Abstract
The conformations of supercoiled (sc) DNA and linear DNA bound to polylysine (PL)-coated mica were investigated by scanning force microscopy (SFM) in solution. From the polymer statistical analysis of linear DNA, we could distinguish between re-arrangements or trapping of the DNA on the surface. Conditions of re-arrangements to an almost equilibrated state can be achieved at appropriate PL surface concentrations. We could show that the ability of re-arrangements depends on the salt concentration of the adsorption/imaging buffer. Comparing the statistical analysis of the linear DNA with SFM images of scDNA suggested that irregular scDNA conformations are formed under conditions of trapping, whereas plectonemic structures are favoured under conditions of surface re-arrangements. Salt-dependent changes in the scDNA conformation over the range of 10–100 mM NaCl, as characterised by the parameters writhe and the superhelix radius r, are observable only under conditions that enable surface re-arrangements. The measured values of writhe suggest that the scDNA loses approximately one-half of the supercoils during the binding to the surface. At the same time r increases systematically with decreasing writhe, thus the scDNA topology remains determined by the constraints on supercoiling during the binding to PL-coated mica.
INTRODUCTION
DNA supercoiling by a helical winding deficit is a general feature in all organisms and is essential for the three-dimensional organisation of the genome. The tertiary structure of supercoiled (sc) DNA can be important for protein–DNA interactions (1,2) or for specific interactions of distant sites on the DNA by DNA looping (3–6). In order to assess the biological role of supercoiling, small circular scDNA is frequently studied as a model system, e.g. as for analysis of transcription activation by enhancer (5,7). Also, scDNA can help to study the mode of action of DNA-binding proteins altering the DNA conformation (8,9).
scDNA adopts predominantly a plectonemic conformation, in which the DNA double strand is wound around itself (10–14). The helical winding deficit in such a molecule, given by the linking number difference ΔLk, distributes into writhe (Wr), being proportional to the number of supercoils, and twist (Tw), the number of windings of the DNA double helix around its local axis (15). The ratio Wr/Tw depends on the salt concentration, as shown by electron microscopy (EM) (12,14,16). Lowering the ionic strength reduces the Wr/Tw ratio from ∼3:1 at moderate to high ionic strengths (≥50 mM) to ∼1:1 at 10 mM. This change in geometry is connected with an increased electrostatic repulsion between opposing DNA double strands, such that the superhelix radius r increases from ∼5 nm in 100 mM NaCl to ∼8.5 nm in 10 mM NaCl (17,18).
While the tertiary structure of scDNA can be imaged by EM and by scanning force microscopy (SFM) at a high resolution, both techniques require an immobilisation of the molecules on a flat surface, resulting in a transition from three-dimensional to a two-dimensional conformation. Only in cryo-EM, the DNA is immobilised within a vitrified layer and the molecules approximately maintain a three-dimensional state, which is, however, limited by the thickness of the layer. For conventional EM and SFM, the DNA is generally adsorbed on a surface via ionic interactions, so that the salt dependence of the scDNA conformation can critically influence the structure during the adsorption process.
An advantage of SFM over EM is that a sample can be prepared without staining the DNA. Moreover, rinsing with water and drying the sample can be avoided by imaging in solution (13,19). Mica, an atomically flat and negatively charged surface, is usually used in SFM to bind DNA in a buffer supplied with Mg2+ or other divalent ions, which mediate the interaction between the phosphate groups of the DNA and the substrate (20). Mica can also be modified with chemicals carrying positively charged groups, like 3-aminopropyltriethoxy silane (APTES), to make the surface adhesive for DNA (functionalised mica) (21,22). In such cases, the DNA can be bound to the substrate in a wide range of buffer conditions, thereby avoiding the requirement of millimolar amounts of magnesium ions in the adsorption buffer. However, despite the advantages of SFM, enabling a soft sample preparation, scDNA conformations imaged with this technique appear to depend rather strongly on the preparation conditions. While systematic changes in superhelix parameters (e.g. Wr, Tw and the superhelix radius) at varying salt conditions have been observed by cryo-EM and conventional EM (12,14,16), no corresponding SFM measurements are reported. scDNA prepared with the magnesium-mediated adsorption technique for SFM in air tends to lose supercoils and can even assume open circular conformations, similar to relaxed DNA (16,23–25). This depends on the incubation time before rinsing the sample with water and could in part be caused by water rinsing (16,24). Cherny and Jovin (16) have also demonstrated that the addition of KCl to the deposition buffer (10 mM HEPES, pH 7.5, 5 mM MgCl2) can lead to even more open conformations, against the expected salt effect in scDNA. In contrast, scDNAs deposited on mica treated with APTES (AP-coated mica) and imaged in solution or in air, displayed the expected extended, tightly interwound conformation in 20 mM Tris, pH 7.6, 161 mM NaCl, but the majority of molecules tended to adopt folded shapes with no defined superhelix axis on this substrate in 10 mM NaCl (21). Although these latter conformations clearly reflect an expected irregularity and a more loose interwinding at low ionic strength (18), they may be anomalous, since they lack the plectomemic appearence. Fujimoto and Schurr (26) conducted Monte Carlo computer simulations of the surface binding of scDNA with the same length and at the same ionic conditions as in Lyubchenko and Shlyakhtenko (21). They found a good agreement with the SFM experiment at 161 mM ionic strength; also, at low salt concentrations, plectonemically interwound conformations were predicted. In these simulations, reversible DNA–surface interactions were assumed, and the molecules could achieve an equilibrated two-dimensional state. The segmental mobility of scDNA imaged on AP-coated mica in solution (21) together with the qualitative agreement between experiment and simulation at 161 mM ionic strength, could indicate a capability of surface re-equilibration as well. Otherwise, it has been argued that the tendency to form open circular conformations on untreated mica could be a result of surface re-equilibration, such that plectonemic scDNA can be imaged only under conditions of trapping on pre-treated mica (25).
In the present work, the relationship between the DNA–surface interaction strength, the salt concentration and the two-dimensional geometry of scDNA is investigated by SFM in liquid using polylysine (PL) to functionalise mica. In EM, glow-discharged carbon surfaces coated with PL are routinely used to adsorb DNA (10,16,27).
The ability of surface re-equilibration can be characterised quantitatively by means of linear DNA, whose conformation can be described statistically in both three or two dimensions by the worm-like chain (WLC) model (28–30). Equilibrated linear DNA can move freely on the surface and should behave like an ideal two-dimensional coil, which can be verified by determining the mean square end-to-end distance <R2> of imaged DNA paths. Kinetic trapping is the opposite extreme case of an adsorption process and is caused by irreversible DNA–surface interactions. This results in a collapsed conformation similar to a projection of the three-dimensional structure.
Here, PL-coated mica was used to adsorb and image comparatively scDNA and linear DNA by SFM in solutions containing different concentrations of NaCl. We investigated whether the adhesive force between the surface and the DNA could be controlled by treating the mica with different concentrations of the polypeptide PL. To test for surface equilibration or kinetic trapping, the linear DNA was analysed assuming the WLC model. These data were used to discriminate between structural alterations in scDNA due to varying salt concentrations in the adsorption/imaging buffer and due to the strength of interaction with the surface. The superhelix parameters Wr and the superhelix radius r were measured according to Boles et al. (10) to analyse quantitatively changes in the scDNA conformation.
MATERIALS AND METHODS
Analysed DNA
In all experiments, a derivative of pUC18 [pUC18-1A-H1/2 (4)] with a size of 2497 bp was examined. The plasmid DNA was amplified in Escherichia coli Nova Blue cells and isolated using the Plasmid Maxi Kit (Qiagen, Hilden, Germany). A major fraction of open circular DNA was removed by HPLC using a reverse-phase column (RP-18; Merck, Darmstad, Germany) (31). The scDNA was analysed by electrophoresis in a 1% agarose gel containing 1 µg ml–1 chloroquine to determine the distribution of topoisomers and their ΔLk values relative to a pUC18 topoisomer standard (32). Band quantification was done with the gel analysis option of NIH-Image (version 1.62). The scDNA had an average ΔLk = –12.3 (calculated by fitting a Gaussian function to the observed topoisomer distribution) corresponding to a superhelical density of σ = –0.052. The prepared plasmid DNA was also used to produce linear DNA fragments by cleavage with PvuI restriction endonuclease. This enzyme converts the DNA into two fragments, 627 and 1870 bp in size.
SFM imaging
SFM images were acquired with a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) in tapping mode. For imaging in solution, sharpened silicon nitride tips (type NP-S; Digital Instruments) were used at drive frequencies of 8.0–9.5 kHz. SFM was also done in air using etched silicon tips (type NHC; Nanosensors, Neuchatel, Switzerland), operated at drive frequencies of ∼280 kHz. The images were recorded both in solution and in air at a scan diameter of 2 × 2 µm, a scan rate of 2–3 Hz and a resolution of 512 × 512 pixels.
Preparation of PL-coated mica
A mixture of PLs with molecular masses of 500–2000 (Sigma, Deisenhofen, Germany) corresponding to ∼4–15 lysine residues per polypeptide was used for coating mica. Twenty microlitres of an aqueous PL solution (1 µg ml–1–1 mg ml–1) were dropped on freshly cleaved mica. After incubation for 30 s, the mica was rinsed with 4 ml of millipore purged water, the water was soaked up, and finally, the mica was dried in a nitrogen stream. The dried PL-coated mica was immediately used for an SFM experiment.
Sample preparation
For imaging in solution, the DNA in its supercoiled or linearised form was dissolved at a concentration of 0.5 nM in 10 mM HEPES, pH 8.0, supplemented with 5–100 mM NaCl (buffer I). The PL-coated mica was pre-incubated with a 30 µl drop of the corresponding buffer without DNA. Subsequently, the SFM liquid cell (Digital Instruments) with a mounted SFM tip and wet with another 30 µl of the buffer, was placed in the instrument. The cell was approached to the surface manually, until a water column between the mica and the liquid cell was established. Then, the solution was substituted with 60 µl of the DNA solution. After a few minutes, depending on the PL surface concentration, stable images were obtained.
In a control experiment, the linear DNA was imaged on untreated mica initially with the same procedure now using 10 mM HEPES, pH 8.0, 10 mM NaCl, 2 mM MgCl2 (buffer II) as a deposition buffer. Linearised DNA was allowed to bind and equilibrate on the surface for 20 min, after which the solution in the liquid cell was substituted with fresh buffer II. Then, the solution was substituted by 10 mM HEPES, pH 8.0, 10 mM NaCl, 2 mM NiSO4 to fix the DNA on the mica and image it in solution.
The linear DNA was also imaged in air by dropping 20 µl of ∼5 nM DNA in buffer I, 50 mM NaCl on PL-coated mica. After 5 min of incubation, the sample was rinsed with water and dried carefully.
Image analysis
Linear DNAs. Polymer chain statistical analysis was used to distinguish between trapping and equilibration of linear DNA fragments on the surfaces, essentially as described (29). The mean square end-to-end distance <R2> is related to the DNA persistence length P and the contour length L according to equation 9 in Rivetti et al. (29). Kinetically trapped DNA features a reduction of <R2> as compared with the theoretical value for a complete equilibration according to equations 10 and 11 in Rivetti et al. (29).
Using the NanoScope IIIa software (version 3.0) the images were flattened and zoomed to a size of 500 × 500 nm, such that one pixel corresponded to ∼1 nm. The DNA paths were converted to xy coordinates by manually tracing the DNA contours using NIH-Image (version 1.62). Using a self-written program [Thetascan (version. 1.0) run in Origin (version 7.0)] the digitised paths were smoothed twice by calculating a correction for each xy coordinate Pi, given by the weighted average of five contiguous coordinates:
Picorrected = (1Pi–2 + 2Pi–1 + 4Pi + 2Pi+1 + 1Pi+2) / 10 1
It was shown by analysing simulated DNA paths that this smoothing procedure is suitable for removing the curvatures due to pixelisation while keeping levelling of the real curvatures to a minimum (Mücke,N., Kreplak,L., Kirmse,R., Wedig,T., Herrmann,H., Aebi,U. and Langowski,J., manuscript in preparation). It follows that two pixels at the ends of the paths are removed in each smoothing step, leading to a shortening of the evaluated paths. These were analysed to determine <R2> using the program Thetascan. Only molecules with a contour length within two standard deviations of the average length of all fragments were included, where different preparation methods were treated separately.
scDNA. The scDNAs were analysed according to the procedure reported in Boles et al. (10) to determine the superhelix parameters Wr, the superhelix pitch angle α and the superhelix radius r. The described model considers an idealised superhelix with a uniform winding along the surface of a cylinder with radius r and the end loops represented by semi-circles with the same radius r. α and r are calculated from the measurable quantities: the superhelix axis length l, the number of visible crossover points defined as the node number n, and the number of end loops E . l was measured by manually tracing the superhelix axis using the Nanoscope IIIa software (version 3.0) after flattening and zooming. Only those molecules were analysed which display a well distinguishable superhelix axis and number of nodes.
RESULTS
General properties of PL-coated mica
The flatness of mica treated with PL, as described in Materials and Methods, was the same as that of untreated mica, independently of the amount of PL, as verified by measuring roughness values using the NanoScope IIIa software on images recorded at constant scanning parameters (data not shown). A stable probing of the DNA in solution required at least ∼3 or ∼5 µg ml–1 PL in deposition buffers containing 10 or 25–100 mM NaCl, respectively (incubating mica with PL for 30 s before rinsing and drying). For probing DNA in air, the minimal PL concentration was ∼1 µg ml–1 (analysed by deposition from buffer I, 50 mM NaCl). A comparatively small PL size (a mixture of 4–15 lysine residues per polypeptide) was chosen in order to ensure an even charge density on the surface, in particular at low PL concentrations. This was expected to minimise possible influences on the DNA structure at the surface–solution interface and to yield a flat surface. On the other hand, the binding of small PL chains to mica is weaker than for longer chains, so that it might be possible that PL dissociates from the surface, binds to DNA and thereby changes its structure. We believe that the following observations indicate that the low molecular mass PL is stably bound to mica, even though a small portion may dissociate slowly. (i) Even when mica was incubated with the lowest PL concentration sufficient for DNA immobilisation and then vigorously rinsed under a continuous flow of water for ∼1 min, DNA still binds to the surface and can be imaged. This was verified by SFM in solution using 5 µg ml–1 PL and deposition from buffer I, 100 mM NaCl. (ii) When dried mica treated with a high PL concentration (1 mg ml–1) was pre-incubated with deposition buffer I for ∼1 h before DNA addition, we found a few intermolecularly linked scDNAs, although crowding of the surface with DNA was low. In this case, some PL released into free solution might have caused these aggregates. However, when we used the preparation procedure described in Materials and Methods, where the PL-coated mica was pre-incubated only for the time required to make the instrument settings, intermolecularly linked molecules were not observed at similar DNA surface densities.
Analysis of linear DNAs
Figure 1 shows SFM micrographs of the 627 and 1870 bp fragments imaged in solution at different conditions. When fragments were deposited on mica treated with the lowest PL concentrations that still enabled stable scanning at a given salt concentration, we observed extended conformations without self crossings (Fig. 1B). Such conformations indicate a pronounced capability of surface re-arrangements, comparable with fragments deposited on bare mica with Mg2+ and imaged by SFM in air, where surface equilibration was shown (29). Qualitatively, we observed similar conformations by binding the DNA to bare mica in the same ionic environment as in Rivetti et al. (29) for 20 min (buffer II), subsequent fixation with Ni2+ and imaging in solution (Fig. 1C). On the other hand, the fragments adopted more bent conformations, which frequently exhibit self crossings, on mica treated with a high PL concentration (1 mg ml–1) (Fig. 1A). In Rivetti et al. (29), such shapes were shown to be a consequence of trapping on differently pre-treated mica surfaces, i.e. the DNA becomes unable to move on the surface after the initial contact. Alternatively, looped structures could be stabilised by dissociated PL when it is used at a high concentration. However, strongly bent structures, similar to those shown in Figure 1A, were also observed on mica treated with only 5 µg ml–1 PL and deposition from 10 mM NaCl. In 50 mM NaCl, the fragments were clearly extended at this PL concentration. This difference is explained by enhanced DNA–surface interactions at low salt concentrations, where the negative charges of the DNA are less shielded.
Figure 1.
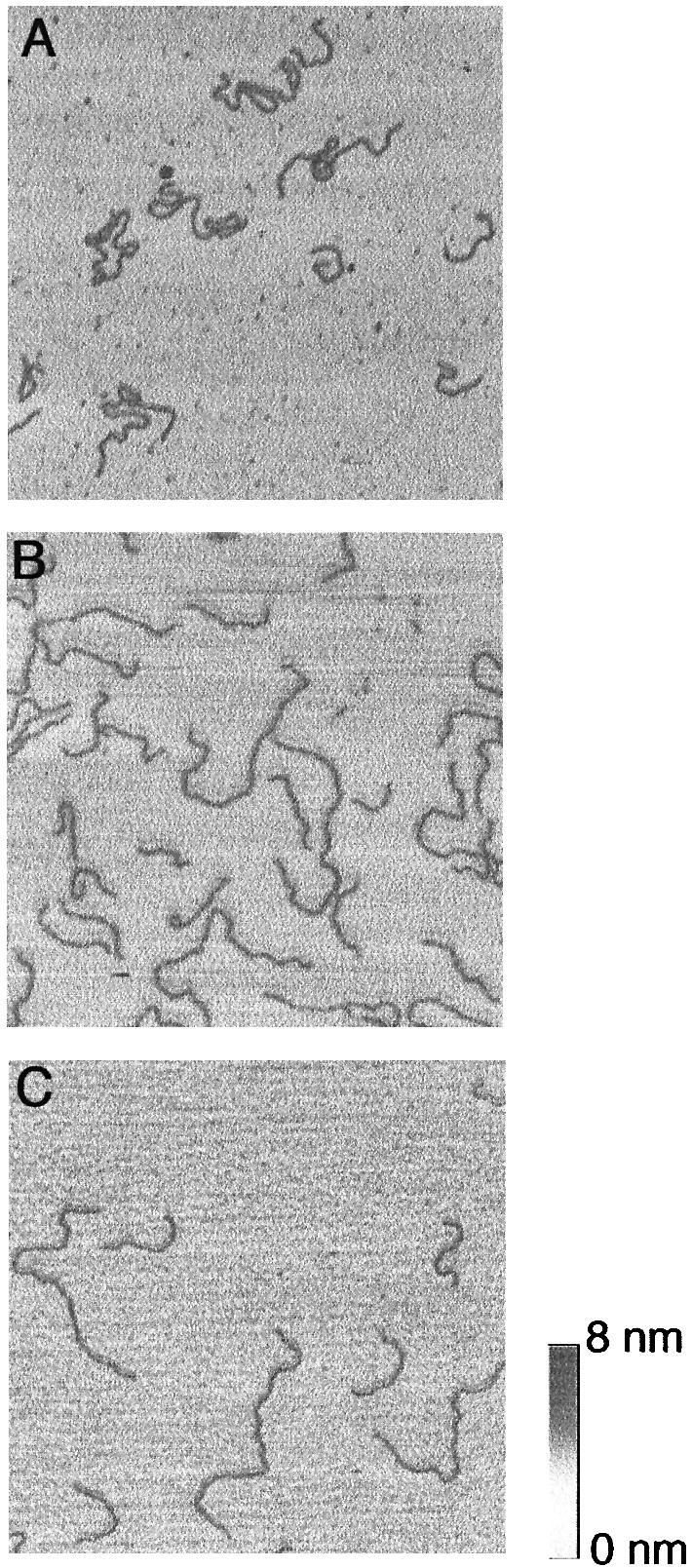
SFM images recorded in liquid of linear DNA fragments with sizes of 627 and 1870 bp. (A) Deposition of the DNA from 10 mM HEPES, pH 8.0, 50 mM NaCl on mica treated with 1 mg ml–1 PL. (B) Deposition of the DNA from 10 mM HEPES, pH 8.0, 10 mM NaCl on mica treated with 3 µg ml–1 PL. (C) Deposition of the DNA from 10 mM HEPES, pH 8.0, 10 mM NaCl, 2 mM MgCl2 on untreated mica, incubation for 20 min and subsequent fixation of the DNA by exchange of the scanning solution with 10 mM HEPES, pH 8.0, 10 mM NaCl, 2 mM NiSO4. The image frames are zooms representing 1 × 1 µm.
Only the shorter 627 bp fragment was analysed statistically in all samples, because the paths and the ends of the larger fragment were not always distinguishable at conditions causing the highly bent conformations. The average contour length of the 627 bp fragment was 203.8 ± 9.2 nm, as measured in the whole set of images on PL-coated mica recorded in solution and corrected for the estimated length of the eight pixels removed after smoothing. This is 4.4% shorter than the expected contour length, assuming a helical rise of 0.34 nm/bp. No systematic length difference among preparations with varying surface concentrations of PL was noticeable, suggesting that PL did not influence the contour length (Table 1). The slight shortening results most probably from the fact that sometimes bends in very short sections in the DNA are not detected due to the thickness of the imaged DNA.
Table 1. Mean square end-to-end distances of the 627 bp fragment at different adsorption/imaging conditions.
| [PL] (mg ml–1) | Deposition conditionsa | L (nm)b | <R2> (nm2)c | Number of molecules | |
|---|---|---|---|---|---|
| TIF | 1.0 | 10 mM NaCl | 200 | 6300 | 75 |
| 1.0 | 50 mM NaCl | 195 | 7500 | 135 | |
| 0.005 | 10 mM NaCl | 197 | 8300 | 97 | |
| 0.005 | 50 mM NaCl | 188 | 16 400 | 68 | |
| 0.003 | 10 mM NaCl | 197 | 16 900 | 94 | |
| 0 | 10 mM NaCl, 2 mM MgCl2 | 203 | 19 600 | 77 | |
| TIA | 0.001 | 50 mM NaCl | 198 | 18 600 | 50 |
TIF, tapping in fluid; TIA, tapping in air.
aThe DNA was bound to the surfaces in 10 mM HEPES, pH 8.0 plus the given salt conditions.
bThe contour lengths L represent the average length of the DNAs traced and smoothed as described in Materials and Methods (note that eight pixels are removed from the original paths by smoothing).
cThe theoretical <R2> for a complete surface equilibration of the 627 bp fragment is equal to 22 200 nm2 [calculated using equation 9 in Rivetti et al. (29) for the average contour length of the smoothed DNA paths of all samples (197 nm) and a DNA persistence length of 50 nm]. The theoretical value for trapping is 9600 nm2 [calculated using equations 10 and 11 in Rivetti et al. (29)].
The measured mean square end-to-end distances <R2> (Table 1) reflect the visually apparent differences in the shape of the imaged fragments under different adsorption conditions (Fig. 1). The <R2> value from the control experiment with untreated mica and deposition with Mg2+ is 1.1-fold smaller than the theoretical value for perfect equilibration [22 200 nm2, based on a DNA persistence length of 50 nm (29,33)]. An analysis of the longer 1870 bp DNA imaged in this experiment yielded an <R2> value of 123 200 nm2, which is higher than the theoretical value for equilibrated DNA of this length (105 100 nm2). This is probably due to excluded volume effects, which are more pronounced in longer fragments. Such effects should appear only in the case of free movements on the surface, expected for the chosen adsorption conditions (29). Therefore, the <R2> value of the 627 bp fragment represents most likely the correct reference value for a surface equilibration. The measured <R2> values correlate with the salt dependence of the DNA conformation on mica at low PL concentration (5 µg ml–1): in 50 mM NaCl, <R2> was slightly reduced compared with the measured reference value (1.2-fold). This confirms surface re-arrangements, since <R2> is significantly higher than expected for a trapping mechanism: from equations 10 and 11 in Rivetti et al. (29), a 2.3-fold reduction would be expected. In 10 mM NaCl, on the other hand, re-arrangements are completely inhibited, as is indicated by an <R2> value 2.4-fold lower than the reference value. Thus, under these adsorption conditions, trapping prevails. Further, re-arrangements are also seen in 10 mM NaCl when the PL concentration is reduced to 3 µg ml–1. When mica treated with a high PL concentration (1 mg ml–1) was used, <R2> was reduced even more than theoretically expected for trapping (e.g. 3.1-fold in 10 mM NaCl). At these extreme conditions, other influences in addition to trapping must be taken into account; one possibility could be that noticeable amounts of dissociated PL lead to an increased compaction of the DNA.
In summary, the measurements show that (i) surface re-arrangements of the DNA are possible even at the lowest PL concentrations necessary for imaging the DNA in solution and (ii) changing from higher to low salt concentration can cause a switch between surface re-arrangements and complete trapping at a constant PL concentration. It is important to note that the persistence length of DNA does not change in the analysed range of salt concentrations (33). Finally, we assume in the following that the degree of re-arrangements is similar for all moderate salt concentrations (25–100 mM NaCl), since the lowest PL concentration sufficient for a stable DNA immobilisation was approximately the same in this concentration range (5 µg ml–1, only in 25 mM NaCl could loosely bound DNA be observed when using 3 µg ml–1 PL).
The DNA could be imaged by SFM in air at an even lower PL concentration of 1 µg ml–1. This yielded an <R2> value of the 627 bp fragment exceeding the highest values obtained by SFM in solution. However, the value was still below the theoretical value for equilibration (1.2-fold) but in good agreement with the reference value measured on naked mica. The apparent persistence length derived from the <R2> value obtained on naked mica using equation 9 of Rivetti et al. (29) is 36 nm.
Characterisation of the imaged scDNA
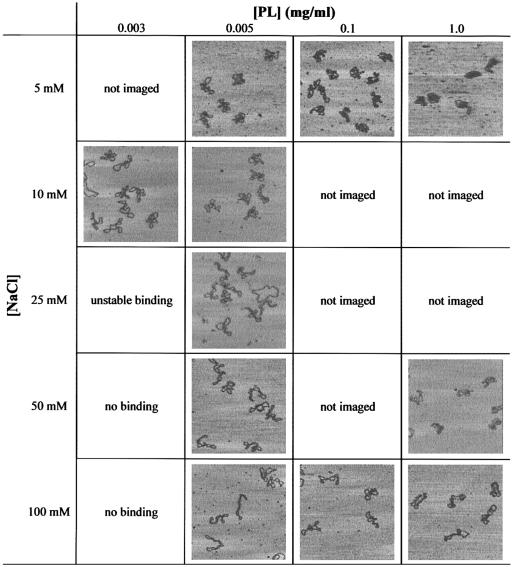
The characteristics of the 2497 bp scDNA as a function of salt concentration and surface concentration of PL are shown in Table 2. When using 5 µg ml–1 PL at 25–100 mM NaCl, the molecules assumed predominantly extended, interwound conformations. Lowering the salt concentration to 5 or 10 mM NaCl at the same PL amount showed an obvious change in the scDNA geometry, since the molecules became almost invariably irregular and folded with no defined superhelix axis. This change coincides with the inhibition of re-arrangements at low salt concentration, as shown by the analysis of the linear DNAs at the same PL concentration (see Table 1). A significant number of the scDNAs (a total of 55%) became plectonemic, thereby more loosely interwound in 10 mM NaCl, when the PL concentration was reduced to 3 µg ml–1. This structural alteration also parallels the behaviour of the linear DNA, since re-arrangements could be re-enabled in 10 mM NaCl by reducing the PL concentration from 5 to 3 µg ml–1. Increasing the PL concentration up to 1 mg ml–1 had the effect of a stronger compaction of the scDNAs at low ionic strength, while they were also irregular but less compacted in 50 mM NaCl. In 100 mM NaCl, extended conformations were retained at up to 1 mg ml–1 PL, but were not regularly interwound.
Table 2. scDNA conformations as imaged by SFM in solution at different NaCl concentrations and different surface concentrations of PL.
The image frames shown correspond to a scan size of 1 × 1 µm. The height scale corresponds to Figure 1.
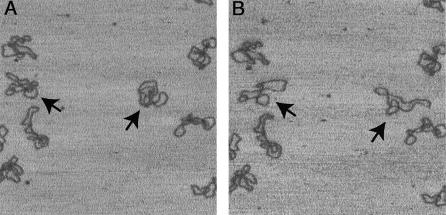
The time-lapse experiment shown in Figure 2 may confirm the salt-dependent ability of surface re-arrangements of scDNA on PL-coated mica. scDNA was adsorbed in buffer I, 10 mM NaCl on mica treated with 5 µg ml–1 PL, i.e. under conditions causing folded and compacted scDNA conformations. After ∼10 min, the imaging buffer was changed to 100 mM NaCl, during which the same area as directly before the buffer exchange was scanned continuously. Two molecules in the shown image frame underwent a global conformational change to a more extended shape and others showed a segmental mobility. The DNA does not appear smeared, indicating that it was not displaced by the scanning tip. However, the resulting conformations are not typical for higher salt concentrations such as those shown in Table 2, presumably because they originate from the initially trapped molecules.
Figure 2.
Observation of conformational changes of scDNA after buffer exchange. In (A) the scDNA was adsorbed and imaged in 10 mM HEPES, pH 8.0, 10 mM NaCl on mica treated with 5 µg ml–1 PL. In (B) the same area as in (A) was recorded directly after the scanning solution was exchanged with 10 mM HEPES, pH 8.0, 100 mM NaCl. The molecules marked by arrows show pronounced conformational changes. The scan size of the images and the height scale correspond to Figure 1.
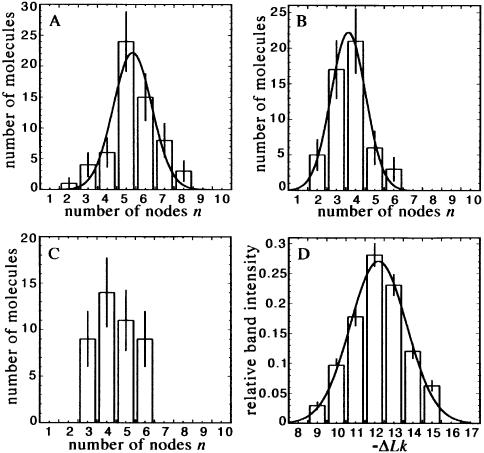
Next, the imaged superhelix conformations were analysed quantitatively in order to characterise changes in the two-dimensional geometry in terms of Wr and the superhelix radius r. Here, those molecules were not taken into account which either appeared too irregular or exhibited regions of close contacts of the opposing DNA strands where the nodes could not always be distinguished. The latter feature is reported in other SFM studies of scDNA as well (13,21). Under conditions of surface re-arrangements, the observed distribution of node number n of the evaluated molecules was nearly Gaussian and comparable with the topoisomer distribution determined by gel electrophoresis (Fig. 3). Therefore, we assumed that the occurrence of close contacts is not correlated with the superhelical density, so that an exclusion of some molecules from the analysis will affect the measurement of the superhelix parameters Wr and r only to a small degree.
Figure 3.
Distribution of counted node number n compared with the distribution of topoisomers, as determined by gel electrophoresis. The node numbers were counted in scDNA imaged at (A) 100 mM NaCl, 5 µg ml–1 PL, (B) 10 mM NaCl, 3 µg ml–1 PL and (C) 100 mM NaCl, 1.0, 0.1 mg ml–1 PL (combined). (D) Relative band intensities of the plasmid topoisomers calculated with a gel quantification option in NIH-Image. Data fitting was done assuming a Gaussian function (results for the node number distributions are given in Table 3, the average ΔLk determined by fitting to the measured band intensities is –12.3 with a standard deviation of 1.4).
Table 3 reports the superhelix parameters, showing that under conditions of surface re-arrangements, Wr and r change systematically with the salt concentration. Thus, binding to PL-coated mica reproduced the expected change of the scDNA conformation from tightly to more loosely interwound shapes, when the salt concentration was reduced from 100 to 10 mM. On the other hand, the absolute values differ significantly from expectation: e.g. at 100 mM NaCl, Wr was only approximately one-half that expected for the analysed plasmids [Wr = –8.2 for ΔLk = –12.3 (10,12,14)] and r was 2.6-fold higher than the value of 5 nm predicted by simulations or measured by solution scattering (17,18). The measured superhelix pitch angles α were 54° (50 and 100 mM NaCl), 50° (25 mM NaCl) and 51° (10 mM NaCl), in good agreement with other SFM or EM measurements (10,13). The average superhelix axis lengths l were 39% (100 and 50 mM NaCl) and 38% (25 and 10 mM NaCl) of the total contour length L of the plasmid, which is close to 41%, measured previously by EM (10).
Table 3. Superhelix parameters measured by SFM in liquid.
| [NaCl] (mM)a | [PL] (mg ml–1) | Superhelix radius r (nm)b | –Wrc | Average number of nodes <n> | Number of molecules |
|---|---|---|---|---|---|
| 100 | 0.005 | 12.9 ± 3.8 | 4.4 ± 1.1 | 5.4 ± 1.0 | 61 |
| 50 | 0.005 | 13.7 ± 3.5 | 4.1 ± 0.8 | 5.0 ± 0.9 | 62 |
| 25 | 0.005 | 15.0 ± 3.9 | 3.9 ± 1.1 | 5.0 ± 1.4 | 92 |
| 10 | 0.003 | 17.6 ± 4.7 | 2.9 ± 0.8 | 3.7 ± 0.9 | 52 |
| 100 | 0.1, 1.0 (combined) | 14.9 ± 4.6 | 3.6 ± 1.0 | 4.5 ± 1.0 | 46 |
aThe scDNA was adsorbed and imaged in 10 mM HEPES, pH 8.0 plus the given salt concentration.
bCalculated using equation 4 in Boles et al. (10) using the measured values n, l and E.
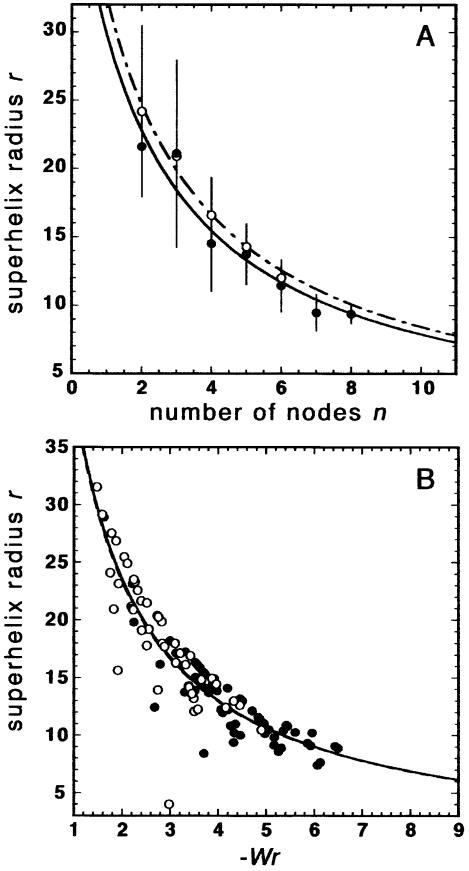
In Figure 4A it is visible that the average r, calculated separately for all molecules with a given node number n, depends on n in a systematic manner. A similar dependence is observed when the r values of individual molecules are plotted against their Wr values (Fig. 4B). Following figure 12 in Boles et al. (10), where r determined by EM is plotted against the average superhelical density σ, regression lines were fitted to the data points in Figure 4A and B. These lines are derived from the linear dependence of 1/r against σ (10). It is noticeable that the curves for 10 and 100 mM NaCl are very close in Figure 4A and overlay each other in Figure 4B (also the curves for 25 and 50 mM, data not shown). Thus, the data points for each salt concentration are located within a certain region along the same curve (this is less obvious in Fig. 4A, since the information about the frequency of molecules with a distinct node number is hidden). These observations point to a systematic coupling of Wr and r in the scDNAs bound to PL-coated mica.
Figure 4.
Dependence of the superhelix radius r on the counted node number n (A) and Wr (B). (A) For all molecules with a given node number n, the average r was calculated and plotted against n. (B) Plot of r against Wr of individual molecules. The fitting curves are derived from a linear function given by a plot of 1/r against n or Wr. In (A) and (B), measurements in 100 and 10 mM NaCl are indicated by closed circles, continuous line and open circles, dashed line, respectively.
For conditions inhibiting re-arrangements (PL concentrations of 0.1 and 1.0 mg ml–1 at all salt concentrations or of 5 µg ml–1 at 10 mM NaCl), a quantitative analysis was not possible due the irregular shapes. However, a few molecules could be evaluated that were bound to mica treated with 0.1 and 1.0 mg ml–1 PL in 100 mM NaCl. The data for both PL concentrations were pooled for better statistics (a separate analysis yielded similar results with poorer statistics). Wr and r differed more from the expected values (Table 3) and the distribution of node number was not Gaussian (Fig. 3C), indicating significant distortions of the scDNA conformation. It is clear that a quantitative evaluation under these conditions is artificial because the few selected molecules appearing regularly interwound are not representative for the majority of structures.
DISCUSSION
Equilibration and trapping of DNA on PL-coated mica
For SFM in liquid, a controlled adsorption of DNA on pre-treated mica can be critical, since probing DNA in liquid requires relatively strong DNA–surface interactions as compared with SFM of dried samples. For analysis of DNA by SFM, the flatness of the surface support is an additional critical factor, which limits the number of usable surfaces. We observed that PL treatment does not increase the surface roughness. PL-coated mica enabled a controllable DNA adsorption and imaging in solution in such a way that conditions of pronounced surface re-arrangements can be adjusted. Furthermore, it was shown that the ability of re-arrangements can depend on the ionic strength of the deposition buffer. In controls using either untreated mica for scanning in solution or using a very low PL concentration (1 µg ml–1) and scanning in air, we expected a complete equilibration. This is known for untreated mica at the deposition conditions used (29). For SFM in air, a surface equilibration is probable, because at the PL concentration used no DNA can be detected by SFM in solution. This suggests that the surface mobility is sufficiently high before final immobilisation by water rinsing and drying. The <R2> values obtained in both experiments were still below the theoretical value for a complete equilibration. Whether specific sequence properties of the 627 bp contribute to this reduction of <R2> could be the basis of a more systematic study but falls outside the subject of this work. A comparative analysis of the longer 1870 bp fragment imaged on untreated mica indicated excluded volume effects, since the <R2> value was higher than the corresponding theoretical value for equilibrated DNA. Excluded volume effects require free movements on the surface, so that the <R2> value measured for the 627 bp DNA in this experiment is accepted as the reference for a surface equilibration. An <R2> value smaller than theoretical expectation might be interpreted as a deviation from the ideal two-dimensional random walk assumed in the WLC model, provided that the assumed DNA persistence length of 50 nm is correct. This deviation could be caused by sequence- dependent features, such as unusual local flexibility or curvature. Further, temporary circularisation events between the AT overhangs produced by the restriction endonuclease used to cleave the plasmid DNA may have affected the equilibrium conformation of the DNA. On the other hand, the persistence length value of 36 nm calculated from the measured <R2> agrees with a reported SFM measurement where a similar protocol for imaging DNA in solution on naked mica was used (34). This agreement suggests an influence of Ni2+ used to fix the DNA after the surface equilibration. Ni2+ may cause an apparent increase of the DNA flexibility. PL on the surface could affect the DNA in a similar way, which would explain the slightly reduced <R2> value measured by scanning in air.
The <R2> values observed from images recorded in solution at minimal PL and appropriate salt concentration are slightly lower (∼1.2-fold) than those from the control experiments. This could be due to an incomplete surface equilibration and/or to the PL interacting with the DNA. A surface equilibration is suggested by frequently observed movements of DNA molecules between scans, including changes of the overall shape of molecules, as shown in Figure 2. The DNA mobility is expected to be enabled by reversible interactions between DNA and PL at low density on the mica. Nevertheless, an additional mobility of PL with bound DNA is conceivable, which could contribute to DNA movements to some degree.
We also observed that under conditions inhibiting surface re-arrangements, the <R2> values of the 627 bp DNA could become smaller than theoretically expected for the trapping mechanism according to equations 10 and 11 in Rivetti et al. (29). Taking the measured reference value of 19 600 nm2 for equilibration, the measurement in 10 mM NaCl and a low PL concentration of 5 µg ml–1 (see Table 1) agrees well with the theoretical 2.3-fold reduction of <R2> by trapping. The values measured at a very high PL concentration (1 mg ml–1) are lower, suggesting that PL at a high surface concentration influences the deposition process beyond simple trapping. Here, it is possible that a noticeable fraction of PL was released into free solution and lead to more compacted conformations. We cannot distinguish at the moment quantitatively such an influence from the possibility that the adsorption proceeds in an unexpected way because of a very high DNA–surface interaction energy. An effect of PL at a high concentration is also suggested by comparing the superhelix structures at low salt concentration in Table 2. The DNA appears to become increasingly compacted by increasing the PL concentration, although re-arrangements are already completely inhibited when 5 µg ml–1 PL is used. The following characterisation of the scDNA will focus on structures observed at low PL concentrations.
Alterations in scDNA structure
Like linear DNA, the conformation of the two-dimensional scDNA also changed significantly depending on the adsorption conditions. By comparing the polymer statistical analysis with SFM images of the scDNA, it can be concluded that plectonemic conformations are favoured under conditions of surface re-arrangements, while trapping leads to more irregular or folded conformations. Using a PL surface concentration of 5 µg ml–1, most molecules were plectonemic at moderate salt concentrations which enable re-arrangements, whereas they were irregular at ≤10 mM NaCl, i.e. under trapping conditions. The irregular conformations at low salt concentration are a consequence of trapping: a significant number of molecules could be converted to loosely interwound structures with a defined superhelix axis by reducing the PL concentration such that re-arrangements were possible. From scattering experiments, sedimentation analysis and computer simulations, loosely interwound scDNAs in low salt concentrations were postulated (17,18). Such structures may be less rigid against deformations during the surface adsorption compared with the more tightly interwound structures in higher salt concentrations, which agrees with their higher tendency of forming folded two-dimensional conformations under all conditions analysed. Furthermore, our findings agree with those reported in Lyubchenko and Shlyakhtenko (21), where scDNA deposited on AP-coated mica assumed plectonemic conformations in 161 mM NaCl and, to a major part, irregular and folded conformations in 10 mM NaCl. Possibly, a difference in the degree of surface equilibration by changing the ionic strength applies also to the AP-coated mica. The salt-dependent capability of the scDNA to re-arrange on PL-coated mica was directly observable by increasing the salt concentration during scanning from conditions of trapping to re-arrangements. scDNAs, initially trapped in compacted conformations in 10 mM NaCl, assumed extended conformations after the change to 100 mM NaCl, demonstrating that large-scale movements on the surface are possible. A speculative conclusion from this observation is that extended two-dimensional scDNA conformations could often emerge via more compacted intermediary states formed during the flattening process.
The conformational change of scDNA with varying salt concentrations was analysed by a quantitative evaluation of plectonemic structures imaged under conditions of re-arrangements. The relative changes in Wr (and correspondingly in the average number of nodes <n>) and r are somewhat weaker than determined by EM, solution measurements and simulations (14,16–18). This could be explained by an overestimate of Wr in low salt concentrations, since in the more loosely interwound structures some of the crossings may represent positive writhe. Additionally, the partial exclusion of molecules containing close DNA–DNA contacts may lead to a slight overestimate of r and a corresponding underestimate of Wr at higher salt concentrations, where such structures were more frequent. The major determinant of Wr is the node number n [given by Wr = –n·sin α, where sin α = 2l/L (10,35)], whose distribution reflected the topoisomer distribution observed by gel electrophoresis. Therefore, the ∼2-fold decrease in the absolute values of Wr (or <n>) compared with the expected value given by the superhelical density of the plasmids is probably a consequence of the adsorption to the surface. Monte Carlo simulations of the surface binding of scDNA, which took into account reversible DNA–surface interactions and a complete surface equilibration, predict that Wr does not decrease by the transition from free solution to a surface (Wr even increased slightly in higher ionic strength) (26). The comparison with the simulations may indicate that the observed drop of Wr, corresponding to a partial unwinding of the superhelix, is associated with the strength of the DNA–surface interaction. scDNA adsorbed to glow-discharged carbon surfaces coated with PL and imaged by EM exhibited a less pronounced unwinding in terms of the node number n (16). An explanation for this difference might be that significantly weaker DNA–surface interactions are used for EM, which is possible, since also DNA loosely attached to the surface would be completely fixed afterwards by drying the sample.
A plot of r as a function of node number n or a plot of r as function of Wr of individual molecules indicates a systematic coupling of r with the degree of supercoiling (see Fig. 4A and B). The dependence of r on Wr (or n) resembles that of r on the average superhelical density σ, as measured by EM, i.e. a hyperbolic decrease of r with increasing σ (10). This dependence is consistent over high and low salt concentrations, showing that the inter-dependence of Wr and r persists, even if the degree of supercoiling is modulated by different mechanisms. For instance, varying the salt concentration, which changes the electrostatic interaction between nearby DNA segments, also changes the Wr/Tw ratio (12,14). This ratio is, in turn, constant over different superhelical densities (10). Interestingly, the extrapolation of the fitting curves in Figure 4 to the expected average n (7.6 and 10.7 in 10 and 100 mM NaCl, respectively) or Wr (–6.2 and –8.6 in 10 and 100 mM NaCl, respectively) for the analysed plasmid yields values of r close to those determined by simulations and solution scattering [∼8.5 and ∼5 nm in 10 and 100 mM NaCl, respectively (17,18)]. This suggests that the inter-dependence of Wr and r is only slightly disturbed during the adsorption to PL-coated mica.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Laurent Kreplak, Thomas Weidemann and Katalin Tóth for helpful advice and comments on the manuscript.
REFERENCES
- 1.Bates A.D. and Maxwell,A. (1993) DNA Topology. Oxford University Press, Oxford, UK. [Google Scholar]
- 2.Negri R., Costanzo,G., Buttinelli,M., Venditti,S. and Dimauro,E. (1994) Effects of DNA topology in the interaction with histone octamers and DNA topoisomerase I. Biophys. Chem., 50, 169–181. [DOI] [PubMed] [Google Scholar]
- 3.Vologodskii A.V., Levene,S.D., Klenin,K.V., Frank-Kamenetskii,M. and Cozzarelli,N.R. (1992) Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol., 227, 1224–1243. [DOI] [PubMed] [Google Scholar]
- 4.Bussiek M., Klenin,K. and Langowski,J. (2002) Kinetics of site–site interactions in supercoiled DNA with bent sequences. J. Mol. Biol., 322, 707–718. [DOI] [PubMed] [Google Scholar]
- 5.Schulz A., Langowski,J. and Rippe,K. (2000) The effect of the DNA conformation on the rate of NtrC activated transcription of E. coli RNA polymerase σ54 holoenzyme. J. Mol. Biol., 300, 709–725. [DOI] [PubMed] [Google Scholar]
- 6.Rippe K., von Hippel,P.H. and Langowski,J. (1995) Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci., 20, 500–506. [DOI] [PubMed] [Google Scholar]
- 7.Santero E., Hoover,T.R., North,A.K., Berger,D.K., Porter,S.C. and Kustu,S. (1992) Role of integration host factor in stimulating transcription from the σ54-dependent nifH promoter. J. Mol. Biol., 227, 602–620. [DOI] [PubMed] [Google Scholar]
- 8.Zivanovic Y., Goulet,I., Révet,B., Le Bret,M. and Prunell,A. (1988) Chromatin reconstitution on small DNA rings. II. DNA supercoiling on the nucleosome. J. Mol. Biol., 200, 267–290. [DOI] [PubMed] [Google Scholar]
- 9.Jett S.D., Cherny,D.I., Subramaniam,V. and Jovin,T.M. (2000) Scanning force microscopy of the complexes of p53 core domain with supercoiled DNA. J. Mol. Biol., 299, 585–592. [DOI] [PubMed] [Google Scholar]
- 10.Boles T.C., White,J.H. and Cozzarelli,N.R. (1990) Structure of plectonemically supercoiled DNA. J. Mol. Biol., 213, 931–951. [DOI] [PubMed] [Google Scholar]
- 11.Langowski J. and Giesen,U. (1989) Configurational and dynamic properties of different length superhelical DNAs measured by dynamic light scattering. Biophys. Chem., 34, 9–18. [DOI] [PubMed] [Google Scholar]
- 12.Adrian M., ten Heggeler-Bordier,B., Wahli,W., Stasiak,A.Z., Stasiak,A. and Dubochet,J. (1990) Direct visualization of supercoiled DNA molecules in solution. EMBO J., 9, 4551–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rippe K., Mücke,N. and Langowski,J. (1997) Superhelix dimensions of a 1868 base pair plasmid determined by scanning force microscopy in air and in aqueous solution. Nucleic Acids Res., 25, 1736–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bednar J., Furrer,P., Stasiak,A., Dubochet,J., Egelman,E.H. and Bates,A.D. (1994) The twist, writhe and overall shape of superhelical DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. Possible implications for DNA structure in vivo.J. Mol. Biol., 235, 825–847. [DOI] [PubMed] [Google Scholar]
- 15.Cozzarelli N.R., Boles,T.C.B. and White,J.H. (1990) Chapter 4. In Wang,J.C. (ed.), DNA Topology and Its Biological Effects. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, pp. 139–184. [Google Scholar]
- 16.Cherny D.I. and Jovin,T.M. (2001) Electron and scanning force microscopy studies of alterations in supercoiled DNA tertiary structure. J. Mol. Biol., 313, 295–307. [DOI] [PubMed] [Google Scholar]
- 17.Rybenkov V.V., Vologodskii,A.V. and Cozzarelli,N.R. (1997) The effect of ionic conditions on the conformations of supercoiled DNA.1. Sedimentation analysis. J. Mol. Biol., 267, 299–311. [DOI] [PubMed] [Google Scholar]
- 18.Hammermann M., Brun,N., Klenin,K.V., May,R., Toth,K. and Langowski,J. (1998) Salt-dependent DNA superhelix diameter studied by small angle neutron scattering measurements and Monte Carlo simulations. Biophys. J., 75, 3057–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansma H.G., Bezanilla,M., Zenhausern,F., Adrian,M. and Sinsheimer,R.L. (1993) Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res., 21, 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustamante C., Vesenka,J., Tang,C.L., Rees,W., Guthold,M. and Keller,R. (1992) Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry, 31, 22–26. [DOI] [PubMed] [Google Scholar]
- 21.Lyubchenko Y.L. and Shlyakhtenko,L.S. (1997) Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl Acad. Sci. USA, 94, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyubchenko Y.L., Gall,A.A., Shlyakhtenko,L.S., Harrington,R.E., Jacobs,B.L., Oden,P.I. and Lindsay,S.M. (1992) Atomic force microscopy imaging of double stranded DNA and RNA. J. Biomol. Struct. Dyn., 10, 589–606. [DOI] [PubMed] [Google Scholar]
- 23.Pfannschmidt C. and Langowski,J. (1998) Superhelix organization by DNA curvature as measured through site-specific labeling. J. Mol. Biol., 275, 601–611. [DOI] [PubMed] [Google Scholar]
- 24.Nagami F., Zuccheri,G., Samori,B. and Kuroda,R. (2002) Time-lapse imaging of conformational changes in supercoiled DNA by scanning force microscopy. Anal. Biochem., 300, 170–176. [DOI] [PubMed] [Google Scholar]
- 25.Tanigawa M. and Okada,T. (1998) Atomic force microscopy of supercoiled DNA structure on mica. Anal. Chim. Acta, 365, 19–25. [Google Scholar]
- 26.Fujimoto B.S. and Schurr,J.M. (2002) Monte Carlo simulations of supercoiled DNAs confined to a plane. Biophys. J., 82, 944–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams R.C. (1977) Use of polylysine for adsorption of nucleic acids and enzymes to electron microscope specimen films. Proc. Natl Acad. Sci. USA, 74, 2311–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frontali C., Dore,E., Ferrauto,A., Gratton,E., Bettini,A., Pozzan,M.R. and Valdevit,E. (1979) An absolute method for the determination of the persistence length of native DNA from electron micrographs. Biopolymers, 18, 1353–1373. [DOI] [PubMed] [Google Scholar]
- 29.Rivetti C., Guthold,M. and Bustamante,C. (1996) Scanning force microscopy of DNA deposited onto mica: equilibration vs. molecular kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol., 264, 919–932. [DOI] [PubMed] [Google Scholar]
- 30.Kratky O. and Porod,G. (1949) Röntgenuntersuchung gelöster Fadenmoleküle. Rec. Trav. Chim., 68, 1106–1113. [Google Scholar]
- 31.Kapp U. and Langowski,J. (1992) Preparation of DNA topoisomers by RP-18 high-performance liquid chromatography. Anal. Biochem., 206, 293–299. [DOI] [PubMed] [Google Scholar]
- 32.Keller W. (1975) Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc. Natl Acad. Sci. USA, 72, 4876–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagerman P.J. (1988) Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem., 17, 265–286. [DOI] [PubMed] [Google Scholar]
- 34.Lysetska M., Knoll,A., Boehringer,D., Hey,T., Krauss,G. and Krausch,G. (2002) UV light-damaged DNA and its interaction with human replication protein A: an atomic force microscopy study. Nucleic Acids Res., 30, 2686–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuller F.B. (1971) The writhing number of a space curve. Proc. Natl Acad. Sci. USA, 68, 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]