Abstract
The 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase isoenzymes 1 and 2 (HSD3B1 and HSD3B2) are membrane-bound enzymes that play essential roles in the biosynthesis of steroid hormones. Therefore, variation in the HSD3B1 and HSD3B2 genes might play a role in the pathophysiology of steroid hormone-related disease. We set out to systematically identify common polymorphisms and haplotypes in human HSD3B1 and HSD3B2. We identified 17 single nucleotide polymorphisms (SNPs) in HSD3B1 and 9 in HSD3B2 – the majority of which were not present in public databases – by resequencing human HSD3B1 and HSD3B2 using 240 DNA samples from four different ethnic groups (60 samples per group). Functional genomic studies of the 5 nonsynonymous cSNPs in HSD3B1 and the one observed in HSD3B2 showed that two of these polymorphisms resulted in significant decreases in the quantity of enzyme protein expressed. However, none of the 3 nonsynonymous SNPs located in areas encoding putative membrane-binding domains altered subcellular localization of the enzyme as determined by immunofluorescence microscopy. Finally, common variant haplotypes in the 5′-flanking regions of these genes showed significant cell line-dependent variation in their ability to drive transcription. In aggregate, these results provide a basis for study of the possible role in human disease of common genetic variation in HSD3B1 and HSD3B2.
Keywords: 3β-hydroxysteroid dehydrogenase 1, HSD3B1, 3β-hydroxysteroid dehydrogenase 2, HSD3B2, genetic polymorphisms, single nucleotide polymorphisms, SNPs, haplotypes, gene resequencing, functional genomics
1. Introduction
The 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase 1 and 2 isoenzymes (HSD3B1 and HSD3B2) are responsible for the oxidation and isomerization of Δ5-3β-hydroxysteroid precursors to form Δ4-ketosteroids [1–3]. HSD3B1 and HSD3B2 are NAD+-dependent membrane-bound enzymes that localize to the endoplasmic reticulum and mitochondrion [4–6]. HSD3B-catalyzed oxidation and isomerization are essential steps in the formation of all classes of active steroid hormones [7]. In humans, HSD3B1 is expressed in the placenta and peripheral tissues, while HSD3B2 is expressed predominantly in the adrenal gland, ovary and testis [8–11]. Deficiency of HSD3B2 is associated with a rare form of congenital adrenal hyperplasia [12]. The two human HSD3B isoforms are encoded by HSD3B1 and HSD3B2, genes that encode proteins consisting of 372 and 371 amino acids, respectively. Both genes map to a 7.8 kb region in chromosome 1p13.1 [13–15].
Because of the essential role of these two enzymes in the formation of steroid hormones, it is possible that common genetic variation in the human HSD3B1 and HSD3B2 genes might influence steroid biosynthesis and, as a result, the pathogenesis of steroid hormone-related diseases such as breast cancer or prostatic hyperplasia [7, 16]. In the present study, we set out to systematically identify common polymorphisms and haplotypes in the genes encoding human HSD3B1 and HSD3B2 and to begin to study the functional implications of that gene sequence variation. Specifically, we resequenced both genes using 60 DNA samples each from four different ethnic groups and identified common polymorphisms and haplotypes in the coding regions, 5′-flanking regions (5′-FRs) and 5′- and 3′-untranslated regions (UTRs), followed by characterization of the functional implications of nonsynonymous cSNPs and common 5′-FR haplotypes in both genes. These studies represent a step toward determining the possible relationship of common DNA sequence variation in these two important genes with the pathophysiology of steroid hormone-dependent disease in humans.
2. Materials and methods
2.1. DNA samples
DNA samples from 60 Caucasian-American (CA), 60 African-American (AA), 60 Han Chinese-American (HCA) and 60 Mexican-American (MA) subjects (sample sets HD100CAU, HD100AA, HD100CHI and HD100MEX) were obtained from the Coriell Cell Repository (Camden, NJ). All of these DNA samples had been anonymized by the National Institute of General Medical Sciences prior to deposit, and all subjects had provided written consent for the use of their DNA for experimental purposes. These DNA samples have been widely used for human gene resequencing studies [17–20]. The present study was reviewed and approved by Mayo Clinic Institutional Review Board.
2.2. HSD3B1 and HSD3B2 gene resequencing
Each of the 240 DNA samples studied was used to perform PCR amplifications of the areas of the two genes to be resequenced. Primer sequences are listed in the Supplemental Material, Table 1. Because there are several HSD3B pseudogenes with high homology to HSD3B1 and HSD3B2, a long PCR was performed for the 5′-FRs and exons 1 and 2 of both HSD3B1 and HSD3B2. Specifically, a long PCR was performed, followed by two nested amplifications to obtain 5′-FR sequence as well as the sequences of exons 1 and 2. The same strategy was used, for the same reason, to amplify exon 4 in both genes. All amplifications were performed with AmpliTaq Gold DNA polymerase (Perkin-Elmer, Foster City, CA), and the 25 µl reaction mixtures contained 0.75 U of DNA polymerase, 0.5 µl of a 10-fold diluted DNA sample (16–19 ng DNA per reaction), 5 to 10 pmol of each primer, 0.08 mM dNTP (Boehringer Mannheim, Indianapolis, IN), 0 or 2% DMSO and 2.5 µl of 10X PCR buffer containing 15 mM MgCl2 (Perkin-Elmer). Amplicons were sequenced on both strands in the Mayo Molecular Biology Core Facility with an ABI 377 DNA sequencer using BigDye (Perkin-Elmer) dye primer sequencing chemistry. To exclude PCR-induced artifacts, independent amplifications were performed for samples in which a SNP was observed only once or for any sample with an ambiguous chromatogram. Chromatograms were analyzed using the University of Washington PolyPhred 3.0 and Consed 8.0 programs [21, 22] as well as with Mutation Surveyor (SoftGenetics, LLC, State College, PA).
2.3. HSD3B1 and HSD3B2 COS-1 cell transient expression
The wild type (WT) HSD3B1 and HSD3B2 cDNA sequences were cloned into the expression vector pcDNA4/HisMax-TOPO® TA (Invitrogen, Carlsbad, CA). These WT constructs were then used as template for site-directed mutagenesis performed using circular PCR to create variant allozyme expression constructs. The HSD3B1 Thr367 variant construct was used as template to create the Phe96, Thr367 and the Leu286, Thr367 double variant constructs. In a similar fashion, the HSD3B1 Val79 variant sequence was used as template to create the Val79, Leu286 double variant construct. This latter construct was then used to create a triple variant Val79, Leu286, Thr367 construct. Sequences of all constructs were confirmed by sequencing inserts in both directions. All DNA constructs were isolated using the Qiagen Plasmid Maxi Kit (Qiagen, Valencia, CA). In order to maximize construct DNA quality and to minimize variation during transient transfection as a result of DNA quality, DNA preparations all had concentrations of over 1.0 mg/ml and A260/A280 ratios of ≥ 1.8. These expression constructs were then transfected into COS-1 cells with the TransFast reagent (Promega, Madison, WI) at a charge ratio of 1:3. Specifically, 10 µg of HSD3B1 and HSD3B2 expression construct DNA was cotransfected with 2 µg of pSV-β-galactosidase DNA (Promega) to make it possible to correct for transfection efficiency. Cells were lysed after 48 hr in culture with 0.1% NP40 lysis buffer, followed by centrifugation at 3000×g for 10 min. Supernatant preparations after centrifugation were used to perform Western blot analyses.
2.4. HSD3B1 and HSD3B2 Western blot analyses
Supernatant lysates of COS-1 cells transfected with WT and variant allozyme expression constructs were used to perform Western blot analyses. After correction, on the basis of β-galactosidase activity for possible variation in transfection efficiency, cell lysates were loaded on 12% SDS gels. After electrophoresis, proteins were transferred to PVDF membranes (BioRad, Hercules, CA), followed by blotting with monoclonal anti-His antibody (Sigma, St Louis, MO). Western blot results were quantified with the AMBIS Radioanalytic Imaging System, Version 4.31 (Ambis, Inc., San Diego, CA), and the data were expressed as a percentage of the intensity of the recombinant WT HSD3B1 or HSD3B2 protein band on the same gel.
2.5. HSD3B1 and HSD3B2 in vitro translation and degradation
Transcription and translation of HSD3B1 and HSD3B2 allozymes were performed with the TnT® coupled rabbit reticulocyte lysate (RRL) System (Promega), as described by Wang et al. [23–25]. Specifically, 1 µg of expression construct DNA, together with 2 µL T7 buffer, 1 µL T7 polymerase, 1 µL of a mixture of amino acids that lacked methionine, 1 µL RNasin (Promega) and 2 µL of 35S-methionine (1000 Ci/mm, 10 mCi/mL, 0.4 µm final concentration) (Amersham Pharmacia Biotech) were added to 25µL RRL that had been “treated” to inhibit protein degradation. With the exception of the RNasin and 35S-methionine, all reagents were included in the Promega kit. The reaction volume was increased to 50 µL with nuclease-free water (Promega), and the mixture was incubated at 30°C for 90 min. A 5 µL aliquot was then used to perform SDS-PAGE, followed by autoradiography.
For the protein degradation experiments, 10 µL of the in vitro-translated 35S-methionine-labeled protein described in the preceding paragraph was added to 50 µL of an adenosine 5'-triphosphate (ATP) generating system and 50 µL of “untreated” RRL. The ATP generating system consisted of 100 µL each of 1 M Tris-HCl (pH 7.8), 160 mM MgCl2, 120 mM KCl, 100 mM dithiothreitol, 100 mM ATP, 200 mM creatine phosphate and 2 mg/mL creatine kinase (all from Sigma), plus 300 µL of nuclease-free water (Promega). This mixture was incubated at 37°C, and aliquots were removed at 0, 4 and 8 hr to perform SDS-PAGE, followed by autoradiography.
2.6. HSD3B1 and HSD3B2 quantitative RT-PCR
mRNA was isolated according to the manufacturer’s instructions with the RNeasy Mini Kit (Qiagen) from COS-1 cells transfected with expression constructs for HSD3B1 WT and Phe96 allozymes. These cells had been cotransfected with β-galactosidase to make it possible to correct for transfection efficiency [18, 20, 26]. RT-PCR was then performed with primers for HSD3B1 and, as an internal standard, for β-galactosidase.
2.7. HSD3B1 and HSD3B2 reporter gene studies
Luciferase reporter gene constructs were created for the most common human HSD3B1 and HSD3B2 5′-FR sequences with frequencies of ≥ 3% in any of the populations studied. Specifically, approximately 1000 bp of HSD3B1 or HSD3B2 5′-FR sequence was amplified from human genomic DNA samples that contained the desired haplotypes. The forward and reverse primers for those amplifications contained ACC65I and XhoI restriction sites, respectively, to make it possible to subclone the amplicons into pGL-3 Basic (Promega), upstream of the firefly luciferase gene open reading frame (ORF). The forward and reverse primers for the amplifications of HSD3B1 5′-FRs were GGTAACG GAACCCAGGGCTCTCCAGGGGCAAAT and CTCGAGGGCCATCCA AAGTAGCAGGGAT. The primers used to amplify the HSD3B2 5′-FRs were GGTACCCAACTACAAATCCCCACACTTGTA and CTCGAGCGTGGCCAATCCAAAGTAG CAGGAA, respectively. Underlined areas in the primers contained the restriction sites. Each of the inserts was sequenced in both directions to ensure that the correct sequence had been amplified. The same precautions were taken to establish and maintain construct quality and to minimize possible variation during construct DNA isolation as were used for the expression constructs. The luciferase reporter gene constructs were designated HSD3B1 pGL3WT, HSD3B1 pGL3(−236T/C), and HSD3B2 pGL3WT, HSD3B2 pGL3(−194G/A) and HSD3B2 pGL3(−145T/G), based on the location of the variant nucleotide. These constructs were used to transiently transfect MCF-7, DU-145 and JEG-3 cell lines. All of these cell lines have been shown to express HSD3B1 or HSD3B2 [7].
Specifically, 5 µg of purified plasmid DNA was transfected into the cell lines together with 50 ng pRL-TK DNA (Promega). The Renilla luciferase activity expressed by pRL-TK was used as a control for transfection efficiency. As an additional control, cells were also transfected with pGL-3 Basic that lacked insert. All transfections were performed using the TransFast reagent (Promega). Cells were lysed after 48 hr in culture, and reporter gene activity was measured with the Promega dual-luciferase assay system. Results were reported as the ratio of firefly luciferase light units to Renilla luciferase light units, and all values were expressed as a percentage of the activity of the WT 5′-FR construct. Assays in all three cell lines were performed in triplicate.
2.8. HSD3B1 and HSD3B2 confocal microscopy
Fluoresceine isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin and tetramethylrhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit immunoglobulin were purchased from Southern Biotech (Birmingham, AL). COS-1 cells were subcultured to 50–70% confluence on coverslips and were transfected with HSD3B expression constructs, followed by culture for an additional 48 hr. The cells were washed with phosphate buffered saline (PBS), fixed at room temperature for 12 min with 3% paraformaldehyde and –finally – were washed and incubated at room temperature for 5 min with buffer containing 0.5% Triton X-100. The coverslips were then incubated with the primary antibodies – rabbit polyclonal anti-human antibody against calnexin, an endoplasmic reticulum marker [26], and mouse monoclonal anti-His antibody – followed by incubation with FITC-conjugated goat anti-mouse or TRITC-conjugated goat antirabbit IgG antibody. The COS-1 cells were then viewed by Zeiss LSM 510 confocal microscopy with 488 or 570 nm filters for green or red fluorochrome excitation, respectively [26].
2.9. Data analysis
DNA sequence obtained during the gene resequencing studies was compared with HSD3B1 and HSD3B2 genomic and cDNA genomic consensus sequences (GenBank Accession numbers NM_000862, NM_000198, BC031999 and BC038419). Average levels of recombinant allozyme immunoreactive protein from the expression studies and luciferase activities obtained during the reporter gene studies were compared by ANOVA performed with the Prism program. Linkage disequilibrium among HSD3B1 and HSD3B2 polymorphisms was determined by calculating D' values for all possible pairwise combinations of polymorphisms [27, 28]. This method for reporting linkage data is independent of allele frequency. Haplotypes were inferred computationally as described by Schaid et al. [29].
3. Results
3.1. Human HSD3B1 and HSD3B2 resequencing
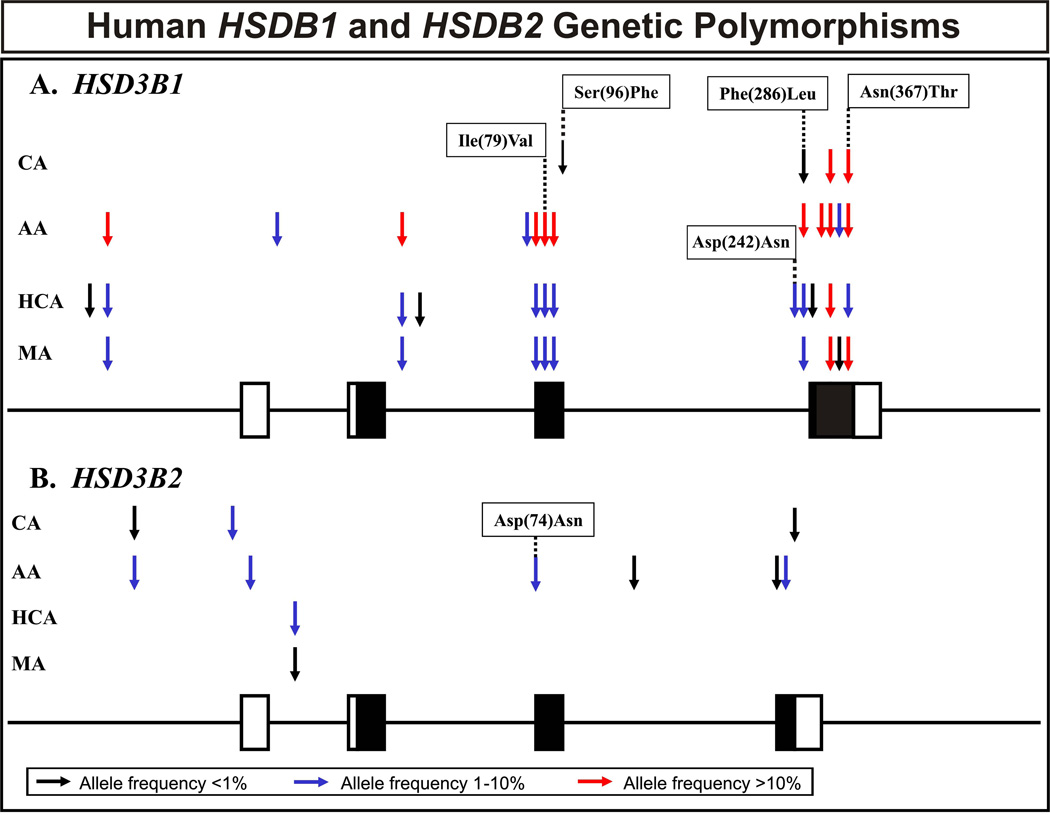
The HSD3B1 and HSD3B2 genes were resequenced using 240 DNA samples obtained from 60 AA , 60 CA, 60 HCA and 60 MA subjects. The areas resequenced included all exons, exon-intron splice junctions and approximately 1–2 kb of 5′-FR for both genes. A schematic representation of SNP locations in the two genes is shown graphically in Fig. 1, and polymorphism locations and frequencies are listed in Table 1. Seventeen SNPs were identified in HSD3B1, including 5 non-synonymous cSNPs (Ile79Val, Ser96Phe, Asp242Asn, Phe286Leu and Asn367Thr) (Table 1). Nine SNPs were identified in HSD3B2, including one nonsynonymous cSNP (Asp74Asn) (Table 1). All polymorphisms in both genes were in Hardy-Weinberg equilibrium (p > 0.05). We also calculated “nucleotide diversity”, a quantitative measure of genetic variation, adjusted for the number of alleles studied. Two standard measures of nucleotide diversity are π, average heterozygosity per site, and θ, a population mutation measure that is theoretically equal to the neutral mutation parameter [30]. Table 2 contains a list of these values for HSD3B1 and HSD3B2. In addition, values for Tajima’s D, a test of the neutral mutation hypothesis [31], were estimated for each population (Table 2). For both genes, the DNA from AA subjects showed greater apparent “diversity” in sequence than that for the other populations, probably reflecting the greater relative antiquity of these sequences [32].
Figure 1.
Human HSD3B1 and HSD3B2 genetic polymorphisms. The figures show a schematic representation of the (A) HSD3B1 and (B) HSD3B2 gene structures, with arrows indicating the locations of polymorphisms. Exons are depicted as rectangles, with black rectangles representing the open reading frame and open rectangles representing untranslated regions. Red arrows represent frequencies greater than 10%; dark blue arrows frequencies from 1 to 10% and black arrows polymorphisms with frequencies of less than 1%. “AA” is African-American; “CA” Caucasian-American; “HCA” Han Chinese-American and “MA” Mexican-American subjects. Alterations in encoded amino acids as a result of nonsynonymous cSNPs are also indicated.
Table 1.
Human HSD3B1 and HSD3B2 genetic polymorphisms. Polymorphism locations, alterations in nucleotide and amino acid sequences and frequencies of the polymorphisms observed are listed for each of the 4 ethnic groups studied. Data for exons are boxed. The table also indicates whether the polymorphism is presently represented in the HapMap or dbSNP. Polymorphisms in exons and UTRs have been numbered with respect to the “A” in the “ATG” translation initiation codon, with positive numbers located 3′- and negative numbers 5′to that position. The numbering scheme for nucleotides located within introns 5′and 3′ of the exons is based on their distance from splice junctions, using negative or positive numbers, respectively.
| Frequency of Variant Allele | Present in | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Location | Nucleotide | Sequence Change | Amino Acid Change | AA | CA | HCA | MA | Databases |
| HSD3B1 | |||||||||
| 1 | 5'FR | (−386) | C→T | 0.000 | 0.000 | 0.008 | 0.000 | no | |
| 2 | 5'FR | (−236) | T→C | 0.308 | 0.000 | 0.042 | 0.033 | yes | |
| 3 | Intron 1 | 48 | C→A | 0.033 | 0.000 | 0.000 | 0.000 | no | |
| 4 | Intron 2 | 56 | G→T | 0.175 | 0.000 | 0.042 | 0.025 | yes | |
| 5 | Intron 2 | (−22) | G→C | 0.000 | 0.000 | 0.008 | 0.000 | no | |
| 6 | Exon 3 | 166 | C→T | 0.026 | 0.000 | 0.000 | 0.000 | no | |
| 7 | Exon 3 | 228 | C→T | 0.319 | 0.000 | 0.042 | 0.025 | yes | |
| 8 | Exon 3 | 235 | A→G | Ile (79) Val | 0.331 | 0.000 | 0.042 | 0.033 | yes |
| 9 | Exon 3 | 270 | T→A | 0.314 | 0.000 | 0.042 | 0.025 | yes | |
| 10 | Exon 3 | 287 | C→T | Ser (96) Phe | 0.000 | 0.009 | 0.000 | 0.000 | no |
| 11 | Exon 4 | 724 | G→A | Asp (242) Asn | 0.000 | 0.000 | 0.017 | 0.000 | no |
| 12 | Exon 4 | 856 | T→C | Phe (286) Leu | 0.358 | 0.008 | 0.033 | 0.033 | yes |
| 13 | Exon 4 | 933 | T→C | 0.000 | 0.000 | 0.008 | 0.000 | no | |
| 14 | Exon 4 | 939 | G→A | 0.142 | 0.000 | 0.000 | 0.000 | no | |
| 15 | Exon 4 | 1012 | C→T | 0.117 | 0.388 | 0.608 | 0.608 | yes | |
| 16 | Exon 4 | 1017 | G→A | 0.017 | 0.000 | 0.000 | 0.008 | no | |
| 17 | Exon 4 | 1100 | A→C | Asn (367) Thr | 0.117 | 0.310 | 0.085 | 0.175 | yes |
| HSD3B2 | |||||||||
| 18 | 5'FR | (−541) | C→T | 0.025 | 0.008 | 0.000 | 0.000 | yes | |
| 19 | 5'UTR | (−194) | G→A | 0.000 | 0.033 | 0.000 | 0.000 | no | |
| 20 | 5'UTR | (−145) | G→A | 0.017 | 0.000 | 0.000 | 0.000 | no | |
| 21 | Intron 1 | 58 | T→G | 0.000 | 0.000 | 0.050 | 0.008 | yes | |
| 22 | Exon 3 | 220 | G→A | Asp (74) Asn | 0.042 | 0.000 | 0.000 | 0.000 | yes |
| 23 | Intron 3 | 31 | G→A | 0.008 | 0.000 | 0.000 | 0.000 | no | |
| 24 | Exon 4 | 549 | G→A | 0.008 | 0.000 | 0.000 | 0.000 | no | |
| 25 | Exon 4 | 777 | G→A | 0.034 | 0.000 | 0.000 | 0.000 | no | |
| 26 | Exon 4 | 1029 | C→T | 0.000 | 0.008 | 0.000 | 0.000 | no | |
Table 2.
Estimates of values for π, θand Tajima’s D for HSD3B1 and HSD3B2 in four ethnic groups. Values are parameter estimate means ± SE. P values refer to Tajima’s D.
| Genetic Diversity and Neutrality Test Values | ||||
|---|---|---|---|---|
| Population | π | θ | Tajima's D | P Values |
| HSD3B1 | ||||
| AA | 12.5 ± 7.13 | 8.44 ± 3.10 | 1.24 | 0.22 |
| CA | 3.60 ± 2.73 | 2.83 ± 1.52 | 0.52 | 0.62 |
| HCA | 4.54 ± 3.23 | 8.41 ± 3.07 | −1.20 | 0.24 |
| MA | 4.24 ± 3.10 | 6.30 ± 2.51 | −0.79 | 0.44 |
| HSD3B2 | ||||
| AA | 1.15 ± 1.45 | 5.19 ± 2.39 | −1.70 | 0.09 |
| CA | 0.46 ± 0.87 | 2.60 ± 1.57 | −1.43 | 0.15 |
| HCA | 0.44 ± 0.86 | 0.86 ± 0.86 | −0.54 | 0.61 |
| MA | 0.08 ± 0.35 | 0.86 ± 0.86 | −1.01 | 0.33 |
Many rare mutations in the HSD3B2 gene have been reported previously, mainly because they were observed in DNA obtained from patients suffering from classical HSD3B2 deficiency [7, 33]. However, none of those mutations was observed during our resequencing studies. When we compared our resequencing data with data in the HapMap [34] and in dbSNP, we found that only 8 of 17 HSD3B1 SNPs, and only 3 of the 9 SNPs that we observed in HSD3B2 were represented in the most recent version of the HapMap or dbSNP (see Table 1). This observation suggested that indepth gene resequencing efforts, such as that described here, are still required in order to obtain haplotype and haplotype tag SNP (htSNP) information for use in association and functional genomic studies.
Structural analysis and mutagenesis studies have indicated that HSD3B1 and HSD3B2 contain two characteristic Y-X-X-X-K sequence motifs that are located between Tyr154 and Lys158 and between Tyr 269 and Lys273 [7, 33, 35]. These motifs are thought to be associated with the catalytic sites of the enzymes. In addition, affinity labeling of purified human HSD3B1 resulted in the identification of two substrate binding sites, Asn176 to Arg186 and Gly251 to Lys274 [36]. None of the HSD3B1 nonsynonymous cSNPs observed during our gene resequencing studies were located in any of these regions. In addition, two putative membrane-binding domains have been identified in these enzymes that are located between residues 72 and 89 in the N-terminus of the proteins and between resides 283 and 310 in the C-terminal region [11, 37]. Two of the nonsynonymous cSNPs in HSD3B1 (Ile79Val and Phe286Leu), and one in HSD3B2 (Asp74Asn), were located in these putative membrane-binding domain regions. Therefore, as described subsequently, we performed confocal microscopy subcellular localization studies with variant allozymes including these polymorphisms to determine whether the observed alterations in amino acid sequence might disrupt membrane binding.
3.2 HSD3B1 and HSD3B2 haplotype and linkage disequilibrium analyses
Haplotype and pairwise linkage disequilibrium analyses were performed with both genes because it has become increasingly clear that the determination of haplotype may be more helpful than the assay of individual SNPs for use in association studies [38]. In all, nine common (frequency greater than 1%) unequivocal and inferred haplotypes for HSD3B1, and 6 unequivocal haplotypes for HSD3B2 were present in the 240 DNA samples that we resequenced (Table 3). We have also listed in Table 3 any haplotype that encoded a variant amino acid sequence, even if that haplotype did not meet our 1% frequency criterion. Among HSD3B1 haplotypes, 7 were observed in the AA population, with 3 unique to that group; 4 were present in CA subjects, with one unique haplotype; 4 haplotypes were present in MA and 5 were present in HCA subjects, with one unique to HCA DNA. All 5 HSD3B2 haplotypes were observed in the AA population, with only 2 in the CA population subjects and only the *1 (WT) haplotype in HCA and MA subjects (Table 3). Haplotype “designations” listed in Table 3 were based on the encoded amino acid sequence of the allozyme, with the WT sequence designated as *1. Subsequent number designations refer to haplotypes containing nonsynonymous cSNPs, beginning at the N- and proceeding to the C-terminus of the protein. In the case of HSD3B1, no haplotypes containing only the Val79 or Phe96 alterations in amino acid sequence were observed. Instead, combinations of Val79 with Leu286 or Phe96 with Thr367 were observed. Therefore, we designated these two double variant sequences as haplotypes *5 and *6, respectively (Table 3). Letter designations were then made in the order of descending haplotype frequencies, based on data for the AA population. For example, *1A was more frequent than *1B in AA subjects, and both were more frequent than *1C.
Table 3.
HSD3B1 and HSD3B2 haplotypes. Nucleotide positions are numbered as described in Table 1. Variant nucleotides as compared to the “reference sequence”, i.e., the most common sequence in AA subjects, are highlighted as white on black. Initial haplotype designations (*1, *2, *3, etc.) were made on the basis of amino acids that vary, with the WT sequence designated *1. Subsequent assignments (letter designations) were made within ethnic groups, based on decreasing frequencies. For HSD3B1, combinations of Ile79Val and Phe286Leu, as well as combinations of Ser96Phe and Asn367Thr were observed. We have designated those combination as *5 and *6 in Table 3. “o” indicates unequivocal haplotypes, while “i” indicates inferred haplotypes.
| Frequencies | Nucleotide Locations | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Designation | AA | CA | HCA | MA | 5'-FR (−236) |
Intron 2 (56) |
Exon 3 (166) |
Exon 3 (228) |
Exon 3 (235) |
Exon 3 (270) |
Exon 3 (287) |
Exon 4 (724) |
Exon 4 (856) |
Exon 4 (939) |
Exon 4 (1012) |
Exon 4 (1100) |
|
| HSD3B1 | |||||||||||||||||
| o | *1A | 0.415 | 0.297 | 0.245 | 0.192 | T | G | C | C | A | T | C | G | T | G | C | A |
| o | *1B | 0.103 | 0.381 | 0.586 | 0.608 | T | G | C | C | A | T | C | G | T | G | T | A |
| o | *4 | 0.014 | T | G | C | C | A | T | C | A | T | G | T | A | |||
| o | *5 | 0.011 | T | G | C | C | A | T | C | G | C | G | C | A | |||
| o | *6 | 0.108 | 0.295 | 0.085 | 0.167 | T | G | C | C | A | T | C | G | T | G | C | C |
| o | *7A | 0.133 | 0.042 | 0.025 | C | T | C | T | G | A | C | G | C | G | C | A | |
| o | *7B | 0.117 | C | G | C | T | G | A | C | G | C | A | C | A | |||
| i | *7C | 0.025 | C | T | T | T | G | A | C | G | C | G | C | A | |||
| i | *8 | 0.008 | T | G | C | C | A | T | T | G | T | G | C | C | |||
| Designation | AA | CA | HCA | MA | 5'-FR (−541) |
5'-UTR (−194) |
5'-UTR (−145) |
Exon 3 (220) |
Exon 4 (777) |
||||||||
| HSD3B2 | |||||||||||||||||
| o | *1A | 0.866 | 0.958 | 1.000 | 1.000 | C | G | G | G | G | |||||||
| o | *1B | 0.034 | C | G | G | G | A | ||||||||||
| o | *1C | 0.025 | T | G | G | G | G | ||||||||||
| o | *1D | 0.017 | C | G | A | G | G | ||||||||||
| o | *1E | 0.025 | C | A | G | G | G | ||||||||||
| o | *2A | 0.042 | C | G | G | A | G | ||||||||||
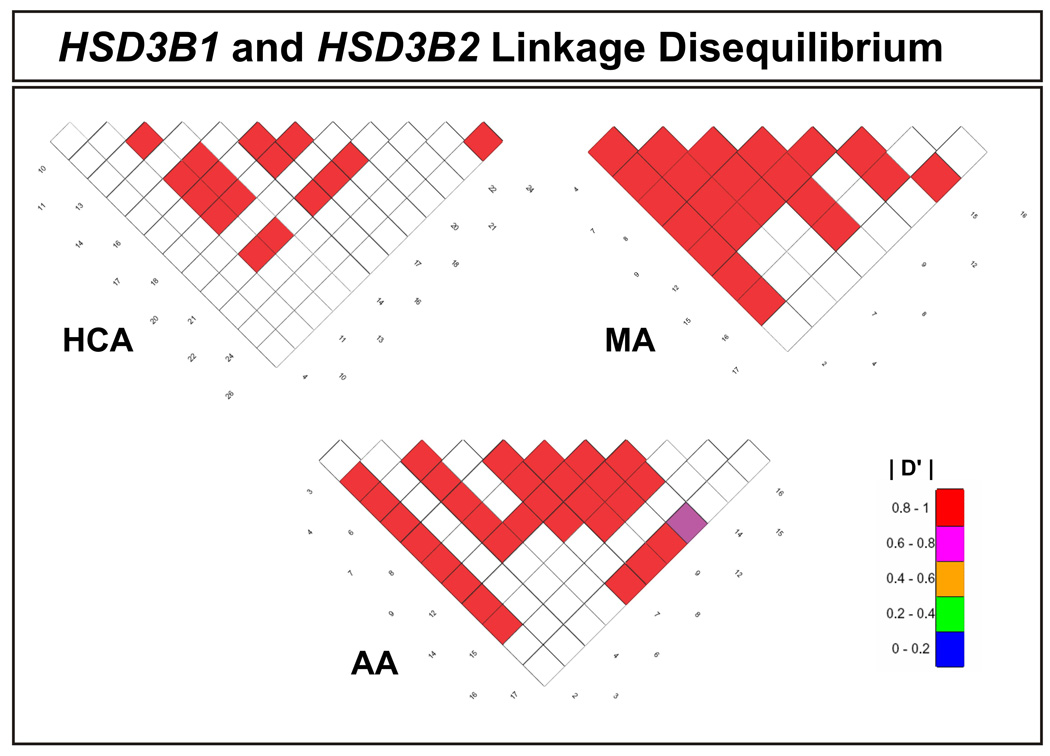
Linkage disequilibrium analysis was also performed for all pairwise combinations of the SNPs observed in these two genes. To make this possible, D' values were calculated [27, 28]. D' values can range from +1.0 when two polymorphisms are maximally associated, to zero when they are randomly associated. The linkage disequilibrium data for the two genes together are displayed graphically in Fig. 2.
Figure 2.
Human HSD3B1 and HSD3B2 linkage disequilibrium. The extent of population-based LD within the area resequenced, shown as pairwise D′ values, is depicted graphically for both genes compared for three ethnic groups. There were inadequate polymorphisms of high enough frequency to make it possible to create a similar figure for CA DNA. The values shown were all significantly correlated, with P < 0.05. The numbers along diagonals represent the polymorphisms listed in Table 1.
3.3. HSD3B1 and HSD3B2 quantitative Western blot analyses
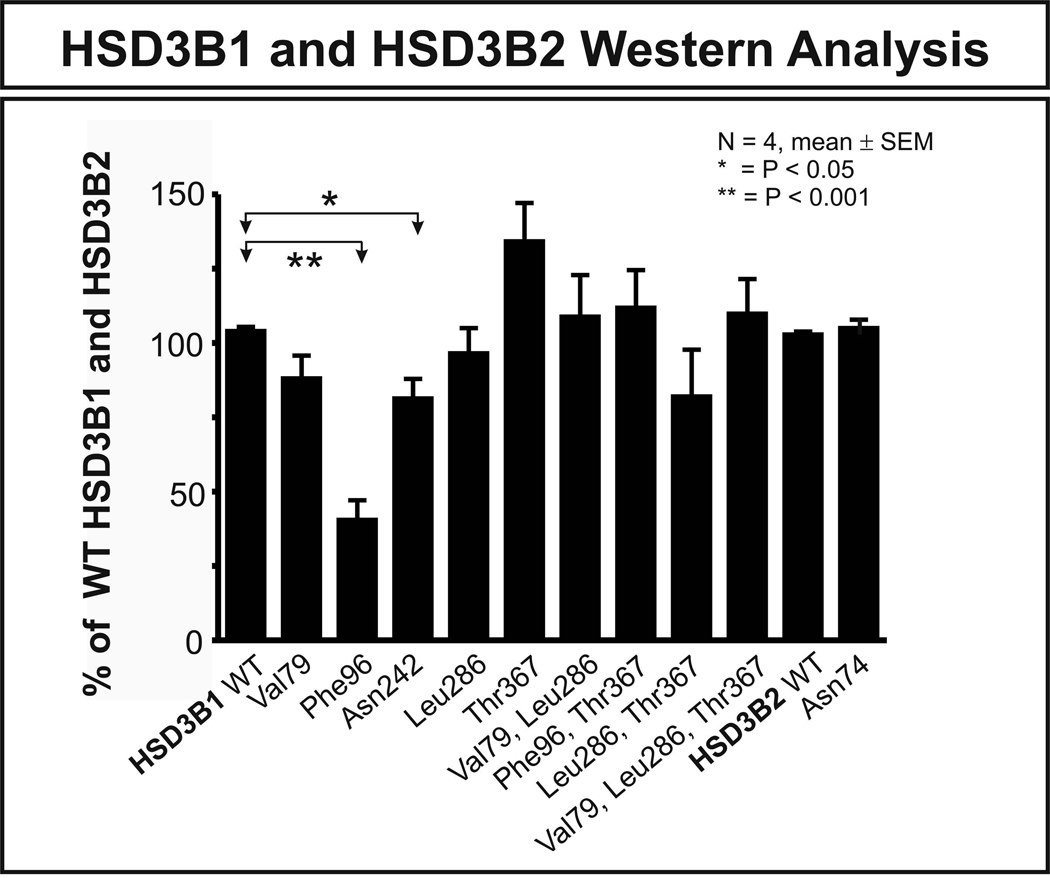
To study the functional effects of common nonsynonymous cSNPs, expression constructs were created for WT HSD3B1 and HSD3B2, as well as constructs containing variant nonsynonymous cSNPs, including those encoding HSD3B1 Val79, Phe96, Asn242, Leu286 and Thr367 as well as HSD3B2 Asn74. We also created several double and triple variant HSD3B1 constructs on the basis of the results of the haplotype analysis. They included (Val79, Leu286), (Phe96, Thr367), (Leu286, Thr367) and (Val79, Leu286, Thr367) constructs. All of these constructs were transiently expressed in COS-1 cells and, after correction for the β-galactosidase activity, quantitative Western blot analysis was performed. COS-1 cells were used to perform these experiments rather than bacterial cells to assure the presence of mammalian mechanisms for post-translational modification and protein degradation. Results of the Western blot studies are shown in Fig. 3. HSD3B1 Phe96 showed a greater than 50% decrease in protein level when compared with WT (p < 0.001), and the Asn242 variant allozyme showed a decrease of approximately 30% (p < 0.05) (Fig. 3). Several mechanisms might be responsible for these decreases in level of enzyme protein. However, the most common mechanism by which nonsynonymous cSNP affect protein quantity is accelerated degradation [23, 24, 39]. Therefore, in vitro degradation studies were performed with the HSD3B1 Phe96 variant allozyme that displayed a greater than 50% decrease in protein quantity.
Figure 3.
HSD3B1 and HSD3B2 quantitative Western blot analysis. Each bar represents the average of 4 independent transfections (mean ± SEM). The results are shown as a percentage of HSD3B1 and HSD3B2 WT allozyme on the same gel. The HSD3B1 Phe96 allozyme showed a greater than 50% decrease in protein level when compared with the WT (p < 0.001), and the HSD3B1 Asn242 variant allozyme showed a decrease of approximately 30% (p < 0.05).
3.4. HSD3B1 in vitro degradation study
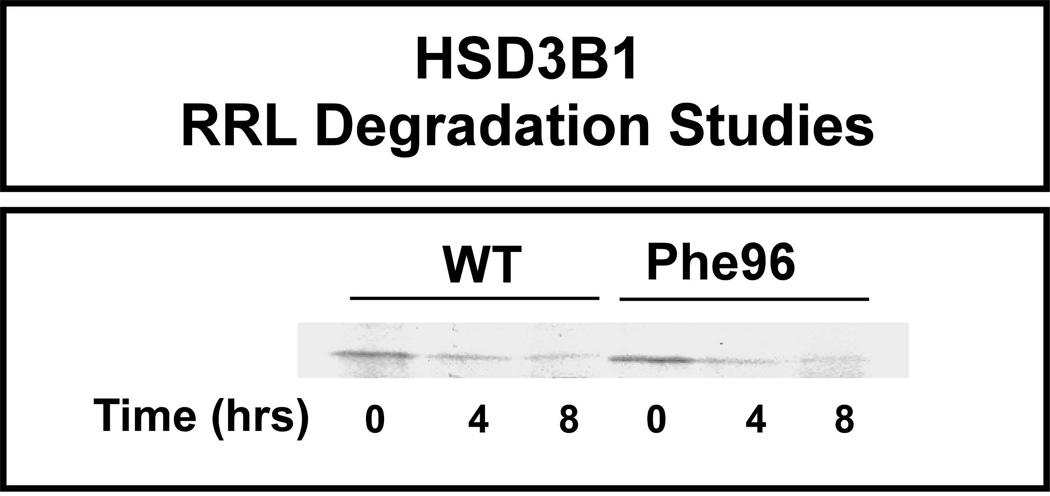
In vitro-translated 35S-methionine-labeled HSD3B1 WT and Phe96 were mixed with an ATP generating system and “untreated” RRL as described in the Materials and Methods. Degradation assays were then performed at 37°C for 0, 4 and 8 hr, followed by SDS-PAGE (Fig. 4). However, there was no apparent difference between WT and the Phe96 variant HSD3B1 allozymes with regard to rate of degradation (Fig. 4). Since we had not observed a striking difference in rate of degradation, we next compared mRNA levels for these allozymes to test the hypothesis that the differences in protein level in the cells might be related to differences in mRNA levels.
Figure 4.
Human HSD3B1 rabbit reticulocyte lysate (RRL) degradation. Recombinant human HSD3B1 allozymes were translated using the TnT® RRL system. The figure shows representative autoradiographs for 35S-methionine radioactively labeled WT and Phe96 allozymes at different time points in the degradation experiment.
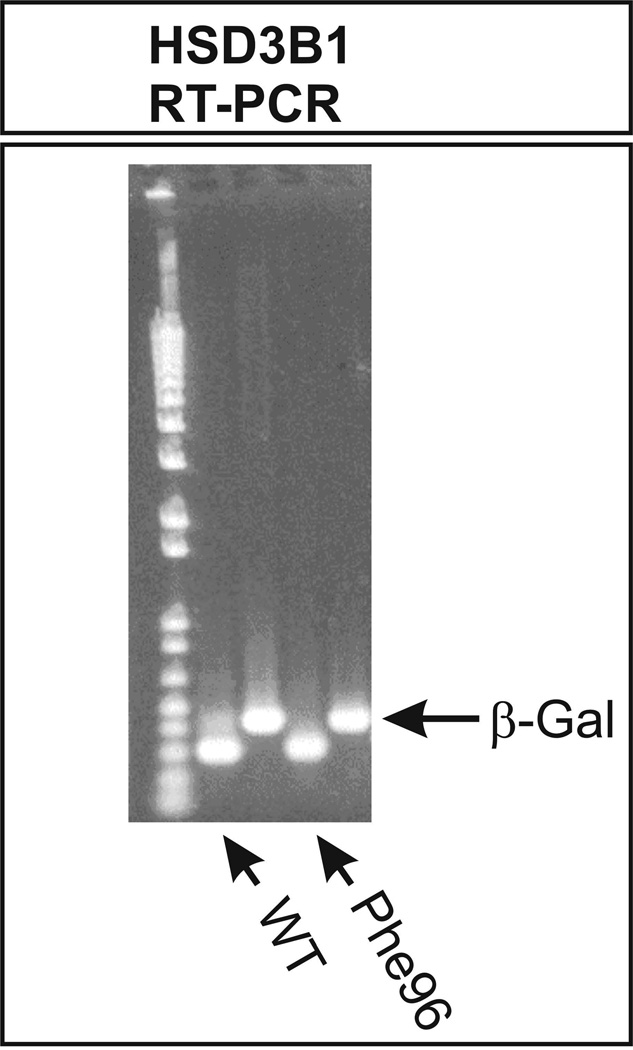
3.5. HSD3B1 RT-PCR
To compare mRNA levels after the transient expression of WT and the HSD3B1 Phe96 variant allozyme, RT-PCR was performed with 30 thermal cycles as well as 15 cycles with mRNA isolated from COS-1 cells transfected with constructs encoding these allozymes. β-Galactosidase assays were used to correct for transfection efficiency, with the β-galactosidase mRNA also serving as an internal control for the RT-PCR assays. Fig. 5 shows the results obtained with 30 cycles. Similar results were obtained at 15 cycles. After correction for the internal control, levels of mRNA for these two allozymes did not appear to differ. Therefore, we had no obvious explanation for the striking decrease in expression of the HSD3B1 Phe96 variant allozyme after transient expression in COS-1 cells.
Figure 5.
Human HSD3B1 RT-PCR of WT and Phe96 mRNA after the transfection of COS-1 cells. The cells were cotransfected with β-galactosidase, both as a control for transfection efficiency and for the RT-PCR. The figure shows results obtained after 30 amplification cycles, but similar results were also observed after 15 cycles (data not shown).
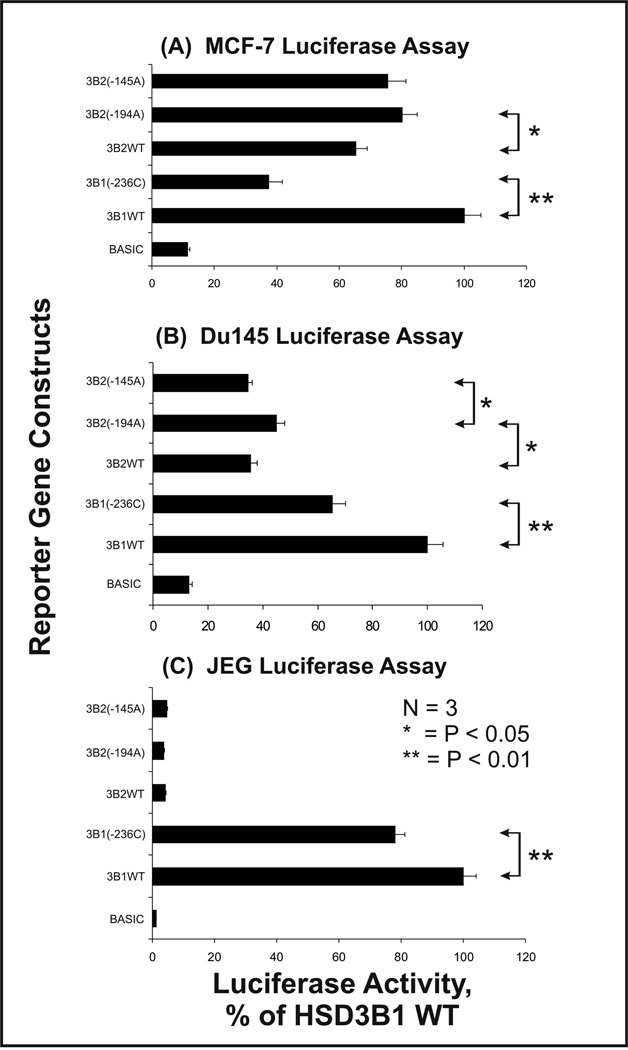
3.6. HSD3B1 and HSD3B2 5’-FR reporter gene studies
Our resequencing studies have resulted in the identification of 5 sequences in the HSD3B1 and HSD3B2 5′-FR with frequencies greater than 3%. Reporter gene constructs were created for these 5′-FR sequences – each of which included only a single SNP. Each construct included the initial 1 kb upstream of the translation initiation codon, and these constructs were used to transfect three cell lines, a breast cancer cell line, MCF-7; a prostate cancer cell line, DU-145; and a placental cell line, JEG-3. HSD3B1 is the major isoform expressed in placenta and peripheral tissues [7], whereas HSD3B2 is predominantly expressed in the adrenal gland, ovary and testis [7]. Luciferase activity measured after transfection of these three cell lines with the reporter gene constructs showed very similar patterns for HSD3B1, in all cell lines, except that overall activity was much higher in JEG-3 than in the other two cell lines, consistent with the fact that HSD3B1 is the major isoform expressed in the placenta (Fig. 6). The change from T to C at nucleotide (−236) in the HSD3B1 5′-FR was associated with a significant decrease in luciferase activity in all three cell lines (p < 0.01 in all cases) (Fig. 6). As anticipated, luciferase activity for the HSD3B2 5′-FR sequences was very low in JEG-3 cells when compared with that for the HSD3B1 constructs (Fig. 6). Patterns of luciferase activity for HSD3B2 5′-FRs were similar in MCF-7 and DU145 cell lines, but the nucleotide change at position (−194) in HSD3B2 significantly increased luciferase activity in both cell lines (p < 0.05). There was also a statistically significant difference between the HSD3B2 (−194) variant and the (−145) variant sequence in DU145, but not in MCF-7 cells, although the directional trend was similar in both cell lines (Fig. 6). Transcription factors such as STAT5, STAT6, AP-1, and SF-1 have been reported to participate in the regulation of human HSD3B1 and HSD3B2 expression [7, 40–42]. The T→C alteration at position (−236) of the HSD3B1 5′-FR created a new AP-2 binding site on the basis of the results of a TFSEARCH search. That alteration might account for the decrease in luciferase activity for the (−236C) construct that is shown in Fig. 6.
Figure 6.
Human HSD3B1 and HSD3B2 luciferase reporter gene studies. Activities of luciferase reporter gene constructs containing different 5′-FR SNPs in the two genes are reported as a percentage of the HSD3B1 WT construct activity. Each bar represents the mean ± SEM of 3 experiments.
3.7. HSD3B1 and HSD3B2 confocal microscopy
Since the HSD3B1 Val79 and Leu286 and the HSD3B2 Asn74 alterations in amino acid sequence were located within putative membrane-binding domains [11, 37], confocal microscopy was performed to determine the possible effects of these polymorphisms on the subcellular localization of these isoforms. Specifically, with calnexin as an endoplasmic reticulum marker, immunofluorescent studies were performed using COS-1 cells transiently transfected with constructs encoding HSD3B1 WT, Val79, Leu286 and a combination of the codon 79 and 286 variants, as well as HSD3B2 WT and Asn74 constructs. All of these allozymes, including those with variant sequence, colocalized with calnexin (Fig. 7), indicating that the polymorphisms did not appear to disrupt the subcellular localization of the variant allozymes.
Figure 7.
Subcellular localization of recombinant HSD3B1 and HSD3B2 allozymes. The red strain indicates the endoplasmic reticular marker calnexin. Green represents the location of HSD3B1 or HSD3B2. The “merge” images show the colocalization of calnexin with HSD3B isoforms.
4. Discussion
Steroid hormones play a crucial role in differentiation, development, growth and the physiologic function of most human tissues [43]. Major pathways for steroid hormone biosynthesis have been well characterized, and one important activity within those pathways is catalyzed the NAD+-dependent membrane-bound HSD3B enzymes. The two HSD3B isoforms expressed in human tissues are encoded by the HSD3B1 and HSD3B2 genes. The enzymes encoded by these genes catalyze sequential 3β-hydroxysteroid dehydrogenation and the Δ5 to Δ4-isomerization of the Δ5-steroid precursors pregnenolone, 17α-hydroxypregnenolone, dehydroepiandrosterone and androst-5-ene-3β,17β-diol to form their respective Δ4-ketosteroids, progesterone, 17α-hydroxyprogesterone, Δ4-androstenedione and testosterone, respectively [1–3]. Therefore, these bifunctional dimeric enzymes are required for the biosynthesis of all major classes of steroid hormones, including glucocorticoids, mineralocorticoids, progesterones, androgens and estrogens. In addition, enzymes of the HSD3B family also catalyze the formation and/or biotransformation of 5α-androstanes and 5α-pregnanes such as dihydrotestosterone and dihydroprogesterone [1–3]. Therefore, the two HSD3B isoenzymes control critical steroidogenic reactions in the adrenal cortex, gonads, placenta and a variety of peripheral tissues [44]. Mutations in the HSD3B2 gene result in classical HSD3B deficiency, a rare form of congenital adrenal hyperplasia that accounts for approximately 1% of all cases of that disease [45, 46]. Inherited variation in these enzymes has also been reported to represent a risk factor for other steroid hormone-related diseases [7, 16]. However, there have been no systematic studies of common genetic variation in these genes. Because of the medical and biological importance of the reactions catalyzed by HSD3B1 and HSD3B2, we set out to identify common genetic polymorphisms in HSD3B1 and HSD3B2 and to begin the process of determining whether that gene sequence variation might influence function.
We began by resequencing HSD3B1 and HSD3B2 using 240 DNA samples from four ethnic groups. That effort resulted in the identification of 17 SNPs in HSD3B1 and 9 in HSD3B2 (Fig. 1, Table 1), the majority of which were not present in the HapMap or dbSNP. Linkage disequilibrium and haplotype analyses were then performed for both genes. There were significant differences among the four ethnic groups studied with regard to polymorphism and haplotype frequencies, raising the possibility that the genetic variation that we observed might have ethnic-specific effects on function. We then performed functional genomic studies with the nonsynonymous cSNPs observed, as well as luciferase reporter gene assays with common sequence variation within the 5′-FRs of both genes. None of the nonsynonymous cSNPs were located within substrate binding or catalytic sites for these enzymes. When we performed Western blot analyses for allozymes expressing the nonsynonymous cSNPs, we found that the Phe96 variant allozyme for HSD3B1 displayed a greater than 50% decrease in protein level (Fig. 3). The most common mechanism by which nonsynonymous cSNPs affect function is an alteration in protein level [39], and decreased protein quantity under these circumstances is most often due to accelerated degradation of the variant allozyme [25, 39]. Therefore, we tested the possibility that this HSD3B1 Phe96 variant allozyme might be degraded more rapidly than the WT. However, we failed to observe a striking alteration in RRL degradation for the variant allozyme (Fig. 4), nor did we observe striking differences in mRNA levels after the transient expression of this allozyme in COS-1 cells (Fig. 5). Therefore, the underlying mechanism responsible for this observation will have to be pursued in the course of future studies.
Our resequencing experiments also resulted in the identification of SNPs in the HSD3B1 and HSD3B2 5′-FRs. Luciferase reporter gene assays for constructs containing these 5′-FR SNPs were performed in 3 different cell lines. Patterns of reporter gene expression were similar in these cell lines, with the HSD3B1 (−236C) polymorphism showing a significant decrease in luciferase activity when compared with the WT sequence in all cell lines studied and the HSD3B2 (−194) SNP showing a significant increase in two cell lines (Fig. 6). Detailed studies of molecular mechanisms responsible for these observations can now be pursued in the course of future experiments.
In summary, we have resequenced and determined common sequence variation in the human HSD3B1 and HSD3B2 genes in 4 ethnic groups. That effort resulted in the identification and functional characterization of a series of common polymorphisms and haplotypes in these two important steroid hormone biosynthetic genes. It will now be possible to use this information to determine whether common DNA sequence variation in these genes might provide insight into the role of these enzymes in hormone-dependent disease or in variation in response to the treatment of that disease.
Supplementary Material
Acknowledgements
We thank Mrs. Luanne Wussow for her assistance with the preparation of this manuscript.
This work was supported in part by National Institutes of Health (NIH) grants R01 GM28157, R01 GM35720 and U01 GM61388 (The Pharmacogenetics Research Network), as well as a PhRMA Foundation Center of Excellence in Clinical Pharmacology Award.
Appendix A. Supplemental data
Supplementary Material Table 1 data associated with this article can be found in the online version at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The gene sequence data reported here have been deposited in the NIH database PharmGKB, with accession numbers PS204303, PS204998, PS205002, PS205003 and PS20517 for HSD3B1 and with accession numbers PS204304, PS205035, PS205036, PS205037 and PS205038 for HSD3B2.
References
- 1.Simard J, Durocher F, Mebarki F, Turgeon C, Sanchez R, Labrie Y, Couet J, Trudel C, Rheaume E, Morel Y, Luu-The V, Labrie F. Molecular biology and genetics of the 3 beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. J. Endocrinol. 1996;(150 Suppl):S189–S207. [PubMed] [Google Scholar]
- 2.Payne AH, Abbaszade IG, Clarke TR, Bain PA, Park CH. The multiple murine 3 beta-hydroxysteroid dehydrogenase isoforms: structure, function, and tissue-and developmentally specific expression. Steroids. 1997;62:169–175. doi: 10.1016/s0039-128x(96)00177-8. [DOI] [PubMed] [Google Scholar]
- 3.Mason JI, Keeney DS, Bird IM, Rainey WE, Morohashi K, Leers-Sucheta S, Melner MH, Simard J, Durocher F, Mebarki F, Turgeon C, Sanchez R, Labrie Y, Couet J, Trudel C, Rheaume E, Morel Y, Luu-The V, Labrie F, Payne AH, Abbaszade IG, Clarke TR, Bain PA, Park CH. The regulation of 3 beta-hydroxysteroid dehydrogenase expression. Steroids. 1997;62:164–168. doi: 10.1016/s0039-128x(96)00176-6. [DOI] [PubMed] [Google Scholar]
- 4.Cherradi N, Defaye G, Chambaz EM. Dual subcellular localization of the 3 beta-hydroxysteroid dehydrogenase isomerase: characterization of the mitochondrial enzyme in the bovine adrenal cortex. J. Steroid Biochem. Mol. Biol. 1993;46:773–779. doi: 10.1016/0960-0760(93)90318-q. [DOI] [PubMed] [Google Scholar]
- 5.Cherradi N, Defaye G, Chambaz EM. Characterization of the 3 beta-hydroxysteroid dehydrogenase activity associated with bovine adrenocortical mitochondria. Endocrinology. 1994;134:1358–1364. doi: 10.1210/endo.134.3.8119176. [DOI] [PubMed] [Google Scholar]
- 6.Sauer LA, Chapman JC, Dauchy RT. Topology of 3 beta-hydroxy-5-ene-steroid dehydrogenase/delta 5-delta 4-isomerase in adrenal cortex mitochondria and microsomes. Endocrinology. 1994;134:751–759. doi: 10.1210/endo.134.2.8299570. [DOI] [PubMed] [Google Scholar]
- 7.Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr. Rev. 2005;26:525–582. doi: 10.1210/er.2002-0050. [DOI] [PubMed] [Google Scholar]
- 8.Lachance Y, Luu-The V, Labrie C, Simard J, Dumont M, de Launoit Y, Guerin S, Leblanc G, Labrie F. Characterization of human 3 beta-hydroxysteroid dehydrogenase/delta 5-delta 4-isomerase gene and its expression in mammalian cells. J. Biol. Chem. 1992;267:3551. [PubMed] [Google Scholar]
- 9.Gingras S, Moriggl R, Groner B, Simard J. Induction of 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase type 1 gene transcription in human breast cancer cell lines and in normal mammary epithelial cells by interleukin-4 and interleukin-13. Mol. Endocrinol. 1999;13:66–81. doi: 10.1210/mend.13.1.0221. [DOI] [PubMed] [Google Scholar]
- 10.Gingras S, Simard J. Induction of 3beta-hydroxysteroid dehydrogenase/isomerase type 1 expression by interleukin-4 in human normal prostate epithelial cells, immortalized keratinocytes, colon, and cervix cancer cell lines. Endocrinology. 1999;140:4573–4584. doi: 10.1210/endo.140.10.7038. [DOI] [PubMed] [Google Scholar]
- 11.Thomas JL, Mason JI, Blanco G, Veisaga ML. The engineered, cytosolic form of human type I 3beta-hydroxysteroid dehydrogenase/isomerase: purification, characterization and crystallization. J. Mol. Endocrinol. 2001;27:77–83. doi: 10.1677/jme.0.0270077. [DOI] [PubMed] [Google Scholar]
- 12.Simard J, Moisan AM, Morel Y. Congenital adrenal hyperplasia due to 3beta-hydroxysteroid dehydrogenase/Delta(5)-Delta(4) isomerase deficiency. Semin. Reprod. Med. 2002;20:255–276. doi: 10.1055/s-2002-35373. [DOI] [PubMed] [Google Scholar]
- 13.Lorence MC, Corbin CJ, Kamimura N, Mahendroo MS, Mason JI. Structural analysis of the gene encoding human 3 beta-hydroxysteroid dehydrogenase/delta 5-4-isomerase. Mol. Endocrinol. 1990;4:1850–1855. doi: 10.1210/mend-4-12-1850. [DOI] [PubMed] [Google Scholar]
- 14.Lachance Y, Luu-The V, Labrie C, Simard J, Dumont M, de Launoit Y, Guerin S, Leblanc G, Labrie F. Characterization of human 3 beta-hydroxysteroid dehydrogenase/delta 5-delta 4-isomerase gene and its expression in mammalian cells. J. Biol. Chem. 1990;265:20469–20475. [PubMed] [Google Scholar]
- 15.Lachance Y, Luu-The V, Verreault H, Dumont M, Rheaume E, Leblanc G, Labrie F. Structure of the human type II 3 beta-hydroxysteroid dehydrogenase/delta 5-delta 4 isomerase (3 beta-HSD) gene: adrenal and gonadal specificity. DNA Cell. Biol. 1991;10:701–711. doi: 10.1089/dna.1991.10.701. [DOI] [PubMed] [Google Scholar]
- 16.Roberts RO, Bergstralh EJ, Farmer SA, Jacobson DJ, Hebbring SJ, Cunningham JM, Thibodeau SN, Lieber MM, Jacobsen SJ. Polymorphisms in genes involved in sex hormone metabolism may increase risk of benign prostatic hyperplasia. Prostate. 2006;66:392–404. doi: 10.1002/pros.20362. [DOI] [PubMed] [Google Scholar]
- 17.Ji Y, Salavaggione OE, Wang L, Adjei AA, Eckloff B, Wieben ED, Weinshilboum RM. Human phenylethanolamine N-methyltransferase pharmacogenomics: gene resequencing and functional genomics. J. Neurochem. 2005;95:1766–1776. doi: 10.1111/j.1471-4159.2005.03453.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin YN, Salavaggione OE, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. Human methylenetetrahydrofolate reductase pharmacogenomics: gene resequencing and functional genomics. Pharmacogenet. Genomics. 2006;16:265–277. doi: 10.1097/01.fpc.0000194423.20393.08. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee B, Salavaggione OE, Pelleymounter LL, Moon I, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Glutathione S-transferase omega 1 and omega 2 pharmacogenomics. Drug Metab. Dispos. 2006;34:1237–1246. doi: 10.1124/dmd.106.009613. [DOI] [PubMed] [Google Scholar]
- 20.Wood TC, Salavaggione OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. J. Biol. Chem. 2006;281:7364–7373. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- 21.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 22.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics. 2003;13:555–564. doi: 10.1097/01.fpc.0000054124.14659.99. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM. Human thiopurine S-methyltransferase (TPMT) pharmacogenetics: variant misfolding and aggresome formation. Proc. Natl. Acad. Sci. USA. 2005;102:9394–9399. doi: 10.1073/pnas.0502352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Weinshilboum RM. Thiopurine S-methyltransferase (TPMT) pharmacogenetics: insights, challenges and future directions. Oncogene Rev. 2006;25:1629–1938. doi: 10.1038/sj.onc.1209372. [DOI] [PubMed] [Google Scholar]
- 26.Ma CX, Adjei AA, Salavaggione OE, Coronel J, Pelleymounter L, Wang L, Eckloff BW, Schaid D, Wieben ED, Adjei AA, Weinshilboum RM. Human aromatase: gene resequencing and functional genomics. Cancer Res. 2005;65:11071–11082. doi: 10.1158/0008-5472.CAN-05-1218. [DOI] [PubMed] [Google Scholar]
- 27.Hartl DL, Clark AG. Organization of genetic variation, Principles of Population Genetics. Chapter 3. Sunderland, MA: Sinauer Associates, Inc.; 2000. pp. 95–107. [Google Scholar]
- 28.Hendrick PW. Genetics of Populations. Sudbury, MA: Jones and Bartlett Publ.; 2000. pp. 396–405. [Google Scholar]
- 29.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengard JH, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaessmann H, Heissig F, von Haeseler A, Paabo S. DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat. Genet. 1999;22:78–81. doi: 10.1038/8785. [DOI] [PubMed] [Google Scholar]
- 33.Morel Y, Mebarki F, Rheaume E, Sanchez R, Forest MG, Simard J. Structure-function relationships of 3 beta-hydroxysteroid dehydrogenase: contribution made by the molecular genetics of 3 beta-hydroxysteroid dehydrogenase deficiency. Steroids. 1997;62:176–184. doi: 10.1016/s0039-128x(96)00178-x. [DOI] [PubMed] [Google Scholar]
- 34.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas JL, Mason JI, Brandt S, Spencer BR, Norris W., Jr. Structure/function relationships responsible for the kinetic differences between human type 1 and type 2 3beta-hydroxysteroid dehydrogenase and for the catalysis of the type 1 activity. J. Biol. Chem. 2002;277:42795–42801. doi: 10.1074/jbc.M208537200. [DOI] [PubMed] [Google Scholar]
- 36.Thomas JL, Nash WE, Myers RP, Crankshaw MW, Strickler RC. Affinity radiolabeling identifies peptides and amino acids associated with substrate binding in human placental 3 beta-hydroxy-delta(5)-steroid dehydrogenase. J. Biol. Chem. 1993;268:18507–18512. [PubMed] [Google Scholar]
- 37.Thomas JL, Evans BW, Blanco G, Mason JI, Strickler RC. Creation of a fully active, cytosolic form of human type I 3beta-hydroxysteroid dehydrogenase/isomerase by the deletion of a membrane-spanning domain. J. Mol. Endocrinol. 1999;23:231–239. doi: 10.1677/jme.0.0230231. [DOI] [PubMed] [Google Scholar]
- 38.Brodde OE, Leineweber K. Beta2-adrenoceptor gene polymorphisms. Pharmacogenet. Genomics. 2005;15:267–275. doi: 10.1097/01213011-200505000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Weinshilboum R, Wang L. Pharmacogenetics: inherited variation in amino acid sequence and altered protein quantity. Clin. Pharmacol. Ther. 2004;75:253–258. doi: 10.1016/j.clpt.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Kazansky AV, Raught B, Lindsey SM, Wang YF, Rosen JM. Regulation of mammary gland factor/Stat5a during mammary gland development. Mol. Endocrinol. 1995;9:1598–1609. doi: 10.1210/mend.9.11.8584036. [DOI] [PubMed] [Google Scholar]
- 41.Leers-Sucheta S, Morohashi K, Mason JI, Melner MH. Synergistic activation of the human type II 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase promoter by the transcription factor steroidogenic factor-1/adrenal 4-binding protein and phorbol ester. J Biol Chem. 1997;272:7960–7967. doi: 10.1074/jbc.272.12.7960. [DOI] [PubMed] [Google Scholar]
- 42.Feltus FA, Groner B, Melner MH. Stat5-mediated regulation of the human type II 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene: activation by prolactin. Mol. Endocrinol. 1999;13:1084–1093. doi: 10.1210/mend.13.7.0314. [DOI] [PubMed] [Google Scholar]
- 43.Payne AH, Hales DB. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 2004;25:947–970. doi: 10.1210/er.2003-0030. [DOI] [PubMed] [Google Scholar]
- 44.Mason JI, Naville D, Evans BW, Thomas JL. Functional activity of 3beta-hydroxysteroid dehydrogenase/isomerase. Endocr. Res. 1998;24:549–557. doi: 10.3109/07435809809032644. [DOI] [PubMed] [Google Scholar]
- 45.Bois E, Mornet E, Chompret A, Feingold J, Hochez J, Goulet V. [Congenital adrenal hyperplasia (21-OH) in France. Population genetics] Arch. Fr. Pediatr. 1985;42:175–179. [PubMed] [Google Scholar]
- 46.Thilen A, Larsson A. Congenital adrenal hyperplasia in Sweden 1969–1986. Prevalence, symptoms and age at diagnosis. Acta Paediatr. Scand. 1990;79:168–175. doi: 10.1111/j.1651-2227.1990.tb11434.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.