Abstract
The process of salvaging sugars released from extracellular matrix, during plant cell growth and development, is not well understood, and many molecular components remain to be identified. Here we identify and functionally characterize a unique Arabidopsis gene encoding an α-d-galacturonic acid-1-phosphate kinase (GalAK) and compare it with galactokinase. The GalAK gene appeared to be expressed in all tissues implicating that glycose salvage is a common catabolic pathway. GalAK catalyzes the ATP-dependent conversion of α-d-galacturonic acid (d-GalA) to α-d-galacturonic acid-1-phosphate (GalA-1-P). This sugar phosphate is then converted to UDP-GalA by a UDP-sugar pyrophosphorylase as determined by a real-time 1H NMR-based assay. GalAK is a distinct member of the GHMP kinase family that includes galactokinase (G), homoserine kinase (H), mevalonate kinase (M), and phosphomevalonate kinase (P). Although these kinases have conserved motifs for sugar binding, nucleotide binding, and catalysis, they do have subtle difference. For example, GalAK has an additional domain near the sugar-binding motif. Using site-directed mutagenesis we established that mutation in A368S reduces phosphorylation activity by 40%; A41E mutation completely abolishes GalAK activity; Y250F alters sugar specificity and allows phosphorylation of d-glucuronic acid, the 4-epimer of GalA. Unlike many plant genes that undergo duplication, GalAK occurs as a single copy gene in vascular plants. We suggest that GalAK generates GalA-1-P from the salvaged GalA that is released during growth-dependent cell wall restructuring, or from storage tissue. The GalA-1-P itself is then available for use in the formation of UDP-GalA required for glycan synthesis.
d-Galacturonic acid (d-GalA)3 is a quantitatively major glycose present in numerous plant polysaccharides including pectins and arabinogalactan proteins (1, 2). The synthesis of these polysaccharides requires a large number of glycosyltransferases and diverse nucleotide-sugar (NDP-sugar) donors (1, 3). Some of these NDP-sugars are formed by interconversion of pre-existing NDP-sugars and by salvage pathways. For example, the main pathway for UDP-GalA formation is the 4-epimerization of UDP-GlcA, a reaction catalyzed by UDP-GlcA 4-epimerase (4–6). However, in ripening Fragaria fruit d-GalA is incorporated into pectin (7). It is likely that a sugar kinase converts the d-GalA to GalA-1-P (8), which is then converted to UDP-GalA by a nonspecific UDP-sugar pyrophosphorylase (9). Myo-inositol may also be a source of GalA for polysaccharide biosynthesis (10).
Galacturonic acid is likely to be generated by enzyme-catalyzed hydrolysis of pectic polysaccharides in plant tissues. Polysaccharide hydrolase activities are present in germinating seeds (11, 12), in germinating and elongating pollen (13–15), and in ripening fruit (14). Thus, monosaccharide salvage pathways may be required for normal plant growth and development.
Numerous sugar-1-P kinases, including d-Gal-1-P kinase (16), l-Ara-1-P kinase (17), and l-Fuc-1-P kinase (18), have been described (19), but no d-GalA-1-P kinase has been identified in any species to account for the hydrolysis and recycle of pectic polymers. The subsequent pyrophosphorylation of UDP-sugars could be carried out by UDP-sugar pyrophosphorylases (20).
Here, we report the identification and characterization of a functional galacturonic acid kinase (GalAK). We compared the activity of GalAK with a previously uncharacterized Arabidopsis GalK and discussed the evolution of these sugar kinase members of the GHMP kinase.
EXPERIMENTAL PROCEDURES
cDNA Cloning and Site-directed Mutagenesis of Arabidopsis GalAK
Total RNA was extracted from the stems of 6-week-old Arabidopsis plants and used as a template to reverse-transcribe cDNA with oligo(dT) as primer (4). The coding sequence of Arabidopsis GalAK was amplified by PCR using 1 unit of high-fidelity proofreading Platinum DNA polymerase (Invitrogen), and 0.2 μm of each forward and reverse primers: 5′-AC atg tct tgg cct acg gat tct gag-3′ and 5′-GG TAC CTC gag aag aac acg agc agc gtc-3′. The reverse transcription-PCR product was cloned to generate plasmid pCR4-topoTA:At3g10700#11; and sequenced (GenBankTM FJ439676). The PciI-KpnI fragment (1276 bp) containing the full-length AtGalAK gene without the stop codon was subcloned into an Escherichia coli expression vector derived from pET28b (4), generating GalAK with an extension of six histidines at its C-terminal. GalK (At3g06580.1) and UDP-sugar pyrophosphorylase (At5g52560.1 “Sloppy”) were cloned using primer sets (GalK, S#1 5′-GCC atg gcg aaa ccg gaa gaa gta tca gtc-3′ + AS#2 5′-GAT ATC TCG AGg agg ttg aag atg gca gca c-3′) and (Sloppy, S#1 5′-atg gct tct acg gtt gat tcc-3′ + AS#2 5′-gaa gag aag tcc att tgt atc ttg-3′), respectively, and subsequently used to generate expression plasmids GalK with a His6 extension at the C-terminal (pET28a:At3g06580#3.1) and Sloppy with a His6 fusion at its N-terminal (pET28b:At5g52560# a73f/2#2).
Site-directed Mutagenesis
For mutagenesis, pET28b:At3g10700#11.3 encoding wild-type AtGalAK was used as template in the PCR-mediated QuikChange® XL site-directed mutagenesis kit (Stratagene). The primer pairs 5′-gt cct tta gga gAG cac att gat cac cag gg-3′ and 5′-cc ctg gtg atc aat gtg CTc tcc taa agg ac-3′ (mutagenized nucleotide residues are indicated in uppercase) were used to mutate codon 41A to E of GalAK (abbreviated as A41E); for mutation of Y250F of GalAK, the mutagenized primers were 5′-c aac cca gga tTt aat ctg cga gtt tct gag tg-3′ and 5′-c tcg cag att aAa tcc tgg gtt ggt ggt caa cg-3′; A368S mutation of GalAK was generated with the primer pairs 5′-ga ttc agc ggt Tca ggt ttc agg gga tgt tg-3′ and 5′-ct gaa acc tgA acc gct gaa tct agc tcc-3′.
For site-directed mutagenesis of GalK, pET28a:At3g06580#3.1 was used as template. The mutagenized primer pairs 5′-ctg ata gga gCg cac att gac tat gaa gga tac-3′ and 5′-gtc aat gtg cGc tcc tat cag att cac tct tcc-3′ were used to mutate codon 62E to A of GalK; for mutation of Y262F in GalK, the mutagenized primers were 5′-cg gct gct aag aat tTc aat aac agg gtc gtt g-3′ and 5′-cct gtt att gAa att ctt agc agc cgt gac cg-3′; to generate GalK A437S mutation, the primers were 5′-ga ctg acc gga Tct gga tgg ggc ggt tgc-3′ and 5′-gcc cca tcc agA tcc ggt cag tct tgc tcc-3′; to generate GalK S206G mutation, the primers were 5′-t gga aca caa GGt ggt ggg atg gac cag gc-3′ and 5′-g gtc cat ccc acc aCC ttg tgt tcc aat gtg tc-3′. Each 25-μl site-directed mutagenesis reaction consists of 10 ng of plasmid, 65 ng of primer pairs, 2.5 units of PfuTurbo DNA polymerase, and Stratagene supplied buffer and proprietary nucleotides. Reactions were carried out in a thermocycler using one cycle of 95 °C for 0.5 min, followed by 12 cycles each (95 °C, 0.5 min; 55 °C, 1 min; and 68 °C for 7 min). Reaction products were treated with 10 units of DpnI, subsequently plasmids were transformed to Dh5α-competent cells, and positive clones were selected on LB agar supplemented with kanamycin (50 μg/ml). DNA sequencing confirmed the intended single- or double-based substitution of each mutant constructs, and the resulting plasmids were named pET28b:AtGalAK#11.3A41E, pET28b:AtGalAK#11.3Y250F, pET28b: AtGalAK#11.3A368S, pET28a:AtGalK#3.1E62A, pET28a: AtGalK#3.1Y262F, pET28a:AtGalK#3.1A437S, and pET28a: AtGalK#3.1S206G. Expression of wild-type and mutant genes is under the T7 promoter; thus each plasmid was transformed to BL21(de3)plysS-derived E. coli strain (Novagen) for gene expression.
Protein Expression and Purification
E. coli cells harboring each plasmid construct or an empty vector were cultured for 16 h at 37 °C in LB medium (20 ml) supplemented with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml). A portion (8 ml) of the cultured cells was transferred into fresh LB liquid medium (250 ml) supplemented with the same antibiotics, and the cells then grown at 37 °C at 250 rpm until the cell density reached A600 = 0.6. The cultures were then transferred to 18 °C, and gene expression was induced by the addition of isopropyl β-d-thiogalactoside to a final concentration of 0.5 mm. After 24-h growth while shaking (250 rpm), the cells were harvested by centrifugation (6,000 × g for 10 min at 4 °C), resuspended in lysis buffer (10 ml of 50 mm Tris-HCl, pH 7.5, containing 10% (v/v) glycerol, 50 mm NaCl, 1 mm MgCl2, 0.5 mm EDTA, 1 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride) and lysed in an ice bath by 24 sonication cycles each (10-s pulse; 20-s rest) using a Misonix S-4000 (Misonix Inc., Farmingdale, NY) equipped with microtip probe. The lysed cells were centrifuged at 4 °C for 30 min at 20,000 × g, and the supernatant (termed s20) was recovered and kept at −20 °C.
His-tagged proteins were purified on a column (10-mm inner diameter × 150 mm long) containing Ni-Sepharose (2 ml, Qiagen) equilibrated with 50 mm sodium phosphate, pH 7.5, containing 0.3 m NaCl. The bound His-tagged protein was eluted with the same buffer containing increasing concentrations of imidazole. The fractions containing GalK or GalAK activities were supplemented with equal volume of 50% (v/v) glycerol and stored in aliquots at −80 °C, whereas Sloppy was stored in 40% glycerol (v/v). The concentration of proteins was determined using the Bradford reagent using bovine serum albumin as standard.
The molecular weights of the recombinant proteins were estimated by size-exclusion chromatography using a Waters 626 LC HPLC system equipped with a photo diode array detector (PDA 996) and a Waters Millennium32 work station. Separate solutions (0.5 ml) of GalK, GalAK, and Sloppy or a mixture of standard proteins (10 mg each of alcohol dehydrogenase (150 kDa), ovalbumin (48.9 kDa), ribonuclease A (15.6 kDa), and cytochrome c (12.4 kDa)) were separately chromatographed at 1 ml min−1 on a Superdex75 column (10 mm inner diameter × 300 mm long, Amersham Biosciences) equilibrated with 0.1 m sodium phosphate, pH 7.6, containing 0.1 m NaCl. The eluant was monitored at A280 nm, and fractions were collected every 20 s. Fractions containing enzyme activity were pooled and kept at −80 °C. To confirm the amino acid sequence of each recombinant protein, the purified recombinant proteins were fragmented with trypsin, and the resulting peptides were sequenced as described (4) by electrospray tandem mass spectrometry analyses (data not shown).
Enzyme Assays and NMR Spectroscopic Analysis
The following three assays were performed.
Formation of a Sugar-1-P from a Sugar and ATP
GalAK reactions (50-μl final volume) were performed in 100 mm Tris-HCl, pH 7.6 (or other buffer when indicated), containing 5 mm MgCl2, 2 mm ATP, 2 mm d-GalA, and 1.5 μg of recombinant GalAK. Reactions were kept at 37 °C for up to 10 min, and then terminated by adding an equal volume of chloroform. After vortexing (30 s) and centrifugation (12,000 rpm for 5 min, at room temperature), the upper aqueous phase was collected and chromatographed on a Q15 anion-exchange column (2 mm inner diameter × 250 mm long, Amersham Biosciences) or a TSK-DEAE-5-pw column (7.5 mm inner diameter × 75 mm long, Bio-Rad) using an Agilent Series 1100 HPLC system equipped with an autosampler, diode-array detector, and a Corona (ESA, Chelmsford, MA), and ChemStation software as described (4).
Formation of UDP-sugars from Monosaccharides Using a Kinase, UDP-sugar Pyrophosphorylase (Sloppy), and Yeast PPase
The coupled assays were performed in conjunction with HPLC, to easily detect the formation of reaction products by UV. The coupled assay has two steps. Unless otherwise noted see below for GalAK enzyme characterization, the kinase reaction (50-μl volume) conditions were as described above for recombinant GalAK for up to 10 min. The reactions were terminated in a boiling bath (1 min at 100 °C). After cooling, 2 mm UTP, 1 μg of purified recombinant UDP-sugar pyrophosphorylase (Sloppy, At5g52560), and 1 unit of yeast PPase (Sigma) were added (final volume, 60 μl). Sloppy is a reversible enzyme (i.e. can also convert PPi + UDP-sugar to sugar-1-P + UTP), thus to measure kinase activity, we included yeast PPase to drive the coupled-reaction forward toward formation of UDP-GalA by depleting the PPi to 2 × Pi. After 15 min at 37 °C, the reactions were terminated by adding chloroform, and the products were analyzed by anion-exchange chromatography as described above. Nucleotides and nucleotide sugars were detected by their UV absorbance using a Waters or Agilent photodiode array detector. The maximum absorbance for uridine-nucleotides and UDP-GalA was 261.8 nm in ammonium formate. The peak area of analytes was compared with calibration curves of internal standard (UDP-Gal or UDP-GalA).
Real-time 1H NMR Analysis of Sugar Phosphorylation
Two NMR-based assays were performed: phosphorylation assay and coupled assay. Phosphorylation reactions were performed in 50 mm sodium phosphate, pH 7.5, in a mixture of D2O:H2O (8:1 v/v, 180 μl) containing 5 mm MgCl2, 2 mm ATP, 2 mm GalA, and 1.5 μg of recombinant GalAK supplied in H2O buffer. In early experiments GalA was incubated without enzyme in the reaction buffer to monitor at equilibrium the ratio between the α and β configuration of the anomeric sugar prior adding the kinase and ATP. In the coupled assay, 2 mm UTP, 1 μg of recombinant Sloppy, and 1 unit of yeast PPase were also added. Immediately upon addition of enzyme, the reaction mixture was transferred to a 3-mm NMR tube. Real-time 1H NMR spectra were obtained using a Varian Inova 600-MHz spectrometer equipped with a cryogenic probe. Data acquisition was not started until ∼2 min after the addition of enzyme to the reaction mixture due to spectrometer set-up requirements (shimming). Sequential one-dimensional proton spectra were acquired over the course of the enzymatic reaction. All spectra were referenced to the water resonance at 4.765 ppm downfield of 2,2-dimethyl-2-silapentane-5-sulfonate. Processing of the data as covariance matrices was performed with Matlab (The Mathworks, Inc.).
Enzyme Properties of GalK and GalAK
To characterize the properties of recombinant GalK and GalAK, the kinase activities were tested under a variety of conditions: with various buffers, at different temperatures, different ions, or with different potential inhibitors. For the optimal pH experiments, 1.5 μg of recombinant enzyme (GalK or GalAK) was first mixed with 5 mm MgCl2, 2 mm ATP, and 100 mm of each individual buffer (Tris-HCl, phosphate, MES, MOPS, or HEPES). The optimal pH assays were initiated after the addition of specific sugar (Gal or GalA). Inhibitor assays were performed under standard assay conditions except for the addition of various additives (sugars and nucleotides) to the reaction buffer. These kinase assays were incubated for 10 min at 37 °C and were subsequently terminated by heat (1 min at 100 °C). After cooling, a coupled assay was used to detect the relative activity of the kinase by adding 2 mm UTP, 1 μg of purified recombinant UDP-sugar pyrophosphorylase (Sloppy), and 1 unit of yeast PPase (Sigma). The amount of UDP-sugar formed was calculated from HPLC UV spectra. Early work in our laboratory has shown that the amount of Sloppy used in the assays is sufficient to provide full conversion of sugar-1-P to UDP-sugar under the variety of pH and inhibitors tested. Similarly, the yeast PPase is fully active under these broad experimental conditions. For the experiments aimed at defining the optimal temperature the GalK or GalAK assays were performed under standard assay conditions except that reactions were incubated at different temperature for 10 min. Subsequently, the kinase activities were terminated (100 °C), and the relative kinase activity was measured by the coupled assay at 37 °C. For the experiments aimed at determining if the kinases required metals, the GalK or GalAK assays were performed with ATP, specific sugar (GalA or Gal) with a variety of ions. After a 10-min incubation at 37 °C the kinase assay was terminated by heat. After cooling, the amount of Mg2+ was adjusted to 5 mm, Sloppy, UTP, and yeast PPase were added, and the coupled assay was carried out at 37 °C. The amount of UDP-sugar formed was calculated from HPLC UV spectra.
For the experiments aimed at determining the ability of the kinases to utilize other sugars, GalK or GalAK assays were performed under standard assay conditions except for substitute the sugar (Gal or GalA) with different sugars (for example, mannose, fucose, and others). These kinase assays were incubated for 60 min (unless otherwise mentioned) at 37 °C and were subsequently terminated by heat (1 min at 100 °C). After cooling, reactions were separated by Q15 anion-exchange HPLC column (as above), and sugar or sugar-1-P peaks were monitored using Corona detector. The amount of sugar-1-P formed was calculated from HPLC Corona spectra.
Kinetics
The catalytic activity of GalAK was determined using the enzyme-coupled assay at 37 °C for 5 min using Tris-HCl, pH 7.6, containing MgCl2 (5 mm), variable concentrations of ATP (40 μm to 2 mm) with a fixed concentration of sugar (2 mm), and 0.27 μg of recombinant GalAK (6 pmol). In a separate series of experiments reactions were performed with a fixed amount of ATP (2 mm) and variable concentrations of sugar (20–400 μm). GalK kinetic assays were performed for 5 min in the same buffer as above, using 0.5 μg of recombinant enzyme (10 pmol), with variable concentrations of ATP (40 μm to 8 mm) and sugar (2 mm) or with ATP (2 mm) and variable concentrations of sugar (40 μm to 8 mm). Enzyme velocity data of the amount (μm) of phosphorylated sugar produced per second per microgram of enzyme, as a function of substrate concentrations, were plotted. The Solver tool (Excel version 11.5 program) was used to generate best-fit curve calculated by nonlinear regression analyses, and for calculation of Vmax and apparent Km.
Quantitative PCR of AtGalAK and AtGalK Transcripts in Different Tissues
RNA was extracted from roots, leaves, flowers, young siliques, and various sections of the stem (the lower region non-elongating bottom stems, the elongating middle stem region, and the upper stem regions), of 6-week-old Arabidopsis plants using RNeasy Plant Mini Kit (Qiagen). Two micrograms of total RNA was reverse transcribed using oligo(dT) as primer in 20-μl reactions (4), and the resulting cDNA was then diluted with sterile water to 200 μl prior for qPCR.
qPCR (final volume of 20 μl) was carried out using 2 μl of diluted cDNA, 0.5 μm each primer, and 10 μl of 2 × iQTM SYBR Green Supermix (Bio-Rad 170-8880) consisted of polymerase, dNTPs, company propriety buffer, and SYBR Green I. The qPCRs, in 96-well plate format, were carried using iCycler Real-Time PCR System (Bio-Rad). The thermal cycle conditions were: 95 °C for 3 min, and 40 cycle repeats of (95 °C for 10 s and 56 °C for 30 s), followed by 95 °C for 1 min, and 55 °C for 1 min. The GalAK primers for qPCR reactions were: qS#1 5′-gta tct ggg tct gcg gaa tg-3′ and qAS#1 5′-caa gct cgt ggt cca aag tc-3′; for GalK, qS#2 5′-ggt gct tct ccc caa ctc tt-3′ and qAS#2 5′-gaa tag cca tcg gca aca ct-3′; the qPCR primes for actin, used as reference gene control, were qS#3 5′-ggt aac att gtg ctc agt ggt gg-3′ and qAS#3 5′-aac gac ctt aat ctt cat gct gc-3′.
RESULTS
Identification, Cloning, and Characterization of Arabidopsis GalAK
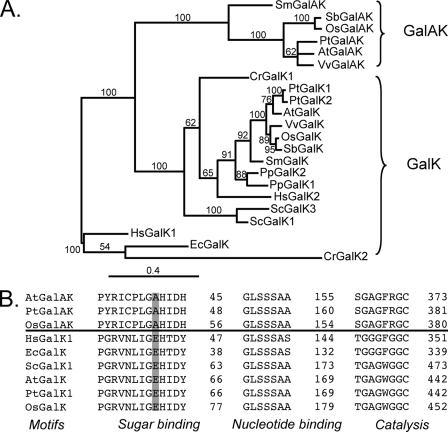
GalK from E. coli, yeast, human, and Pyrococcus furiosus (21–24) has conserved folds and amino acid domains. GalK is a member of the GHMP superfamily that includes galactokinase (G), homoserine kinase (H), mevalonate kinase (M), and phosphomevalonate kinase (P). All of these proteins contain three conserved motifs. The motif, PGRVNLIG(AE)HXDY, at the N-terminal region of the protein, is involved in sugar binding. The motif GL(GS)SSA, which is located ∼100 aa downstream of the N-terminal motif, is involved in binding the α- and β-phosphate groups of ADP (21), whereas the motif, GAGXG, located at the C-terminal region is involved in catalysis and transfer of the γ phosphate to the sugar (23).
To identify plant enzymes that contribute to the flux of monosaccharides into nucleotide-sugar metabolism, we searched the Arabidopsis genomic data base (www.ncbi.nlm.nih.gov/) for candidate GlcA-1-P and GalA-1-P kinases. BLAST analysis and sequence alignment of human, E. coli, and yeast GalK identified two Arabidopsis genes (At3g06580 and At3g10700).
At3g06580 encodes a protein (abbreviated GalK) that has 27 and 29% aa identity with the human and E. coli GalK, respectively, and has been shown to compliment a galk yeast mutant (16). However, the substrate specificities of the Arabidopsis enzyme were not determined. By contrast, At3g10700 encodes a protein (abbreviated herein GalAK) that has 26 and 27% aa sequence identity with human and E. coli GalK, respectively. Phylogenetic analysis (Fig. 1A) indicated that the GalAKs from different plant species such as Arabidopsis, rice (OsGalAK, Oryza sativa Os04g0608100), and poplar GalAK (PtGalAK, Populus trichocarpa JGI:427630) are distinguished from GalK in all other species.
FIGURE 1.
Sequence alignments and phylogenetic relationships of GalAK and GalK from different organisms. A, phylogenetic relationships of GalAK and GalK in different species. Protein sequences (see name and gene accession numbers below) were aligned and analyzed using Muscle 3.7 software (38) and the phylogenetic trees were created using MrBayes 3.1.2 software (39). Branch support values (more than 50%) are shown. The bar represents 0.4 protein substitutions per site. B, the conserved motifs of the putative GalAK sugar-binding, ATP-binding, and catalytic domains were aligned with GalK from different species. Note: the conserved AA Glu in the sugar binding motif of GalK is altered in GalAK to Ala. A. thaliana GalA-kinase (AtGalAK, At3g10700), Oryza sativa GalAK (OsGalAK, Os4g51880), Populus trichocarpa GalAK (PtGalAK, JGI:427630), Vitis vinifera GalAK (VvGalAK, GSVIVT00007137001), Sorghum bicolor GalAK (SbGalAK, Sb06g027910), Selaginella moellendorffii GalAK (SmGalAK, JGI:82393), A. thaliana Gal kinase (AtGalK, At3g06580), GalK from Saccharomyces cerevisiae (ScGalK1, 2AJ4 and ScGalK3, NP_010292), GalK from Homo Sapiens (HsGalK1, 1WUU and HsGalK2, 2A2D), E. coli (EcGalK, NP_308812), Chlamydomonas reinhardtii (CrGalK1, XP_001701527 and CrGalK2, XP_001691828), Physcomitrella patens GalK (PpGalK1, XP_001767042 and PpGalK2, XP_001756018), O. sativa (OsGalK, Os03g0832600), Populus trichocarpa (PtGalK1, XP_002314964 and PtGalK2, XP_002312315), V. vinifera (VvGalK, GSVIVT00033170001), S. bicolor (SbGalK, Sb01g002480), and S. moellendorffii (SmGalK, JGI:144964).
Arabidopsis GalK and GalAK have only 22% aa identity. The conserved glutamate (Glu-62) found in GalK proteins (Fig. 1B), is replaced by alanine in Arabidopsis, rice, and poplar GalAKs. We reasoned that in GalAK the non-polar alanine residue (Ala-41) may accommodate charged glycoses such as GalA or GlcA, whereas a glutamate residue would repulse the carboxyl group of these uronic acids (Fig. 1B). We cloned and expressed the Arabidopsis proteins in E. coli to determine if At3g10700 encodes a uronic acid kinase and to determine if At3g06580 encodes a protein with only GalK activity.
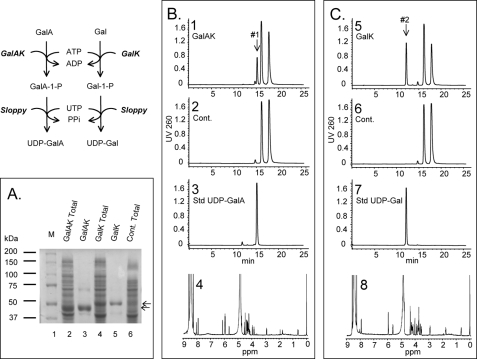
A highly expressed protein band (50 kDa) was detected after SDS-PAGE analysis of E. coli cells expressing GalK (Fig. 2A, lane 4, marked by the upper arrow). Similarly a band (46 kDa) was observed in cell overexpressing GalAK (Fig. 2A, lane 2). GalK and GalAK were column-purified and gave distinct 50 and 46 kDa protein bands, respectively (Fig. 2A, lanes 5 and 3), which agree with the calculated mass of the translated gene products.
FIGURE 2.
Expression, enzymatic activities of recombinant Arabidopsis GalA-kinase (GalAK), Gal-kinase (GalK), and identification of reaction products. A, SDS-PAGE of total soluble protein isolated from E. coli cells expressing Arabidopsis recombinant GalAK (lane 2), control empty vector (lane 6), or Arabidopsis GalK (lane 4), and of nickel-column purified recombinant GalAK (lane 3) and GalK (lane 5). B, HPLC chromatogram of GalAK coupled-based assay. A distinct UDP-GalA peak (marked by arrow #1, in panel 1) is detected in GalAK reaction but not in control (empty vector control cells, panel 2). The peak eluted at 15.2 min (arrow #1, in panel 1) was collected and confirmed after analysis by 1H NMR (panel 4) as UDP-α-GalA. For detailed proton NMR spectra of GalAK enzymatic product, see supplemental Fig. S1, A and B. The HPLC peaks in B (panels 1–3) are ATP (18 min), UTP+ADP (16.3 min), and UDP-GalA (15.2 min). The minor peak at 14.8 min is AMP contamination. C, HPLC chromatogram of GalK coupling-based assay. A UDP-Gal peak (marked by arrow #2 in panel 5) is detected in reaction contained GalK but not in empty vector control cells (panel 6). The peak eluted at 12.2 min (arrow #2, in panel 5) was collected and confirmed after analysis by 1H NMR (panel 8) as UDP-α-Gal. For detailed proton NMR spectra of the enzymatic product, see supplemental Fig. S2, A and B. The HPLC peaks in C (panels 5–7) are ATP (18 min), UTP+ADP (16.3 min), and UDP-Gal (12.2 min). The minor peak at 14.8 min is AMP contamination.
Preliminary experiments demonstrated that GalAK converts GalA and ATP to GalA-1-P and ADP (data not shown). We then developed a sensitive two-step coupled assay for the quantification of GalAK activity. First, the GalAK was reacted with ATP and GalA, and the enzyme was then heat-inactivated. The amount of GalA-1-P produced was quantified by measuring the amount of UDP-GalA formed in the presence of UDP-sugar pyrophosphorylase (abbreviated herein “Sloppy”). The enzymatic products of this second reaction were analyzed by anion-exchange chromatography (Fig. 2B, panel 1), which showed the presence of two new products with retention time characteristic of ADP (16.3 min overlap with UTP) and UDP-GalA (15.2 min). The peak marked #1 (Fig. 2B, panel 1) was collected, and its structure was confirmed by 1H NMR spectroscopy as UDP-α-d-GalA (Fig. 2B, panel 4, and supplemental Fig. S1, A and B) using authentic standards.
Characterization and Properties of GalAK
Crude recombinant GalAK is stable when stored at −20 °C. However, the purified enzyme lost >90% of its initial activity within 1 week of storage at this temperature. We found that GalAK could be stabilized for several months at −80 °C when diluted with glycerol (to 25% v/v) and flash-frozen in liquid nitrogen.
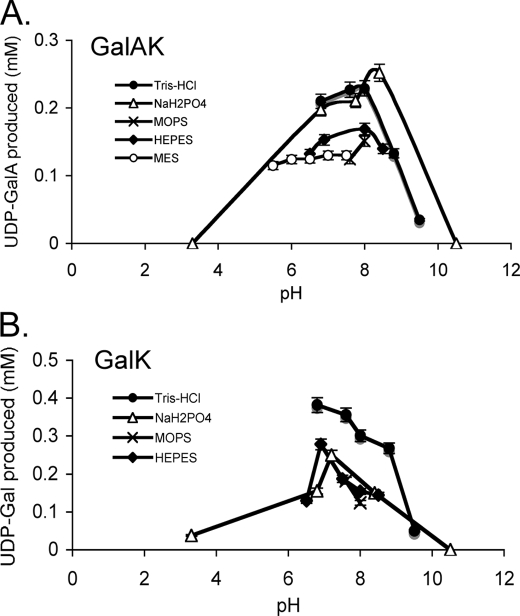
GalAK requires Mg2+, although other divalent cations, including Mn2+ and Ca2+, can substitute for magnesium. GalAK activity was 61% in the presence of Ca2+, but reduced by ∼32% in the presence of Mn2+ (Table 1). GalAK activity was completely abolished in the presence of EDTA. The recombinant GalAK was active between pH 3.3 and pH 9.5 (Fig. 3A), with maximum activity at pH 7.5–7.8 in Tris, or phosphate buffers. Activity was reduced when reactions were performed in HEPES, MOPS, or MES buffers.
TABLE 1.
GalAK and GalK require metals for activity
GalAK or GalK enzyme was mixed with additive (metal, EDTA, or water control) for 10 min on ice. Subsequently, ATP and appropriate sugar were added and kinase-assay was carried out under standard conditions. Each value is the mean of duplicate reactions, and the values varied by no more than ±5%.
| Metal | Relative GalAK activity | Relative GalK activity |
|---|---|---|
| 5 mm | % | |
| MgCl2 | 100 | 100 |
| MnCl2 | 32 | 42 |
| CaCl2 | 61 | 13 |
| CuCl2 | 0 | 0 |
| ZnSO4 | 0 | 0 |
| EDTA | 0 | 0 |
FIGURE 3.
The effects of buffer and pH on GalAK and GalK activities. The activity of GalAK (A) and GalK (B) were analyzed at different buffers (Tris-HCl, phosphate, MES, MOPS, and HEPES) at different pH levels. Except for the buffer, the kinase reactions were performed under standard conditions. Each value is the mean of triplicate reactions, and the values varied by no more than 5%.
GalAK was active between 4 °C and 42 °C, with maximum activity at 37 °C (Table 2). No GalAK activity was obtained when assays were performed at 55 °C. Preincubating the recombinant GalAK with GalA or ATP at different temperatures (55–75 °C, for 5 min), prior to performing the activity assays at 37 °C did not stabilize GalAK enzymatic activity.
TABLE 2.
The effect of temperature on GalAK and GalK activities
The enzymatic reactions were performed under standard conditions for each enzyme activity except for the reaction temperature. Each value is the mean of duplicate reactions, and the values varied by no more than ±5%.
| Temperature | Relative GalAK activity | Relative GalK activity |
|---|---|---|
| °C | % | |
| 4 | 24 | 26 |
| 25 | 53 | 75 |
| 30 | 88 | 101 |
| 37 | 100 | 100 |
| 42 | 80 | 100 |
| 55 | 0 | 1 |
| 65 | 0 | 0 |
To investigate the sugar specificity of GalAK, the enzyme was reacted for up to 60 min with ATP and different monosaccharides. No phosphorylation occurred with d-Gal, d-Glc, d-Fru, d-Man, d-gluconic acid, d-Xyl, d-GalNac, d-GlcNac, d-glucosamine, d-Ara, l-Ara, l-Rha, or l-Fuc. These monosaccharides (at 4 mm) also had no effect on GalAK activity in the presence of GalA. Thus we concluded that the recombinant GalAK is specific for GalA.
We next investigated the nucleotide triphosphate phosphorylation donor specificity of GalAK. Neither CTP, GTP, ITP, TTP, nor UTP are substrates for GalAK. The recombinant GalAK was also not inhibited by these tri-phosphate-nucleotides (4 mm). By contrast, enzyme activity was reduced by 60% in the presence of 4 mm ADP (Table 3). Other nucleotide diphosphates tested had no effect on activity. The specificity for GalA as substrate and ATP as nucleotide phosphate donor indicates the unique function for this GalA kinase. It is therefore likely that free GalA residues would be recycled back into the nucleotide sugar pool by GalAK and Sloppy.
TABLE 3.
The effect of potential inhibitors on GalAK and GalK activities
Inhibitors (at 4 mm), or control (water) were mixed with GalAK or GalK in 100 mm Tris-HCl, pH 7.6, for 10 min on ice prior to performing the enzymatic reaction under standard conditions for each enzyme activity. Each value is the mean of triplicate reactions, and the values varied by no more than ±10%.
| Nucleotide inhibitor | Relative GalAK activity | Relative GalK activity |
|---|---|---|
| 4 mm | % | |
| NAD+ | 100 | 111 |
| UTP | 105 | 112 |
| CTP | 100 | 112 |
| GTP | 104 | 107 |
| PPi | 109 | 110 |
| NADP+ | 93 | 95 |
| ITP | 104 | 110 |
| ADP | 60 | 77 |
| UDP | 105 | 111 |
| TTP | 101 | 96 |
| AMP | 101 | 95 |
| Control | 100 | 100 |
Real-time NMR Analysis of GalAK
Guyett et al. (25) have demonstrated the ability of 1H NMR spectroscopy to monitor enzymatic reactions in real-time. This procedure can be used to monitor the dynamics of an enzymatic assay, the appearance and disappearance of intermediates, and the detection of unstable products. Thus we used real-time 1H NMR spectroscopy to follow the reactions catalyzed by GalAK (Fig. 4A and supplemental Fig. S5, A–C).
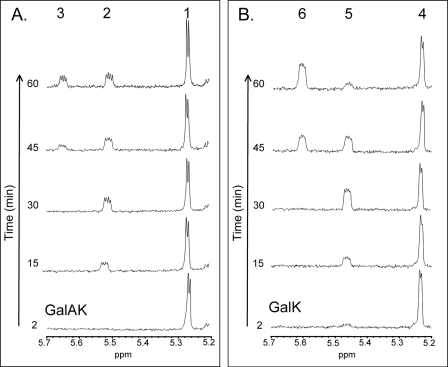
FIGURE 4.
Real- t ime 1 H NMR - based GalAK and GalK assays. Individual enzyme (GalK or GalAK) was mixed with sugar (Gal or GalA), buffer, and ATP, and the reaction was placed in an NMR tube. Approximately 2 min after enzyme addition and NMR shimming, NMR data were collected (bottom trace). Progressions of enzyme activity at 15 and 30 min are shown. At 30 min, UTP and Sloppy were added to the NMR tube, and the NMR spectra were continued to be collected. The sugar anomeric region of the 1H NMR spectrum is shown. A: NMR peak annotations of coupled GalAK-Sloppy assay are: peak 1, α-GalA at 5.27 ppm; peak 2, α-GalA-1-P at 5.52 ppm; and peak 3, UDP-α-GalA at 5.65 ppm. B: NMR peak annotations of coupled GalK-Sloppy assay are: peak 4, α-Gal, at 5.24 ppm; peak 5, α-Gal-1-P at 5.47 ppm; and peak 6, UDP-α-Gal at 5.61 ppm.
Before enzyme was added, two NMR signals of GalA could be observed corresponding to the anomeric protons α-d-GalA (5.27 ppm) and β-form of GalA (4.55 ppm) (see supplemental Fig. S5A). The ratio of α- to β-GalA form was constant (∼30%/70%). GalAK did not have mutarotase activity (data not shown). When GalAK was supplemented with ATP an immediate conversion of the α-d-GalA (5.27 ppm) into α-d-GalA-1-P (5.52 ppm) was observed (see time course of enzymatic progression in Fig. 4A and supplemental Fig. S5C). Addition of UTP followed by recombinant UDP-sugar pyrophosphorylase (Sloppy), resulted in the conversion of GalA-1-P to UDP-α-d-GalA indicated by the shift (5.65 ppm) (Fig. 4A). During the kinase reaction GalAK converted ATP to ADP indicating that the γ-phosphoryl of ATP is transferred to galacturonic acid.
Arabidopsis At3g06580 Is Galactokinase (GalK)
To determine the activity and specificity of recombinant Arabidopsis GalK we performed experiments similar to those described for GalAK. The HPLC-based assays show that recombinant GalK in the presence of ATP readily converts d-Gal to Gal-1-P and in the present of Sloppy to UDP-Gal (see Fig. 2C, panel 5, arrow #2). The UDP-Gal peak (12.2 min) was collected, and the product was confirmed by 1H NMR spectroscopy (Fig. 2C, panel 8, and supplemental Fig. S2, A and B) as UDP-α-d-Gal. GalK requires divalent metals for activity. However, unlike GalAK, 42 and 13% GalK activity were observed with Mn2+ and Ca2+, respectively, when compared with Mg2+ (Table 1). GalK has highest activity between 30 °C and 42 °C (Table 2), with an optimal pH of 7 to 8 at phosphate and Tris buffers. GalK activity was reduced somewhat when Tris was replaced by HEPES or MOPS buffer (Fig. 3B).
The GalK sugar specificity was also examined. In addition to d-Gal, the recombinant enzyme efficiently converts 2-deoxy-d-Gal to 2-deoxy-d-Gal-1-phosphate and ADP. GalK did not phosphorylate GalA, GlcA, d-Glc, d-Fru, d-Man, d-Xyl, d- GalNc, d-GlcNAc, d-glucosamine, d-Ara, l-Ara, l-Rha, or l-Fuc. Thus, we suggest that Arabidopsis At3g06580 encodes a GalK.
Purified GalK is less stable than GalAK, and within 2 days at −20 °C the enzyme lost all its activity. However, the purified recombinant protein was stable and retained activity when stored at −80 °C in the presence of glycerol (final 25%v/v). Interestingly, E. coli cultures containing the GalK expression plasmid and producing large amounts of GalK protein did not grow to cell densities above A600 = 0.8. This phenomenon was not observed with E. coli expressing GalAK or Sloppy.
Comparing Kinetic and Catalytic Properties of GalK and GalAK
Kinetics analyses of GalAK and GalK are summarized in Table 4. The apparent Km values for GalAK were 71 μm (GalA) and 195 μm (ATP), with a Vmax of 1 μm s−1μg−1 (GalA) and 0.5 μm s−1μg−1 (ATP), and kcat/Km values (s−1 mm−1) were 36 (GalA) and 6.3 (ATP). The apparent Km values for GalK were 701 μm (Gal) and 701 μm (ATP), with Vmax values of 3.5 μm s−1μg−1 (Gal) and 3.3 μm s−1μg−1 (ATP), and kcat/Km (s−1 mm−1) were 12.3 (Gal) and 11.8 (ATP). The Km for human GalK is 120 μm (Gal) and 350 μm (ATP). Thus, our kinetic data are comparable with kinetic analyses of other sugar kinases.
TABLE 4.
Enzyme kinetics of GalAK and GalK
GalAK activity was measured with varied concentrations of d-GalA (0.02–0.4 mm) and ATP (0.04–2.0 mm), after 5 min at standard conditions. GalK activity was measured with varied concentrations of d-Gal (0.04–8 mm) and ATP (0.04–8.0 mm), after 5 min at standard conditions. Enzyme velocities were plotted, and Solver software was used to generate best-fit curve and for calculation of Vmax and apparent Km. Each value is the mean of quadruple reactions, and the values varied by no more than ±10%. The kinetics of GalK obtained from H. sapiens, E. coli, S. cerevisiae, and P. furiosus ((40)), are displayed for comparison.
| Km(sugar) | Km (ATP) | Vmax (sugar) | Vmax (ATP) | kcat/Km | kcat/Km (ATP) | |
|---|---|---|---|---|---|---|
| μm | μm | μm s−1 μg−1 | μm s−1 μg−1 | s−1mm−1 | s−1mm−1 | |
| AtGalAK | 70.8 | 195 | 1.0 | 0.5 | 36 | 6.3 |
| AtGalK | 701 | 701 | 3.5 | 3.3 | 12.3 | 11.8 |
| HsGalK | 120 | 350 | 81.2 | 81.2 | 568 | 195 |
| EcGalK | 700 | 100 | 14 | 14 | 13.8 | 96.7 |
| ScGalK | 600 | 150 | 55.8 | 55.8 | 89.9 | 360 |
| PfGalK | 270 | 8 | 43.2 | 41.9 | 105 | 3439 |
Substrate Specificity of Site-directed Mutated GalAK and GalK Kinases
Sequence comparisons of functional GalK and homologous proteins in different species indicated that these proteins all have a glutamic acid residue (E62), that forms a hydrogen bond with the C6 hydroxyl group of galactose as depicted by structural studies (26). On the other hand, in genes encoding GalAK proteins from Arabidopsis and other plant species, the glutamate at position 41 is alanine (Fig. 1B). Thus, to investigate if this aa is critical for sugar binding we altered these aa residues in both GalAK and GalK using site-directed mutagenesis (Table 5). GalAKA41E did not phosphorylate either d-GalA or d-GlcA, suggesting that the Glu substitution is not compatible with the carboxyl group of uronic acid. Somewhat unexpectedly GalAKA41E did not phosphorylate Gal or any other sugar tested even with extended reaction times.
TABLE 5.
The effect of selective mutation on GalAK and GalK activities
Selected amino acids of wild-type GalA and Gal proteins were altered by site-directed mutagenesis (“Experimental Procedures”). The recombinant proteins (wt and mutants) were purified and GalAK and GalK activities were measured under standard conditions but for 15 min. The relative activities of mutant were compared to wt (100%), and the value is the mean of duplicate reactions, and the values varied by no more than ±5%.
| Enzyme mutation site | Sugar substrate | Relative enzyme activity |
|---|---|---|
| % | ||
| GalAKWT | d-GalA | 100 |
| d-GlcA | 0 | |
| GalAKA41E | d-GalA | 0 |
| d-GlcA | 0 | |
| GalAKY250F | d-GalA | 90 |
| d-GlcA | 50 | |
| GalAKA368S | d-GalA | 90 |
| d-GlcA | 0 | |
| GalKWT | d-Gal | 100 |
| 2-d-Gal | 100 | |
| GalKE62A | d-Gal | 0 |
| 2-d-Gal | 0 | |
| GalKY262F | d-Gal | 80 |
| 2-d-Gal | 57 | |
| GalKA437S | d-Gal | 60 |
| 2-d-Gal | 40 | |
| GalKS206G | d-Gal | 95 |
| 2-d-Gal | 85 | |
| GalNac | 30 |
Analyses of the human galk gene of GALK-deficient individuals with galactosemia has provided information on those aa residues that have a critical role in catalysis (26). Based on these studies we selected Ala-368 and Ala-437 for mutation. GalAKA368S and GalKA437S reduced phosphorylation activities by only ∼40 and 10%, respectively, when compared with native enzyme.
We next sought to convert GalAK to a GlcAK by changing aa residues we believed to interact with the C-4 hydroxyl of GalA. This assumption was based on mutation of GalK of E. coli (24), which showed that a tyrosine mutation significantly changed enzyme substrate specificity. The tyrosine mutation of GalAK to phenylalanine (GalAKY250F) resulted in phosphorylation of GlcA with kcat/Km values of 0.76 s−1 mm−1. Interestingly, the Y250F-mutated enzyme was still active toward GalA but with reduced catalytic efficiency kcat/Km decreased to 24 s−1 mm−1 (GalA), and increase Km 395 μm (GalA).
The ability of AtGalAKY250F to phosphorylate GlcA led us to explore how similar mutations affect Arabidopsis GalK (Table 5). AtGalKY262F failed to phosphorylate d-Glc but was still fully active toward Gal suggesting that other amino acids are involved in the interaction with the C-4 hydroxyl of monosaccharides. Based on mutation studies of E. coli GalK (24), and our GalAKY250F, we identified Ser-206 of AtGalK as a potential aa to be altered in accepting sugars with C-2 modifications. Indeed, converting Ser-206 to Gly allowed AtGalKS206G to phosphorylate GalNac. However, converting the conserved Glu-62 to Ala in AtGalK (GalKE62A) did not result in the phosphorylation of GalA.
Gene Expression of GalAK and GalK in Arabidopsis
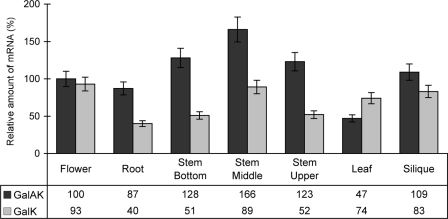
GalAK and GalK were shown by qPCR (see Fig. 5) to be expressed in roots, stems, leaves, flowers, and young siliques. Stem and floral tissue had the highest expression of the mRNAs of GalK and GalAK. Interestingly, GalAK expression was almost 1.5-fold higher in the elongating middle stem region than in the lower or upper stem region.
FIGURE 5.
The expression of GalAK and GalK genes in different Arabidopsis tissues. Total RNA obtained from different tissues was reverse transcribed, and the relative amounts of GalAK and GalK mRNAs compared with Actin mRNA were quantified by real-time qPCR. Each value is the mean of triplicate reactions, and the values varied by no more than 10%.
DISCUSSION
We have described the cloning and biochemical characterization of a plant sugar kinase (GalAK) that in the presence of ATP specifically phosphorylates α-d-GalpA to α-d-GalpA-1-P and ADP. GalA kinase did not phosphorylate the β-anomer of GalA under our assay conditions (supplemental Fig. S5C). The selectivity of the GalAK for the α-anomeric configuration of monosaccharides is not unique to this kinase. The GalK from human (23, 27), bacteria (24), and yeast (22), as well as the Arabidopsis GalK, are also selective for the α-configuration of the sugar (see Fig. 4B and supplemental Fig. S6C). At equilibrium in solution ∼30% of galactose exists in the α configuration and ∼64% in the β-form (supplemental Fig. S6A); a similar anomeric ratio was observed with GalAK (supplemental Fig. S5A). In humans, a galactose mutarotase converts the thermodynamically more stable β-Galp to the bioactive α-form (27). Thus, galactose mutarotase may be required to rapidly generate and maintain a sufficient pool of α-galactose for GalK-catalyzed conversion of Gal to Gal-1-P. To date no Gal or GalA mutarotase activities have been identified in plants. Nevertheless, the possibility cannot be discounted that such mutarotases do exist. Indeed, plant genes with “mutarotase-like” motifs exist, but their functions remain to be determined. There is also the possibility that the chelation of cations such as Ca2+ or Mg2+ by free GalA could non-enzymatically shift the anomeric equilibrium to the α-form. For example, Angyal et al. (28) have shown that the α-anomeric configuration of GalA is stabilized in the presence of selected metal ions that form a complex with O5 and a carboxylate oxygen of the monosaccharide. Thus, plant cells may not require a GalA mutarotase if the Ca-α-GalA complex is indeed more stable than the β-anomer of GalA. The fact that GalAK is active in the presence of calcium ion could explain that, if such a GalA-cation complex is made, it will serve a substrate for the kinase.
Genes encoding GalK are highly conserved in Eukaryote, Bacteria, and Archaea. By contrast, genes encoding GalAK, appear to be restricted to land plants. Using a Blast search of the Arabidopsis GalAK sequence with a translated genomic data base we found other closely related sequences (Fig. 1). A GalAK homolog was also found in the lycopodiophyte Selaginella moellendorffii (a member of one of the oldest extant vascular plant groups) but is not present in the moss Physcomitrella patens, a plant that lacks a vascular system. Thus, the appearance of GalAK may have been one of the factors required for the development of the plant vascular system. However, we have also identified a sequence with low identity (36%) with GalAK in one of the two recently sequenced Micromonas (www.ncbi.nlm.nih.gov/). A full understanding of the origins and evolution of GalAK will require complete genomic sequences from a wide range of embryophytes, streptophytes, and chlorophytes.
Many plant genes have undergone rounds of duplications thereby generating large gene families (29). However, GalAK exists as a single copy gene even in those plant species (e.g. poplar, maize, and legumes) that are recent polyploids (30). Interestingly, GalK has two gene copies in Populus, Physcomitrella, and Chlamydomonas, but in Arabidopsis and rice genome for example, only one copy of GalK exist. The reason that GalAK exists as a single copy gene is not known. However, the possibility cannot be discounted that additional copies of the gene interferes with as yet unidentified aspects of plant metabolism. Such a hypothesis can now be investigated by determining whether overexpression of GalAK has an adverse effect on plant growth and development.
UDP-GlcA 4-epimerases (4–6) are believed to be the predominant enzymes involved in formation of the UDP-GalA that is required in the Golgi for the synthesis of plant cell wall glycans. Here we have provided biochemical evidence showing that UDP-GalA is also generated from GalA in a series of reactions involving GalAK and Sloppy (a UDP-sugar PPase). Such data are consistent with early metabolic labeling studies demonstrating that plants can use exogenous GalA for the synthesis of wall polysaccharides (8).
The observation that the GalAK gene is transcribed in all plant tissues suggests that most if not all plant cells have the ability to recycle GalA. Hydrolytic enzymes involved in pectin depolymerization, including endo- and exogalacturonases, have been identified in numerous plant tissues, including pollen tubes (31), fruits (32), seeds (12), floral and leaf tissues (14), and stem (33). Thus, free GalA formed by depolymerization of pectic polysaccharides present in plant organs and storage tissues may provide an immediate non-photosynthetic source of carbon that enters the sugar nucleotide pathway. The newly formed UDP-GalA can then be used for cell wall synthesis by the rapidly developing seedlings and during pollen tube germination and growth.
The primary wall of growing plant cells is itself a potential source of free GalA. The assembly and restructuring of this wall is believed to be required to allow plant cells to expand and grow. These processes may result in the generation of GalA that could then be recycled by specific GalA transporter into the cell as a substrate for GalAK (see a model in Fig. 6). Although no plasmamembrane-localized transporters that import GalA into the cytosol have been identified, a large gene family with over 50 monosaccharide transporter-like genes exist in Arabidopsis (34), perhaps one of them is GalA transporter. Baluska et al. (35) have suggested that the pectic fragment can be endocytosed. Previous studies indicate that exogalacturonates are present in the cytosol of pollen (36). Thus hydrolyzes of pectins may provide an alternative route for the production of cytosolic GalA.
FIGURE 6.
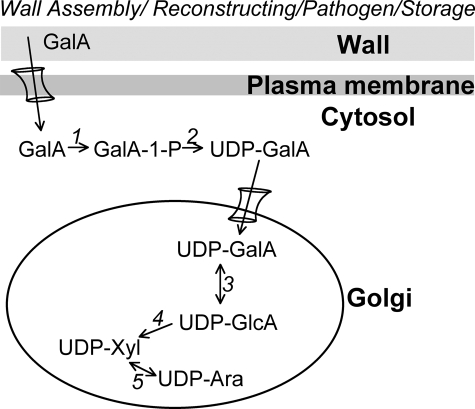
A model for GalA recycling during wall assembly and restructuring. GalA hydrolytically released from wall polymers during wall assembly and restructuring or from storage tissues is transferred to the cytosol by plasmamembrane sugar transporter(s). GalA is subsequently phosphorylated by GalAK (1, At3g10700) and then converted to UDP-GalA in the cytosol by Sloppy (2, a UDP-sugar PPase, At5g52560). UDP-GalA can then be transferred by a UDP-GalA transporter into the Golgi, where it can be directly incorporated by glycosyltransferases to glycans or enter the nucleotide-sugar interconversion pathway. UDP-GlcA 4-epimerase (3, e.g. At2g45310) interconverts UDP-GalA and UDP-GlcA. UDP-GlcA is converted to UDP-xylose by UDP-GlcA decarboxylase (4, Uxs, e.g. At3g53520). UDP-xylose 4-epimerase (5, Uxe, e.g. At1g30620) interconverts UDP-xylose and UDP-arabinose.
The combined activity of GalAK and Sloppy results in the formation of UDP-GalA in the cytosol. Thus, a Golgi-localized UDP-GalA transporter (Fig. 6) will be required for this nucleotide sugar to be used in glycan synthesis. Previous studies have suggested that such a UDP-GalA transporter exists, because intact microsomes are able to take up exogenous UDP-[14C]GalA (37).
In summary, our results demonstrate unambiguously that vascular plants have enzymes that convert GalpA to UDP-GalpA. This salvage pathway is likely common in every cell and has an important role in recycling GalA released from cell wall glycans during plant growth. Elucidating the metabolic pathways that involve GalAK and the evolutionary origins of this plant-specific enzyme are major new challenges for plant scientists.
Supplementary Material
Acknowledgments
We thank Andy Martin and Ben Mullenbach, the undergraduate students who helped in the project, and Dr. John Glushka and Yingnan Jiang for their guidance and assistant with the early NMR experiments, and Haibao Tang who helped with the phylogeny analysis. Lastly, we especially thank Malcolm O'Neil of the Complex Carbohydrate Research Center for his constructive comments on the manuscript.
This work was supported by National Science Foundation Grant IOB-0453664 (to M. B.-P.), by U.S. Dept. of Agriculture Grant 2002-35318-12620 (to M. B.-P.), and by the BioEnergy Science Center, which is supported by the Office of Biological and Environmental Research in the DOE Office of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- d-GalA
- α-d-galacturonic acid
- GalAK
- α-d-galacturonic acid-1-phosphate kinase
- HPLC
- high-performance liquid chromatography
- PPase
- pyrophosphorylase
- MES
- 4-morpholineethanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid
- qPCR
- quantitative PCR
- aa
- amino acid(s)
- GalA-1-P
- α-d-galacturonic acid-1-phosphate
- GalK
- α-d-galactose-1 phosphate kinase.
REFERENCES
- 1.Mohnen D. (2002) in Pectins and Their Manipulation (Knox J. P., Seymour G. B. eds) pp. 52–98, CRC Press/Blackwell Publishing, Oxford [Google Scholar]
- 2.Yates E. A., Valdor J. F., Haslam S. M., Morris H. R., Dell A., Mackie W., Knox J. P. (1996) Glycobiology 6, 131–139 [DOI] [PubMed] [Google Scholar]
- 3.Mohnen D., Bar-Peled M., Somerville C. R. (2008) in Biosynthesis of Plant Cell Walls (Himmel M. ed) pp. 94–187, Blackwell Publishing, Oxford [Google Scholar]
- 4.Gu X., Bar-Peled M. (2004) Plant Physiol. 136, 4256–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Usadel B., Schlüter U., Mølhøj M., Gipmans M., Verma R., Kossmann J., Reiter W. D., Pauly M. (2004) FEBS Lett. 569, 327–331 [DOI] [PubMed] [Google Scholar]
- 6.Mølhøj M., Verma R., Reiter W. D. (2004) Plant Physiol. 135, 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loewus F. A. (1961) Ann. N.Y. Acad. Sci. 92, 57–78 [DOI] [PubMed] [Google Scholar]
- 8.Neufeld E. F., Feingold D. S., Ilves S. M., Kessler G., Hassid W. Z. (1961) J. Biol. Chem. 236, 3102–3105 [PubMed] [Google Scholar]
- 9.Litterer L. A., Schnurr J. A., Plaisance K. L., Storey K. K., Gronwald J. W., Somers D. A. (2006) Plant Physiol. Biochem. 44, 171–180 [DOI] [PubMed] [Google Scholar]
- 10.Young L. C., Stanley R. G., Loewus F. A. (1966) Nature 209, 530–531 [Google Scholar]
- 11.Ren C., Kermode A. R. (2000) Plant Physiol. 124, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sitrit Y., Hadfield K. A., Bennett A. B., Bradford K. J., Downie A. B. (1999) Plant Physiol. 121, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes-Davis R., Tanaka C. K., Vensel W. H., Hurkman W. J., McCormick S. (2005) Proteomics 5, 4864–4884 [DOI] [PubMed] [Google Scholar]
- 14.Hadfield K. A., Bennett A. B. (1998) Plant Physiol. 117, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch M., Cheung A. Y., Hepler P. K. (2005) Plant Physiol 138, 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan C. P., Tugal H. B., Baker A. (1997) Plant Mol Biol. 34, 497–506 [DOI] [PubMed] [Google Scholar]
- 17.Sherson S., Gy I., Medd J., Schmidt R., Dean C., Kreis M., Lecharny A., Cobbett C. (1999) Plant Mol Biol. 39, 1003–1012 [DOI] [PubMed] [Google Scholar]
- 18.Kotake T., Hojo S., Tajima N., Matsuoka K., Koyama T., Tsumuraya Y. (2008) J. Biol. Chem. 283, 8125–8135 [DOI] [PubMed] [Google Scholar]
- 19.Neufeld E. F., Feingold D. S., Hassid W. Z. (1959) Arch Biochem. Biophys 83, 96–100 [DOI] [PubMed] [Google Scholar]
- 20.Kotake T., Yamaguchi D., Ohzono H., Hojo S., Kaneko S., Ishida H. K., Tsumuraya Y. (2004) J. Biol. Chem. 279, 45728–45736 [DOI] [PubMed] [Google Scholar]
- 21.Hartley A., Glynn S. E., Barynin V., Baker P. J., Sedelnikova S. E., Verhees C., de Geus D., van der Oost J., Timson D. J., Reece R. J., Rice D. W. (2004) J. Mol. Biol. 337, 387–398 [DOI] [PubMed] [Google Scholar]
- 22.Thoden J. B., Sellick C. A., Timson D. J., Reece R. J., Holden H. M. (2005) J. Biol. Chem. 280, 36905–36911 [DOI] [PubMed] [Google Scholar]
- 23.Thoden J. B., Timson D. J., Reece R. J., Holden H. M. (2005) J. Biol. Chem. 280, 9662–9670 [DOI] [PubMed] [Google Scholar]
- 24.Hoffmeister D., Thorson J. S. (2004) Chembiochem 5, 989–992 [DOI] [PubMed] [Google Scholar]
- 25.Guyett P., Glushka J., Gu X., Bar-Peled M. (2009) Carbohydr. Res. 344, 1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoden J. B., Holden H. M. (2003) J. Biol. Chem. 278, 33305–33311 [DOI] [PubMed] [Google Scholar]
- 27.Timson D. J., Reece R. J. (2003) BMC Biochem. 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angyal S. J., Greeves D., Littlemore L. (1988) Carbohydr. Res. 174, 121–131 [Google Scholar]
- 29.Tang H., Wang X., Bowers J. E., Ming R., Alam M., Paterson A. H. (2008) Genome Res. 18, 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sémon M., Wolfe K. H. (2007) Curr. Opin. Genet. Dev. 17, 505–512 [DOI] [PubMed] [Google Scholar]
- 31.Dubald M., Barakate A., Mandaron P., Mache R. (1993) Plant J. 4, 781–791 [DOI] [PubMed] [Google Scholar]
- 32.Pressey R. (1984) Eur J Biochem. 144, 217–221 [DOI] [PubMed] [Google Scholar]
- 33.Pressey R., Avants J. K. (1977) Plant Physiol 60, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Büttner M. (2007) FEBS Lett. 581, 2318–2324 [DOI] [PubMed] [Google Scholar]
- 35.Baluska F., Hlavacka A., Samaj J., Palme K., Robinson D. G., Matoh T., McCurdy D. W., Menzel D., Volkmann D. (2002) Plant Physiol. 130, 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barakate A., Martin W., Quigley F., Mache R. (1993) J. Mol. Biol. 229, 797–801 [DOI] [PubMed] [Google Scholar]
- 37.Sterling J. D., Quigley H. F., Orellana A., Mohnen D. (2001) Plant Physiol. 127, 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huelsenbeck J. P., Ronquist F. (2001) Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- 40.Verhees C. H., Koot D. G., Ettema T. J., Dijkema C., de Vos W. M., van der Oost J. (2002) Biochem. J. 366, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.