Abstract
Detection and identification of microbial pathogens are important for disease diagnosis, treatment and prophylaxis measurements. By introducing an innovative technique, we show a robust, reliable and accurate microarray-based method for identification of microbial pathogens. The technique utilizes a unique combination of multiplex competitive hybridization, which enhances hybridization accuracy of oligonucleotides to the specific target, and apyrase-mediated allele-specific extension, which improves specific extension. As a model system, different clinically relevant human papillomaviruses were selected for this study. The method generated accurate results and proves to be promising for specific and correct microbial and viral typing.
INTRODUCTION
Accurate and specific typing of microbial and viral pathogens is of utmost importance for clinical diagnosis and for characterizing particular types/species. Many microorganisms usually lack adequate morphological detail for easy identification. Furthermore, the development of specific therapies/vaccines requires the implementation of sufficient parameters for microbial and viral detection, and a reliable and robust genotyping method is necessary for accurate follow-up during clinical trials and monitoring of treatment (1). In addition to reliability and robustness, high throughput genotyping approaches are required. To meet this need, DNA microarrays have been applied for detection of microbial pathogens in some recent studies (2–5). In this work we present a new microarray-based method for microbial and viral identification, using multiplex competitive hybridization (MUCH) in combination with an apyrase-mediated allele-specific primer extension (AMASE) assay (6,7) followed by hybridization to generic tag arrays. MUCH–AMASE was evaluated for detection and genotyping of human papillomaviruses (HPVs) as a model system.
HPVs are considered as an important contributing factor in the etiology of certain benign and malignant lesions in humans such as cervical cancer (8). At present, more than 100 different HPV genotypes have been identified based on DNA sequence differences (9,10). HPVs are classified in low- and high-risk genotypes based on their oncogenic potential. High-risk HPVs have been shown to be present in 99.7% of cervical cancer worldwide. Therefore, high-risk HPV testing has implications for the clinical management of women with cervical lesions and for primary screening for cervical cancer. In addition, it is necessary to identify individual HPV genotypes to investigate the epidemiology and clinical behavior of particular types (11). Furthermore, it is not an unusual phenomenon for an HPV carrier to be infected by more than one HPV genotype with the varying rate of multiple HPV infections depending on the characteristics of the population tested.
HPV cannot be sufficiently cultured in vitro and cytological and histological examinations do not allow direct assessment of HPV but basically diagnose the consequences of viral infection (1). Therefore, detection and genotyping of HPV-DNA can provide more accurate diagnosis of infection and the related risk for development of cervical neoplasia. There are a number of different techniques for detection and genotyping of HPV with advantages and disadvantages. Hybrid Capture II system (HCII, Digene Corp., USA) is a signal amplification method, based on the hybridization of the target HPV-DNA to labeled RNA probes. A disadvantage of the HCII system is that the technique is less sensitive than PCR (1,12,13) and does not allow specific genotyping but only differentiates between high- and low-risk groups. In addition, the accuracy of the test is restrained by cross-hybridizations. PCR is the most widely used target amplification method and two approaches are most common in HPV testing. One approach utilizes type-specific amplification and the second approach uses general amplification. The former specifically amplifies a single HPV genotype by the use of type-specific primers. However, the major disadvantage of this approach lies in the fact that many PCRs have to be performed. The latter utilizes consensus or general PCR primers. There are several different consensus PCR primer sets for amplification of HPV genotypes. A number of different techniques can be performed to score the genotypes of these PCR amplified HPV-DNA. These techniques include restriction fragment length polymorphism (RFLP), hybridization assays and DNA sequencing technologies (14). RFLP is a low throughput, time-consuming and labor intensive method. Hybridization techniques, on the other hand, are generally high throughput but hold the risk for cross-hybridizations and unspecific bindings. In recent years, relatively rapid DNA sequencing methods have been developed for application in routine analysis of clinical samples. Nevertheless, it should be noted that direct DNA sequencing technologies are not capable of detecting simultaneously multiple infections in a specimen (10). In addition, sequences only representing a minority of the PCR products may remain unnoticed. Another drawback is the presence of non-specific PCR products that can give rise to background signals when one of the PCR primers is used as sequencing primer. In a recent work, these problems have been met by utilization of multiple type-specific sequencing primers covering clinically important HPV genotypes (15).
To address the limitations of the existing methods, we introduce MUCH–AMASE, a novel approach based on MUCH of target specific oligonucleotides followed by specific extension of hybridized oligonucleotides. This unique combination has provided an accurate and microarray-based method for viral and microbial detection using HPV as a model system. The method uses triple accuracy control for specific hybridization and specific extension for each sample, minimizing entirely the risk of false results. Furthermore, the sample preparation, annealing and extension steps are automated and data analysis can be obtained in a user-friendly format.
MATERIALS AND METHODS
Microarray preparation and spatial separation of samples
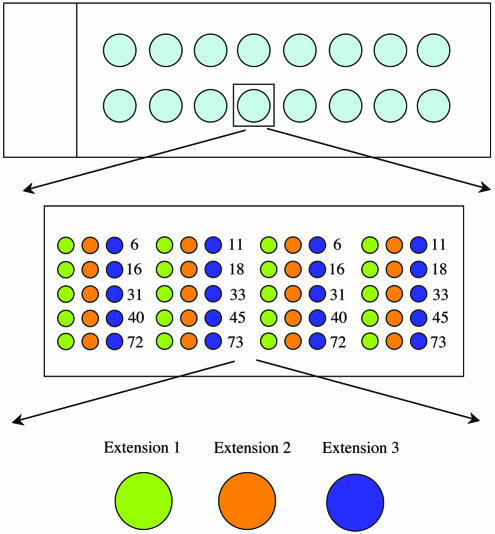
Thirty sequence tags were selected from www.genome.wi.mit.edu (16). The sequences of these unique tags are listed in Table 1. The amino linked oligonucleotides suspended to a concentration of 20 µM in 150 mM sodium phosphate (pH 8.5) and 0.06% sarkosyl were spotted by a GMS 417 arrayer (Affymetrix, USA) on 3D-link activated Motorola slides (Motorola Life Sciences, IL). The oligonucleotides were printed in 16 sub-arrays on the slides (an array of arrays). The 5′-terminus of the 30 sequence tags was linked with an amino group. Each array contained one duplicate of each oligonucleotide. The distance between the spots was 250 µm and the spot diameter was ∼200 µm. The array of arrays was positioned in two columns of eight rows with 9 mm distance between array centers. The printed arrays were incubated overnight in a humid chamber followed by post coupling according to the manufacturer’s instructions. The sub-arrays on the microarray slide were separated during hybridization by using a silicon mask (Elastosile 601 A/B Wacker Chemie GmbH, Munich, Germany) molded in a 96 well plate and excised to fit the slide. A custom made rack was used to press the silicon firmly to the slide during the hybridization.
Table 1. List of sequence tags (5′→3′) used for this study.
| LT2 (T)15-CGCAGGTATCGTATTAATTGATCTGC |
| LT3 (T)15-CCTCATGTCAACGAAGAACAGAACC |
| LT4 (T)15-ATTGAAGCCTGCCGTCGGAGACTAA |
| LT5 (T)15-AGACTGCGTGTTGGCTCTGTCACAG |
| LT6 (T)15-TTATGGTGATCAGTCAACCACCAGG |
| LT7 (T)15-GAGACACCTTATGTTCTATACATGC |
| LT8 (T)15-TCCATGCGCTTGCTCTTCATCTAGC |
| LT9 (T)15-GCCTTACATACATCTGTCGGTTGTA |
| LT10 (T)15-CACAAGGAGGTCAGACCAGATTGAA |
| LT11 (T)15-GCCACAGATAATATTCACATCGTGT |
| LT12 (T)15-ACACATACGATTCTGCGAACTTCAA |
| LT13 (T)15-TTACAGGATGTGCTCAACAGACGTT |
| LT14 (T)15-GCTCACAATAATTGCATGAGTTGCC |
| LT15 (T)15-CTGCACTGCTCATTAATATACTTCTGG |
| LT16 (T)15-TTCACGCACTGACTGACAGACTGCTT |
| LT17 (T)15-CAACATCATCACGCAGAGCATCATT |
| LT18 (T)15-GCATCAGCTAACTCCTTCGTGTATT |
| LT19 (T)15-GGCGTTATCACGGTAATGATTAACAGC |
| LT20 (T)15-ACATCAATCTCTCTGACCGTTCCGC |
| LT21 (T)15-GCCTTATGCTCGAACTGACCATAAC |
| LT22 (T)15-CGGATATCACCACGATCAATCATAGGTAA |
| LT23 (T)15-CCTTAATCTGCTGCAATGCCACAGC |
| LT24 (T)15-TAGCTCTCCGCCTACAATGACGTCA |
| LT25 (T)15-AGGAACGCCTTACGTTGATTATTGA |
| LT26 (T)15-GAGTCAGTACCGATGTAGCCGATAA |
| LT27 (T)15-ACTCGAATGAACCAGGCGATAATGG |
| LT28 (T)15-ATTATATCTGCCGCGAAGGTACGCC |
| LT29 (T)15-GGACAGACAGTGGCTACGGCTCAGTT |
| LT30 (T)15-CGGTATTCGCTTAATTCAGCACAAC |
| LT31 (T)15-GCTCTTACCTGTTGTGCAGATATAA |
5′-terminus tagged extension primers
The DNA sequence of the L1 region of HPV-6, 11, 16, 18, 26, 31, 32, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 61, 66, 67, 68, 69, 70, 71 (CP8061), 72 (LVX100), 73 (MM9), 81 (CP8304), 82 (MM4), 83 (MM7), 84 (MM8), CP6108 and IS324 genotypes were aligned for specific primer design. These genotypes can be amplified by consensus primer sets. Three regions (within a 150 bp long PCR amplified sequence) with high heterogeneity between different HPV types were selected to design extension primers. The distance between these oligonucleotides was 10 to 11 bases to allow elongation by DNA polymerase. A database search (BLAST from National Center for Biotechnology Information, NCBI) showed specific matches with the corresponding HPV types. A total of 30 extension primers (three primers/HPV type) were designed for the 10 HPV types investigated in this study (HPV-6, 11, 16, 18, 31, 33, 40, 45, 72 and 73). Each oligonucleotide contained a tag (barcode) at its 5′-terminus. These HPV extension primers with 5′-terminus tags are listed in Table 2. The oligonucleotides were synthesized by MWG-biotech (Ebersberg, Germany). A comparison of Tables 1 and 2 shows that each oligonucleotide tag on the glass slide is complementary to one of the tags on the 5′-terminus of the extension primers.
Table 2. List of HPV genotype-specific extension primers (5′→3′) with complementary 5′-terminus tags.
| HPV6(1)LT2 GCAGATCAATTAATACGATACCTGCG-ACGCAGTACCAACATGACAT |
| HPV6(2)LT3 GGTTCTGTTCTTCGTTGACATGAGG-CGTAACTACATCTTCCACA |
| HPV6(3)LT4 TTAGTCTCCGACGGCAGGCTTCAAT-TGATTATAAAGAGTACATGC |
| HPV11(1)LT5 CTGTGACAGAGCCAACACGCAGTCT-ACGCAGTACAAATATGACAC |
| HPV11(2)LT6 CCTGGTGGTTGACTGATCACCATAA-TGTGTCTAAATCTGCTACA |
| HPV11(3)LT7 GCATGTATAGAACATAAGGTGTCTC-AGATTATAAGGAATACATGC |
| HPV16(1)LT8 GCTAGATGAAGAGCAAGCGCATGGA-ACGCAGTACAAATATGTCAT |
| HPV16(2)LT9 TACAACCGACAGATGTATGTAAGGC-CATATCTACTTCAGAAACT |
| HPV16(3)LT10 TTCAATCTGGTCTGACCTCCTTGTG-TACTAACTTTAAAGAGTACC |
| HPV18(1)LT11 ACACGATGTGAATATTATCTGTGGC-TCGCAGTACCAATTTAACAA |
| HPV18(2)LT12 TTGAAGTTCGCAGAATCGTATGTGT-TACACAGTCTCCTGTACCT |
| HPV18(3)LT13 AACGTCTGTTGAGCACATCCTGTAA-TGCTACCAAATTTAAGCAGT |
| HPV31(1)LT14 GGCAACTCATGCAATTATTGTGAGC-ACGTAGTACCAATATGTCTG |
| HPV31(2)LT15 CCAGAAGTATATTAATGAGCAGTGCAG-AATTGCAAACAGTGATACT |
| HPV31(3)LT16 AAGCAGTCTGTCAGTCAGTGCGTGAA-TAGTAATTTTAAAGAGTATT |
| HPV33(1)LT17 AATGATGCTCTGCGTGATGATGTTG-TCGCAGTACTAATATGACTT |
| HPV33(2)LT18 AATACACGAAGGAGTTAGCTGATGC-AGTAACTAGTGACAGTACA |
| HPV33(3)LT19 GCTGTTAATCATTACCGTGATAACGCC-GAATTTTAAAGAATATATAA |
| HPV40(1)LT20 GCGGAACGGTCAGAGAGATTGATGT-TCGTAGCACTAATTTAACCT |
| HPV40(2)LT21 GTTATGGTCAGTTCGAGCATAAGGC-CACACAGTCCCCCACACCA |
| HPV40(3)LT22 TTACCTATGATTGATCGTGGTGATATCCG-TAACAGTAATTTCAAGGAAT |
| HPV45(1)LT23 GCTGTGGCATTGCAGCAGATTAAGG-CCGCAGTACTAATTTAACAT |
| HPV45(2)LT24 TGACGTCATTGTAGGCGGAGAGCTA-TACACAAAATCCTGTGCCA |
| HPV45(3)LT25 TCAATAATCAACGTAAGGCGTTCCT-TCCTACTAAGTTTAAGCACT |
| HPV72(1)LT26 TTATCGGCTACATCGGTACTGACTC-TCGCAGTACTAATGTAACTA |
| HPV72(2)LT27 CCATTATCGCCTGGTTCATTCGTGA-CACAGCGTCCTCTGTATCA |
| HPV72(3)LT28 GGCGTACCTTCGCGGCAGATATAAT-TTCTAATTTTCGTGAGTATC |
| HPV73(1)LT29 AACTGAGCCGTAGCCACTGTCTGTCC-TAGAAGCACTAATTTTTCTG |
| HPV73(2)LT30 GTTGTGCTGAATTAAGCGAATACCG-TACACAGGCTAGTAGCTCT |
| HPV73(3)LT31 TTATATCTGCACAACAGGTAAGAGCTGCCAACTCTAATTTTAAGG |
HPV patient samples and DNA plasmids
The material for the study consisted of cervical scrapes taken from the ecto-endo cervix by CytoBrush® of 15 women diagnosed with either primary invasive cervix cancer (Radiumhemmet, Karolinska Hospital) or abnormal cytology diagnosed through the Swedish national cervix cancer screening program (Gynecology Clinic, Karolinska Hospital). The samples were collected in sterile phosphate buffered saline (pH 8.0). DNA extraction was performed using the QIAmp® System (Qiagen Inc., Valencia, CA) according to the instructions described in the kit. The DNA was dissolved in 200 µl of Tris–EDTA buffer (pH 7.4). HPV whole genomic plasmids for types 6, 11, 16, 18, 33, 40, 45, 72 and 73 (kindly donated by Dr E.-M. de Villiers of the Deutsches Krebsforschungszentrum) were also used in this study.
PCR amplification
Samples were amplified with general primer sets GP5+/6+ (GP6+ biotin-labeled) and MY09/11 (MY09 biotin-labeled). The PCRs were performed as previously described (10,17). The PCR products were 150 (GP5+/6+-derived) and 450 bp (MY09/11-derived) in size.
Sample preparation and extension reaction
Five microliters of the PCR products were immobilized onto streptavidin-coated super paramagnetic beads (Dynabeads M280; Dynal, Oslo, Norway) and single-stranded DNA was obtained by alkali elution of the non-biotinylated strand. This and other procedures described in this section were automatically performed by a Magnetrix 1200 robotic system (Magnetic Biosolutions, Stockholm, Sweden). The supernatant was then discarded and the capture strand was resolved in 60 µl of a solution containing annealing buffer [10 mM Tris-acetate (pH 7.75), 2 mM Mg-acetate] and a pool of 30 HPV type-specific/extension-specific primers (0.1 µM of each primer). The primers were allowed to anneal to the captured DNA template by heating the solution to 70°C for 1 min and then cooling to room temperature for 5 min. As mentioned above, each HPV oligonucleotide contained a specific tag (a barcode) at its 5′-terminus. The primers with 5′-terminus tags are listed in Table 1. After the annealing, the unbound oligonucleotides and the excess of perfectly matched oligonucleotides were removed and the immobilized single-stranded DNA with annealed primers were resolved in 20 µl solution of annealing buffer [10 mM Tris-acetate (pH 7.75), 2 mM Mg-acetate] and extension buffer [42 mM Tris–HCl (pH 8), 5 mM MgCl and 1 mM DTT]. To this mixture 20 µl of a solution containing 0.5% (v/v) BSA (Biothema, Dalarö, Sweden), extension buffer [42 mM Tris–HCl (pH 8), 5 mM MgCl and 1 mM DTT], 1 µg SSB, 8 U exonuclease-deficient (exo-) Klenow DNA polymerase and 40 mU apyrase was added. To start the extension reaction, 20 µl of a mixture containing 0.5 µM of each dNTP [50% Cy5-labeled dCTP and 50% Cy5-labeled dTTP (Amersham Biosciences)], 0.5% (v/v) BSA and extension buffer [42 mM Tris–HCl (pH 8), 5 mM MgCl and 1 mM DTT] was injected to the primed immobilized DNA and mixed gently. After polymerization, the enzymes and dNTPs were discarded and immobilized DNA was washed by annealing buffer [10 mM Tris-acetate (pH 7.75), 2 mM Mg-acetate]. The immobilized DNA was then treated with NaOH (0.1 M) and the supernatant (the AMASE product) was neutralized with HCl (0.1 M). The AMASE products (each containing a specific tag at the 5′-terminus) were then hybridized to the generic tag-arrays for 30 min.
Data acquisition
Following the hybridization, data were obtained by scanning slides with an Agilent scanner (Agilent, Palo Alto, CA). Data were then analyzed by GenePix 4.0 software (Axon instruments, USA). The fluorescence intensities (medians after background subtraction) were calculated by the GenePix 4.0 software and the data were analyzed in Microsoft Excel. As mentioned above, one slide was divided into 16 sub-arrays (an array of arrays) and each array contained one duplicate spot of each oligonucleotide tag and thus the mean value of these spots was used to analyze the data.
Pyrosequencing technology and Sanger dideoxy sequencing
All the genotyping results were confirmed by DNA sequencing with both the Pyrosequencing technology and the Sanger method (cycle sequencing) as described previously (10,18,19). Pyrosequencing was performed on a PSQ 96 (Pyrosequencing AB, Uppsala, Sweden) and conventional Sanger DNA sequencing was performed on an ABI 3700 DNA Analyzer (Applied Biosystems, Foster City, CA) using the BigDye terminator chemistry according to the manufacturer’s manual.
RESULTS AND DISCUSSION
There is an imperative need for cost-effective, high throughput and extremely accurate genotyping technologies for adequate monitoring of biological and clinical impacts of viral and microbial infections. Many of today’s available genotyping technologies fulfil one or two of these genotyping criteria, but at the expense of other factor(s). In this work, we present a technology that does not compromise on accuracy and is performed on a microarray format. This technology takes advantage of a MUCH combined with base-specific extension by exploiting the AMASE technology (6,7) followed by high throughput hybridization and analysis on an array of microarrays using generic tag-arrays (Figs 1 and 2). We have evaluated the MUCH–AMASE technique for genotyping of HPVs. Although HPV is a model system, this technique represents a general approach and is applicable for genotyping of other viral and microbial species/genotypes. Nevertheless, in this study, the choice of HPV genotypes was based on the availability of DNA plasmids and patient specimens. HPVs are DNA viruses that are believed to be an important contributing factor in the etiology of certain tumor lesions in humans, such as cervical cancer. The HPV-16, HPV-18, HPV-31, HPV-33 and HPV-45 are clinically important high-risk types and HPV-6, HPV-11 and HPV-40 are clinically relevant low risk types. HPV-72 and HPV-73 were also included in this study.
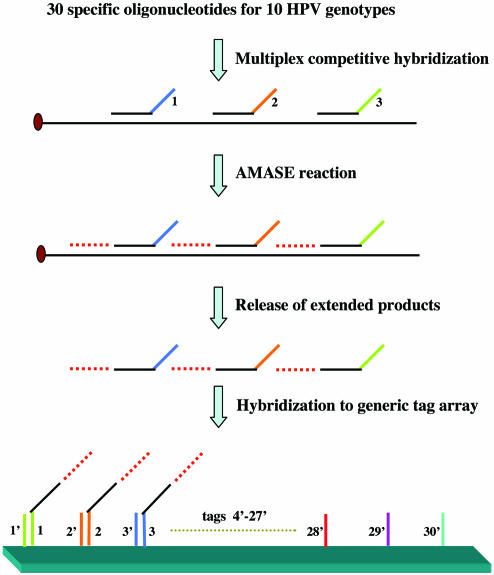
Figure 1.
MUCH–AMASE technique. A schematic principle illustration of the method. The experimental conditions were as described in Materials and Methods. See text for details.
Figure 2.
A schematic illustration of the microarray slide demonstrating the arrangements of the arrays. The slide was arrayed for 16 samples and every spot contained an array of 30 complementary tags (three for every genotype) in duplicate.
Multiplex competitive hybridization (MUCH)
The HPV genome contains extensive genetic heterogeneity and consequently these heterogeneous sequences require specific genotyping tools. There are different techniques for HPV detection and genotyping, all with advantages and disadvantages. Many of these techniques rely on type-specific hybridization assays. Nevertheless, type-specific hybridization (often, when the species has two chromosomes, referred as allele-specific hybridization) is hampered by cross-reactivity (non-specific hybridization) giving false results. This non-specific hybridization is especially a problem in high throughput microarray based analysis. In the microarray-based approaches, a small amount of discriminating probes are spotted and immobilized on the glass slides and an excess of mobile target DNA is allowed to hybridize to these probes. Theoretically, the target DNA will hybridize to perfectly matched probe(s) and by using stringent temperature conditions, hybridization to mismatched probes is avoided. However, in practice, there are extremely small differences in the duplex stability between a perfect match and a mismatch at one base, which is a critical limitation of this technology. Furthermore, since the discriminating probes are immobilized, a perfect match probe cannot influence hybridization of the mobile target DNA to the mismatch probes.
The first step in our approach utilizes specific hybridization of oligonucleotides to the target DNA. However, as will be discussed below, this specific hybridization has been modified to achieve greater discrimination between genotypes without the need for stringent temperature conditions. For this matter, oligonucleotides were designed for the L1 region of 10 investigated HPV types (HPV-6, 11, 16, 18, 31, 33, 40, 45, 72 and 73). The sequences of oligonucleotides were based on alignment of the above-mentioned HPV genotypes, amplified by MY09/11 (450 bp product) and GP5+/6+ (150 bp product) primer sets. Three regions with high heterogeneity between different HPV types were selected to design specific oligonucleotides. The distance between these oligonucleotides was 10 to 11 bases, allowing extension by DNA polymerase in the next phase of the method. Thus, a total of 30 type-specific oligonucleotides (three oligonucleotides/HPV type) were designed. Briefly, one-tenth of biotin-labeled PCR product (5 µl) was immobilized onto streptavidin-coated super paramagnetic beads and single-stranded DNA was obtained by alkali elution of the non-biotinylated strand. A relatively high concentration (6 pmol) of each oligonucleotide was then added in a multiplex fashion to anneal to the captured single-stranded target DNA. The solution was heated to 70°C and then cooled to room temperature. The main difference between this approach and conventional type-specific hybridization assays is that the mobile type-specific probes (with high concentrations compared to the target DNA) are competing to hybridize to the same DNA sequence. Obviously, when the temperature drops from 70°C, the oligonucleotides (or the probes) that are completely matched to the target DNA will have favorable hybridization kinetics compared to the mismatch probes and thus the mismatch probes are out-competed in the hybridization process. Subsequently, at lower temperature (as low as room temperature) the mismatch probes would have the opportunity to hybridize non-specifically. However, this non-specific hybridization will be hindered because the matched probes already occupy the specific regions. After the annealing, the unbound oligonucleotides and the excess of perfectly matched oligonucleotides are removed by a washing step. The washing is a significant step, first to reduce the background signals that predominantly are caused by non-specific primer–primer hybridization, and secondly to enhance the signal intensity by eliminating hybridization of excessive unbound and non-extended oligonucleotides to the tagged array. In order to be able to distinguish which of the multiplexed annealed type- specific oligonucleotides have hybridized to the target DNA, each oligonucleotide contains a specific and unique tag (barcode) at its 5′-terminus functioning as the signature of the oligonucleotide.
Apyrase-mediated allele-specific primer extension (AMASE)
In spite of improvement of type (allele)-specific hybridization assays by employing a multiplex competitive approach, this method may still give rise to false positive signals. In a study performed on HIV-1 as a model system for investigation of the effects of internal primer-template mismatches (18), the authors concluded that the presence of two to four mismatches in the primer-template did not have a significant effect on PCR. However, they observed that the presence of five or more mismatches reduced the PCR product yields by at least 20-fold. This may indicate that despite the kinetic nature of MUCH, some non-specific hybridization may still occur. However, DNA-modifying enzymes such as DNA polymerases and DNA ligases have successfully been employed for genotyping of especially single nucleotide polymorphisms (SNPs). Examples of such methods are allele-specific primer extension assays (19), single base extension (or mini-sequencing) (20) and ligation assays (21). Thus, we sought a combination of MUCH with an enzymatic approach to provide an accurate viral and microbial genotyping technique. For this purpose, allele-specific primer extension assay was employed.
Allele-specific extension methods requiring only a single detection reaction are based on extension of allele-specific primers that differ at their 3′-terminus nucleotide, defining the allelic variants. Despite its simplicity, the use of allele-specific extension technologies has been greatly hampered by poor discrimination property of the DNA polymerases, resulting in certain mismatches being poorly discriminated. However, we have previously shown that DNA polymerases extend the mismatched primer-templates with slower reaction kinetics in comparison to extension of the matched primer-template configurations (22). This kinetic difference was exploited by addition of apyrase (a nucleotide degrading enzyme) in the extension reaction. The AMASE protocol allows incorporation of nucleotides when the reaction kinetics is fast (matched 3′-terminus primer-template) but degrades the nucleotides before extension when the reaction kinetics are slow (mismatched 3′-terminus primer-template). Obviously, since viruses do not carry two or more alleles, the term allele-specific appears to be an incorrect expression. Nevertheless, from a methodological point of view, the discrimination factor of the enzymatic part of the assay is based on the ability of the DNA polymerase to extend a 3′-terminus matched base but not a 3′-terminus mismatched base and this is recognized as allele-specific extension. Thus, we will continue to use the well-known technological term of allele-specific extension for microbial typing with reservation.
In silico evaluation
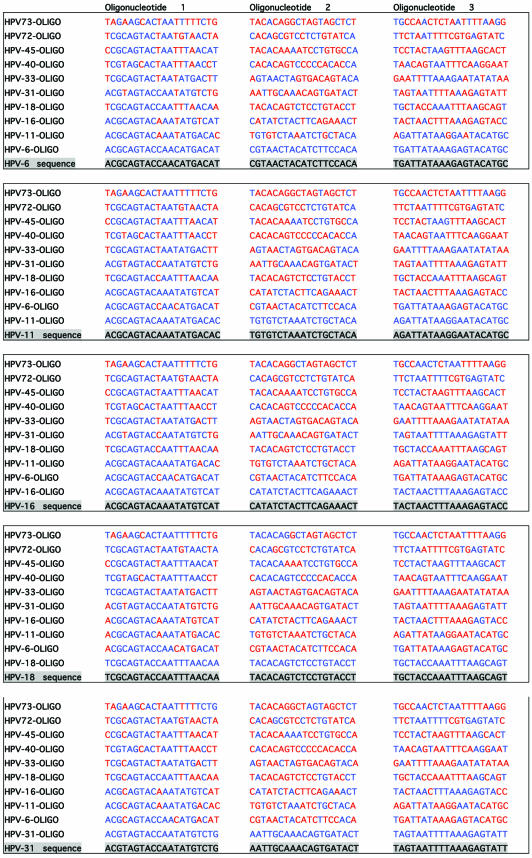
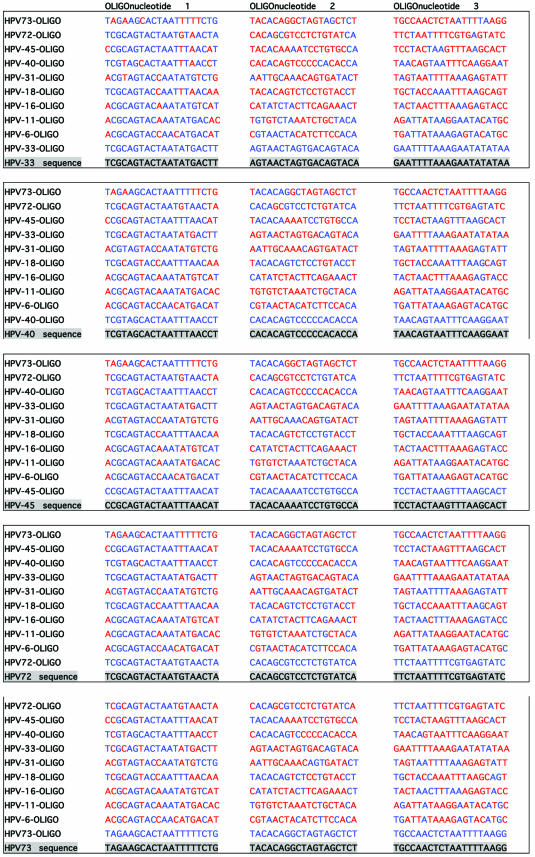
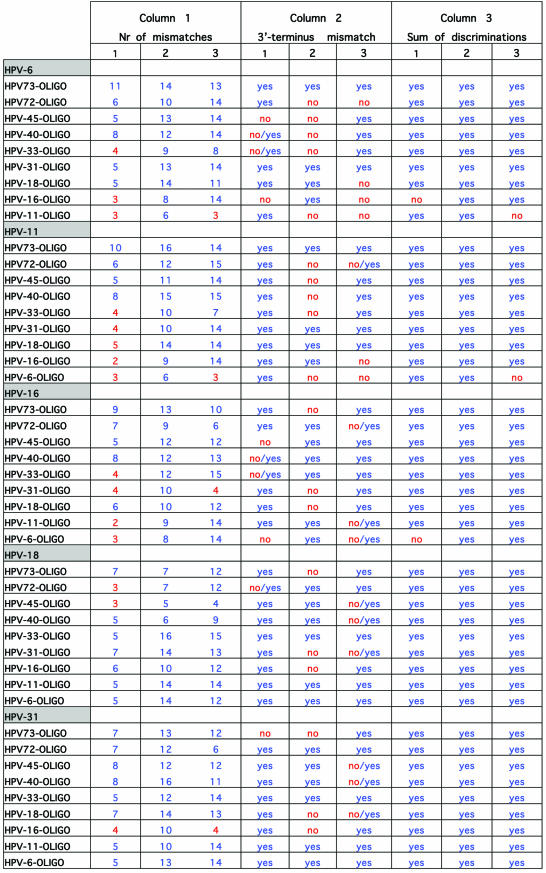
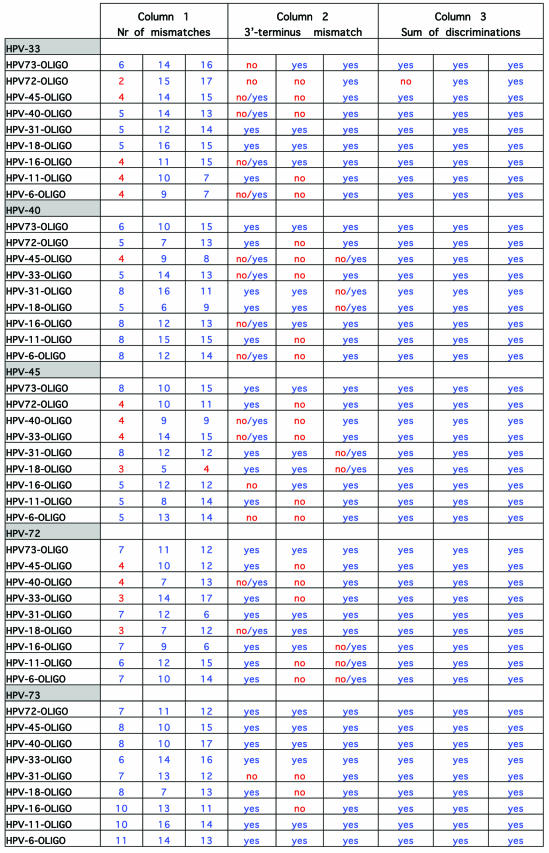
To investigate the accuracy of the method and the probability of false results, DNA sequences of L1 region of 39 HPV genotypes (that are liable to be amplified with consensus primer sets) were aligned for primer design. Specific primers for 10 HPV types in three regions with high heterogeneity on L1 region were designed. The first step in the experimental design of extension primers was a theoretical simulation of the events. For evaluation of the designed primers, all the 39 HPV genotypes were evaluated for cross-hybridizations and mismatch extensions by an in-house developed script written in Perl, but for practical and space reasons only alignments of the 10 selected HPV genotypes (in this study) are shown. Table 3 illustrates the simulation for primer design of three regions with high heterogeneity on DNA sequence of the L1 region of the 10 HPV types. As shown, the consensus sequence of the three regions of the investigated genotypes is highlighted and just above these, the sequence of 10 oligonucleotides is indicated (oligonucleotide 1 for the first region, oligonucleotide 2 for the second region and oligonucleotide 3 for the third region). The distances between oligonucleotides 1 and 2 and oligonucleotides 2 and 3 are 10 and 11 bases, respectively. One of these oligonucleotides (for each region) forms a complete match to the consensus sequence and is always indicated as the first oligonucleotide above the consensus sequence. Nine oligonucleotides that correspond to the other HPV types are listed on top of the complete matched oligonucleotide and contain different numbers of mismatches when compared to the consensus sequence. The mismatching nucleotides are indicated in red. In Table 4, which is actually a continuation of Table 3, columns 1–3 correspond to the number of mismatches to the template, presence or absence of 3′-terminus mismatched primer-template and the sum of the discriminations, respectively. Obviously, a mismatch at or near the 5′- and 3′-ends of an oligonucleotide will have much less consequences on hybridization efficiency than one in the middle. And as the numbers of mismatches to the template differ and occur in different positions, these may have a completely different influence on competitive hybridization. Thus, in this simulation, irrespective of position of the mismatches, when the number of mismatches was ≤4 the effect of mismatches in the competitive hybridization was assumed to be minute. This assumption is partially based on the study performed on the effect of internal mismatches on PCR (18). These numbers are flagged by red in the first column of Table 4. Realistically, in a competitive hybridization, four mismatches should be sufficient to discriminate a complete match oligonucleotide from mismatching oligonucleotides, nevertheless this assumption was kept to challenge the technique. The second discriminating factor in this technique is the AMASE assay and in the second column of Table 4, the presence or absence of 3′-terminus mismatching base(s) is indicated by ‘yes’ or ‘no’, respectively. However, we have previously noticed that in some cases presence of a mismatch in a penultimate base hinders the primer extension and therefore in such cases the discrimination by AMASE was cautiously indicated by ‘no/yes’. When AMASE is not able to influence the process of type discriminations (a consequence of lack of 3′-terminus mismatches), ‘no’ is used. As shown, a high number of flagged discriminations is indicated in the first two columns. Nevertheless, in the third column when the sum of discriminations is evaluated, only five combinations are flagged by ‘no’. The evaluation has been based on combining the discrimination factor of MUCH (the first column) with AMASE (the second column). Four scenarios were possible by this combination. First, when the number of mismatches in MUCH is indicated in blue (more than four internal oligonucleotide-template mismatches) and AMASE results by ‘yes’ (presence of 3′-terminus mismatches in primer-template) the sum of discriminations will be ‘yes’. Second, when one of the discriminating factors fails and is flagged but the other one fulfils the criteria for a correct genotyping, the sum will be ‘yes’. Third, when MUCH is flagged (four or less internal mismatches are present) and AMASE is ‘no/yes’ the resulting sum has been assumed to be ‘yes’. The argument here was that the presence of a mismatch in a penultimate base hinders a proper primer extension (at least in some cases) and this in combination with two to four other internal mismatches should provide adequate discrimination. The fourth scenario is when both factors fail to fulfill correct genotyping criteria and consequently the sum of discriminations will also fail and is notified by a ‘no’ flag. Nevertheless, MUCH–AMASE is performed on three selected regions and this triple oligonucleotide control provides an extreme accuracy in the genotyping.
Table 3. Oligonucleotide sequence alignments of three variable regions of 10 HPV genotypes on L1 region, for evaluation of false hybridizations and extensions.
See text for more details.
Table 4. Theoretical results of false hybridization and extension possibilities of HPV oligonucleotides.
See text for more details.
Experimental evaluation
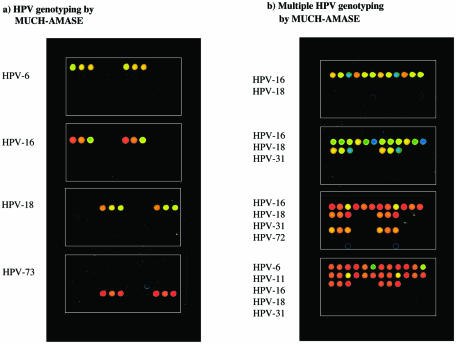
Nine HPV whole genomic DNA plasmids of different genotypes (HPV-6, 11, 16, 18, 31, 33, 40, 45, 72 and 73) and 15 clinical patient specimens were used in this study. The samples were amplified according to the procedures mentioned earlier. In brief, after MUCH of the 30 specific oligonucleotides and removal of the unbound oligonucleotides, the primed templates were subjected to specific extensions (AMASE). After extension, the microarray was scanned and fluorescent intensities for each spot was measured and compared. As mentioned, MUCH–AMASE was performed for genotyping of HPV-amplicon from both whole plasmid and patient samples. Figure 3a shows array scans of MUCH–AMASE on four PCR amplified materials (HPV-6, HPV-16, HPV-18 and HPV-73). In order to evaluate the technique for identification and genotyping multiple HPV infections, PCR products of whole genome plasmids were mixed and analyzed. Figure 3b shows this simulated multiple infections of HPV-16/18, HPV-16/18/31, HPV-16/18/31/72 and HPV-6/11/16/18/31. These results strongly indicate the possibility of typing multiple HPV infections, which is a common phenomenon. As illustrated in Figure 3, the positive HPV spots on the slide show considerably higher signal intensities compared to the negative spots. The in silico evaluation of the sum of the discrimination factors indicated failure of five oligonucleotides (see Table 4); however, the experimental analysis showed only one of these in HPV-11 (the third oligonucleotide corresponding to HPV-6) giving some background signal. The genotyping results here demonstrate fluorescence signal intensities of 10 different HPV genotypes with the lowest positive signal intensity 14-fold over the negative (background) signal. All the HPV DNA plasmid samples and clinical samples analyzed by MUCH–AMASE were analyzed for statistical evaluation. For each sample, the ratio of the highest signal was compared to the other negative signals on the array and a ratio range of 14 to 1648 was obtained with the mean value of 335 and a median of 267. These results indicate highly significant values in themselves and especially with regard to the existing microarray formats. The assay worked on fragments both derived from GP5+/6+ (150 bp) and MY09/11 (450 bp). However, we will here point out that a drawback of the technique is that point mutations (appearance of a sub-type within a specific type) cannot be detected. As mentioned, nine HPV whole genomic DNA plasmids and 15 clinical samples were genotyped by MUCH–AMASE. The clinical samples were scored as HPV-16 (11 samples), HPV-31, HPV-33 and HPV-73 (two samples). In addition, in order to investigate the sensitivity of the method and the possibility to quantify multiple HPV-DNA, two additional patient samples with multiple HPV infections were analyzed. One sample contained triple infections with a frequency of ∼57% HPV-16, 10% HPV-18 and 33% HPV-72. The second sample had a double infection with a frequency of 90–95% HPV-16 and 5–10% HPV-18. However, we will emphasis that these frequencies are approximate (relative) quantifications and not absolute quantifications but since the detection system is very sensitive we are able to easily detect minor types (as low as 5% and probably less). Moreover, high throughput genotyping in combination with detection of multiple infections with low frequencies requires automated scoring. For this reason, a script has been developed that facilitates automated scoring of single and multiple infections and calculates a relative quantification when the sample exhibits multiple infections. All genotyping results were confirmed by DNA sequencing.
Figure 3.
Demonstration of scanned fluorescent images of HPV samples genotyped by MUCH–AMASE demonstrating (a) single infection and (b) multiple infections in each sample.
In conclusion, the HPV detection results obtained by MUCH–AMASE technique have been more than satisfactory with the lowest signal intensity with significant order of magnitude over the negative control. The assay is automated and does not require optimizations or stringent reaction/temperature conditions. To enhance the discriminatory power and accuracy of MUCH–AMASE we utilized three specific extension oligonucleotides for each genotype, and accurate results were achieved. Moreover, multiple infections/variants of HPV were readily detected, which is of clinical importance as a tangible number of HPV carriers are multiple infected. This study presents development of an accurate microarray-based HPV-genotyping method using fluorescently labeled nucleotides suitable for large-scale clinical settings and the technique is applicable for detection and typing of other microorganisms and viruses.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Swedish Research Council and Cancerfonden.
REFERENCES
- 1.van Doorn L.J., Kleter,B. and Quint,W.G. (2001) Molecular detection and genotyping of human papillomavirus. Expert. Rev. Mol. Diagn., 1, 394–402. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Lopez M.A., Mendez-Tenorio,A., Maldonado-Rodriguez,R., Doktycz,M.J., Fleming,J.T. and Beattie,K.L. (2003) Fingerprinting of prokaryotic 16S rRNA genes using oligodeoxyribonucleotide microarrays and virtual hybridization. Nucleic Acids Res., 31, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Coscoy,L., Zylberberg,M., Avila,P.C., Boushey,H.A., Ganem,D. and DeRisi,J.L. (2002) Microarray-based detection and genotyping of viral pathogens. Proc. Natl Acad. Sci. USA, 99, 15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chizhikov V., Wagner,M., Ivshina,A., Hoshino,Y., Kapikian,A.Z. and Chumakov,K. (2002) Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol., 40, 2398–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson W.J., Strout,C.L., DeSantis,T.Z., Stilwell,J.L., Carrano,A.V. and Andersen,G.L. (2002) Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes, 16, 119–127. [DOI] [PubMed] [Google Scholar]
- 6.Ahmadian A., Gharizadeh,B., O‘Meara,D., Odeberg,J. and Lundeberg,J. (2001) Genotyping by apyrase-mediated allele-specific extension. Nucleic Acids Res., 29, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Meara D., Ahmadian,A., Odeberg,J. and Lundeberg,J. (2002) SNP typing by apyrase-mediated allele-specific primer extension on DNA microarrays. Nucleic Acids Res., 30, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godfroid E., Heinderyckx,M., Mansy,F., Fayt,I., Noel,J.C., Thiry,L. and Bollen,A. (1998) Detection and identification of human papilloma viral DNA, types 16, 18 and 33, by a combination of polymerase chain reaction and a colorimetric solid phase capture hybridisation assay. J. Virol. Methods, 75, 69–81. [DOI] [PubMed] [Google Scholar]
- 9.Vernon S.D., Unger,E.R. and Williams,D. (2000) Comparison of human papillomavirus detection and typing by cycle sequencing, line blotting and hybrid capture. J. Clin. Microbiol., 38, 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharizadeh B., Kalantari,M., Garcia,C.A., Johansson,B. and Nyren,P. (2001) Typing of human papillomavirus by pyrosequencing. Lab. Invest., 81, 673–679. [DOI] [PubMed] [Google Scholar]
- 11.van den Brule A.J., Pol,R., Fransen-Daalmeijer,N., Schouls,L.M., Meijer,C.J. and Snijders,P.J. (2002) GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol., 40, 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cope J.U., Hildesheim,A., Schiffman,M.H., Manos,M.M., Lorincz,A.T., Burk,R.D., Glass,A.G., Greer,C., Buckland,J., Helgesen,K. et al. (1997) Comparison of the hybrid capture tube test and PCR for detection of human papillomavirus DNA in cervical specimens. J. Clin. Microbiol., 35, 2262–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smits H.L., Bollen,L.J., Tjong,A.H.S.P., Vonk,J., Van Der Velden,J., Ten Kate,F.J., Kaan,J.A., Mol,B.W. and Ter Schegget,J. (1995) Intermethod variation in detection of human papillomavirus DNA in cervical smears. J. Clin. Microbiol., 33, 2631–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard H.U., Chan,S.Y., Manos,M.M., Ong,C.K., Villa,L.L., Delius,H., Peyton,C.L., Bauer,H.M. and Wheeler,C.M. (1994) Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence and phylogenetic algorithms. J. Infect. Dis., 170, 1077–1085. [DOI] [PubMed] [Google Scholar]
- 15.Gharizadeh B., Ghaderi,M., Donnelly,D., Amini,B., Wallin,K.L. and Nyren,P. (2003) Multiple-primer DNA sequencing method. Electrophoresis, 24, 1145–1151. [DOI] [PubMed] [Google Scholar]
- 16.Hirschhorn J.N., Sklar,P., Lindblad-Toh,K., Lim,Y.M., Ruiz-Gutierrez,M., Bolk,S., Langhorst,B., Schaffner,S., Winchester,E. and Lander,E.S. (2000) SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc. Natl Acad. Sci. USA, 97, 12164–12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravitt P.E., Peyton,C.L., Alessi,T.Q., Wheeler,C.M., Coutlee,F., Hildesheim,A., Schiffman,M.H., Scott,D.R. and Apple,R.J. (2000) Improved amplification of genital human papillomaviruses. J. Clin. Microbiol., 38, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christopherson C., Sninsky,J. and Kwok,S. (1997) The effects of internal primer-template mismatches on RT–PCR: HIV-1 model studies. Nucleic Acids Res., 25, 654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton C.R., Graham,A., Heptinstall,L.E., Powell,S.J., Summers,C., Kalsheker,N., Smith,J.C. and Markham,A.F. (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res., 17, 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syvanen A.C. (1999) From gels to chips: minisequencing primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat., 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 21.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 22.Ahmadian A., Gharizadeh,B., Gustafsson,A.C., Sterky,F., Nyren,P., Uhlen,M. and Lundeberg,J. (2000) Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem., 280, 103–110. [DOI] [PubMed] [Google Scholar]