Abstract
Wnt signaling plays important roles in various physiological and pathophysiological processes. The pathway that leads to β-catenin stabilization is initiated by Wnt binding to its cell surface receptors, which induces the formation of phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) via activation of phosphatidylinositol 4-phosphate 5-kinase (PIP5K) type I. Here, we show that Wnt also stimulated the production of phosphatidylinositol 4-phosphate (PtdIns(4)P), which depended on Frizzled (Fz), Dishevelled (Dvl), and phosphatidylinositol 4-kinase (PI4K) type IIα in HEK293T cells. Dvl directly interacted with and activated PI4KIIα by increasing its Vmax for ATP and PtdIns. In addition, Dvl regulated PI4KIIα and PIP5KI via different domains. Moreover, Dvl, PI4KIIα, and PIP5KI appeared to form a ternary complex upon Wnt3a stimulation. This complex may allow efficient production of PtdIns(4,5)P2 from PtdIns, which is far more abundant than PtdIns(4)P in cells. Therefore, this study provides new insights into the mechanism by which Wnt3a regulates the production of PtdIns(4,5)P2.
The Wnt family of secretory glycoproteins plays important roles in regulation of embryonic development and tumorigenesis. They also regulate many other physiological and pathophysiological processes, including bone development, neuronogenesis, adipogenesis, myogenesis, organogenesis, and lipid and glucose metabolism (1–5). Studies using Drosophila and Xenopus embryos as well as mammalian cells have established a canonical Wnt signaling pathway that leads to stabilization of β-catenin. In the absence of Wnt, a number of proteins, including Axin, adenomatous polyposis coli (APC), casein kinase 1 (CK1), glycogen synthase kinase-3β (GSK3β),3 form a complex that facilitates β-catenin phosphorylation by CK1 and GSK3β. This phosphorylation targets β-catenin for ubiquitination and proteasome-mediated proteolytic degradation (3, 6). Some of the Wnt proteins bind to two cell surface receptors Fz and low density lipoprotein receptor-related protein (LRP) 5/6 and initiate a signaling cascade that eventually leads to the suppression of β-catenin phosphorylation by GSK3β and stabilization of β-catenin.
Because the finding that the canonical Wnt proteins transduce signals by inducing the interaction between LRP5/6 and Axin (7), more has been learned about the mechanisms by which this interaction is regulated by Wnt proteins. Studies have indicated that two phosphorylation events at the C-terminal intracellular domain of LRP5/6, the phosphorylation of Thr1479 by CKIγ (8, 9) and of Ser1490 by GSK3 (10, 11), were required for the interaction. We recently showed that Wnt3a stimulated the production of PtdIns (4,5)P2, which in turn regulated the phosphorylation of LRP5/6 at Thr1479 and Ser1490 (12). We also showed that Wnt3a regulated phosphatidylinositol 4-phosphate 5-kinase type I (PIP5KI) activity by inducing the interaction between Dvl and PIP5KI (12). Moreover, Dvl could directly stimulate the lipid kinase activity of PIP5KI (12).
PtdIns(4,5)P2 plays important roles in various cellular functions, including membrane trafficking, cytoskeletal reorganization, migration, ion channel activation, and signal transduction (13). It, however, represents less than 1% of plasma membrane phospholipids and is primarily synthesized in most cells by sequential phosphorylation of PtdIns on the D4 and D5 positions of the inositol ring by two PtdIns kinases, PI4K and PIP5KI, respectively (14, 15). While PtdIns(4)P, the substrate for PIP5KI, is also accounted for around 1% of plasma membrane phospholipids, PtdIns, the substrate for PI4K, is very abundant. Thus, Wnt3a may have to stimulate PI4K activity to provide enough substrate for PIP5KI in PtdIns(4,5)P2 production.
Two types of PI4K (PI4KI and PI4KII) have been characterized in mammalian cells. There are two isoforms of PI4KII (PI4KIIα and PI4KIIβ) and two isoforms of PI4KI (PI4KIα and PI4KIβ) (16). In our previous study, we demonstrated the involvement of PI4KIIα in Wnt signaling. siRNA-mediated knockdown in mammalian cells and morpholino-mediated suppression in Xenopus embryos of PI4KIIα inhibited LRP6 phosphorylation and Wnt signaling. In this report, we examined whether Wnt3a regulates the lipid kinase activity of PI4KIIα and found that Wnt3a could induce an increase in the level of PtdIns(4)P in a Dvl- and Fz-dependent manner. In addition, the Dvl protein was found to directly interact with and activate PI4KIIα. Moreover, different domains of Dvl appeared to be involved in the regulation of PI4KIIα and PIP5KI, and Wnt3a induced the formation of a complex of Dvl, PI4KIIα, and PIP5KI possibly for more efficient production of PtdIns (4,5)P2 in cells.
MATERIALS AND METHODS
Constructs, siRNAs, Ligands, Chemicals, and Antibodies
Human PI4KIIα and PIP5KIβ cDNAs were subcloned into CMV promoter-based mammalian cell expression vector and confirmed by DNA sequencing. Expression plasmids for Dvl and its mutants have previously been described (17). siRNAs were designed using the siRNA Design Program (Dharmacon) and synthesized by Applied Biosystems. The control siRNA was provided by Ambion. The sequences of all the siRNAs have been described (12). Recombinant purified Wnt3a proteins were purchased from R&D Systems.
Mouse anti-Dvl3 antibodies were kindly provided by D. Sussman. Rabbit anti-PI4KIIα antibody has previously been described (18). Rabbit anti-PIP5KIβ (Abgent), mouse anti-Flag, mouse anti-HA, mouse anti-Myc, and mouse anti-His (Covance) antibodies were acquired commercially.
Cell Culture and Transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's medium in the presence of 10% fetal bovine serum and transfected with DNA using Lipofectamine Plus or with siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The LacZ plasmid was added to make the total amount of DNA equals 0.25 μg/well in a 24-well plate. For transfection of siRNAs, cells were transfected with 10 pmol/well siRNAs, the total amounts remained same when siRNA mixtures were used.
Lipid Extraction and PtdIns(4)P Content Determination by ELISA
Lipid extraction was carried out as previously described (19). Briefly, cells were washed once with ice-cold PBS and lysed in 3.75 volumes of methanol/chloroform/HCl (40:20:1) mixture on ice, followed with 1 volume of chloroform and 2.25 volumes of water. After vortexing for 1 min, samples were centrifuged at 3000 rpm for 2 min at 4 °C, and the lower organic phase was collected and dried under a nitrogen stream.
The PtdIns(4)P ELISA was carried out as previously described with some modifications (19). Lipid extracts were dissolved directly in ethanol at room temperature, loaded into a microplate, and dried under vacuum. The microplate was incubated with 2% bovine serum albumin in PBS at room temperature for 30 min, followed by a mouse anti-PtdIns(4)P IgM antibody (Echelon Biosciences Inc.) for 1 h and goat anti-mouse IgG-HRP (Santa Cruz Biotechnology) for 25 min. The microplate was washed three times with PBS and added with chemiluminescence substrate (SuperSignal West Pico, Pierce). The luminescence intensity was determined by a luminometer.
Immunoprecipitation, Protein Purification, in Vitro Pull-down Assay, and PI4KIIα Kinase Assay
For overexpression assays, HEK293T cells were transiently transfected with the indicated constructs for 24 h. Coimmunoprecipitation was carried out as described previously (17). Preparation of purified recombinant His-PI4KIIα, GST-PI4KIIα-K(N-terminal 92 amino acid truncation), GST-mDvl1, His-mDvl1, His-hDvl3, GST-DIX, GST-PDZ, GST-DEP, and GST-PIP5KIβ proteins was carried out following the procedures described previously (12, 20, 21).
For the in vitro pull-down assay, recombinant proteins were incubated in 250 μl of binding buffer (1× PBS, 0.1% Nonidet P-40, 0.5 mm dithiothreitol, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride) for 3 h at 4 °C. After the proteins were captured by glutathione beads, the pull-down complexes were analyzed by Western blotting.
For the kinase assay, proteins were incubated in the kinase buffer (30 mm Hepes, pH 7.4, 100 mm NaCl, 2 mm MgCl2, 1 mm EDTA, and 0.2% Triton X-100) at room temperature for 2 h. The samples were then incubated with 10 μCi of [32P]ATP, 50 μm cold ATP, 20 μg of PtdIns (Echelon Biosciences Inc.) vesicle for 15 min at 37 °C in a final volume of 50 μl. The reaction was stopped by adding the lipid extraction solution as described above. Lipids were separated by TLC, visualized, and quantified by a phosphoimager.
RESULTS AND DISCUSSION
Wnt3a Increases PtdIns(4)P Content in HEK293T Cells via Fz and Dvl
We previously demonstrated that treatment of HEK293T cells with Wnt3a resulted in an increase in the cellular PtdIns(4,5)P2 content using an ELISA (12). In the present study, we examined whether Wnt3a treatment could change the level of PtdIns(4)P in cells using an ELISA with an antibody specific to PtdIns(4)P (Fig. 1A). Treatment of HEK293T cells with 50 ng/ml Wnt3a increased PtdIns(4)P contents by more than 70% compared with those in mock-treated cells (Fig. 1A).
FIGURE 1.
Effect of Wnt3a on the content of PtdIns(4)P in HEK293T cells. HEK293T cells were transfected with siRNAs as indicated for 48 h and then stimulated with Wnt3a (50 ng/ml) for 30 min before lipid extraction. The contents of PtdIns(4)P were determined by an ELISA. Data are presented as means ± S.D. (Student's t-test). The mock control has 920942 relative light units for A and 400648 relative light units for B.
There are four PtdIns 4-kinases in mammalian cells that catalyze the production of PtdIns(4)P from PtdIns (22). We have previously shown that knocking down of PI4KIIα, but not any of the other three PtdIns 4-kinases, inhibited Wnt3a-induced β-catenin accumulation, LRP6 phosphorylation, and PtdIns(4,5)P2 formation in HEK293T cells (12), suggesting that PI4KIIα could be involved in Wnt3a-induced PtdIns(4)P accumulation in these cells. To test this hypothesis, we transfected cells with the PI4KIIα siRNA and found that it abolished Wnt3a-induced formation of PtdIns(4)P (Fig. 1A). This result indicates that Wnt3a-induced PtdIns(4)P formation depends on PI4KIIα in these cells.
Because Fz and Dvl are required for Wnt3a-induced accumulation of PtdIns(4,5)P2 (12), we examined whether they were also involved in PtdIns(4)P formation. In the previous study we have shown that transfection of a mixture of siRNAs targeting Dvl1–3 or Fz2/4/5 was able to inhibit Wnt3a-induced PtdIns(4,5)P2 formation (12). We tested the same mixtures of the siRNAs and found that they could also inhibit Wnt3a-induced production of PtdIns(4)P (Fig. 1B). This result suggests that Fz and Dvl are also required for Wnt3a-indcued PtdIns(4)P formation in these cells.
Dvl Directly Interacts with and Activates PI4KIIα
Because both Dvl and PI4KIIα are required for Wnt3a-induced PtdIns(4)P formation, we tested whether Dvl may interact with and activate PI4KIIα as it did with PIP5KI (12). We first tested whether Dvl could interact with PI4KIIα using coimmunoprecipitation in HEK293T expressing Flag-Dvl1 and HA-PI4KIIα and found that these two proteins coimmunoprecipitated (Fig. 2, A and B). To determine whether PI4KIIα could directly interact with Dvl, we carried out a pull-down assay using recombinant proteins prepared in Escherichia coli., and found that GST-tagged Dvl1, but not GST, interacted with His-tagged PI4KIIα (Fig. 2C). It seems that N-terminal 92 amino acids of PI4KIIα are not required for the interaction (Fig. 2D).
FIGURE 2.
Interaction between Dvl and PI4KIIα. A and B, Dvl1-Flag and PI4KIIα-HA or GSK3β-HA were coexpressed in HEK293T cells for 24 h, and PI4KIIα-HA or GSK3β-HA was immunoprecipitated with an anti-HA (A) or anti-Flag (B) antibody. The immunocomplexes were detected by indicated antibodies. GSK3β-HA was used as a control. *, Ig. C, purified His-tagged PI4KIIα was incubated with GST or Dvl1-GST proteins bound on glutathione beads for 3 h at 4 °C. After washing, proteins associated with beads were analyzed by Western blotting using anti-His or anti-GST antibody. D, pulldown of His-tagged Dvl1 by GST or PI4KIIα-K-GST proteins was detected by Western blotting using anti-His or anti-GST antibody. PI4KIIα-K lacks the N-terminal 92 amino acids.
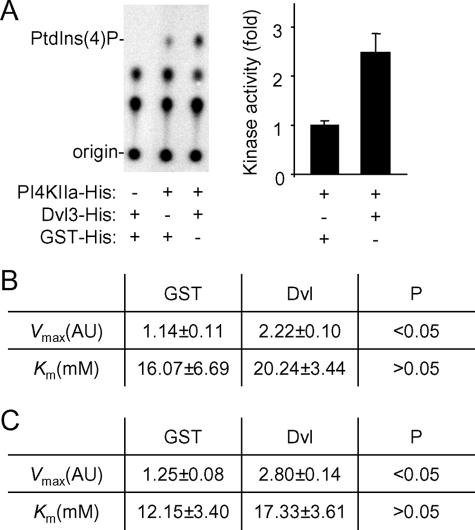
Knowing that purified recombinant Dvl3 protein could stimulate the PIP5KI lipid kinase activity (12), we tested whether recombinant Dvl3 protein could also stimulate the lipid kinase activity of recombinant PI4KIIα prepared from E. coli. In an in vitro kinase assay, we found that recombinant Dvl3 protein was able to stimulate PI4KIIα activity by ∼2.5 fold (Fig. 3A), and recombinant Dvl1 could also stimulate its activity (data not shown). We further characterized the activation of PI4KIIα by Dvl3 by determining the Km and Vmax values of PI4KIIα for its substrates, PtdIns and ATP, in the presence or absence of Dvl3. Using GST protein as a control, Dvl3 increased the Vmax values for both substrates by at least 2-fold without significantly changing the Km values (Fig. 3, B and C and supplemental Fig. S1). These results indicate that Dvl accelerates the rate of phosphorylation reaction, but does not significantly alter the affinities of PI4KIIα for its substrates.
FIGURE 3.
Regulation of PI4KIIα lipid kinase activity by Dvl protein. A, lipid kinase activity of purified PI4KIIα (50 nm) was determined by an in vitro kinase assay followed by TLC analysis in the presence of purified GST and Dvl3 proteins (100 nm). A representative TLC image is shown. The experiment was repeated three times. The data are presented as means ± S.D. p < 0.05 (Student's t-test). B and C, effects of Dvl on the kinetics of PI4KIIα catalysis. Vmax and Km values for ATP (A) and PtdIns (B) were calculated based on dose-dependent curves shown in supplemental Fig. S2. Values are presented as means ± S.D. from three independent experiments (Student's t-test).
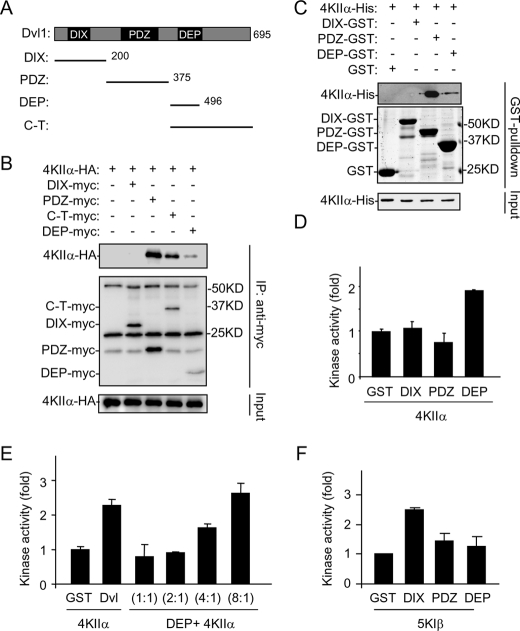
The Dvl proteins are composed of three highly conserved domains: an N-terminal DIX, a PDZ, and a C-terminal DEP domain (Fig. 4A). Coimmunoprecipitation experiments indicated that both PDZ and DEP domains could interact with PI4KIIα (Fig. 4B), and these interactions were direct because purified recombinant proteins interacted in in vitro pull-down assays (Fig. 4C). Although the PDZ domain appeared to have a higher affinity than the DEP domain for PI4KIIα (Fig. 4, B and C), the DEP protein, but not PDZ protein, stimulated the lipid kinase activity of PI4KIIα in an in vitro kinase assay (Fig. 4D). A titration experiment revealed that close to 8 times more DEP protein than the full-length Dvl protein was needed to achieve the same level of PI4KIIα activation as the full length (Fig. 4E). We interpret these results to suggest that the interaction between DEP and PI4KIIα is involved in activating the kinase, whereas the PDZ and PI4KIIα interaction may contribute to the affinity of Dvl for PI4KIIα.
FIGURE 4.
Characterization of Dvl domains in interaction and activation of PI4KIIα. A, schematic representation of Dvl1 and its truncated forms used in the study. B, delineation of Dvl1 domains involved in interaction with PI4KIIα by immunoprecipitation with proteins transiently expressed in HEK293T cells. Three background bands existing in all of the lanes are Ig. C, in vitro pull-down assay using recombinant Dvl1 fragments and PI4KIIα. Samples were analyzed by immunoblotting with the indicated antibodies or Coomassie Blue staining. D and E, effects of Dvl1 fragments on lipid kinase activity of PI4KIIα. Purified PI4KIIα (50 nm) was assayed for its lipid kinase activity in the presence of 200 nm (D) or varying amounts of recombinant proteins (E). The ratio indicates the amount of DEP domain protein to PI4KIIα protein. The amounts of GST and Dvl used in E are 50 nm. The data are presented as means ± S.E. F, effects of Dvl1 fragments on lipid kinase activity of PIP5KIβ. Purified PIP5KIβ (50 nm) was assayed for its lipid kinase activity in the presence of 50 nm Dvl domain proteins.
Wnt Promotes the Formation of a Dvl, PI4KIIα, and PIP5KIβ Complex
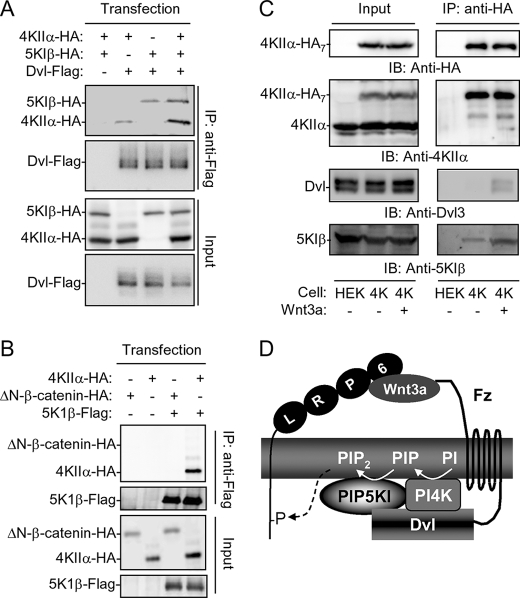
Although Dvl DEP appears to be responsible for PI4KIIα activation, it had little effect on activation of PIP5KIβ (Fig. 4F). The N-terminal DIX domain seems to be primarily responsible for activation of PIP5KIβ (Fig. 4F). While Dvl PDZ was able to bind to both PI4KIIα and PIP5KIβ (12), PI4KIIα and PIP5KIβ did not appear to compete for Dvl binding. In a coimmunoprecipitation experiment, more rather than less PI4KIIα and PIP5PIβ proteins were pulled down with Dvl in cells coexpressing Dvl, PIP5KIβ, and PI4KIIα than those coexpressing Dvl and PI4KIIα or PIP5KIβ (Fig. 5A). One possible explanation to the enhanced interactions is that PI4KIIα may interact with PIP5KIβ. When these two proteins were expressed in HEK293T cells, they coimmunoprecipitated (Fig. 5B and supplemental Fig. S2A). The interaction between these two proteins appeared to be direct, because purified recombinant proteins of PI4KIIα and PIP5KIβ interacted in an in vitro pull-down assay (supplemental Fig. S2, B and C). Consistent with the non-competitive nature of these interactions, expression of Dvl did not reduce, but rather increased, the amount of PI4KIIα pulled down by PIP5KIβ (supplemental Fig. S2D).
FIGURE 5.
Interactions of Dvl, PI4KIIα and PIP5KIβ. A, immunoprecipitation of Dvl-Flag in HEK293T cells overexpressing PI4KIIα and/or PIP5KIβ. B, coimmunoprecipitation of PI4KIIα and PIP5KIβ in transfected HEK293T cells. ΔN-β-catenin-HA was used as a control. C, effect of Wnt3a on Dvl, PIP5KIβ and PI4KIIα complex formation. HEK293T cells stably expressing PI4KIIα-HA7 (4K), whose expression level is lower than that of endogenous PI4KIIα as detected by an anti-PI4KIIα antibody, were treated with or without 50 ng/ml Wnt3a protein for 30 min. Immunoprecipitation was carried out using an anti-HA antibody. HEK is the parental cell. D, hypothetical model depicting the regulation of PtdIns(4,5)P2 production by Wnt3a through Fz, Dvl and phosphatidylinositol kinases.
Next, we wanted to know whether Wnt3a stimulation regulates these interactions. To do this, we need to examine the interactions using proteins that are expressed at near endogenous levels. Antibodies specific for PI4KIIα, PIP5KIβ, and Dvl were not suitable for immunoprecipitation. To circumvent this difficulty, we established a HEK293 cell line that stably expressed PI4KIIα carrying seven HA tags at a level that is lower than that of endogenous PI4KIIα (compare the upper bands with the lower bands in the second left panel of Fig. 5C). The seven HA tags markedly increases immunoprecipitation efficiency. In the immunocomplexes pulled down by anti-HA antibodies, we detected a low level of PIP5KIβ, but not Dvl (Fig. 5C). However, when the cells were treated with Wnt3a, Dvl3 was readily detected (Fig. 5C). In addition, an increased amount of PIP5KIβ was detected (Fig. 5C). All of these results together indicate that Wnt3a is able to induce the binding of Dvl to the PIP5KIβ and PI4KIIα complex, leading to more stable interaction between PIP5KIβ and PI4KIIα.
In this report, we describe our findings that Wnt3a stimulates the production of PtdIns(4)P via one of the PI4Ks, namely PI4KIIα in HEK293 cells. It is likely that the same PI4K is also regulated by Wnt in Xenopus embryos because its suppression by morpholino oligos inhibited Wnt signaling in the embryos (12). In overexpression systems, we found that Dvl could also interact with PI4KIIβ (data not shown). In our previous studies, we presented evidence to indicate that Dvl regulates all of the three PIP5KI isoforms even though PIP5KIβ appeared to be the major isoform in HEK293T cells (12). The lack of significant effects of siRNAs to other PIP5KI or PI4K isoforms on Wnt signaling in these cells may be due to their expression levels or subcellular localization. In HEK293 cells, Wnt3a appears to regulate PI4KIIα via Fz and Dvl, probably through its activation by Dvl. It is intriguing and logical for Dvl to directly stimulate both PI4K and PIP5K, both of which constitute the major pathway for PtdIns(4,5)P2 synthesis in cells. Given that PtdIns(4)P (the substrate of PIP5KI) is as scarce as PtdIns(4,5)P2 (the product of PIP5KI), activation of PI4K would allow Wnt to utilize PtdIns, a more abundant source of substrate on the plasma membrane, for rapid production of PtdIns(4,5)P2. Although we have not visualized the ternary complex of endogenous Dvl, PI4KIIα and PIP5KI proteins, the evidence presented in Fig. 5 and supplemental Fig. S2 suggests that these three proteins are most likely to form a complex. It is also reasonable to postulate that this complex would allow more efficient production of PtdIns(4,5)P2 since PtdIns(4), the product of PI4KIIα, would not have to diffuse far before it is used by PIP5KI to produce PtdIns(4,5)P2 as depicted in Fig. 5D. Therefore, this study provides further insights into the regulation of PtdIns(4,5)P2 production by Wnt and Dvl.
Supplementary Material
Acknowledgments
We thank Michelle Orsulak and other members of the Wu laboratory for assistance and Pietro De Camilli for the anti-PI4KIIα antibody.
This work was supported, in whole or in part, by National Institutes of Health grants (to D. W.). This work was also supported by NSFC grants (to L. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- GSK3β
- glycogen synthase kinase-3β
- LRP
- lipoprotein receptor-related protein
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase
- HA
- hemagglutinin.
REFERENCES
- 1.Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20,781–810 [DOI] [PubMed] [Google Scholar]
- 2.Reya T., Clevers H. (2005) Nature 434,843–850 [DOI] [PubMed] [Google Scholar]
- 3.Clevers H. (2006) Cell 127,469–480 [DOI] [PubMed] [Google Scholar]
- 4.Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5,691–701 [DOI] [PubMed] [Google Scholar]
- 5.Mani A., Radhakrishnan J., Wang H., Mani A., Mani M. A., Nelson-Williams C., Carew K. S., Mane S., Najmabadi H., Wu D., Lifton R. P. (2007) Science 315,1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusse R. (2005) Cell Res. 15,28–32 [DOI] [PubMed] [Google Scholar]
- 7.Mao J., Wang J., Liu B., Pan W., Farr G. H., 3rd, Flynn C., Yuan H., Takada S., Kimelman D., Li L., Wu D. (2001) Mol. Cell 7,801–809 [DOI] [PubMed] [Google Scholar]
- 8.Davidson G., Wu W., Shen J., Bilic J., Fenger U., Stannek P., Glinka A., Niehrs C. (2005) Nature 438,867–872 [DOI] [PubMed] [Google Scholar]
- 9.Bilic J., Huang Y. L., Davidson G., Zimmermann T., Cruciat C. M., Bienz M., Niehrs C. (2007) Science 316,1619–1622 [DOI] [PubMed] [Google Scholar]
- 10.Tamai K., Zeng X., Liu C., Zhang X., Harada Y., Chang Z., He X. (2004) Mol. Cell 13,149–156 [DOI] [PubMed] [Google Scholar]
- 11.Zeng X., Tamai K., Doble B., Li S., Huang H., Habas R., Okamura H., Woodgett J., He X. (2005) Nature 438,873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan W., Choi S. C., Wang H., Qin Y., Volpicelli-Daley L., Swan L., Lucast L., Khoo C., Zhang X., Li L., Abrams C. S., Sokol S. Y., Wu D. (2008) Science 321,1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Paolo G., De Camilli P. (2006) Nature 443,651–657 [DOI] [PubMed] [Google Scholar]
- 14.Clarke J. H., Richardson J. P., Hinchliffe K. A., Irvine R. F. (2007) Biochem. Soc. Symp. 74,149–159 [DOI] [PubMed] [Google Scholar]
- 15.Michell R. H. (2008) Nat. Rev. 9,151–161 [DOI] [PubMed] [Google Scholar]
- 16.De Matteis M. A., Di Campli A., Godi A. (2005) Biochim. Biophys. Acta 1744,396–405 [DOI] [PubMed] [Google Scholar]
- 17.Li L., Yuan H., Weaver C. D., Mao J., Farr G. H., 3rd, Sussman D. J., Jonkers J., Kimelman D., Wu D. (1999) EMBO J. 18,4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J., Wenk M. R., Pellegrini L., Onofri F., Benfenati F., De Camilli P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100,3995–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie F., Okuno M., Pasquale E. B., Yamaguchi Y. (2005) Nat. Cell Biol. 7,501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Zhu J., Yang G. Y., Wang Q. J., Qian L., Chen Y. M., Chen F., Tao Y., Hu H. S., Wang T., Luo Z. G. (2007) Nat. Cell Biol. 9,743–754 [DOI] [PubMed] [Google Scholar]
- 21.Krauss M., Kukhtina V., Pechstein A., Haucke V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balla A., Balla T. (2006) Trends Cell Biol. 16,351–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.