Abstract
Hemojuvelin (HJV) is a glycosylphosphatidylinositol-linked protein and binds both bone morphogenic proteins (BMPs) and neogenin. Cellular HJV acts as a BMP co-receptor to enhance the transcription of hepcidin, a key iron regulatory hormone secreted predominantly by liver hepatocytes. In this study we characterized the role of neogenin in HJV-regulated hepcidin expression. Both HJV and neogenin were expressed in liver hepatocytes. Knockdown of neogenin decreased BMP4-induced hepcidin mRNA levels by 16-fold in HJV-expressing HepG2 cells but only by about 2-fold in cells transfected with either empty vector or G99V mutant HJV that does not bind BMPs. Further studies indicated that disruption of the HJV-neogenin interaction is responsible for a marked suppression of hepcidin expression. Moreover, in vivo studies showed that hepatic hepcidin mRNA could be significantly suppressed by blocking the interaction of HJV with full-length neogenin with a soluble fragment of neogenin in mice. Together, these results suggest that the HJV-neogenin interaction is required for the BMP-mediated induction of hepcidin expression when HJV is expressed. Combined with our previous studies, our results support that hepatic neogenin possesses two functions, mediation of cellular HJV release, and stimulation of HJV-enhanced hepcidin expression.
Iron is an indispensable nutrient required for a variety of biochemical processes such as respiration, metabolism, and DNA synthesis. Iron homeostasis in the body is regulated primarily by the rate of iron absorption from the intestine.
Mutations in the key iron homeostatic proteins result in either hereditary hemochromatosis or iron-deficient anemia (1–4). Hereditary hemochromatosis is a heterogeneous group of inherited iron overload disorders linked to mutations in several genes including HFE, HFE2, HAMP (the hepcidin gene), and TFR2. HFE2 is a recently cloned gene encoding the protein hemojuvelin (HJV)2 (5). Both the HFE2 mRNA and protein are highly expressed in skeletal muscles and the heart and at lower levels in the liver (5, 6).
HJV plays a pivotal role in iron homeostasis. Homozygous or compound heterozygous mutations in HFE2 are the major cause of juvenile hemochromatosis (5), a particularly severe form of hereditary hemochromatosis (7, 8). The marked suppression of hepatic hepcidin expression observed in juvenile hemochromatosis patients with HFE2 mutations as well as in HJV knock-out (Hjv−/−) mice has implicated HJV as a critical upstream regulator of hepcidin transcription (5, 9, 10). Hepcidin, the key iron regulatory hormone, is predominantly expressed in liver hepatocytes, and iron levels in the body positively regulate its expression (11, 12). In the liver HJV is found predominantly in hepatocytes (10), where it regulates hepcidin expression.
HJV is a glycosylphosphatidylinositol-linked membrane-bound protein and is either associated with cells or released by proteolytic cleavage in a soluble form. Cellular HJV acts as a co-receptor for bone morphogenic proteins (BMPs), BMP2, BMP4, BMP5, and BMP6 to enhance the transcription of hepcidin through the BMP-signaling cascade (13, 14). Binding of BMP ligands to BMP receptor complexes on the cell surface triggers the phosphorylation of Smad1, Smad5, and Smad8 (Smad1/5/8) in the cytoplasm. The phosphorylated Smads (pSmad1/5/8) form heteromeric complexes with Smad4 and then translocate into the nucleus where they induce the transcription of target genes (15). Liver-specific disruption of Smad4 in mice markedly decreases hepcidin expression and causes severe iron accumulation in different organs, further supporting the pivotal role of BMP signaling in the regulation of hepcidin expression (16).
HJV cleavage and subsequent release constitutes the major pathway of cellular HJV turnover in HepG2 cells transfected to express HJV (17). Functional studies reveal that soluble HJV suppresses BMP-induced hepcidin expression both in vitro and when injected into mice, likely through competition with membrane-associated HJV for limited BMPs (14, 18). Previous studies suggest that HJV release may involve retrograde trafficking of HJV from the plasma membrane to the Golgi/trans-Golgi network compartment, where it may be subjected to cleavage by the proprotein convertase furin, followed by rapid release from cells (17, 19, 20). Interaction of HJV with neogenin, a type I membrane protein expressed in most tissues including the liver (21), is required for HJV release from different cell lines (6, 17).
In this study we characterized the role of neogenin in HJV-regulated hepcidin expression. Our results indicated that HJV and neogenin are co-expressed in liver hepatocytes. Surprisingly, the HJV-neogenin interaction is required for the induction of hepcidin expression by BMP4 in addition to its role of mediating HJV release from cells.
EXPERIMENTAL PROCEDURES
Quantitative Real-time RT-PCR (qRT-PCR)
qRT-PCR was used to analyze the mRNA levels of HFE2, neogenin, and GAPDH in isolated rat liver hepatocytes, Kupffer cells, sinusoidal endothelial cells, and hepatic stellate cells (HSC) as well as the mRNA levels of hepcidin and GAPDH in HepG2 cells and mouse livers. Total RNA isolation and cDNA preparation were previously described (22). qRT-PCR analysis was performed using primers specific for rat genes and human GAPDH as previously reported (6, 22, 23). The sequences of other primers are 5′-ggctctgttttcccacaacag-3′ (forward, human hepcidin), 5′-tccttcgcctctggaacatgg-3′ (reverse, human hepcidin), 5′-aaatatgacaactcactcaagattgtca-3′ (forward, mouse GAPDH), 5′-cccttccacaatgccaaagt-3′ (reverse, mouse GAPDH), 5′-ctgagcagcaccacctatctc-3′ (forward, mouse hepcidin), and 5′-tggctctaggctatgttttgc-3′ (reverse, mouse hepcidin). The results for each gene of interest are expressed as the amount relative to that of GAPDH in each sample.
Cell Culture and Transfection
HepG2 cells were purchased from ATCC and maintained in MEM, 10% FCS, 1 mm pyruvate/1× nonessential amino acids (complete medium). HepG2 cells stably expressing G99V HJV (G99V-HepG2) were generated using the Nucleofector kit V (Amaxa Biosystems) as previously described (6). HepG2 cells stably transfected with wild type HFE2 (HJV-HepG2) or pcDNA3 empty vector (control-HepG2) were generated previously (6, 24). The stably transfected cells were maintained in complete medium with 800 μg/ml G418.
HepG2 cells stably transfected with the tetracycline repressor (tTA-HepG2) were obtained from Dr. Gregory Longmore at Washington University, St. Louis (25). tTA-HepG2-HJV and tTA-HepG2-G99V HJV cells were generated by subcloning HFE2 or G99V HFE2 cDNA into a tetracycline-inducible pcDNA4 vector, respectively, followed by a stable transfection into tTA-HepG2 cells. Transfected cells were maintained in complete medium with 800 μg/ml G418 and 5 μg/ml blasticidin and induced to express HJV using 2 μg/ml doxycycline (dox), a tetracyline homolog. tTA-HepG2 cells transfected with empty pcDNA4 vector (tTA-HepG2-control) were also generated and used as controls.
Mutagenesis
The G99V mutant HFE2 cDNA was generated using the QuikChangeTM XL site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. pcDNA3-HFE2 construct was used as a template. The primers used to introduce the mutation were 5′-ccgcacctgccgcgtggacctcgccttcc-3′ (forward) and 5′-ggaaggcgaggtccacgcggcaggtgcgg-3′ (reverse). The mutation in the resulting construct was confirmed by DNA sequencing. No other sequence change was detected.
BMP4 Treatment
tTA-HepG2-HJV, -G99V HJV, or -control cells were subcultured into 12-well plates. After 48 h of incubation, cells at about 60% confluence were incubated with MEM, 1 mm pyruvate, 1× nonessential amino acids (NEAA), 1% FCS plus 2 μg/ml dox for 6 h to serum starve the cells and to induce HJV expression. Cells were then incubated in the same medium with BMP4 (R&D Systems, Inc.) at the indicated concentrations for the time intervals described in each figure legend. In some studies 40 nm soluble neogenin fibronectin type III 5–6 domain (Neo FNIII 5–6) or 5 μm decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone (Alexis Biochemicals), a furin convertase inhibitor (FCI), was also included. For Western blot analysis, the conditioned medium (CM) was collected, and cell lysate was prepared for the immunodetection of HJV in both CM and lysate as well as for the immunodetection of phosphorylated Smad1/5/8 (pSmad) and β-actin in the lysates. For qRT-PCR analysis of hepcidin and GAPDH mRNA, total cellular RNA was isolated as described under “Quantitative Real-time RT-PCR.” Neo FNIII 5–6 fragment specifically binds HJV with a KD in the subnanomolar range. Purified Neo FNIII 5–6 was generated from baculovirus as previously described (26).
Knockdown of Endogenous Neogenin
SMARTpool siRNA specific for human neogenin (Dharmacon) was used to knock down the endogenous neogenin in control-, HJV-, and G99V HJV-HepG2 cells as previously described (6). RNAiMAX reagent (Invitrogen) was used for the transfection. The negative control siRNA was previously described (6). siRNA transfection was conducted in 12-well plates in complete medium. About 48-h after transfection, cells were serum-starved with MEM, 1 mm pyruvate, 1× NEAA, 1% FCS for 6 h followed by incubation with the same medium (1 ml per well) supplemented with 0, 0.5, 1, and 5 ng/ml BMP4 for 16 h. CM was then collected, and cell lysate was prepared for immunodetection. Total cellular RNA was isolated for qRT-PCR analysis.
For neogenin rescue studies HJV-HepG2 cells were first transfected with either control or neogenin siRNA. On the following day cells were transfected with either human neogenin cDNA or pcDNA3 empty vector using FuGENE HD transfection reagent (Roche Applied Science). On day 3, cells were serum-starved with MEM, 1 mm pyruvate, 1× NEAA, 1% FCS for 6 h followed by incubation with the same medium (1 ml per well) supplemented with 5 ng/ml BMP4 for 16 h. About 72 h after the siRNA transfection, CM was collected, cell lysate was prepared for immunodetection, and total RNA was isolated for qRT-PCR analysis.
Flow Cytometry
Flow cytometry was used to quantify cell surface HJV in HJV-HepG2 cells after treatment with or without 40 nm Neo FNIII 5–6 for 16 h as well as in G320V HJV, D172E HJV, and G99V HJV-HepG2 cells. Cells were detached from flasks with cell dissociation buffer (Invitrogen). Cells were then incubated with affinity-purified rabbit anti-HJV antibody (4 μg/ml) in Hanks' buffer supplemented with 3% fetal bovine serum for 30 min at 4 °C, washed, and incubated with phycoerythrin-conjugated goat anti-rabbit IgG (1:500 dilution) (Caltag, Burlingame, CA) in the same buffer for 30 min at 4 °C. Flow cytometry analysis was performed on a BD Biosciences FACSCalibur flow cytometer. Rabbit IgG and control-HepG2 cells were used as negative controls. The levels of cell surface HJV are expressed as arbitrary units (6).
Immunodetection
Cell lysates from the isolated rat liver cells and HepG2 cells were prepared using NET-Triton (150 mm NaCl, 5 mm EDTA, 10 mm Tris (pH 7.4), and 1% Triton X-100) supplemented with 1× protease inhibitor mixture (Roche Applied Science), 1 mm sodium fluoride (Sigma), and 1 mm sodium orthovanadate (Sigma). Proteins from the isolated liver cells (250 μg), whole cell extract from 1 well of a 12-well plate, and conditioned medium (120 μl) were separated using 11% SDS-PAGE under reducing conditions followed by transfer onto nitrocellulose membrane. Membranes were probed with affinity-purified rabbit anti-HJV antibody (0.22 μg/ml) (21), rabbit anti-neogenin antibody (0.4 μg/ml, Santa Cruz Biotechnology), rabbit anti-phosphorylated Smad1/5/8 (1:1,000; Cell Signaling Technology), or mouse anti-β-actin antibody (1:10,000; Chemicon International, Temecula, CA) followed by immunodetection using corresponding horseradish peroxidase-conjugated secondary antibodies (Chemicon International). The chemiluminescent bands were exposed to x-ray film (Super Signal, Pierce).
Neo FNIII 5–6 Injection
Eight-week-old male 129EvSv mice were injected intraperitoneally with purified Neo FNIII 5–6 at 3 mg/kg body weight or carrier buffer twice on the same day at 8:30 a.m. and 3:30 p.m. Injected mice had free access to the regular rodent diet. At 24 h after the first injection, animals were euthanized while under anesthesia. Liver tissues were collected for qRT-PCR analysis of hepcidin and GAPDH mRNA. Each group consisted of three mice. All the procedures were approved by the Institutional Animal Care and Use Committee of Oregon Health and Science University.
Statistical Analysis
The S.D. and the paired two-tailed Student's t test were used to compare two sets of data. The one-way analysis of variance and Tukey's post-test were used to compare three or more sets of data.
RESULTS
HJV and Neogenin Are Co-expressed in Hepatocytes
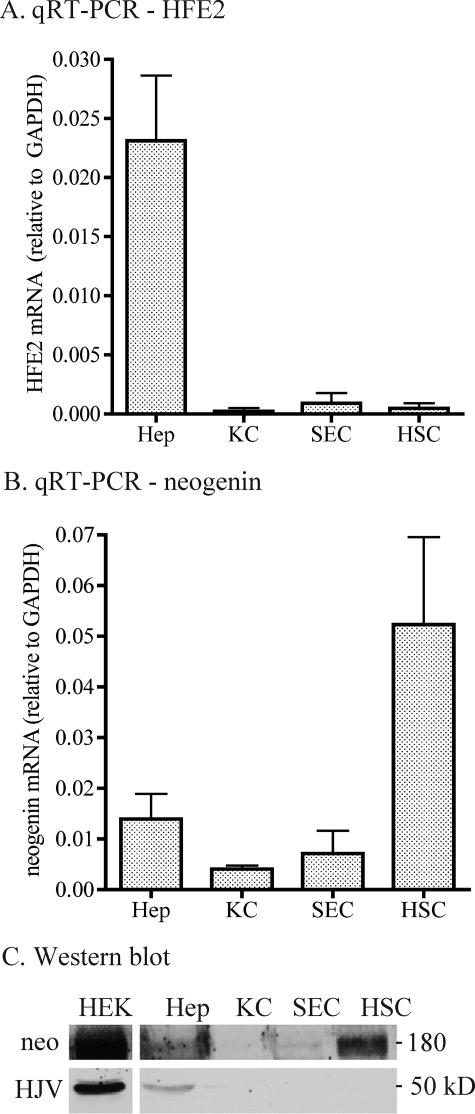
We sought to explore the role of neogenin in HJV-regulated hepcidin expression in hepatocytes in vivo because previous studies demonstrated that HJV binds neogenin and that the HJV-neogenin interaction is required for HJV release (11, 21, 22). We first examined the expression profiles of HFE2 and neogenin mRNA and proteins in the liver using isolated rat liver hepatocytes, Kupffer cells, sinusoidal endothelial cells, and HSC. Consistent with findings in a previous report (10), both HFE2 mRNA and protein were detected predominantly in hepatocytes (Fig. 1, A and C). In contrast, both neogenin mRNA and protein were detected in all liver cell populations (Fig. 1, B and C). The highest level of neogenin mRNA was found in HSC, ∼3.7-, ∼12.9-, and ∼7.3-fold greater than in hepatocytes, Kupffer cells, and sinusoidal endothelial cells, respectively (Fig. 1B). Immunoblots revealed a similar profile of neogenin protein expression (Fig. 1C, upper panel). In the liver, hepatocytes account for approximate 65% in cell number and 80% in volume, but HSCs only account for approximate 5–8% in cell number and 1.4% in volume (27). Therefore, these results suggest that hepatocytes are also the major site of neogenin expression in the liver.
FIGURE 1.
HFE2 and neogenin expression profiles in isolated rat liver cells. A, qRT-PCR analysis of HFE2 mRNA in isolated rat liver hepatocytes (Hep, n = 5), Kupffer cells (KC, n = 7), sinusoidal endothelial cells (SEC, n = 4), and hepatic stellate cells (HSC, n = 6). Results are expressed as the amounts relative to GAPDH. The mean values and the S.D. for each cell population are presented. B, qRT-PCR analysis of neogenin mRNA in isolated rat liver cell populations. The analysis was performed as described in A. C, Western blot analysis of HJV and Neo proteins in isolated rat liver cells. Cell extract protein (250 μg) was separated in SDS-PAGE under reducing conditions. Equal protein loading was confirmed by Ponceau S staining of the membrane (not shown). Membranes were probed with antibodies against HJV and neogenin. Cell lysate from HEK293 cells stably expressing both HJV and neogenin (HEK) was included as a positive control. Experiments were performed using two different sets of isolated cells and showed consistent results.
In situ hybridization and immunohistochemistry analysis revealed that both HFE2 mRNA and protein were evenly detected in hepatocytes in rat liver tissues with no evidence of a zonal distribution (supplemental Fig. 1, A and B). Neogenin mRNA and protein were observed in hepatocytes as well as in non-parenchymal cells throughout the liver tissues (supplemental Fig. 1, A and B). Together, the above results indicate that HJV and neogenin are co-expressed in hepatocytes.
BMP4 Induces Hepcidin Expression in HepG2 Cells
Hepcidin expression is induced via the BMP-signaling cascade (13, 16). We wanted to use the hepatoma cell lines HepG2 cells and tTA-HepG2 cells, where proteins can be expressed in a tetracycline-dependent manner, as model systems to determine the role of neogenin in HJV-regulated hepcidin expression. They are relatively differentiated hepatoma cell lines that express hepcidin, neogenin (24), and many other hepatocyte-specific genes but have no detectable HJV protein by Western blot analysis (data not shown). HFE2 mRNA levels are close to the limit of detection by qRT-PCR and are about 1300-fold lower than in human liver tissue (data not shown).
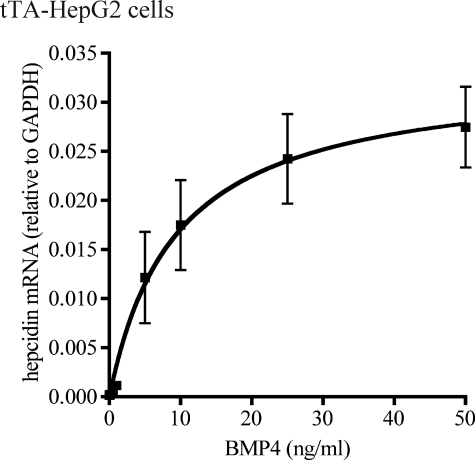
We first titrated the response of tTA-HepG2 cells to BMP4, a ligand for both HJV and BMP receptors that is expressed in the liver (14, 28). Induction of hepcidin could be detectable at as low as 0.1 ng/ml BMP4 as measured by qRT-PCR. Increases in hepcidin mRNA of 1.8-, 3.5-, 13.4-, 130-, 212-, 310-, and 371-fold were detected with the addition of 0.1, 0.5, 1, 5, 10, 25, and 50 ng/ml BMP4, respectively. The induction was nearly linear up to 5 ng/ml BMP4 when cells were treated with BMP4 for 18 h (Fig. 2). On the basis of these observations, a concentration of 5 ng/ml BMP4 was chosen for our studies. At 5 ng/ml BMP4, hepcidin mRNA levels were increased by 130-fold in tTA-HepG2 cells, indicating that BMP4 can induce hepcidin transcription independently of HJV. These results are consistent with previous studies showing that BMP2 induces hepcidin expression in the primary hepatocytes isolated from Hjv−/− mice (13). HepG2 cells are, thus, an appropriate cell line to study the effects of neogenin and different constructs of HJV on hepcidin expression.
FIGURE 2.
Titration of the response of hepcidin mRNA expression to BMP4. tTA-HepG2 cells were subcultured into 12-well plates. After 48 h of incubation, cells at about 60% confluence were first incubated with MEM, 1 mm pyruvate, 1× NEAA, 1% FCS for 6 h to serum starve the cells. Cells were then incubated in the same medium with BMP4 at 0, 0.1, 0.5, 1, 5, 10, 25, and 50 ng/ml for 18 h followed by total RNA isolation, cDNA preparation, and qRT-PCR analysis of hepcidin and GAPDH mRNA. The hepcidin mRNA levels are expressed as the amount relative to that of GAPDH in each specific sample. The results are from three separate experiments, and the mean values and the S.D. are presented.
Knockdown of Neogenin Suppresses Hepcidin Expression Markedly When HJV Is Expressed
Soluble HJV suppresses BMP-induced hepcidin expression through competition with membrane-associated HJV for a limited BMP supply (14, 18). We wanted to test the hypothesis that neogenin negatively regulates hepcidin expression by facilitating the release of HJV from cells as shown previously (24). Endogenous neogenin was knocked down in HepG2 cells stably transfected with HFE2 (HJV) or empty vector (control). The responses of BMP signaling and hepcidin expression to BMP4 treatment were then examined.
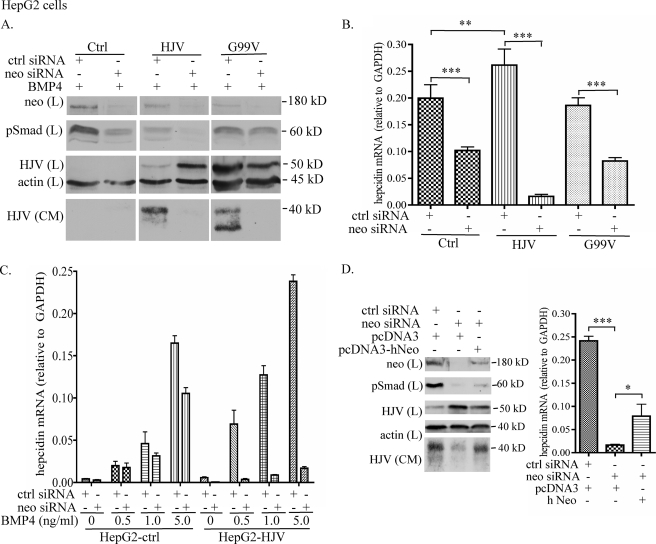
First, we tested the effects of neogenin knockdown on BMP signaling and hepcidin mRNA in the presence of 5 ng/ml BMP4 (Fig. 3, A and B). A single transfection of neogenin siRNA significantly decreased the levels of endogenous neogenin in control and HJV-HepG2 cells (Fig. 3A, top panel). The accumulation of cellular HJV detected in HJV-HepG2 cells with neogenin knockdown (Fig. 3A) is consistent with a previous study, in which a soluble neogenin whole ectodomain that binds HJV was used to block HJV release (17). HJV-HepG2 cells have lower base-line levels of pSmad than control-HepG2 cells (Fig. 3A). Because HepG2 cells were stably transfected with HJV and clonally selected, differences in basal levels in the selected clones could account for this observation.
FIGURE 3.
Knockdown of neogenin suppresses BMP4-induced hepcidin expression. A, knockdown of neogenin decreases pSmad1/5/8. Control-, HJV-, and G99V HJV-HepG2 cells in 12-well plates were transfected with either control (Ctrl) or neogenin (Neo) siRNA using RNAiMAX reagent. After about 48 h, cells were serum-starved for 6 h followed by incubation with the same medium (1 ml/well) supplemented with 5 ng/ml BMP4 for 16 h. CM was then collected, and cell lysate was prepared. About 15% of CM and the total lysate were subjected to SDS-PAGE followed by immunodetection of neogenin (Neo), pSmad1/5/8 (pSmad), HJV, and β-actin in the lysate (L) and HJV in CM. B, knockdown of neogenin decreases the BMP4-induced hepcidin mRNA. Neogenin knockdown and BMP4 treatment in control-, HJV-, and G99V HJV-HepG2 cells were performed in essentially the same manner as described in A. Total RNA was isolated followed by cDNA preparation and qRT-PCR analysis of hepcidin and GAPDH mRNA. The hepcidin mRNA levels are expressed as the amount relative to that of GAPDH in each specific sample. C, knockdown of neogenin decreases hepcidin mRNA in the absence of exogenous BMP4. All the experimental procedures were performed as described in B except that control- and HJV-HepG2 cells were incubated with different concentrations of BMP4 (0, 0.5, 1, and 5 ng/ml). D, neogenin rescue analysis. HJV-HepG2 cells were first transfected with either control (Ctrl) or neogenin (Neo) siRNA on day 1. About 24 h later, cells were introduced with either human neogenin cDNA (hNeo) or pcDNA3 empty vector (pcDNA3). On day 3, cells were serum-starved with MEM, 1 mm pyruvate, 1× NEAA, 1% FCS for 6 h followed by incubation with the same medium (1 ml/well) supplemented with 5 ng/ml BMP4 for 16 h. At about 72 h after the siRNA transfection, CM was collected, and cell lysate was prepared for immunodetection (left panel) as well as for qRT-PCR analysis of hepcidin mRNA (right panel) as described in A and B, respectively. All the experiments were repeated at least three times with consistent results. The qRT-PCR results in B and D were assessed by one-way analysis of variance, and the statistical significant differences relative to the corresponding controls were determined by Tukey's post-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Contrary to our initial hypothesis, neogenin knockdown resulted in a 16-fold decrease, rather than an increase, in hepcidin mRNA in HJV-HepG2 cells and about a 2-fold decrease in control-HepG2 cells compared with their corresponding controls (Fig. 3B). Because hepcidin expression is induced by the BMP-signaling pathway (13, 16), the results of hepcidin mRNA were verified by examining the effects of neogenin knockdown on BMP signaling. Levels of phosphorylated Smad1/5/8 (pSmad) are a direct indicator of BMP receptor activation. In agreement with the reduced induction of hepcidin mRNA, immunoblots revealed decreased pSmad in neogenin-depleted cells (Fig. 3A). Therefore, these results suggest that the suppression of hepcidin expression in both control and HJV-HepG2 cells after neogenin knockdown results from decreased BMP signaling.
To determine whether the inhibition is BMP4-dependent, we next examined the effects of neogenin knockdown on hepcidin expression in HJV-HepG2 cells with the addition of 0, 0.5, and 1 ng/ml concentrations of exogenous BMP4. A similar level of inhibition was obtained under each condition (Fig. 3C). These results indicate that when HJV is expressed, the suppression of hepcidin expression by neogenin knockdown is independent of the addition of exogenous BMP4. Because the levels of HJV mRNA in the control-HepG2 cells are negligible compared with human liver tissues or HJV-HepG2 cells (data not shown), these results suggest that in the presence of no or very low levels of HJV, neogenin acts to maintain the basal levels of BMP signaling, and the resulting hepcidin expression. In contrast, the marked suppression of hepcidin expression detected in HJV-HepG2 cells depleted of neogenin implies that when HJV is expressed, neogenin is required for full extent of the induction of hepcidin expression by BMP4.
To rule out the possibility that the inhibitory effect detected in neogenin-depleted HJV-HepG2 cells results from an off-target effect, we conducted a neogenin rescue study. Transfection of cells with neogenin after endogenous neogenin knockdown was able to partially rescue the HJV release (Fig. 3D, left panel) as well as the hepcidin expression (Fig. 3D, right panel) induced by BMP4 at 5 ng/ml. The lack of a complete rescue might be because of the low transfection efficiency of neogenin cDNA. These results, therefore, suggest that the effect of neogenin knockdown in HJV-HepG2 cells is not caused by an off-target effect.
HepG2 cells have higher basal levels of hepcidin mRNA than tTA-HepG2 cells (Fig. 3B versus Fig. 2). tTA-HepG2 cells were derived by selecting a clone of HepG2 cells stably transfected with the tTA tetracycline repressor (25) and, thus, could account for the lower basal levels of hepcidin than the parent cell line.
Neogenin Knockdown Does Not Markedly Suppress Hepcidin Expression when G99V HJV Is Expressed
HJV can simultaneously bind both BMP2 and neogenin (26). G99V mutant HJV is a non-functional form of HJV. In humans G99V substitution in HJV causes juvenile hemochromatosis (5). G99V HJV binds neogenin (supplemental Fig. 2A) but not BMPs (29). To determine whether or not the suppression of hepcidin expression detected in HJV-HepG2 cells after neogenin knockdown is because of the disruption of the HJV-neogenin interaction, we next examined the effect in HepG2 cells stably expressing the G99V mutant HJV. Because the interaction with neogenin is required for HJV release (6), G99V HJV behaved similarly to wild type HJV in its active release and plasma membrane expression (supplemental Fig. 2, B and C). Similar to HJV-HepG2 cells, neogenin knockdown blocked the release of G99V HJV from G99V HJV-HepG2 cells (Fig. 3A). However, in contrast to the marked suppression of hepcidin expression in HJV-HepG2 cells, knockdown of neogenin knockdown in G99V HJV-HepG2 cells only caused a mild decrease in hepcidin mRNA, which is similar to that in control-HepG2 cells (Fig. 3B). Together with the findings in both control and HJV-HepG2 cells, our results imply that neogenin dictates the extent of the induction of hepcidin expression only when it is associated with a functional HJV that can bind BMPs.
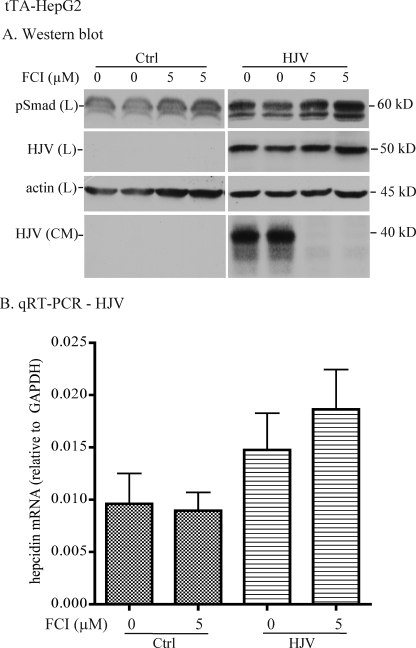
Blockage of HJV Release by a Furin Convertase Inhibitor Does Not Suppress BMP4-induced Hepcidin Expression
Because neogenin is not necessarily required for HJV trafficking to the plasma membrane (24), the dramatic suppression of BMP4-induced hepcidin expression in HJV-HepG2 cells with neogenin knockdown could result from either the inhibition of HJV release or the lack of HJV-neogenin association. To determine which of these processes is required for the induction of hepcidin expression, we first examined the role of HJV release in BMP4-induced hepcidin expression in tTA-HepG2-HJV cells, in which the expression of HJV was induced by the addition of dox. HJV release is mediated via cleavage by the proprotein convertase, furin (19, 20). tTA-HepG2-HJV cells were incubated with BMP4 to induce BMP signaling in the absence or presence of 5 μm FCI, which blocks the release of HJV. Blockage of HJV release by FCI mildly increased the levels of pSmad protein (Fig. 4A) and hepcidin mRNA (Fig. 4B). This is consistent with previous observations that soluble HJV suppresses BMP-induced hepcidin expression (14, 18). Thus, the suppression of hepcidin expression observed in HJV-HepG2 cells when neogenin expression is down-regulated does not result from the inhibition of HJV release.
FIGURE 4.
Inhibition of HJV release by a furin convertase inhibitor does not suppress BMP4-induced hepcidin expression. A, Western blot analysis. tTA-HepG2 cells stably expressing HJV or transfected with empty vector (Ctrl) were maintained in the absence of dox. At about 48 h after subculturing into 12-well plates, cells were first incubated with MEM, 1 mm pyruvate, 1× NEAA, 1% FCS plus 2 μg/ml dox for 6 h to serum starve the cells and to induce HJV expression. Cells were then incubated in the same medium containing 5 ng/ml BMP4 with or without 5 μm decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone, a FCI, for 18 h. CM was collected, and cell lysates were prepared. About 20% of CM and the total lysate were subjected to SDS-PAGE followed by immunodetection of pSmad1/5/8 (pSmad), HJV, and β-actin in the lysate (L) and HJV in CM. This experiment was repeated three times with consistent results. B, qRT-PCR analysis of hepcidin mRNA. Incubation of tTA-HepG2-control and -HJV cells with FCI and BMP4 was performed in essentially the same manner as described in A. Total RNA was isolated followed by cDNA preparation and qRT-PCR analysis of hepcidin and GAPDH mRNA. The hepcidin mRNA levels are expressed as the amount relative to that of GAPDH in each specific sample. The results are from four separate experiments, and the mean values and the S.D. are presented.
Disruption of the HJV-Neogenin Interaction by a Soluble Neogenin Fragment Suppresses BMP4-induced Hepcidin Expression
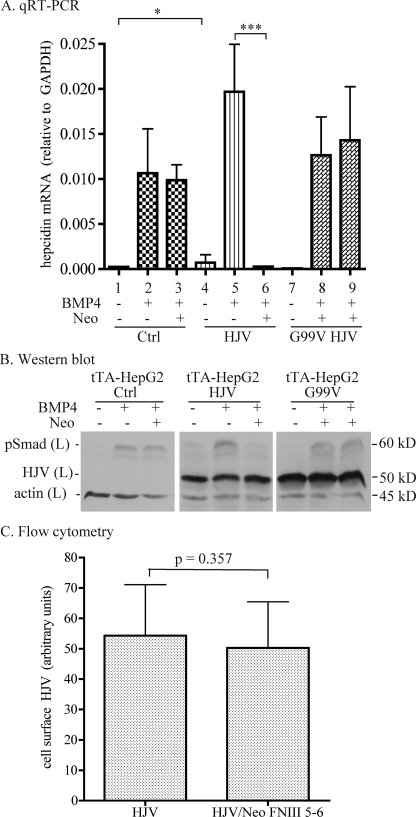
The binding of neogenin to HJV maps to the fibronectin type III 5–6 domain (Neo FNIII 5–6) in the extracellular domain of neogenin (26). HJV can simultaneously bind both BMP2 and neogenin, suggesting that the binding sites do not overlap (26). To determine the effect of the HJV-neogenin interaction on BMP4-induced hepcidin expression, purified Neo FNIII 5–6 fragment was used to disrupt the interaction of HJV with the endogenously expressed full-length neogenin. tTA-HepG2 cells were used as a model system in which the expression of HJV or G99V HJV was under the control of tetracycline-inducible promoter. tTA-HepG2 cells stably transfected with wild type HFE2 (tTA-HepG2-HJV), G99V HFE2 (tTA-HepG2-G99V HJV), or empty vector (tTA-HepG2-control) were maintained in the absence of dox. Because the expression of HJV mRNA in the absence of dox could not be turned off completely in tTA-HepG2-HJV cells, as judged by qRT-PCR, tTA-HepG2-control and tTA-HepG2-G99V HJV cells were used as the controls in this study. The expression of HJV was induced by the addition of dox to the medium 6 h before cells were incubated with Neo FNIII 5–6 and BMP4. Cell lysates were collected 18 h later to analyze the hepcidin mRNA and pSmad protein levels by qRT-PCR and Western blot, respectively.
The addition of Neo FNIII 5–6 to cells suppresses BMP4-stimulated hepcidin expression. Expression of HJV, but not G99V HJV, increased hepcidin mRNA by about 3-fold (1 versus 4, Fig. 5A) and 1.8-fold (2 versus 5, Fig. 5A) without and with addition of 5 ng/ml BMP4, respectively. Upon treatment with both Neo FNIII 5–6 and BMP5, about a 70-fold decrease of BMP4-induced hepcidin mRNA was detected in tTA-HepG2-HJV cells (5 versus 6, Fig. 5A). In contrast, no significant change was observed in tTA-HepG2 control (2 versus 3, Fig. 5A) and G99V HJV (8 versus 9, Fig. 5A) cells under the same conditions. These results are consistent with the finding that HJV is a BMP co-receptor (13) and that Neo FNIII 5–6 disrupts the HJV-neogenin complex that augments BMP signaling.
FIGURE 5.
Soluble neogenin suppresses BMP4-induced hepcidin expression only when HJV is expressed. A, qRT-PCR analysis of hepcidin mRNA. tTA-HepG2 cells stably transfected with empty vector (control), HJV, or G99V HJV were maintained in the absence of dox. At about 48 h after subculturing into 12-well plates, cells were first incubated with MEM, 1 mm pyruvate, 1× NEAA, 1% FCS plus 2 μg/ml dox for 6 h to serum starve the cells and to induce HJV expression. Cells were then incubated in the same medium containing BMP4 at 0 or 5 ng/ml with or without 40 nm Neo FNIII 5–6 (Neo) for 18 h. Total RNA was isolated followed by cDNA preparation and qRT-PCR analysis of hepcidin and GAPDH mRNA. The hepcidin mRNA levels are expressed as the amount relative to that of GAPDH in each specific sample. The results are from four separate experiments and were assessed by one-way analysis of variance. The statistical significant differences relative to the corresponding controls were determined by Tukey's post test. *, p < 0.05; ***, p < 0.001. B, Western blot analysis. Incubation of tTA-HepG2-control, -HJV, and -G99V HJV cells with Neo FNIII 5–6 (Neo) and BMP4 was performed essentially the same as described in A. Total cell lysates (L) were subjected to SDS-PAGE followed by immunodetection of pSmad1/5/8 (pSmad), HJV, and β-actin. This experiment was repeated three times with consistent results. C, flow cytometry analysis of cell surface HJV after incubation with Neo FNIII 5–6. Control- and HJV-HepG2 cells were incubated with or without 40 nm Neo FNIII 5–6 for 18 h. Cells were then detached from flasks with cell dissociation buffer and incubated with affinity-purified rabbit anti-HJV antibody (4 μg/ml) in Hanks' buffer supplemented with 3% fetal bovine serum for 30 min at 4 °C followed by incubation with phycoerythrin-conjugated goat anti-rabbit IgG (1:500 dilution) in the same buffer for 30 min at 4 °C. Flow cytometry analysis was performed on a BD Biosciences FACSCalibur flow cytometer. Rabbit IgG and control-HepG2 cells were used as negative controls. The levels of cell surface HJV are expressed as arbitrary units. The results are from three separate experiments and the mean values and the S.D. are presented. p values are calculated using two-tailed Student's t test to compare the difference between the HepG2-HJV cells incubated with (HJV/Neo FNIII 5–6) and without (HJV) Neo FNIII 5–6.
In agreement with the levels of hepcidin mRNA (Fig. 5A), the addition of Neo FNIII 5–6 also prevented the induction of pSmad signaling by BMP4 in tTA-HepG2-HJV cells but not in tTA-HepG2-control or tTA-HepG2-G99V HJV cells (Fig. 5B). The lack of inhibition in tTA-HepG2-control or tTA-HepG2-G99V HJV cells suggests that Neo FNIII 5–6 does not affect the BMP signaling in the absence of wild type HJV. Given the findings that the HJV-neogenin interaction takes place at the plasma membrane (24), that Neo FNIII 5–6 disrupts the interaction of HJV with full-length neogenin (supplemental Fig. 3), and that HJV can simultaneously bind both BMP2 and neogenin (26), these data suggest that the interaction between HJV and neogenin after it reaches the cell surface is required for the induction of BMP-signaling by BMP4.
To rule out the possibility that the suppressed BMP signaling and hepcidin expression by Neo FNIII 5–6 is caused by the altered HJV trafficking to the plasma membrane, the effect of Neo FNIII 5–6 on cell surface HJV expression was examined. BMP signaling is initiated upon BMP binding to the BMP receptors on the cell surface (15). Our previous studies indicate that knockdown of neogenin does not affect HJV targeting to the plasma membrane (24). HJV-HepG2 cells were incubated in the presence or absence of Neo FNIII 5–6 for 18 h followed by flow cytometry analysis of cell surface HJV (24). Consistent with the neogenin knockdown study (24), Neo FNIII 5–6 did not significantly alter the cell surface HJV levels compared with the corresponding control (Fig. 5C). Together, these results indicate that Neo FNIII 5–6 suppresses BMP signaling and hepcidin expression by disrupting the interaction between full-length neogenin and HJV but not through its participation in HJV cleavage or by affecting its cell surface localization.
Soluble Neogenin Suppresses Hepatic Hepcidin Expression in Vivo
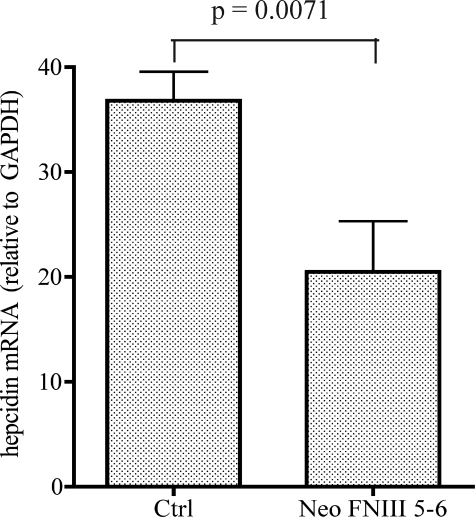
To determine whether the HJV-neogenin interaction is physiologically significant in vivo, we examined the effect of human Neo FNIII 5–6 on hepatic hepcidin expression in mice. Both neogenin and HJV are highly conserved between species. Our previous study showed that knockdown of neogenin in C2C12 cells, a mouse myoblast cell line, suppresses the release of transfected human HJV (6). Because the inhibition of HJV release in HepG2 cells is an immediate process (data not shown), purified Neo FNIII 5–6 at 3 mg/kg body weight was intraperitoneally injected into wild type male 129EvSv mice (8 weeks old) at 16 and 24 h before the collection of liver tissues. Control mice were injected with the same volume of carrier buffer. We used qRT-PCR to examine the levels of hepatic hepcidin mRNA. Injection of Neo FNIII 5–6 significantly suppressed hepcidin mRNA by ∼40% (p = 0.0071) (Fig. 6). These results suggest that interaction of HJV with neogenin is required for the induction of hepcidin expression in vivo.
FIGURE 6.
Soluble neogenin suppresses hepatic hepcidin expression in mice. Eight-week-old male 129EvSv mice were injected intraperitoneally with purified neogenin FNIII 5–6 (Neo FNIII 5–6) at 3 mg/kg body weight or the carrier buffer (Ctrl) twice at 8:30 am and 3:30 pm on the same day. Injected mice had free access to the regular rodent diet. At 24 h after the first injection, animals were euthanized while under anesthesia. Liver tissues were collected for qRT-PCR analysis of hepcidin and GAPDH mRNA. The hepcidin mRNA levels were calculated as the amount relative to that of GAPDH in each specific sample and are expressed as the amount relative to the carrier buffer-injected mice. Each group consisted of three mice. The mean values and the S.D. are presented. p values are calculated using two-tailed Student's t test to compare the difference between the two groups.
DISCUSSION
In this study we characterized the role of neogenin in HJV-regulated hepcidin expression. Results showed that both HJV and neogenin were expressed in liver hepatocytes and evenly detected throughout the liver tissues. Disruption of the interaction between HJV and neogenin by down-regulation of neogenin by siRNA or by the addition of a soluble neogenin fragment that binds HJV markedly decreased BMP4-induced hepcidin mRNA levels in HJV-expressing HepG2 cells. Together with the in vivo study using the HJV binding fragment of neogenin (FNIII 5–6) to disrupt the interaction between HJV and the full-length neogenin, our data support the idea that HJV-neogenin interaction enhances BMP-mediated induction of hepcidin expression.
To determine the role of neogenin in HJV-regulated hepcidin expression, we examined the expression profiles of HJV and neogenin in the liver. A previous report using lacZ protein as an indirect marker showed a selective expression of HJV, a critical upstream regulator of hepcidin transcription (5, 9, 10), in the hepatocytes around the portal triad (10). Here, using more direct methods, we confirmed the predominant expression of HJV in liver hepatocytes. But rather than a selective expression of HJV in hepatocyte cells in the periportal zone (10), our results indicate a uniform localization of both HJV mRNA and protein in hepatocytes throughout the tissue. Distinct from HJV, neogenin mRNA and protein were detected in all of the liver cell populations with the hepatocytes as the predominant source of neogenin. Thus, both HJV and neogenin are co-expressed in liver hepatocytes.
The significant inhibition of hepcidin expression by disrupting the HJV-neogenin interaction in HJV-expressing HepG2 cells was an unexpected result. Previous studies show that the interaction of HJV with neogenin is required for HJV release from hepatoma cell lines (HepG2 and Hep3B cells), a myoblast cell line (C2C12 cells), and HEK293 cells (6, 17, 24). Cellular HJV enhances the transcription of hepcidin through the BMP-signaling cascade by acting as a BMP co-receptor (13, 14), whereas the released soluble HJV is thought to serve as a BMP antagonist to suppress BMP-induced hepcidin expression through competition with membrane-associated HJV for limited BMPs (14, 18, 20). In HepG2 cells, HJV release constitutes the major pathway of cellular HJV turnover (17). Initially, we reasoned that blockage of HJV release by neogenin knockdown or a soluble neogenin would enhance BMP signaling and BMP4-induced hepcidin expression. However, the profound suppression of BMP signaling by both neogenin knockdown and treatment with Neo FNIII 5–6, but not by a furin convertase inhibitor that inhibits HJV release, leads us to conclude that the HJV-neogenin interaction is essential for the induction of BMP signaling and hepcidin expression in hepatocytes. Results from the further studies of G99V HJV, a non-functional form of HJV that causes juvenile hemochromatosis in humans (5), support that neogenin functions to guide the induction of BMP signaling and hepcidin expression only when it is associated with the HJV that binds both full-length neogenin and BMPs.
In contrast to HJV-HepG2 cells, we did not observe any significant effect on hepcidin expression when HepG2 cells transfected with empty vector or G99V HJV were incubated with soluble neogenin. However, we detected a 2-fold decrease of hepcidin expression when the endogenous neogenin in these cells was knocked down. These results imply that neogenin alone mildly enhances BMP signaling. Neogenin is a type I transmembrane protein. Recent studies showed that the neogenin 388-amino acid cytoplasmic domain is involved in various signal transduction pathways when associated with the repulsive guidance molecules a (RGMa) (30–33). The RGM family has three members (RGMa, RGMb, and RGMc). RGMc is the ortholog of HJV in mice. Similar to HJV, both RGMa and RGMb are glycosylphosphatidylinositol-linked proteins, bind neogenin, and are BMP coreceptors. In contrast to HJV, RGMa and RGMb are expressed primarily in the developing and adult central nervous system in distinct, mostly non-overlapping patterns (34–36). On the basis of the results in this study, we speculate that in the tissues that do not express HJV or RGMa or RGMb, neogenin may function to maintain the basal level of BMP signaling through its cytoplasmic domain. In the tissues that express either RGMa or RGMb, neogenin may play a similar role in the induction of BMP signaling as it does in the presence of HJV.
In this and our previous studies (17), we observed an interesting phenomenon. Both neogenin knockdown and treatment with either neogenin whole ectodomain or FNIII5–6 fragments can suppress the HJV release from HJV-HepG2 cells. But only the neogenin knockdown and neogenin whole ectodomain treatment result in the accumulation of cellular HJV. Flow cytometry analysis showed no significant change of cell surface HJV, suggesting that the increased cellular HJV does not result from the HJV accumulation at the cell surface. Immunofluorescence analysis revealed an increased accumulation of HJV in an uncharacterized intracellular compartment when neogenin is knocked down.3 As a result, we speculate that cellular HJV accumulation may result from an altered HJV trafficking.
HJV release is negatively regulated by holo-transferrin and iron (6, 19, 20). The release of HJV from both a myoblast cell line and HepG2 cells depends on the interaction with neogenin (6). Although both HJV and neogenin are expressed in skeletal muscles and the liver (5, 6, 21), only the liver is the predominant site of hepcidin expression (11, 12). On the basis of these observations and the results obtained in this study, we hypothesize that the major function of hepatic neogenin is to augment the HJV-enhanced hepcidin expression through the BMP signaling pathway. We speculate that the major function of neogenin in the skeletal muscles, the major site of HJV expression in the body (5), is to mediate the release of HJV into circulation in response to the body iron status, which indirectly modulates the hepatic hepcidin expression.
In this study we found that the extent of inhibition of hepcidin expression by disrupting the HJV-neogenin interaction is independent of exogenous BMP4 ligand. Our findings are, therefore, not in agreement with the recent report showing that knockdown of neogenin has no evident effect on hepcidin expression in HJV-expressing cells (28). The basis for the difference between our results and theirs is not clear.
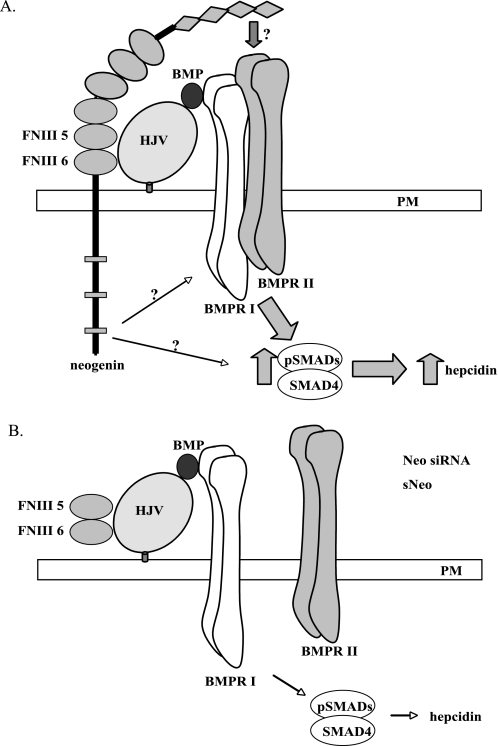
The BMP signaling cascade is initiated by ligand binding to two type I receptors followed by recruitment of two type II receptors to form heterotetramers. Ligand binding induces phosphorylation of the type I receptor by the type II receptor, which leads to phosphorylation of cytoplasmic receptor-activated pSmads. The receptor-activated Smads form heteromeric complexes with Smad4, the central mediator in BMP/Smad signaling, which translocates from the cytoplasm to the nucleus to initiate hepcidin gene transcription (37). We found that the interaction with neogenin is required for cellular HJV processing in our previous studies (17, 24) and that the disruption of the HJV-neogenin interaction suppresses BMP4-induced hepcidin expression in this study. On the basis of these observations, we hypothesize that neogenin may regulate BMP signaling and hepcidin expression through several possible mechanisms. First, neogenin could be directly or indirectly involved in the proper assembly of the BMP receptor tetramer upon the binding of BMP ligand in the presence of HJV. In this scenario, disruption of the HJV-neogenin interactions at the cell surface will lead to the aberrant assembly of BMP receptors with HJV upon the BMP ligand binding and consequently result in the lack of BMP-signaling induction (Fig. 7). Second, neogenin could inhibit the degradation of BMP receptors once associated with HJV. Third, the signal transduction mediated by the neogenin cytoplasmic domain may play an important regulatory role in the induction of hepcidin expression. Understanding the requirement for neogenin in the activation of hepcidin transcription will be the subject of future research.
FIGURE 7.
A model of neogenin in HJV-induced hepcidin expression in hepatocytes. A, HJV-neogenin is required for the proper assembly of HJV-BMP ligand-BMPR I/BMPR II complex to initiate the BMP signaling and to induce hepcidin expression. B, disruption of HJV-neogenin interaction either by neogenin (Neo) knockdown or by soluble neogenin (sNeo) leads to the formation of the aberrant and non-functional HJV-BMP ligand-BMPR I complex. FNIII 5, neogenin fibronectin type III 5 domain. FNIII 6, neogenin fibronectin type III 6 domain. BMPRI, type I BMP receptor. BMPRII, type II BMP receptor.
In addition, two recent studies indicate a critical role of BMP6 in iron homeostasis, presumably through HJV (38, 39). Kautz et al. (40) showed that similar to the hepatic hepcidin expression profile, BMP6 expression in the liver is positively regulated by dietary iron. Therefore, it will also be important to elucidate the role of neogenin in BMP6-regulated hepcidin expression.
In summary, we demonstrated that both HJV and neogenin are uniformly expressed in liver hepatocytes with no distinct gradient of distribution. Our results support the notion that in the presence of HJV, the induction of hepcidin expression by BMP4 requires neogenin.
Supplementary Material
Acknowledgments
We thank Dr. Pamela Bjorkman at Caltech for generously providing us with the neogenin FNIII 5–6 fragment and the neogenin ectodomain, Dr. Gregory Longmore at Washington University for generously giving us tTA-HepG2 cells and a tetracycline-inducible pcDNA4 plasmid, and Julia Maxson, Maja Chloupkova, Gary Reiness, Juxing Cheng, Junwei Gao, and Kristina Nicholson for critical reading of this manuscript and helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants DK080765 (to A.-S. Z.), P50AA11199 (to H. T.), and R24AA12885 (to H. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
A.-S. Zhang and C. A. Enns, unpublished observations.
- HJV
- hemojuvelin
- BMP
- bone morphogenic protein
- CM
- conditioned culture medium
- dox
- doxycycline
- GAPDH
- glyceraldehyde phosphate dehydrogenase
- HSC
- hepatic stellate cells
- neo
- neogenin
- qRT-PCR
- quantitative real-time PCR
- RGM
- repulsive guidance molecule
- tTA
- tetracycline controlled transactivator
- FCI
- furin convertase inhibitor
- MEM
- minimum essential medium
- FCS
- fetal calf serum
- NEAA
- nonessential amino acids
- Neo
- neogenin.
REFERENCES
- 1.Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004) Cell 117,285–297 [DOI] [PubMed] [Google Scholar]
- 2.De Domenico I., McVey Ward D., Kaplan J. (2008) Nat. Rev. Mol. Cell Biol. 9,72–81 [DOI] [PubMed] [Google Scholar]
- 3.Du X., She E., Gelbart T., Truksa J., Lee P., Xia Y., Khovananth K., Mudd S., Mann N., Moresco E. M., Beutler E., Beutler B. (2008) Science 320,1088–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finberg K. E., Heeney M. M., Campagna D. R., Aydinok Y., Pearson H. A., Hartman K. R., Mayo M. M., Samuel S. M., Strouse J. J., Markianos K., Andrews N. C., Fleming M. D. (2008) Nat. Genet. 40,569–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papanikolaou G., Samuels M. E., Ludwig E. H., MacDonald M. L., Franchini P. L., Dubé M. P., Andres L., MacFarlane J., Sakellaropoulos N., Politou M., Nemeth E., Thompson J., Risler J. K., Zaborowska C., Babakaiff R., Radomski C. C., Pape T. D., Davidas O., Christakis J., Brissot P., Lockitch G., Ganz T., Hayden M. R., Goldberg Y. P. (2004) Nat. Genet. 36,77–82 [DOI] [PubMed] [Google Scholar]
- 6.Zhang A. S., Anderson S. A., Meyers K. R., Hernandez C., Eisenstein R. S., Enns C. A. (2007) J. Biol. Chem. 282,12547–12556 [DOI] [PubMed] [Google Scholar]
- 7.De Gobbi M., Roetto A., Piperno A., Mariani R., Alberti F., Papanikolaou G., Politou M., Lockitch G., Girelli D., Fargion S., Cox T. M., Gasparini P., Cazzola M., Camaschella C. (2002) Br. J. Haematol. 117,973–979 [DOI] [PubMed] [Google Scholar]
- 8.Camaschella C., Roetto A., De Gobbi M. (2002) Semin. Hematol. 39,242–248 [DOI] [PubMed] [Google Scholar]
- 9.Huang F. W., Pinkus J. L., Pinkus G. S., Fleming M. D., Andrews N. C. (2005) J. Clin. Invest. 115,2187–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niederkofler V., Salie R., Arber S. (2005) J. Clin. Invest. 115,2180–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pigeon C., Ilyin G., Courselaud B., Leroyer P., Turlin B., Brissot P., Loréal O. (2001) J. Biol. Chem. 276,7811–7819 [DOI] [PubMed] [Google Scholar]
- 12.Frazer D. M., Wilkins S. J., Becker E. M., Vulpe C. D., McKie A. T., Trinder D., Anderson G. J. (2002) Gastroenterology 123,835–844 [DOI] [PubMed] [Google Scholar]
- 13.Babitt J. L., Huang F. W., Wrighting D. M., Xia Y., Sidis Y., Samad T. A., Campagna J. A., Chung R. T., Schneyer A. L., Woolf C. J., Andrews N. C., Lin H. Y. (2006) Nat. Genet. 38,531–539 [DOI] [PubMed] [Google Scholar]
- 14.Babitt J. L., Huang F. W., Xia Y., Sidis Y., Andrews N. C., Lin H. Y. (2007) J. Clin. Invest. 117,1933–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng X. H., Derynck R. (2005) Annu. Rev. Cell Dev. Biol. 21,659–693 [DOI] [PubMed] [Google Scholar]
- 16.Wang R. H., Li C., Xu X., Zheng Y., Xiao C., Zerfas P., Cooperman S., Eckhaus M., Rouault T., Mishra L., Deng C. X. (2005) Cell Metab. 2,399–409 [DOI] [PubMed] [Google Scholar]
- 17.Maxson J. E., Enns C. A., Zhang A. S. (2009) Blood 113,1786–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L., Goldberg Y. P., Ganz T. (2005) Blood 106,2884–2889 [DOI] [PubMed] [Google Scholar]
- 19.Silvestri L., Pagani A., Camaschella C. (2008) Blood 111,924–931 [DOI] [PubMed] [Google Scholar]
- 20.Lin L., Nemeth E., Goodnough J. B., Thapa D. R., Gabayan V., Ganz T. (2008) Blood Cells Mol. Dis. 40,122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang A. S., West A. P., Jr., Wyman A. E., Bjorkman P. J., Enns C. A. (2005) J. Biol. Chem. 280,33885–33894 [DOI] [PubMed] [Google Scholar]
- 22.Zhang A. S., Xiong S., Tsukamoto H., Enns C. A. (2004) Blood 103,1509–1514 [DOI] [PubMed] [Google Scholar]
- 23.Davies P. S., Enns C. A. (2004) J. Biol. Chem. 279,25085–25092 [DOI] [PubMed] [Google Scholar]
- 24.Zhang A. S., Yang F., Meyer K., Hernandez C., Chapman-Arvedson T., Bjorkman P. J., Enns C. A. (2008) J. Biol. Chem. 283,17494–17502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Y., Longmore G. D. (2005) Mol. Cell. Biol. 25,4010–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F., West A. P., Jr., Allendorph G. P., Choe S., Bjorkman P. J. (2008) Biochemistry 47,4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luxon B. A. (2000) in Liver Disease: Diagnosis and Management ( Bacon B. R., DiBisceglie A. M. eds) pp. 3–15, Churchill Livingstone, Philadelphia [Google Scholar]
- 28.Xia Y., Babitt J. L., Sidis Y., Chung R. T., Lin H. Y. (2008) Blood 111,5195–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuns-Hashimoto R., Kuninger D., Nili M., Rotwein P. (2008) Am. J. Physiol. Cell Physiol. 294,C994–C1003 [DOI] [PubMed] [Google Scholar]
- 30.Conrad S., Genth H., Hofmann F., Just I., Skutella T. (2007) J. Biol. Chem. 282,16423–16433 [DOI] [PubMed] [Google Scholar]
- 31.Goldschneider D., Rama N., Guix C., Mehlen P. (2008) Mol. Cell. Biol. 28,4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffar G., Taniguchi J., Brodbeck T., Meyer A. H., Schmidt M., Yamashita T., Mueller B. K. (2008) J. Neurochem. 107,418–431 [DOI] [PubMed] [Google Scholar]
- 33.Matsunaga E., Chédotal A. (2004) Dev. Growth Differ. 46,481–486 [DOI] [PubMed] [Google Scholar]
- 34.Matsunaga E., Tauszig-Delamasure S., Monnier P. P., Mueller B. K., Strittmatter S. M., Mehlen P., Chédotal A. (2004) Nat. Cell Biol. 6,749–755 [DOI] [PubMed] [Google Scholar]
- 35.Babitt J. L., Zhang Y., Samad T. A., Xia Y., Tang J., Campagna J. A., Schneyer A. L., Woolf C. J., Lin H. Y. (2005) J. Biol. Chem. 280,29820–29827 [DOI] [PubMed] [Google Scholar]
- 36.Xia Y., Sidis Y., Mukherjee A., Samad T. A., Brenner G., Woolf C. J., Lin H. Y., Schneyer A. (2005) Endocrinology 146,3614–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samad T. A., Rebbapragada A., Bell E., Zhang Y., Sidis Y., Jeong S. J., Campagna J. A., Perusini S., Fabrizio D. A., Schneyer A. L., Lin H. Y., Brivanlou A. H., Attisano L., Woolf C. J. (2005) J. Biol. Chem. 280,14122–14129 [DOI] [PubMed] [Google Scholar]
- 38.Meynard D., Kautz L., Darnaud V., Canonne-Hergaux F., Coppin H., Roth M. P. (2009) Nat. Genet. 41,478–481 [DOI] [PubMed] [Google Scholar]
- 39.Andriopoulos B., Jr., Corradini E., Xia Y., Faasse S. A., Chen S., Grgurevic L., Knutson M. D., Pietrangelo A., Vukicevic S., Lin H. Y., Babitt J. L. (2009) Nat. Genet. 41,482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kautz L., Meynard D., Monnier A., Darnaud V., Bouvet R., Wang R. H., Deng C., Vaulont S., Mosser J., Coppin H., Roth M. P. (2008) Blood 112,1503–1509 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.