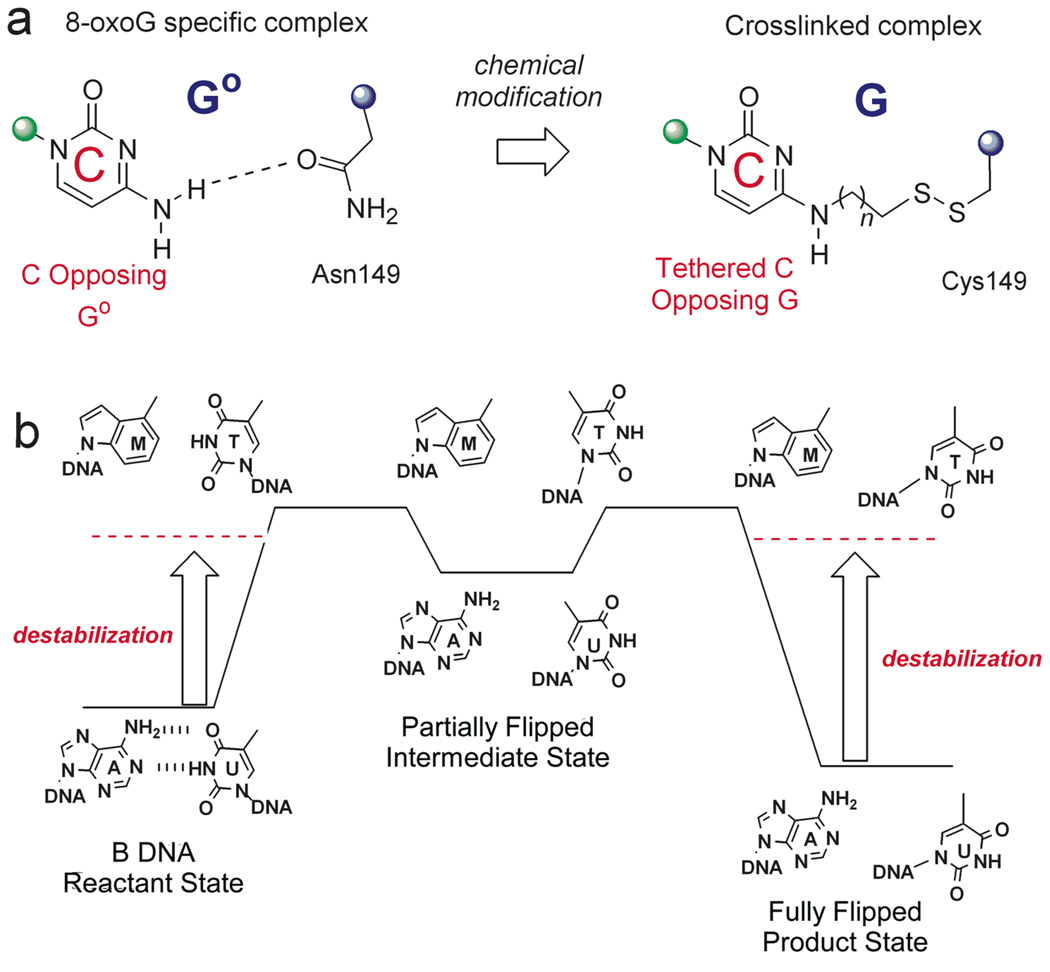
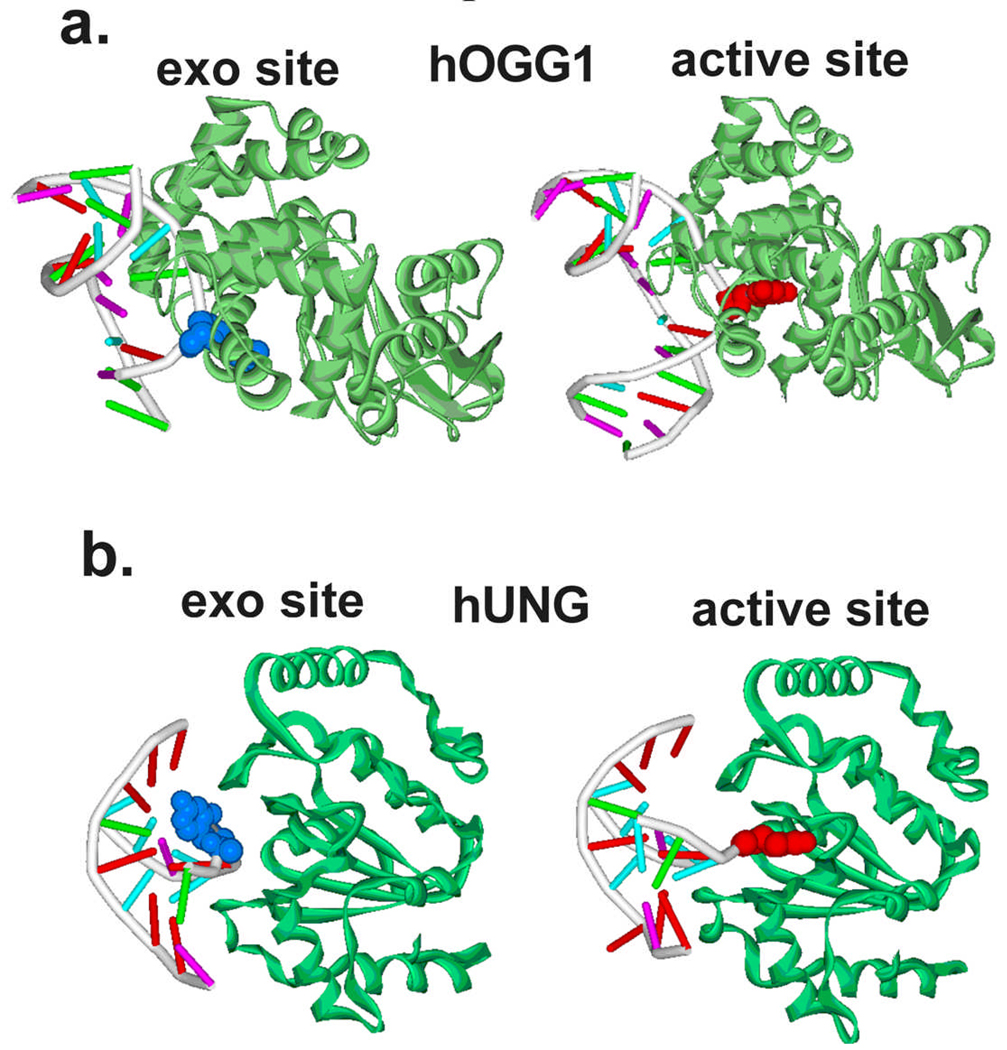
The trapping strategy employed for hOGG1 was to crosslink the DNA to the enzyme thereby locking the enzyme in a single binding register along the DNA duplex (Figure 4a)[28, 29]. This strategy took advantage of structural information provided in a previously solved specific complex where the cognate base 8-oxoG was flipped into the active site[30]. A key interaction in the specific recognition complex was a close contact between the cytosine base on the opposite strand to 8-oxoG and an asparagine side chain of hOgg1 (Figure 4a). This observation prompted the investigators to modify the exocyclic amino group of the cytosine with an alkylthiol linker, and then mutate the asparagine to cysteine to allow a disulfide linkage to form between the DNA and enzyme. The strategy worked, and the structure of a non-specific DNA complex with G in place of 8-oxoG was determined (Figure 5a). The most informative finding in this work was that the guanine base was rotated about 130° from the DNA base stack into an exo-pocket distinct from the active site pocket occupied by 8-oxoG in the specific complex. This important finding suggested that hOGG1 flips both G and 8-oxoG into this transient discrimination pocket, but that only 8-oxoG can proceed further into the active site. Nearly all of the DNA backbone interactions were shared between the non-specific and specific complexes indicating that these interactions form early on the pathway and perhaps drive the reaction towards the final state. In addition, Asn149 which intercalates into the DNA in the final state is observed in the same position in the non-specific complex, suggesting this interaction forms before or during the transition state preceding the exo site intermediate. In the context of the entire flipping coordinate of hOGG1, which involves a 150° rotation of the 8-oxoG nucleotide, the exo intermediate is quite “late” on the trajectory. Elegant kinetic studies on hOGG1 have also provided evidence for several intermediates on the reaction coordinate to the active site[31, 32].
Figure 4.
Strategies for trapping unstable intermediates during base flipping (see text). (a) Disulfide crosslinking. (b) Reaction coordinate tuning.
Figure 5.
Intermediates on the base flipping pathways of hOGG1 and UNG[28, 23]. (a) The exo-site complex of hOGG1 with an extrahelical guanine (blue) obtained by disulfide crosslinking technology (left). The fully extrahelical complex with 8-oxoG is shown on the right for comparison [30]. (b) The early exo-site complex of hUNG with an extrahelical thymine (blue) obtained using the reaction coordinate tuning method. The fully extrahelical complex with uracil is shown on the right [33].
hUNG
An entirely different approach was used to trap an extrahelical thymine intermediate during the UNG reaction (Figure 4b)[23]. The method relied on two effects: energetic destabilization of the thymine base pair in the reactant state (bound B DNA) by using a purine analogue that had no hydrogen bonding groups (M, Figure 4b), and destabilization of the final flipped product state. The latter effect arises because the methyl substituent of thymine sterically precludes binding to the uracil specific active site [33]. Accordingly, if the reactant state and product states are no longer the most stable species on the reaction coordinate, otherwise unstable intermediate flipped states are now populated. For this reason, the approach was called “reaction coordinate tuning (RCT)” [23]. Highly reminiscent of the hOGG1 example, the RCT approach allowed crystallization of a complex of UNG with thymine rotated into a T and U specific exo pocket. The DNA backbone interactions were nearly identical to that observed in the final extrahelical product state, and a leucine intercalative residue was fully inserted in the DNA minor groove, which are interactions that are also seen in the hOGG1 non-specific complex. However, the thymine base was only rotated about 30° from the base stack, and was highly solvent exposed with its Watson-Crick edge docked against an extended loop region of UNG far from the active site pocket. The O4, H3 and O2 hydrogen bond donor and acceptor groups were engaged with a histidine side chain (O4) or backbone amide and carbonyl groups (O2 and H3). As compared to the non-specific intermediate observed in the hOGG1 system, this thymine complex is much earlier on the reaction pathway.
A surprising structural observation with important mechanistic implications was that thymine had an unusual syn configuration around the glycosidic bond in the exo intermediate, as opposed to the standard anti configuration in the B DNA reactant state. This result requires that a full 180° rotation around the glycosidic bond occurs upon moving from the reactant to intermediate state. This dynamic motion has the benefit of allowing the enzyme to read out the hydrogen bond pattern of thymine and uracil (see above). Mechanistically, it is likely that free rotation around the glycosidic bond occurs on the nanosecond time scale after the base has left the restricted confines of the DNA base stack[13]. UNG can then trap the anti configuration that presents the correct hydrogen bond donor acceptor pattern.
Structural Implications
To generalize, the above structural studies suggest some common mechanistic principles that govern enzymatic base flipping. First, the reaction pathway is broken down into bite size pieces by the enzyme. Intermediate docking points are used not only to selectively guide the cognate base into the active site, but to also serve as sieving gates to distinguish between a damaged base and its normal counterpart, which may differ by only a single atom. In between these docking points, the base and its attached sugar likely migrate without interacting directly with the enzyme. This proposal is strongly supported by the anti-syn rotation of the thymine base observed in the UNG system. Second, phosphate backbone interactions and enzyme side chain intercalation occur very early in the process. A reasonable scenario is that the phosphate interactions are used to drive the reaction forward through a series of one or more intermediate states, where each intermediate forms stronger interactions with the backbone than the previous state. This type of downhill energy funnel is supported by rapid kinetic studies with UNG and hOgg1, where two intermediates before the active site have been detected, and the second is more stable than the first, but is not as stable as the final extrahelical state. Finally, measurements indicate that each step is rapid (> 300 s−1) and highly reversible. Thus, the intermediates formed are highly transient, and the reaction is pulled forward solely by the first irreversible step: glycosidic bond hydrolysis.
Insights from NMR Dynamic Studies of UNG
A key mechanistic question in damaged base recognition is whether the initiating event is exposure of the base from dynamic breathing motions of the base pair, or whether the enzyme actively accelerates expulsion of the base by directly interacting with the site. Although it is tempting to infer an initiating recognition event from inspection of these crystal structures, it is fundamentally impossible to deduce a pathway from inspection of a structure alone. For instance, the exo complexes of both hOGG1 and UNG unambiguously reveal an undamaged base bound in a transient binding site with well-developed phosphate backbone interactions and an enzyme residue intercalated into the DNA base stack. These structural observations do not distinguish between a passive mechanism in which the enzyme responds to and traps the base after it has been exposed through base pair breathing motions, or alternatively, the active mechanism in which the enzyme propels the base from the base stack through direct interactions.
A powerful approach to resolve these opposing mechanisms is NMR spectroscopy, where one can ask whether the imino proton exchange rates increase in the presence of the enzyme. This approach has been taken with UNG, where it was intially discovered that UNG did indeed selectively increase the exchange rate of the thymine imino proton in the context of a T:A base pair[34]. The revealing finding was that the exchange rate increase was brought about by increasing the lifetime of the open state by almost 100-fold, and not by increasing the opening rate of the base pair as compared to the free DNA. Thus by definition, UNG uses a passive trapping mechanism to catch T and U bases that have emerged from the duplex due to spontaneous breathing motions. This result was later confirmed for another DNA sequence, and also, for a series of base pair constructs where the partner base had one, two or three hydrogen bond donor acceptor groups (Figure 3a)[19]. The exchangeable state observed in the NMR studies is likely to be the same extrahelical state trapped in the structural studies because removal of the observed phosphate or base interactions by mutagenesis negated the UNG enhancement of the imino proton exchange rate[23].
New Frontiers
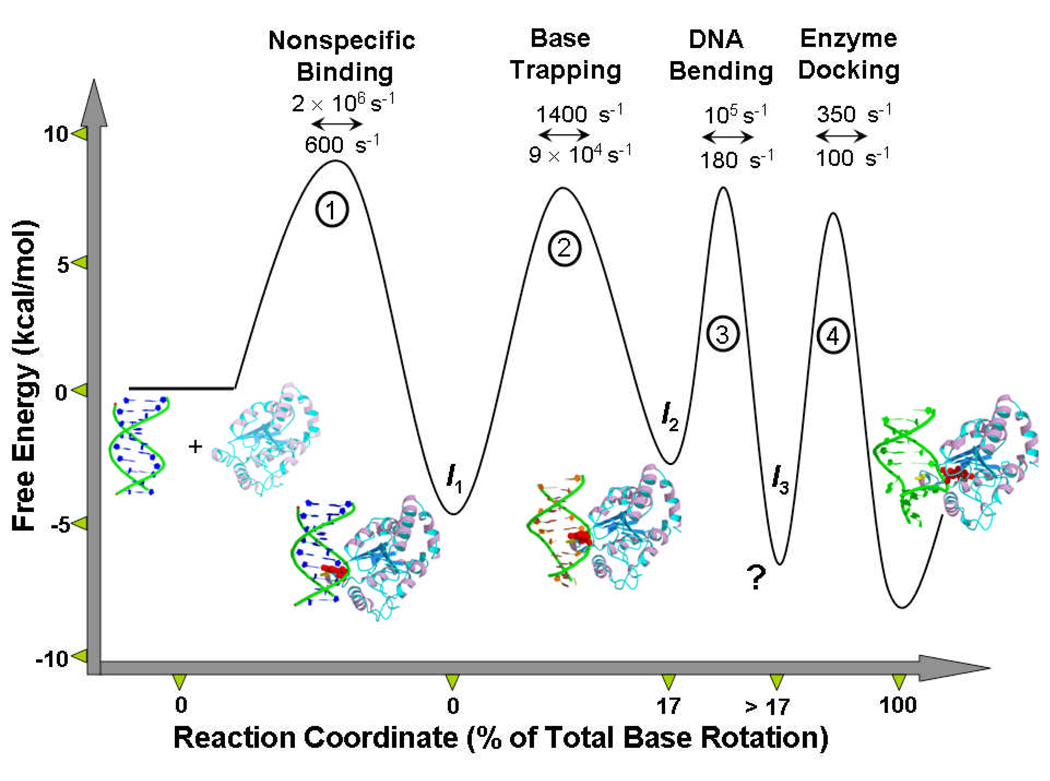
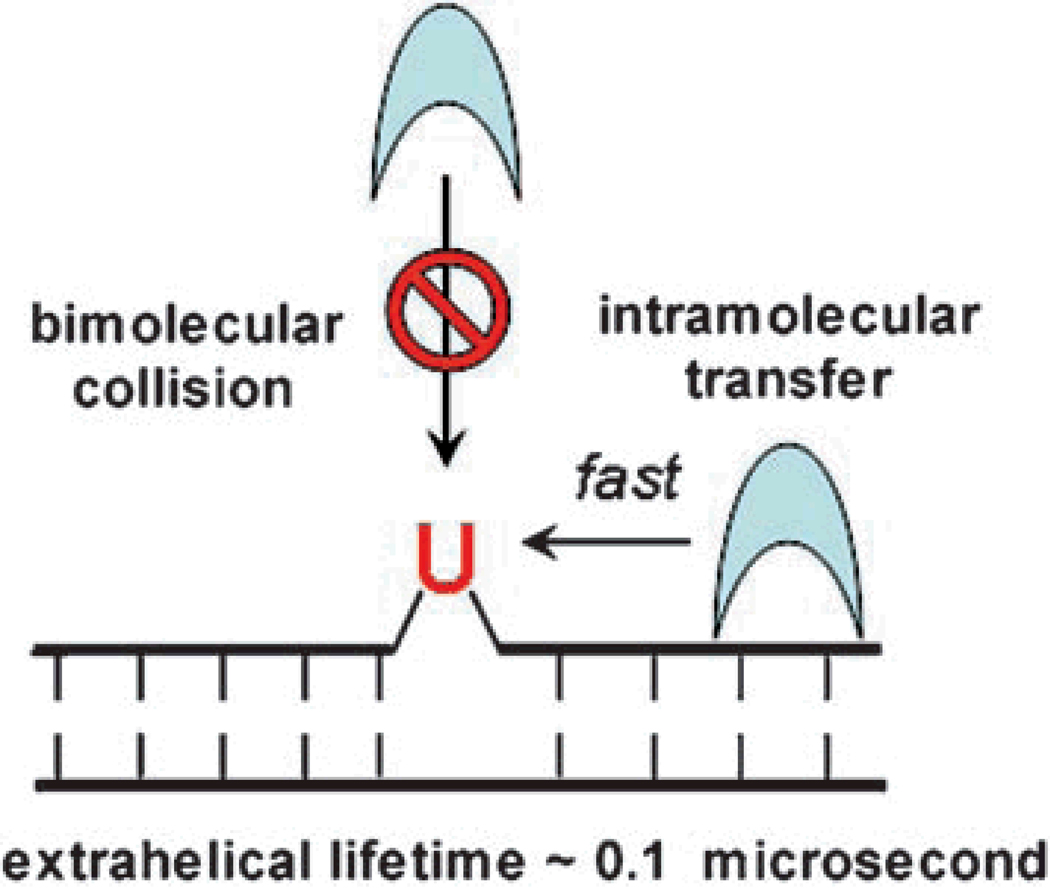
The kinetic, structural and NMR studies of UNG have provided the most complete description of the reaction coordinate for base flipping on an enzyme (Figure 6). Despite this detailed picture of the reaction coordinate, the initial mechanism of recognition is still inscrutable. The enigma arises for two reasons. First, bimolecular encounter with an out state that is present at an equilibrium concentration of only 1/100,000 of the in state, requires a diffusion constant that exceeds physical limits by several orders of magnitude[34]. Second, recognition of a spontaneously flipped base requires a dynamic response from the enzyme that allows trapping of the out state during its ∼1/10 microsecond lifetime. In other words, effective trapping requires dynamic motions of the enzyme exceeding 107 s−1 (1/τout). These considerations force one to embrace a mechanism that solves both aspects of the enigma (Figure 7). Thus UNG must possess rapid dynamic motions that allow it to sample the duplex for extrahelical bases. This essential dynamic flexibility must be combined with an ability to rapidly scan short-lengths of the DNA contour using stochastic one-dimensional sliding to bypass the kinetic limitations of diffusional encounter from bulk solution[35, 36]. Short range sliding back and forth many times over the same DNA segment provides multiple opportunities for capture of a extrahelical uracil base that may be present, and provides an effective scanning mechanism. We anticipate that experimental approaches that can address both the dynamic and diffusional aspects of the initial recognition step will be most valuable in unravelling the remaining mysteries of DNA damaged base recognition.
Figure 6.
The reaction coordinate for uracil flipping by UNG. The microscopic rate constants have been calculated by combining NMR[19,34] and rapid kinetic measurements[24,25]. The profile pertains to 25 °C. The structures are: free human UNG (pdb 1AKZ), intermediate 1 (encounter complex with B DNA, model)[23], intermediate 2 (partially flipped intermediate state, pdb 2OXM), intermediate 3 (detected kinetically, no structural model)[20,25], final flipped state (pdb 1EMH)[33]. Since the rates by neccesity were obtained using different substrates and by extrapolation of the base pair opening rates to 25 °C, the values should only be considered best approximations.
Figure 7.
Possible mechanisms for enzymatic recognition of an extrahelical base with a short extrahelical lifetime. A pathway involving bimolecular collision of the enzyme with the DNA base while it exists in an extrahelical conformation is not kinetically competent[34]. Rapid intramolecular transfer of the enzyme along the DNA bypasses the kinetic problem of diffusion and allows the enzyme to rapidly scan short lengths of the DNA duplex before dissociation.
Acknowledgements
The author’s work was supported by NIH grant RO1-GM56834.
References
- 1.Stivers JT. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:37–65. doi: 10.1016/S0079-6603(04)77002-6. [DOI] [PubMed] [Google Scholar]
- 2.Stivers JT, Jiang YL. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 3.Berti PJ, McCann JA. Chem. Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 4.Cheng X, Roberts RJ. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D'Amare AR. Mol. Cell. 2006;24:535–545. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Jeltsch A. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 7.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 8.Priyakumar UD, MacKerell AD., Jr Chem. Rev. 2006;106:489–505. doi: 10.1021/cr040475z. [DOI] [PubMed] [Google Scholar]
- 9.Huang N, MacKerell AD., Jr Philos. Transact A. Math. Phys. Eng. Sci. 2004;362:1439–1460. doi: 10.1098/rsta.2004.1383. [DOI] [PubMed] [Google Scholar]
- 10.Huang N, Banavali NK, MacKerell AD., Jr Proc. Natl. Acad. Sci. U. S. A. 2003;100:68–73. doi: 10.1073/pnas.0135427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banavali NK, MacKerell AD., Jr J. Mol. Biol. 2002;319:141–160. doi: 10.1016/S0022-2836(02)00194-8. [DOI] [PubMed] [Google Scholar]
- 12.Song K, Hornak V, de los Santos C, Grollman AP, Simmerling C. Biochemistry. 2006;45:10886–10894. doi: 10.1021/bi060380m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Kelso C, Hornak V, de los Santos C, Grollman AP, Simmerling C. J. Am. Chem. Soc. 2005;127:13906–13918. doi: 10.1021/ja052542s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giudice E, Varnai P, Lavery R. Nucleic Acids Res. 2003;31:1434–1443. doi: 10.1093/nar/gkg239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gueron M, Leroy J. Meth. Enz,ymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 16.Gueron M, Kochoyan M, Leroy JL. Nature. 1987;328:89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- 17.Gueron M, Charretier E, Hagerhorst J, Kochoyan M, Leroy L, Moraillon A, Charretier E, Hagerhorst J, Kochoyan M, Leroy JL, Moraillon A. Struct Methods; Applications of imino proton exchange to nucleic acid kinetics and structures. 1990:113–137. Adenine pressGueron, M. [Google Scholar]
- 18.Priyakumar UD, Mackerell AD., Jr J. Am. Chem. Soc. 2006;128:678–679. doi: 10.1021/ja056445a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao C, Jiang YL, Krosky DJ, Stivers JT. J. Am. Chem. Soc. 2006;128:13034–13035. doi: 10.1021/ja062978n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krosky DJ, Song F, Stivers JT. Biochemistry. 2005;44:5949–5959. doi: 10.1021/bi050084u. [DOI] [PubMed] [Google Scholar]
- 21.Krosky DJ, Schwarz FP, Stivers JT. Biochemistry. 2004;43:4188–4195. doi: 10.1021/bi036303y. [DOI] [PubMed] [Google Scholar]
- 22.Moe JG, Russu IM. Biochemistry. 1992;31:8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- 23.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Nature. 2007 doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang YL, Stivers JT. Biochemistry. 2002;41:11236–11247. doi: 10.1021/bi026226r. [DOI] [PubMed] [Google Scholar]
- 25.Stivers JT, Pankiewicz KW, Watanabe KA. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 26.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindahl T. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee A, Yang W, Karplus M, Verdine GL. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 29.Verdine GL, Norman DP. Annu. Rev. Biochem. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- 30.Bruner SD, Norman DP, Verdine GL. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 31.Kuznetsov NA, Koval VV, Nevinsky GA, Douglas KT, Zharkov DO, Fedorova OS. J. Biol. Chem. 2007;282:1029–1038. doi: 10.1074/jbc.M605788200. [DOI] [PubMed] [Google Scholar]
- 32.Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Proc. Natl. Acad. Sci.USA. 2004;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao C, Jiang YL, Stivers JT, Song F. Nat Struct Mol Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Haushalter KA, Lieber CM, Verdine GL. Chem. Biol. 2002;9:345–350. doi: 10.1016/s1074-5521(02)00120-5. [DOI] [PubMed] [Google Scholar]
- 36.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]