Abstract
The degradation of the aromatic compound phenylpropionate (PP) in Escherichia coli K-12 requires the activation of two different catabolic pathways coded by the hca and the mhp gene clusters involved in the mineralization of PP and 3-hydroxyphenylpropionate (3HPP), respectively. The compound 3-(2,3-dihydroxyphenyl)propionate (DHPP) is a common intermediate of both pathways which must be cleaved by the MhpB dioxygenase before entering into the primary cell metabolism. Therefore, the degradation of PP has to be controlled by both its specific regulator (HcaR) but also by the MhpR regulator of the mhp cluster. We have demonstrated that 3HPP and DHPP are the true and best activators of MhpR, whereas PP only induces no response. However, in vivo and in vitro transcription experiments have demonstrated that PP activates the MhpR regulator synergistically with the true inducers, representing the first case of such a peculiar synergistic effect described for a bacterial regulator. The three compounds enhanced the interaction of MhpR with its DNA operator in electrophoretic mobility shift assays. Inducer binding to MhpR is detected by circular dichroism and fluorescence spectroscopies. Fluorescence quenching measurements have revealed that the true inducers (3HPP and DHPP) and PP bind with similar affinities and independently to MhpR. This type of dual-metabolite synergy provides great potential for a rapid modulation of gene expression and represents an important feature of transcriptional control. The mhp regulatory system is an example of the high complexity achievable in prokaryotes.
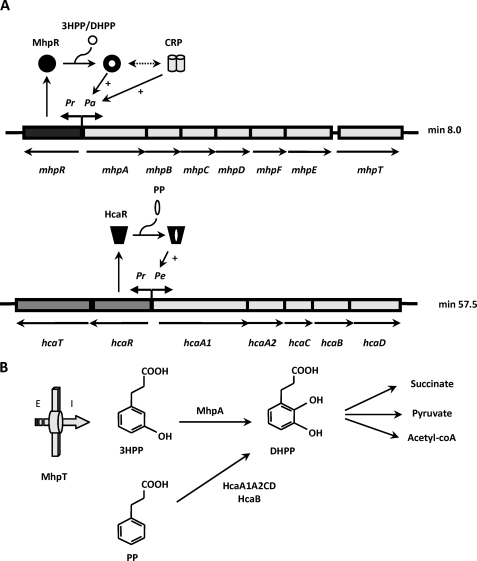
Phenylpropanoic and phenylpropenoic acids and their hydroxylated derivatives are widely distributed in the environment, arising from digestion of aromatic amino acids or as breakdown products of lignin and other plant-derived phenylpropanoids and flavonoids. The bacterial catabolism of these aromatic compounds plays a key role in recycling of such carbon sources in the ecosystem (1, 2). Most Escherichia coli strains are able to degrade these compounds via a meta-fission pathway (3). A scheme of the biochemical pathway for the catabolism of 3-hydroxyphenylpropionate (3HPP)2 and 3-hydoxycinnamate (3HCI) in E. coli K-12 is shown in Fig. 1B. The first step is catalyzed by the MhpA hydroxylase, which inserts one atom of molecular oxygen at the position 2 of the phenyl ring of 3HPP to give 3-(2,3-dihydroxyphenyl)propionic acid (DHPP). This intermediate is then converted to succinate, pyruvate, and acetyl-CoA through the action of a meta-cleavage hydrolytic route whose enzymes are encoded by the mhp cluster located at minute 8.0 of the genome (Fig. 1A), being the first hydroxyphenylpropionate degradation pathway described both at the biochemical and genetic levels (3–6). The mhp cluster is arranged as follows: (i) six catabolic genes encoding the initial monooxygenase (mhpA), the extradiol dioxygenase (mhpB), and the hydrolytic meta-cleavage enzymes (mhpCDFE); (ii) a gene (mhpT) that encodes a potential transporter; (iii) a regulatory gene (mhpR) which is adjacent to the catabolic genes but transcribed in the opposite direction (5). Promoters Pr and Pa control the expression of the divergently transcribed mhpR regulatory gene and mhp catabolic genes, respectively (Fig. 1A).
FIGURE 1.
Regulation and biochemistry of the mhp and hca clusters encoding the pathways for the catabolism of 3HPP and PP, respectively, in E. coli. A, the organization of the catabolic (mhpABCDFE and hcaA1A2CBD), transport (mhpT and hcaT), and regulatory (mhpR and hcaR) genes as well as their regulation by MhpR and HcaR and the global regulator CRP are represented. The thick arrows indicate the direction of gene transcription. Pr, Pa, and Pe are promoter regions. The black circle and the black trapezoid mean the inactive forms of the MhpR activator and the HcaR activator, respectively; the white circle represents the inducer, and + indicates transcriptional activation. The double-headed arrow means synergistic transcription activation by MhpR and CRP. B, scheme of the biochemistry of 3HPP and PP catabolic pathways. The 3HPP transport protein (MhpT) is represented by a thick arrow. E and I indicate outside and inside the cell, respectively. A brief scheme of the pathways including the final products and the first step for the transformation of 3HPP and PP into the common intermediate DHPP by the action of the MhpA monooxygenase and the HcaA1A2CD/HcaB dioxygenase/dihydrodiol-dehydrogenase, respectively, is shown.
Remarkably, the catabolism of 3HPP is connected with degradation of phenylpropionic acid (PP) through the common intermediate DHPP (6). The hca cluster encoding the enzymes responsible for the early steps of PP catabolism is located at minute 57.5 of the genome and contains (i) five genes encoding PP-dioxygenase (hcaEFCD; formerly named as hcaA1A2CD) and PP-dihydrodiol dehydrogenase (hcaB), (ii) a regulatory gene (hcaR), and (iii) a gene (hcaT) that might encode a transporter. The genes hcaR and hcaT are transcribed in the opposite direction from the other genes of cluster (Fig. 1A). The first biochemical step of PP degradation is catalyzed by a PP dioxygenase (HcaA1A2CD), which adds oxygen atoms to positions 2 and 3 of the PP phenyl ring and is subsequently oxidized by the HcaB dehydrogenase to give DHPP (Fig. 1B). Therefore, this compound links the catabolism of PP and 3HPP in E. coli.
As mentioned above, the 3HPP and PP catabolic pathways are regulated by two different regulatory proteins, MhpR and HcaR, respectively. Expression of mhp catabolic genes depends on the transcriptional activator MhpR belonging to the IclR family of transcriptional regulators (Fig. 1A) comprising more than 500 members identified from bacterial and Archaea genomes. IclR regulators typically have an N-terminal helix-turn-helix DNA binding motif and are linked by a long helix to the effector binding domain (the best defining trait of the family) located at the C-terminal domain. Sequence analysis of IclR regulators revealed a very low conservation of the amino acids residues involved in the effector binding, reflecting the chemical diversity of effector molecules recognized by the members of this family. HcaR belongs to the LysR family of transcriptional regulators and positively controls the neighboring genes, hcaA1A2CBD, in the presence of PP and negatively controls its own expression (Fig. 1A) (7, 8). MhpR behaves as a 3HPP-dependent activator of the Pa catabolic promoter by binding to its specific operator sequence centered at position −58 with respect to the transcription start site in Pa promoter. In contrast to HcaR, MhpR does not autoregulate its own expression (9). Expression of Pa promoter is also influenced by the cAMP receptor protein (CRP), which allows expression of the mhp catabolic genes when the preferred carbon source (glucose) is not available but 3HPP is present in the medium (Fig. 1A). MhpR shows a synergistic transcription activation mechanism with CRP (9).
Although transcriptional regulators often respond to one molecule which alters their binding to the promoter region, it has been described that in some particular occasions multiple effectors can regulate gene expression (10–12). The combined effect of these compounds on transcriptional control by a single regulator is in general poorly understood. Such combined effects could play a relevant role in those cases where two or more pathways share common intermediates and must be synchronically regulated, as appears to be the case for the mhp and hca pathways. In this work we have used different in vivo and in vitro experimental approaches to describe the synergistic activation of MhpR in response to different metabolites.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The E. coli strains as well as the plasmids used in this work are listed in Table 1. Unless otherwise stated, bacteria were grown in Luria-Bertani (LB) medium (16) at 37 °C. Growth in M63 minimal medium (20) was achieved at 37 °C using the necessary nutritional supplements and 20 mm glycerol as the carbon source. When required, the appropriate amounts of aromatic compounds were added to the medium. Antibiotics were used if indicated at the following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml), streptomycin (50 μg/ml), spectinomycin (50 μg/ml), and rifampicin (50 μg/ml).
TABLE 1.
Bacterial strains and plasmids with relevant genotype and phenotype
| Strain or plasmid | Relevant genotype and characteristic(s) | Source or reference |
|---|---|---|
| Strain | ||
| S17-1λpir | Tpr SmrrecA thi hsdRM+ RP4 ::2-Tc ::Mu ::Km Tn7 λpir phage lysogen | 13 |
| M15 | Strain for regulated high level expression with pQE vectors | Qiagen |
| MC4100 | F−, araD319 Δ(argF-lac)U169 rpsL150(Smr) relA1 flbB5301 deoC1 ptsF25 rbsR | 14 |
| AFMC | MC4100 spontaneous rifampicin-resistant mutant (Rifr) | 15 |
| AFMCRAL | AFMC with chromosomal insertion mini-Tn5Km mhpR/Pa-lacZ | 9 |
| AFMCAL | AFMC with chromosomal insertion mini-Tn5Km Pa-lacZ | 9 |
| MC1061 | hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi mhpRABCD | 16 |
| ED1061 | MC1061 derivative with chromosomal insertion Km in hcaA1 | 8 |
| ED1061AL | ED1061 with chromosomal insertion mini-Tn5Sm/Sp Pa-lacZ | This work |
| Plasmid | ||
| pUC18 | Apr; oriColE1 high copy number cloning vector, lacZα | 16 |
| pPAL | Apr, pUC18 derivative harboring the mhpR gene | 9 |
| pSJ3 | Apr; lacZ promoter probe vector, lacZ fusion flanked by NotI sites | 17 |
| pAL | Apr; pSJ3 containing a 0.5-kilobase DNA fragment to produce the mhpR/Pa-lacZ fusion | 9 |
| pRAL | Apr; pSJ3 containing a 1.1-kilobase DNA fragment to produce the mhpR/Pa-lacZ fusion | 9 |
| pJCD01 | Apr; oriColE1, polylinker of pUC19 flanked by rpoC and rrnBT1T2 terminators | 18 |
| pJCDAR | pJCD01 derivative containing Pr-Pa promoters | This work |
| pREP4 | kmr, plasmid that expresses the lacI repressor | Qiagen |
| pQE32 | Apr, oriColE1 T5 promoter lac operator, λ to/E. coli rrnB T1 terminators, N-terminal His6 | Qiagen |
| pQMH | Apr, pQE32 derivative containing the His6-mhpR gene | This work |
| pUT miniTn5 Sm/Sp | Apr Smr; delivery plasmid for mini-Tn5 Sm/Sp | 19 |
| pUTAL-2 | Apr Smr, pUTminiTn5 Sm/Sp containing the 4.4-kilobase NotI Pa-lacZ fragment from pAL | This work |
Plasmid DNA was prepared with the High Pure Plasmid Isolation Kit (Roche Applied Science). DNA fragments were purified with Gene-Clean Turbo (Q-BIOgene) and with High Pure PCR Product Purification kit (Roche Applied Science). Oligonucleotides were synthesized on an Oligo-1000 M nucleotide synthesizer (Beckman Instruments). All the cloned inserts and the DNA fragments were confirmed by DNA sequencing through an ABI Prism 377 automated DNA sequencer (Applied Biosystems Inc.). Transformation of E. coli cells was carried out by using the RbCl method or by electroporation (Gene Pulser, Bio-Rad) (16). Proteins were analyzed by SDS-PAGE as described previously (21). The protein concentration of cell extracts was determined by the method of Bradford (22) using bovine serum albumin as standard.
By means of REP4-mediated mobilization, plasmid pUTAL2 (Table 1) containing the mini-Tn5Sm/Sp hybrid transposon that expresses the Pa-lacZ fusion was transferred from E. coli S17–1λpir (Table 1) into kanamycin and streptomycin-resistant E. coli ED1061 through biparental filter mating as described previously (13). The reporter strains were selected among three different candidates with similar expression levels to avoid Pa unrelated lacZ expression rate caused by the position of the insertion of the minitransposon. The relevant genotype of the resulting strain ED1061AL is indicated in Table 1.
Chemicals
DHPP, 2,3-dihydroxycinnamic acid, and 4-hydroxy-2-ketopentanoic acid were provided by T. Bugg from University of Warwick, Coventry, UK. Phenylacetic acid (PA), 3-hydroxyphenylacetic acid (3HPA), 4-hydroxyphenylacetic acid, cinnamic acid (CI), 3-methylphenylpropionic acid, and phenol were purchased from Aldrich. Benzoic acid, 4-hydroxybenzoic acid, PP, 4-hydroxyphenylpropionic acid, 3HCI, 4-hydroxycinnamic acid, and indole were purchased from Sigma. 3HPP, 2-hydroxyphenylpropionic acid, 2-hydroxycinnamic acid, 3-methylcinnamic acid, 3-methoxycinnamic acid, 3-chlorocinnamic acid, 2-carboxycinnamic acid, 3-nitrocinnamic acid, phenylbutyric acid, dl-meta-tyrosine, and 3-methoxyphenylpropionic acid were purchased from Lancaster. 2-Hydroxyphenylacetic acid, 3-hydroxybenzoic acid, 4-fluorophenylacetic acid, and propylbenzene were acquired from Fluka. l-phenylalanine was purchased from Merck.
Overexpression and Purification of MhpR-His
Plasmid pQMH was constructed by cloning into the double-digested pQE32 plasmid a 850-bp BamHI/HindIII fragment harboring the mhpR gene that was PCR-amplified from the plasmid pPAL (Table 1) by using oligonucleotides MhpRhis5′ (5′- CGAGGATCCCGCAGAACAATGAGCAGACGG-3′; the engineered BamHI site is underlined) and MhpRhis3′ (5′-CCCAAGCTTTCAACGTAAATGCATGCCGC-3′; the engineered HindIII is underlined). The recombinant plasmid called pQMH (Table 1) carries the mhpR gene without the ATG start codon and with a His6 tag coding sequence at its 5′-end under control of the T5 promoter and two lac operator boxes. The His tail adds 13 amino acids (MRGSHHHHHHGIP) to the N-terminal end of the MhpR protein (expected mass, 32,712 Da). The His-tagged protein was overproduced in the E. coli M15 strain harboring the plasmid pREP4 (Table 1), which produces the LacI repressor to strictly control gene expression from pQE32 derivatives in the presence of isopropyl-1-thio-β-d-galactopyranoside. E. coli M15 (pREP4, pQMH) cells were grown at 37 °C in ampicillin- and kanamycin-containing LB medium until the cultures reached an absorbance at 600 nm of A0.5. Overexpression of MhpR-His was then induced for 4 h by the addition of 0.2 mm isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested, resuspended in 40 ml of buffer A (50 mm NaH2PO4, 300 mm NaCl, 100 mm imidazole, pH 8.0), and disrupted by passage through a French press (Aminco Corp.) operated at a pressure of 20,000 p.s.i. All purification steps were carried out at 4 °C. The cell lysate was centrifuged at 20,000 × g for 20 min, and the supernatant was ultracentrifuged at 150,000 × g for 60 min. Nucleic acids present in the supernatant were removed by precipitation with 2% streptomycin sulfate (Sigma) and stirred gently for 60 min, and after centrifugation at 23,500 × g for 20 min, the supernatant was dialyzed against buffer A and then applied to a 5-ml nickel-nitrilotriacetic acid-agarose column (Qiagen). The column was washed with 3 volumes of buffer A at a flow rate of 0.5 ml/min. Elution of purified MhpR-His protein was carried out with buffer B (50 mm NaH2PO4, 300 mm NaCl, 500 mm imidazole, pH 8.0) at a flow rate of 0.2 ml/min. The purified protein was dialyzed against buffer C (100 mm Tris-HCl, pH 7.5, 10% glycerol, and 500 mm KCl) and stored at −80 °C. Protein purity and molecular mass were confirmed by SDS/PAGE. MhpR-His concentration was determined spectrophotometrically by using the molar extinction coefficient at 280 nm (ϵ = 30,560 m−1cm−1) calculated on the basis of its amino acid sequence.
β-Galactosidase Assays
β-Galactosidase activities were measured with permeabilized cells as described by Miller (20).
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed at 20 °C with a Beckman Optima XL-A analytical ultracentrifuge (Beckman Coulter) equipped with UV-visible absorbance optics and using double sector Epon-charcoal centerpieces. Concentration gradients were monitored at 200,000 × g by measuring sample absorbance at 280 nm for detection of the concentration gradient. Measurements were performed at 15 μm MhpR-His in the absence and presence of 400 and 800 μm 3HPP. Differential sedimentation coefficients, c(s), were calculated by least-squares boundary modeling sedimentation velocity data using the program SEDFIT (23, 24). The sedimentation coefficients were corrected for buffer composition using the program SEDNTERP (25) to get the corresponding standard values (water and 20 °C).
Gel Retardation Assays (Electrophoretic Mobility Shift Assays)
The DNA fragment Pa-Pr used as a probe was amplified by PCR using 10 ng of plasmid pRAL (Table 1) as a template and the oligonucleotides PP6 (5′-CCGTCTGCTCATTGTTCTG-3′) and LAC57 (5′-CGATTAAGTTGGGTAACGCCAGGG-3′). The DNA fragment was labeled at its 5′-end using the phage T4 DNA polynucleotide kinase (10 units/μl) (Biolabs) and [γ-32P]ATP (3000 Ci/mmol) (Amersham Biosciences). The fragment (274 bp) was purified on a glass fiber column (High Pure PCR purification kit, Roche Applied Science). The reaction mixtures contained 30 mm Tris-HCl, pH 7.5, 100 mm KCl, 2 mm β-mercaptoethanol, 10% glycerol, 1 nm DNA probe, 50 μg/ml bovine serum albumin, 50 μg/ml salmon sperm (competitor) DNA, and purified MhpR-His in a 10-μl final volume. After incubation at 25 °C for 15 min, mixtures were loaded into 4% native polyacrylamide gel with buffer 0.5 × TBE (45 mm Tris borate pH 8.3, 1 mm EDTA). The gels were dried onto Whatman No. 3MM paper and exposed to Hyperfilm MP (Amersham Biosciences).
In Vitro Transcription Assays
Single-round transcription by E. coli RNAp was carried out under standard conditions (18) using as reaction buffer 40 mm Tris-HCl, pH 7.5, containing 10 mm MgCl2, 100 mm KCl, 200 μm cAMP, and 500 μg/ml acetylated bovine serum albumin. Plasmid pJCDAR was used as a DNA template. To construct this plasmid, the oligonucleotides PaPr5 (5′-CGCGAATTCCGGTTTTGTATTCCGTCTGC-3′) and PaPr3 (CGCGGATCCCATTTCAGTACCTCACGACTC-3′) were used for PCR amplification of the mhpR-mhpA intergenic region using the plasmid pRAL as DNA template, and the resulting fragment was cloned into the plasmid pJCD01 which contains the pUC19 polylinker between EcoRI and PstI sites flanked by the divergent terminators rpoCT and rrnBT1T2, respectively (Table 1). The reaction mixtures (9 μl) contained the plasmid DNA (5 nm) with CRP (200 nm) and MhpR (100 nm) or buffer. When required the effector molecules were also added, and the mixture was incubated at 25 °C for 20 min. Finally, 3 μl of RNAp (Epicenter) at 375 nm were added, and the mixture was incubated at 37 °C for 5 min in a final volume of 12 μl. Elongation was started by the addition of 3 μl of a prewarmed mixture containing 1 mm ATP, 1 mm GTP, 1 mm CTP, 50 μm UTP, 1 μCi of [α-32P]UTP and 500 μg/ml heparin in the same buffer to the template-polymerase mix, and the reaction was allowed to proceed for 15 min at 37 °C. Reaction was stopped by the addition of 10 μl of loading buffer containing 1% SDS. After heating at 70 °C, samples were subjected to electrophoresis on 6% sequencing gels, and run-off products were quantified using a Quantity One program (Bio-Rad).
Circular Dichroism (CD) Spectroscopy
Circular dichroism measurements were made with a Jasco J-715 spectropolarimeter using 0.1-cm path length quartz cuvettes. CD spectra were acquired at 20 °C in 20 mm potassium phosphate, 100 mm KCl buffer, pH 7.5, using a protein concentration of 6.5 μm (1-nm bandwidth, 4-s response, and 20-nm/min scan speed). Five spectra were averaged for each sample, and the spectrum of the buffer was subtracted. Thermal denaturation studies were performed with the same samples, overlaid with mineral oil. The CD signal at 220 nm was measured as the temperature was increased from 5 to 90 °C at 50 °C/h using a Peltier temperature control accessory. The content of secondary structure was estimated by analyzing the far-UV CD spectra using three different programs: CONTIN, which implements the ridge regression algorithm of Provencher and Glöckner (26); SELCON, which incorporates a self-consistent method together with the singular value decomposition algorithm to assign protein secondary structure (27), and CDNN based on the use of neural networks (28).
Fluorescence Measurements of Ligand Binding to MhpR-His
Fluorescence spectra of MhpR-His were acquired at 25 °C employing a Fluorolog spectrofluorometer with a 5-mm (excitation) × 10-mm (emission) quartz cuvette, with magnetic stirring to reduce photolysis. Tryptophan residues in the protein were selectively excited at 295 nm (1-nm bandwidth), and the emission was scanned from 305 to 400 nm (5-nm bandwidth). Samples of MhpR-His were diluted to a final concentration of 500 nm in 20 mm Tris-HCl, 100 mm KCl, pH 7.5, and a maximum volume of 10 μl ligand solution (prepared in the same buffer) was added to the protein solution to achieve final ligand concentrations in the range of 10–100 μm (1 ml total reaction volume). To avoid inner filter effects, the absorbance of ligand solutions at 295 and 340 nm was kept below 0.02 and 0.005, respectively. All measurements were corrected for the background emission of the buffer, free ligand solutions, and dilution.
Quenching of protein fluorescence by ligand binding was analyzed with the single-site binding model, employing the following expression,
where F is the fluorescence intensity at 335 nm (relative to the intensity in the absence of ligand), Kb = Kd−1 is the binding equilibrium constant, and Fmin is the minimal relative fluorescence ligand saturating conditions. Equation 1 was fitted to the data employing the Sigmaplot software to obtain the best-fitting binding parameters. Given the large excess of ligand over protein concentration, the total ligand concentration was to a good approximation equal to the free ligand concentration.
RESULTS
Effector Specificity of the MhpR Regulator
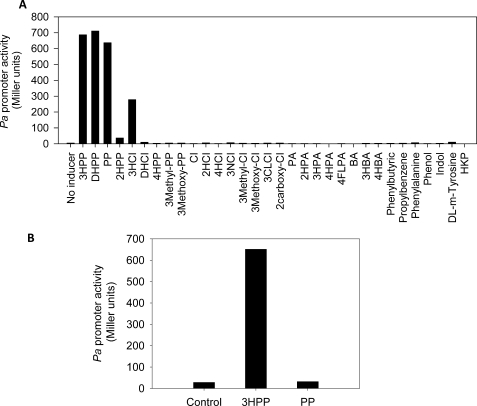
Previous studies showed that 3HPP and 3HCI, the natural substrates of the mhp pathway, induced the expression of the catabolic genes driven by the Pa promoter (8). To investigate other potential effectors of MhpR using lacZ as a reporter gene, we cultured the strain E. coli AFMCRAL (Table 1) in the presence of a large collection of aromatic compounds. The parental strain MC4100 is unable to transform 3HPP into DHPP because it contains a chromosomal deletion spanning the entire lac operon and the first genes of the mhp cluster (29). Fig. 2A shows that 3HPP, DHPP, and PP are the best inducers molecules of MhpR. Concerning the effect of PP, it is important to take into account that PP can be transformed in vivo into DHPP through the hca cluster contained in this strain. Furthermore, we observed that the other substrate of the mhp pathway, 3HCI, induces MhpR 2.5-fold less than 3HPP, whereas its derivative, 2,3-dihydroxycinnamic acid, does not induce the Pa promoter. These results suggest that the presence of a hydroxyl group at positions 2 or 3 of the PP aromatic ring is necessary for MhpR recognition. Nevertheless, the induction by 2-hydroxyphenylpropionic acid is 20-fold lower than by 3HPP, and 2-hydroxycinnamic acid is not an effector of MhpR. Moreover, the hydroxylated derivates at position 4, 4-hydroxyphenylpropionic acid and 4-hydroxycinnamic acid, are not inducers. In addition, the substitution of the hydroxyl group at position 3 of the aromatic ring of PP or CI by methyl (3-methylphenylpropionic acid; 3-methylcinnamic acid), methoxy (3-methoxyphenylpropionic acid; 3-methoxycinnamic acid), chloro (3-chlorocinnamic acid), or nitro (3-nitrocinnamic acid) groups did not allow the expression of mhp genes. Finally, the compound 2-carboxycinnamic acid does not act as an effector molecule. As mentioned above, the inducer DHPP is an intermediate metabolite also produced during the PP degradation (Fig. 1B). Therefore, it was necessary to determine whether the activation of Pa promoter by PP observed in E. coli AFMCRAL was because of the DHPP derived from PP, or the effect was because of a PP direct activation. To this end, β-galactosidase assays were performed using E. coli ED1061AL (pPAL) (Table 1). This is a mutant strain that carries a chromosomal disruption of the hcaA1 gene (8) harboring Pa::lacZ fusion in the chromosome (Table 1). This mutant strain does not transform PP into DHPP, and thus, the putative effect of PP can be tested without interferences. Remarkably, Fig. 2B shows that under these circumstances β-galactosidase activity was not detected in the presence of PP, demonstrating that it is not a real inducer of the mhp pathway.
FIGURE 2.
Inducer profile of mhp genes. A, E. coli AFMCRAL (mhpR/Pa-lacZ) was grown in LB medium in the presence of 1 mm concentrations of the tested compounds until the cultures reached an A600 of 0.8–1.0. 2HPP, 2-hydroxyphenylpropionic acid; 3HCI, 3-hydroxycinnamic acid; DHCI, 2,3-dihydroxycinnamic acid; 4HPP, 4-hydroxyphenylpropionic acid; 3-methyl-PP, 3-methylphenylpropionic acid; 3-methoxy-PP, 3-methoxyphenylpropionic acid; CI, cinnamic acid; 4HCI, 4-hydroxycinnamic acid; 2HCI, 2-hydroxycinnamic acid; 3NCI, 3-nitrocinnamic acid; 3CLCI, 3-chlorocinnamic acid; 3-methyl-CI, 3-methylcinnamic acid; 3-methoxy-CI, 3-methoxycinnamic acid; 2-carboxy-CI, 2-carboxycinnamic acid; 2HPA, 2-hydroxyphenylacetic acid; 3HPA, 3-hydroxyphenylacetic acid; 4HPA, 4-hydroxyphenylacetic acid; 4FlPA, 4-fluorophenylacetic acid; BA, benzoic acid; 4HBA, 4-hydroxybenzoic acid; HKP, 4-hydroxy-2-ketopentanoic acid. B, E. coli ED1061AL (pPAL) was grown in LB medium at an A600 of 0.8–1.0 in the presence of 1 mm 3HPP or PP. Results of one experiment are shown; values were reproducible in three separate experiments.
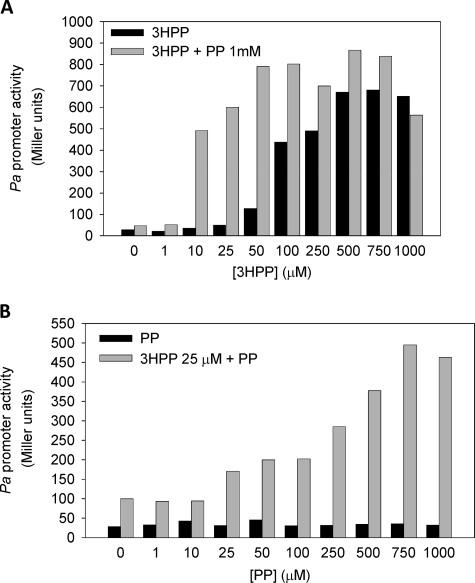
In Vivo Modulation of MhpR Activity by PP
Because degradation of PP in E. coli requires the induction of the mhp genes (Fig. 1), it could be anticipated that PP might also modulate the activity of the MhpR regulator that controls the expression of the mhp genes. To investigate the putative PP effect, we monitored the activity of Pa promoter in E. coli ED1061AL (pPAL) (Table 1). Cells were grown in LB medium in the presence of increasing concentrations of 3HPP and with or without 1 mm PP. Interestingly, the addition of PP increased the activity of MhpR; although it was not system-induced, we observed an additive effect that increased the activity of MhpR (Fig. 3A). This synergistic effect of PP was also observed when 3HPP was replaced in the assay by the other inducer molecules, i.e. 3HCI, and to a lower degree by DHPP (data not shown). These results suggest that PP does not compete with the natural inducers for the effector binding site at MhpR, but on the contrary, it produces a synergistic effect. To assess whether the synergistic effect was PP dosage-dependent, we measured the β-galactosidase activity of E. coli ED1061AL (pPAL) (Table 1) in the presence of 25 μm 3HPP and increasing amounts of PP. Fig. 3B shows that the PP synergistic effect was indeed dosage-dependent, reaching the maximum activation effect (∼15-fold) at 750 μm PP.
FIGURE 3.
Synergistic effect of PP on the induction of mhp genes produced by 3HPP. A, E. coli ED1061AL (pPAL) was grown in LB medium in the presence of increasing concentrations of 3HPP (1–1000 μm) (black bar) or combined with PP (1 mm) (gray bar) until the cultures reached an A600 of 0.8–1.0. Results of one experiment are shown; values were reproducible in three separate experiments. B, concentration-dependent effect of PP on the activation mediated by 3HPP. E. coli ED1061AL (pPAL) was grown in LB medium in the presence of increasing concentrations of PP (1–1000 μm) (black bar) or combined with a non-saturating concentration of 25 μm 3HPP (gray bar). Results of one experiment are shown; values were reproducible in three separate experiments.
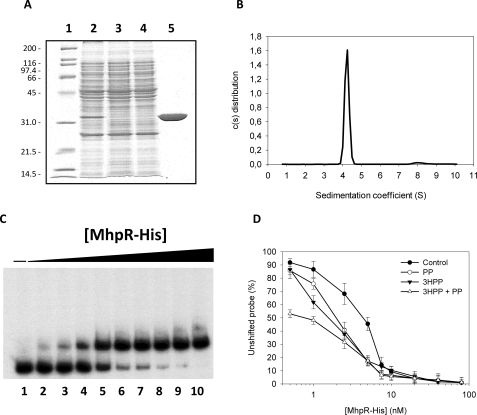
MhpR-His Overexpression and Purification of MhpR-His
To confirm the data obtained in vivo, we also characterized the PP synergistic effect by in vitro studies. For this purpose we cloned the gene mhpR in the plasmid pQE32 to express MhpR-His, as described under “Experimental Procedures.” First, and to test the functionality of the fusion MhpR-His regulator, the strain of E. coli AFMCAL (Table 1) harboring plasmid pQMH, expressing MhpR-His, and E. coli AFMCAL (pPAL), expressing the wild type MhpR regulator, were assayed for β-galactosidase activity. As expected, the levels of β-galactosidase activity were similar in both strains, suggesting that both proteins, MhpR and MhpR-His, were equally functional (data not shown). The MhpR-His protein was then expressed in E. coli M15 (Table 1) and purified to homogeneity as described under “Experimental Procedures.” The purification yield determined by densitometric scanning of SDS-PAGE gels was 85%. The purity and the expected molecular mass of the purified monomer were confirmed by SDS/PAGE (Fig. 4A). Sedimentation velocity experiments were carried out at different concentrations of MhpR in the presence and in the absence of 3HPP, PP, or DHPP and analyzed in terms of distribution of sedimentation coefficients, allowing an evaluation of protein homogeneity and self-association. Fig. 4B shows the sedimentation velocity data for a 15 μm MhpR-His solution in the absence of 3HPP, demonstrating that under these conditions MhpR sediments as a unique species with an s value of 4.4 ± 0.1 S, which is compatible with a globular protein dimer (according to SEDFIT and SEDNTERP calculations). Because the c(s) distributions showed a single major peak, c(s) values were transformed to a molar mass distribution with SEDPHAT program, obtaining a molar mass value also compatible with a protein dimer. The binding of effectors did not alter the oligomeric state of the protein, as demonstrated by sedimentation velocity studies of MhpR (15 μm) performed in the presence of 400 and 800 μm 3HPP (data not shown).
FIGURE 4.
A, overexpression and purification of MhpR-His protein. Analysis on a 12.5% SDS-PAGE of the purification of MhpR-His from E. coli M15 (pREP4, pQMH) cells. Lane 1, molecular mass markers shown in kDa; lane 2, soluble fraction of the crude extract from E. coli M15 (pREP4, pQMH); lane 3, extract that flows through the nickel-nitrilotriacetic acid-agarose column; lane 4, washing step; lane 5, purified MhpR-His protein loaded at 1.5 mg/ml. B, distribution of the sedimentation coefficients for the MhpR-His protein. c(s) sedimentation coefficient distribution was at a protein concentration of 0.5 mg/ml, a rotor temperature of 20 °C, and a rotor speed 200,000 × g. C, gel retardation analyses of MhpR-His binding to the Pa-Pr promoter in absence of aromatic compound. Increasing concentrations of purified MhpR-His were used: lane 1 (0 nm), lane 2 (0.5 nm), lane 3 (1 nm), lane 4 (2.5 nm), lane 5 (5 nm), lane 6 (7.5 nm), lane 7 (10 nm), lane 8 (20 nm), lane 9 (40 nm), and lane 10 (80 nm). Pa-Pr concentration was 1 nm. D, determination of the Kdfor MhpR-His binding to the mhpR-mhpA intergenic region in the absence (closed circles) and presence of 0.5 mm PP (open circles), 0.5 mm 3HPP (closed triangles), or at 0.25 mm both compound (open triangles). The Kd was the MhpR-His concentration at which 50% of the total probe was bound. This value was determined from the curves. The concentration of the probe is significantly lower than the MhpR-His concentration. Errors bars are S.D.
Effect of 3HPP and PP on the in Vitro Binding of MhpR-His to the mhpR-mhpA Intergenic Region
The effect of 3HPP and PP on MhpR-His binding to its target DNA was tested in vitro by electrophoretic mobility shift assay using the purified protein. Data displayed in Fig. 4C, lane 1, show that MhpR-His binds the DNA probe (mhpR-mhpA intergenic region) in the absence of any aromatic compound, with a Kd of 4.5 ± 0.6 nm. The addition of PP or 3HPP to the reaction mix slightly decreased the dissociation constant to a Kd of 2.1 ± 0.2 and 1.7 ± 0.3 nm, respectively (Fig. 4, lanes 2 and 3). Furthermore, when both PP and 3HPP were added together to the reaction mix (Fig. 4C, lane 4), formation of the MhpR-His-DNA complex was observed at even lower protein concentrations (Kd of 0.8 ± 0.3 nm), supporting the synergistic effect observed in vivo.
The Synergistic Activation of MhpR by PP Was Observed in Vitro
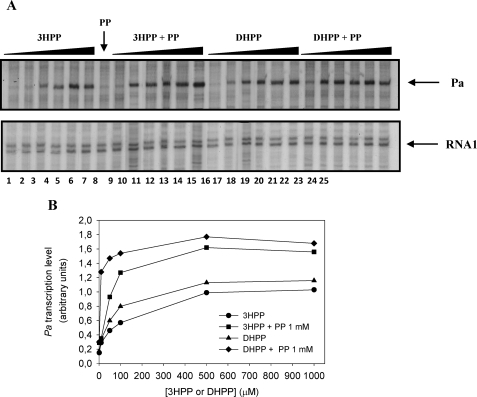
To study the functionality of MhpR-His as a transcriptional activator we designed a single-round in vitro transcription assay. We constructed plasmid pJCDAR that carries the whole mhpR-mhpA intergenic region including both Pr and Pa promoters (Fig. 1A). Although this template allowed us to monitor transcription from Pa and Pr promoters, we focused our analysis in the Pa promoter. As expected, no transcript band was detected in the absence of MhpR-His (data not shown), and there was an absolute requirement for the inducers (3HPP or DHPP) to detect the transcript band (Fig. 5A). The addition of increasing concentrations of 3HPP (1–1000 μm) significantly increased transcription from the Pa promoter (Fig. 5A, lanes 1–6), and the same effect was observed in the presence of DHPP (Fig. 5A, lanes 14–19). Interestingly, DHPP seems to be a better inducer than 3HPP (Fig. 5B).
FIGURE 5.
Effect of 3HPP and DHPP on the transcriptional activation from Pa promoter mediated by MhpR-His. A, single round in vitro transcription was carried out by using the plasmid pJCDAR as template. The concentrations of proteins were 100 nm RNAp, 100 nm MhpR-His, and 200 nm CRP. Lanes 1–6 contained 1, 10, 50, 100, 500, and 1000 μm 3HPP, respectively. Lane 7 contained 1 mm PP. Lanes 8–13 contained the same gradient but in the presence of 1 mm PP. Lanes 14–19 contained 1, 10, 50, 100, 500, and 1000 μm DHPP, respectively. Lanes 20–25 contained the same gradient but in presence of 1 mm PP. Arrows point to the Pa-derived mRNA (181 nucleotides) and the vector-derived RNA1 (108–109 nucleotides). B, Pa transcription levels (in arbitrary units) produced from pJCDAR template in the presence of 1–1000 μm 3HPP or DHPP in the absence and presence of 1 mm PP.
In agreement with the in vivo results, no transcript band was detected in the presence of 1 mm PP (Fig. 5A, lane 7). However, assays performed with MhpR-His and different proportions of PP and either 3HPP or DHPP (Fig. 5A, lanes 8–13 and lanes 20–25, respectively) confirmed the synergistic effect of PP on transcription from the Pa promoter, and the enhancement was even more evident when DHPP instead of 3HPP was used as the inducer molecule (Fig. 5B).
Changes in the MhpR-His Secondary Structure Induced by 3HPP and PP
To investigate the effect of 3HPP and PP on the protein secondary structure, the far-UV CD spectrum of MhpR-His was recorded in the absence and presence of 3HPP and PP. The CD spectrum is characterized by two minima at 210 and 220 nm and a maximum around 200 nm, indicative of α-helix structure (data not shown). The overall secondary structure composition of MhpR-His was estimated by deconvolution of the CD spectrum using three different methods (see “Experimental Procedures”) which provided a consensus average of 36% (±2%) α-helices, 16.0% (±0.6%) β-strands, and 48% (±2%) turns and non-regular structures. The presence of 3HPP induces a small but significant and reproducible decrease of the dichroic signal at 210–224 nm, and the overall change could be consistent with a slightly increase in the α-helix content (∼4%). Similar results were found by simultaneous addition of 3HPP and PP. Variations induced by PP alone were slightly lower but clearly indicated a direct interaction between PP and MhpR in the absence of 3HPP. The stabilization of MhpR-His derived from 3HPP or PP binding was analyzed by following the lost of secondary structure during thermal denaturation in the absence and presence of the ligands. Variations in the CD signal were monitored at 220 nm from 5 to 90 °C (data not shown). MhpR-His shows a single transition between 30 and 55 °C with a midpoint transition temperature, T½, of 47.1 °C, suggesting that MhpR-His denatures as a single cooperative unit. The addition of 1 mm 3HPP increased the T½ by 7.0 °C, whereas the same concentration of PP produced an increase of 5.5 °C. Remarkably, the simultaneous addition of PP and 3HPP, 1 mm each, did not result in a further stabilization of MhpR-His structure, as the T½ was identical to the value measured in the presence of 3HPP alone. Thus, the stabilization induced by the simultaneous presence of both compounds does not seem to be additive.
Spectrofluorometric Analyses of the Interactions between MhpR-His and Effectors
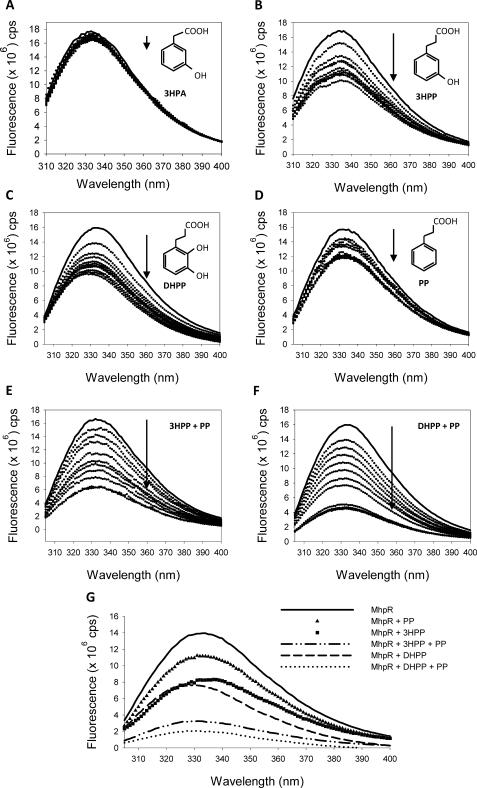
The interactions of MhpR-His with their cognate effectors were studied by fluorescence spectroscopy, a technique that detects local environmental changes in protein aromatic residues (30). MhpR has four tryptophan residues at positions 88, 106, 152, and 204. Residue Trp-88 is located adjacent to the linker region between the N- and C-terminal domains, whereas residues Trp-106, Trp-152, and Trp-204 are located within the C-terminal effector binding domain of MhpR-His (31). The addition of the inducers 3HPP and DHPP caused a significant concentration-dependent quenching of the fluorescence emission spectrum (Fig. 6, B and C, respectively; note as well a ∼5-nm blue-shift in the emission maximum with DHPP). PP also quenched the protein fluorescence in a concentration-dependent manner, corroborating its direct binding to MhpR-His inferred from CD experiments. In contrast, the decrease of the fluorescence intensity observed upon the addition of the non-inducer 3HPA (a 3HPP structural analog employed as a negative control) was very small (Fig. 6A), which suggests that the quenching effects caused by 3HPP, DHPP, and PP are highly specific.
FIGURE 6.
Quenching of intrinsic fluorescence of MhpR-His by different ligands. Fluorescence emission spectra of MhpR-His (0.5 μm) in the presence of increasing concentrations (from 10 to 100 μm) of 3HPA, used as negative control (A), 3HPP (B), DHPP (C), PP (D), 3HPP and PP (E), and DHPP and PP (F). The solid and dotted curves represent the spectra recorded in the absence and presence of increasing concentrations of ligand, respectively. The arrows show the decrease of the fluorescence. G, comparison of the fluorescence decrease caused by the addition of 3HPP, PP, or DHPP in excess (100 μm) with the decrease provided by the previous addition of PP at saturating concentration (100 μm) and the posterior addition of 3HPP or DHPP.
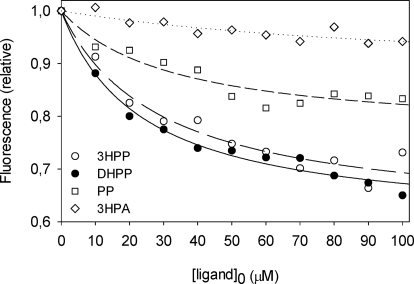
Fluorescence titration data (Fig. 7) were analyzed assuming a single set of binding sites (“Experimental Procedures”) to calculate the affinity constants and the maximum quenching of the three effectors. The best-fitting data yielded similar values for the dissociation constants: 29 ± 1 μm for 3HPP, 24 ± 2 μm for DHPP, and 30 ± 1 μm for PP, whereas the relative fluorescence under saturating conditions (Fmin) of PP (0.76 ± 0.03) was higher than for 3HPP (0.60 ± 0.03) and DHPP (0.59 ± 0.02). The related compound 3HPA, used as the negative control, showed a very weak fluorescence decrease and a higher dissociation constant (∼0.1 μm). Fluorescence studies also allowed us to further investigate the effect of PP on MhpR binding to its natural effectors. When 3HPP or DHPP were added simultaneously with PP, the fluorescence intensity of the protein decreased more than when 3HPP or DHPP was added alone (Fig. 6, E and F). The fluorescence decrease caused by the separate addition of 100 μm 3HPP, PP, or DHPP was compared in Fig. 6G with the decrease provided by the sequential addition of PP and 3HPP or DHPP (100 μm each). In contrast, DHPP addition to MhpR samples containing saturating concentrations of 3HPP does not modify the quenched emission spectra (data not shown). These results altogether are consistent with the notion that DHPP and 3HPP bind to the same site(s), whereas PP has a different binding locus in MhpR, as can also be concluded from the Tm for HhpR in the presence of either or both ligands.
FIGURE 7.
Quenching of MhpR-His (0.5 μm) tryptophan fluorescence as a function of ligand concentration at pH 7.5. Data points represent the decrease at the emission maximum in the presence of 3HPA used as a negative control (open diamonds), PP (open squares), 3HPP (open circles), and DHPP (closed circles). The curves through the data points were generated using Equation 1.
DISCUSSION
The MhpR activator belongs to the IclR family of transcriptional regulators. The characterized members of this family are involved in the regulation of diverse catabolic pathways ranging from the degradation of plant cell polysaccharides in the plant pathogen Erwinia sp (32, 33) to the metabolism of aromatic compounds in E. coli, Acinetobacter, and Pseudomonas (34–39). IclR family members typically have an N-terminal helix-turn-helix DNA binding motif followed by the C-terminal domain adapted to recognize a wide variety of small cellular metabolites as effectors. Sequence analysis of IclR regulators revealed a low conservation of the amino acid residues involved in effector binding. This reflects the large chemical diversity of inducer molecules recognized by members of this family showing the extraordinary structural evolutionary flexibility of these proteins. However, it should be notice that each regulator is able to bind highly specifically only a small number of effectors as shown by MhpR. In fact the in vivo analysis of the potential effectors of MhpR here performed showed that, among all the compounds tested, only the substrates of the mhp pathway (3HPP, 3HCI) and the first intermediate (DHPP) were efficient inducers of the regulator, as 2-hydroxyphenylpropionic acid only produced a very weak induction (Fig. 2A). Very closely related compounds having other substitutions either in the aromatic ring or the lateral chain did not activate the MhpR regulator. This agrees with the narrow effector specificity displayed by other members of the IclR family, with the unique exception of the TtgV regulator, which recognizes a wide range of structurally different effectors (40). The aromatic compounds were tested at 1 mm, as it is generally assumed that at this concentration these molecules can diffuse across biological membranes, making transport theoretically unnecessary (14, 41–45). Nevertheless, we cannot rule out that the absence of induction could be because of the inability of these compounds to enter the cell.
Within the IclR family only a small number of compounds have been formally identified as effectors so far (12, 34, 40, 44, 46–49). Moreover, the regulatory mechanism of these proteins and the structure of the effector binding domain in the free and the effector-bound states have been poorly characterized so far, with the sole exceptions of the regulator AllR and the model system IclR (12, 47).
Previous studies performed using crude cellular extracts showed that MhpR is a regulatory protein that binds to the Pa promoter at position −58 from the transcription initiation site (9). It was also demonstrated by gel retardation assays that the MhpR binding from crude extracts to the mhpR-mhpA intergenic region was improved in the presence of 3HPP, although it was not essential (9). The binding studies here performed by electrophoretic mobility shift assay with the purified MhpR-His protein indicate that 3HPP improved the retardation of the DNA probe, although MhpR-His alone can efficiently bind to the Pa promoter in vitro. These results rule out the possibility that the binding effect previously observed could be because of the presence of a 3HPP-like effector in crude cell extracts. Therefore, our current results demonstrate that MhpR recognizes the Pa operator sequence even in the absence of the effector. However, in vivo assays (9) and in vitro transcription experiments showed that initiation of transcription from Pa promoter is only achieved in the presence of the effectors molecules 3HPP or DHPP (Fig. 5, lanes 1–6 and lanes 14–19, respectively). Several IclR regulators involved in aromatic catabolic pathways bind DNA in the absence of their effector molecules (35, 37, 51, 52). Although no explanation has been put forward so far to account for this phenomenon, it could be hypothesized that these regulators could acquire a conformation that allows them to recognize and interact with the operator region DNA, but only in the presence of effectors might they promote formation of the closed complex between RNAp and the core promoter, which favors contacts with the non-consensus −35 boxes, as occurs in the case of the Pa promoter.
Analytical ultracentrifugation experiments showed that MhpR-His sediments (in the absence and in the presence of 3HPP) as a single species with an s value compatible with a protein dimer. The founding member of the family, IclR, is in a dynamic equilibrium between the dimeric and tetrameric states in solution and binds its operator as a tetramer (53, 54). TM-IclR from Thermotoga maritima is a dimer in solution, but it was proposed to bind as a dimer of dimers to its cognate operator (55). Other IclR members such as Pir from Erwinia chrysanthemi and PcaR and PcaU both from Acinetobacter sp. ADP1 were shown to be dimers in solution (37, 48, 56). However, TtgV is a tetramer in solution both in the presence and absence of effectors and also in its DNA-bound state (57). The addition of ligand drastically increased AllR tetramerization, whereas three species (monomer, dimer, and tetramer) were detected in solution in the absence of effectors (47). Nevertheless, very few data have been reported so far about the interactions of these regulators with aromatic effectors or synergistic regulatory mechanisms. In this sense, the synergistic effect of PP, which is not a MhpR effector, on transcriptional activation by 3HPP or 3HCI is surprising. Moreover, this effect depends on the PP concentration, showing maximum activation at 750 μm in our experimental conditions (Fig. 3B). A similar, but not identical, synergistic response has been described for the BenM regulator, belonging to the LysR family from Acinetobacter sp. strain ADP1, which responds to two metabolites, benzoate and cis,cis-muconate. In this particular case both effectors are true inducers, being as cis,cis-muconate is more effective than benzoate as the sole co-activator (10, 11). On the other hand, it has been described that two different metabolites, glyoxylate and pyruvate, are able to bind to the IclR regulator and show opposite effects. Glyoxylate disrupted the formation of the IclR/operator complex by favoring the dimeric state of the protein, whereas pyruvate increased the binding of the IclR to the target promoter by stabilizing the active tetrameric form of the protein (12).
In this work we have demonstrated for the first time that two compounds metabolized by different degradation pathways, 3HPP and PP, act synergistically to activate gene expression of the mhp operon through a sole regulatory protein. By electrophoretic mobility shift assay we have confirmed that MhpR-His binds its operator more efficiently when 3HPP and PP are present, as the Kd decreased ∼2-fold (Fig. 4C, lanes 2–4). In vitro transcription assays have shown that the level of transcription regulated by MhpR-His in response to both compounds was significantly higher than the sum of the effects because of each compound alone. This synergistic response was more efficient when DHPP instead of 3HPP was used as inducer (Fig. 5, A and B). The in vitro transcription experiments also suggest that DHPP is a more efficient inducer than 3HPP (Fig. 5). This result agrees with the hypothesis that during the catabolism of PP, the mhp pathway must be induced very quickly by low concentrations of DHPP to avoid the accumulation of this catecholic compound, which could became toxic for the cell (58). Nevertheless, this result does not appear to agree with the data obtained in vivo in which the PP synergistic effect was not observed in the presence of 1 mm DHPP (data not shown). However, a plausible explanation for this observation is that DHPP, as occurs with other catecholic compounds, might not enter into the cells in vivo as efficiently as 3HPP or 3HCI (6).
It is well known that ligand binding stabilizes protein structures, and thus, the analytical techniques that measure protein thermostability can be used to study protein-ligand interactions (50, 59–61). The transition temperature of MhpR is increased by 7 and 5.5 °C in the presence of 1 mm 3HPP or PP, respectively. However, the simultaneous addition of 3HPP and PP did not produce any additional increase of the T½, indicating that the presence of both effectors does not results in a further stabilizing of MhpR structure. In the case of the C-terminal domains of AllR and IclR regulators from E. coli, glyoxylate was identified as the strongest thermo-stabilizing inducer (12). The binding of 3HPP, DHPP, and PP to MhpR-His was characterized using fluorescence spectroscopy for monitoring changes in the environment of MhpR tryptophan residues. We also observed that the addition of 3HPP or DHPP to MhpR-His previously saturated with PP induced a further quenching of the fluorescence (Fig. 7), suggesting that the binding site of PP is different from that/those shared by DHPP and 3HPP. In related proteins, the structure of the BenM effector binding domain revealed two different binding sites for benzoate and cis,cis-muconate (11), whereas the IclR crystal structure has confirmed the binding of pyruvate and glyoxylate to a single effector recognition site (12).
The in vitro studies demonstrated that MhpR-His is the protein responsible for the dual compound response. The simultaneous binding of PP and 3HPP/DHPP may alter the conformation of MhpR-His in a way that significantly changes the regulator-DNA and/or regulator-polymerase interaction activating Pa transcription. Therefore, in the presence of both compounds MhpR-His would achieve a unique conformation that favors a higher level of transcription.
The physiological role of PP as a MhpR-His synergic effector can be explained by the convergence of PP and 3HPP catabolic pathways. As shown in Fig. 1B, cells need to activate the mhp pathway to catabolize PP, and such activation is produced as long as DHPP is synthesized. As mentioned above, this can explain why DHPP behaves in vitro as a better inductor than 3HPP, as DHPP should not be accumulated because, like other catechols, it could be highly toxic for the cells (58) and has to be cleaved as quickly as possible. Then, immediately after DHPP begins to be accumulated, the synergism between PP and DHPP on MhpR-mediated activation favors a rapid expression of the mhp genes, and the subsequent synthesis of the MhpB dioxygenase should reduce its toxic effect.
Summarizing, our data reinforce the preliminary assumption that the Pa promoter of the mhp pathway and its MhpR regulator represent an illustrative example of the great complexity achieved by certain bacterial regulatory systems. Such complexity is required to precisely control the expression of a key enzyme in two convergent metabolic pathways using two substrates, 3HPP and PP, which generate the same intermediate DHPP. This tightly regulated system uses two transcription factors for its activation, MhpR and CRP, and maximizes the efficiency of activation by using two synergistic effectors for the same transcription factor.
Acknowledgments
We thank Dr. Douglas V. Laurents for critical reading of the manuscript and revising the English version. We thank T. Bugg for providing DHPP, 2,3-dihydroxycinnamic acid, and 4-hydroxy-2-ketopentanoic acid. The technical work of A. Valencia is greatly appreciated.
This work was supported by Comisión Interministerial de Ciencia y Tecnología Grants GEN2006-27750-C5-3-E, BIO2006-05957, and CSD2007-00005 and Comunidad Autónoma de Madrid Grant P-AMB-259-0505.
- 3HPP
- 3-hydroxyphenylpropionate
- CRP
- cAMP receptor protein
- DHPP
- 3-(2,3-dihydroxyphenyl)propionic acid
- PA
- phenylacetic acid
- 3HPA
- 3-hydroxyphenylacetic acid
- PP
- phenylpropionic acid
- CI
- cinnamic acid
- 3HCI
- 3-hydroxycinnamic acid.
REFERENCES
- 1.Masai E., Harada K., Peng X., Kitayama H., Katayama Y., Fukuda M. (2002) Appl. Environ. Microbiol. 68, 4416–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith M. A., Weaver V. B., Young D. M., Ornston L. N. (2003) Appl. Environ. Microbiol. 69, 524–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burlingame R., Chapman P. J. (1983) J. Bacteriol. 155, 424–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burlingame R. P., Wyman L., Chapman P. J. (1986) J. Bacteriol. 168, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrández A., Garciá J. L., Díaz E. (1997) J. Bacteriol. 179, 2573–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Díaz E., Ferrández A., Prieto M. A., García J. L. (2001) Microbiol. Mol. Biol. Rev. 65, 523–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turlin E., Perrotte-piquemal M., Danchin A., Biville F. (2001) J. Mol. Microbiol. Biotechnol. 3, 127–133 [PubMed] [Google Scholar]
- 8.Díaz E., Ferrández A., García J. L. (1998) J. Bacteriol. 180, 2915–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres B., Porras G., Garcia J. L., Diaz E. (2003) J. Biol. Chem. 278, 27575–27585 [DOI] [PubMed] [Google Scholar]
- 10.Bundy B. M., Collier L. S., Hoover T. R., Neidle E. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7693–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezezika O. C., Haddad S., Clark T. J., Neidle E. L., Momany C. (2007) J. Mol. Biol. 367, 616–629 [DOI] [PubMed] [Google Scholar]
- 12.Lorca G. L., Ezersky A., Lunin V. V., Walker J. R., Altamentova S., Evdokimova E., Vedadi M., Bochkarev A., Savchenko A. (2007) J. Biol. Chem. 282, 16476–16491 [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo V., Timmis K. N. (1994) Methods Enzymol. 235, 386–405 [DOI] [PubMed] [Google Scholar]
- 14.Prieto M. A., García J. L. (1997) Biochem. Biophys. Res. Commun. 232, 759–765 [DOI] [PubMed] [Google Scholar]
- 15.Ferrández A., García J. L., Díaz E. (2000) J. Biol. Chem. 275, 12214–12222 [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17.Ferrández A., Miñambres B., García B., Olivera E. R., Luengo J. M., García J. L., Díaz E. (1998) J. Biol. Chem. 273, 25974–25986 [DOI] [PubMed] [Google Scholar]
- 18.Marschall C., Labrousse V., Kreimer M., Weichart D., Kolb A., Hengge-Aronis R. (1998) J. Mol. Biol. 276, 339–353 [DOI] [PubMed] [Google Scholar]
- 19.de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. (1990) J. Bacteriol. 172, 6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J. H. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 21.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 22.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 23.Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuck P. (2005) in Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., Rowe A. J. eds) The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 25.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S., Rowe A., Horton J. eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 26.Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 27.Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 28.Böhm G., Muhr R., Jaenicke R. (1992) Protein Eng. 5, 191–195 [DOI] [PubMed] [Google Scholar]
- 29.Perkins J. D., Heath J. D., Sharma B. R., Weinstock G. M. (1993) J. Mol. Biol. 232, 419–445 [DOI] [PubMed] [Google Scholar]
- 30.Lakowicz J. R., Kierdaszuk B., Callis P., Malak H., Gryczynski I. (1995) Biophys. Chem. 56, 263–271 [DOI] [PubMed] [Google Scholar]
- 31.Molina-Henares A. J., Krell T., Guazzaroni M. E., Segura A., Ramos J. L. (2006) FEMS Microbiol. Rev. 30, 157–186 [DOI] [PubMed] [Google Scholar]
- 32.Nasser W., Reverchon S., Robert-Baudouy J. (1992) Mol. Microbiol. 6, 257–265 [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., Jiang G., Cui Y., Mukherjee A., Ma W. L., Chatterjee A. K. (1999) J. Bacteriol. 181, 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiMarco A. A., Averhoff B., Ornston L. N. (1993) J. Bacteriol. 175, 4499–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerischer U., Segura A., Ornston L. N. (1998) J. Bacteriol. 180, 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kok R. G., D'Argenio D. A., Ornston L. N. (1998) J. Bacteriol. 180, 5058–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z., Houghton J. E. (1999) Mol. Microbiol. 32, 253–263 [DOI] [PubMed] [Google Scholar]
- 38.Bertani I., Kojic M., Venturi V. (2001) Microbiology 147, 1611–1620 [DOI] [PubMed] [Google Scholar]
- 39.Carmona M., Prieto M. A., Galán B., García J. L., Díaz E. (2008) in Microbial Biology: Genomics and Molecular Biology. (Diaz E. ed) pp. 97–147, Horizon Press, Norfolk, UK [Google Scholar]
- 40.Guazzaroni M. E., Krell T., Felipe A., Ruiz R., Meng C., Zhang X., Gallegos M. T., Ramos J. L. (2005) J. Biol. Chem. 280, 20887–20893 [DOI] [PubMed] [Google Scholar]
- 41.Kashket E. R. (1985) Annu. Rev. Microbiol. 39, 219–242 [DOI] [PubMed] [Google Scholar]
- 42.Olivera E. R., Miñambres B., García B., Muñiz C., Moreno M. A., Ferrández A., Díaz E., García J. L., Luengo J. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cebolla A., Sousa C., de Lorenzo V. (2001) Nucleic Acids Res. 29, 759–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arias-Barrau E., Olivera E. R., Luengo J. M., Fernández C., Galán B., García J. L., Díaz E., Miñambres B. (2004) J. Bacteriol. 186, 5062–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prieto M. A., García J. L. (1997) FEBS Lett. 414, 293–297 [DOI] [PubMed] [Google Scholar]
- 46.Reverchon S., Nasser W., Robert-Baudouy J. (1991) Mol. Microbiol. 5, 2203–2216 [DOI] [PubMed] [Google Scholar]
- 47.Walker J. R., Altamentova S., Ezersky A., Lorca G., Skarina T., Kudritska M., Ball L. J., Bochkarev A., Savchenko A. (2006) J. Mol. Biol. 358, 810–828 [DOI] [PubMed] [Google Scholar]
- 48.Popp R., Kohl T., Patz P., Trautwein G., Gerischer U. (2002) J. Bacteriol. 184, 1988–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H. B., Wang L. H., Zhang L. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pace C. N., McGrath T. (1980) J. Biol. Chem. 255, 3862–3865 [PubMed] [Google Scholar]
- 51.DiMarco A. A., Ornston L. N. (1994) J. Bacteriol. 176, 4277–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eulberg D., Schlömann M. (1998) Antonie van Leeuwenhoek 74, 71–82 [DOI] [PubMed] [Google Scholar]
- 53.Donald L. J., Hosfield D. J., Cuvelier S. L., Ens W., Standing K. G., Duckworth H. W. (2001) Protein Sci. 10, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto K., Ishihama A. (2003) Mol. Microbiol. 47, 183–194 [DOI] [PubMed] [Google Scholar]
- 55.Zhang R. G., Kim Y., Skarina T., Beasley S., Laskowski R., Arrowsmith C., Edwards A., Joachimiak A., Savchenko A. (2002) J. Biol. Chem. 277, 19183–19190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nomura K., Nasser W., Kawagishi H., Tsuyumu S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14034–14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guazzaroni M. E., Gallegos M. T., Ramos J. L., Krell T. (2007) J. Biol. Chem. 282, 16308–16316 [DOI] [PubMed] [Google Scholar]
- 58.Schweigert N., Zehnder A. J., Eggen R. I. (2001) Environ. Microbiol. 3, 81–91 [DOI] [PubMed] [Google Scholar]
- 59.Brandts J. F., Lin L. N. (1990) Biochemistry 29, 6927–6940 [DOI] [PubMed] [Google Scholar]
- 60.Weber P. C., Pantoliano M. W., Salemme F. R. (1995) Acta Crystallogr. D 51, 590–596 [DOI] [PubMed] [Google Scholar]
- 61.Gloss L. M., Matthews C. R. (1997) Biochemistry 36, 5612–5623 [DOI] [PubMed] [Google Scholar]