Abstract
Disruption of Golgi α-mannosidase II activity can result in type II congenital dyserythropoietic anemia and induce lupus-like autoimmunity in mice. Here, we isolated a mutant human embryonic kidney (HEK) 293T cell line called Lec36, which displays sensitivity to ricin that lies between the parental HEK 293T cells, in which the secreted and membrane-expressed proteins are dominated by complex-type glycosylation, and 293S Lec1 cells, which produce only oligomannose-type N-linked glycans. Stem cell marker 19A was transiently expressed in the HEK 293T Lec36 cells and in parental HEK 293T cells with and without the potent Golgi α-mannosidase II inhibitor, swainsonine. Negative ion nano-electrospray ionization mass spectra of the 19A N-linked glycans from HEK 293T Lec36 and swainsonine-treated HEK 293T cells were qualitatively indistinguishable and, as shown by collision-induced dissociation spectra, were dominated by hybrid-type glycosylation. Nucleotide sequencing revealed mutations in each allele of MAN2A1, the gene encoding Golgi α-mannosidase II: a point mutation that mapped to the active site was found in one allele, and an in-frame deletion of 12 nucleotides was found in the other allele. Expression of the wild type but not the mutant MAN2A1 alleles in Lec36 cells restored processing of the 19A reporter glycoprotein to complex-type glycosylation. The Lec36 cell line will be useful for expressing therapeutic glycoproteins with hybrid-type glycans and as a sensitive host for detecting mutations in human MAN2A1 causing type II congenital dyserythropoietic anemia.
Mammalian N-linked glycosylation is characterized by significant chemical heterogeneity generated by an array of competing glycosidases and glycosyltransferases (1). The structural analysis of recombinant glycoproteins, such as human erythropoietin (2, 3), has illustrated the capacity of mammalian expression systems for generating diverse N-linked glycans.
Heterogeneity develops during egress of a glycoprotein through the secretory system (1). N-linked glycosylation is initiated in the rough endoplasmic reticulum (ER)4 by the co-translational transfer of Glc3Man9GlcNAc2 to the asparagine residues of the glycosylation sequon. In the absence of protein misfolding, hydrolysis by ER α-mannosidase I plus α-glucosidase I and II results in the transfer of glycoproteins dominated by the D1,D3 isomer of Man8GlcNAc2 glycans to the Golgi apparatus (4). Further processing by Golgi α-mannosidases IA–C generates Man5GlcNAc2 (5–7), the principle substrate for UDP-N-acetyl-d-glucosamine:α-3-d-mannoside β1,2-N-acetylglucosaminyltransferase I (GnT I). The action of this enzyme yields classic hybrid-type glycans with mannosyl 6-antennae and processed 3-antennae (1). In the absence of the GnT III-mediated addition of bisecting GlcNAc, the two terminal α-mannose residues of the 6-antenna of hybrid-type glycans are cleaved by Golgi α-mannosidase II, forming mono-antennary complex-type glycans. These may then be processed by N-acetylglucosaminyltransferases, generating multiantennary complex-type glycans of enormous potential heterogeneity following the sequential transfer of monosaccharides such as galactose, N-acetylgalactosamine, fucose, and N-acetylneuraminic acid (8).
The importance of this carbohydrate diversity in metazoan biology is illustrated by the disease phenotypes that manifest when the biosynthesis of particular glycoforms is disrupted. In humans, about 12 congenital disorders of glycosylation (CDG) have been identified with defects in the biosynthesis of N-linked glycans (9). One disorder characterized by changes in glycosylation is congenital dyserythropoietic anemia type II (hereditary erythroblastic multinuclearity with a positive acidified serum lysis test (HEMPAS)) (10, 11). HEMPAS is a heterogenous autosomal recessive disorder that renders erythrocytes prone to lysis. Although the precise molecular basis of HEMPAS remains to be determined, it is characterized by either a reduction in β1→4-galactosyltransferase, GnT II, or, in some patients, Golgi α-mannosidase II activity (11, 12). Interestingly, the increase in cell surface terminal mannose in mice deficient in Golgi α-mannosidase II leads to autoimmunity through chronic activation of the innate immune system (13, 14).
Lectin-resistant (Lec) cell lines harboring loss- or gain-of-function mutations affecting the biosynthesis of N-glycans have emerged as powerful tools for the investigation of these disorders (15). For example, genetic complementation using Lec2, containing a mutation in the cytosine monophosphate sialic acid transporter, was used to identify a novel CDG, type IIf (16). Lectin-resistant cell lines can also be used as hosts to study naturally occurring mutations, as in the case of CHO Lec23 cells used to screen α-glucosidase I mutations in CDG, type IIb (17). Other applications of lectin-resistant cell lines include the expression of specific glycoforms of therapeutic glycoproteins. Manipulating the structure of their carbohydrate moieties modulates the pharmacological properties of glycoproteins by altering their bioactivity, serum half-life, and/or tissue tropism (18). For example, β-glucocerebrosidase expressed in CHO Lec1 cells (deficient in GnT I activity) exhibits mannosylation and improved macrophage uptake for the treatment of Gaucher disease (19). Lectin-resistant CHO cell lines have also been used to improve the crystallizability of glycoproteins for structural determination by x-ray crystallography (20–24).
The expression of therapeutic glycoproteins as one or more defined “glycoforms” is essential for their optimization and may even be necessary to obtain regulatory approval (25). To this end, eukaryotic expression systems have been developed that allow glycosylation to be controlled. Recently, Pichia pastoris-based strains with human glycosyltransferases have been established, allowing the expression of glycoforms with oligomannose-, hybrid-, and some complex-type glycans (26, 27) and even sialylated complex-type structures (28). However, mammalian expression remains the dominant technology in industrial settings, presumably because of its reliability for the expression of human secreted glycoproteins.
Although the majority of lectin-resistant cell lines have been generated using CHO cells, no Golgi α-mannosidase II-deficient CHO cell line has been generated thus far (15). Furthermore, only one human lectin-resistant cell line, i.e. GnT I-deficient (Lec1) HEK 293S cells (29), has been produced. Hybrid-type glycosylation has been reported to accumulate in ricin-resistant baby hamster kidney cells (30, 31); however, these cells contain a reduced but detectable level of cell-surface complex-type glycans, consistent with an incomplete ablation of Golgi α-mannosidase II activity (31). Moreover, the hybrids from one of these lines contain a trimannosyl rather than pentamannosyl core and appear to be heavily influenced by GnT II deficiency, resulting in the formation of what are now commonly called monoantennary complex-type glycans (32–34). We now describe the isolation of an HEK 293T cell line mutated at the MAN2A1 locus and deficient in Golgi α-mannosidase II activity via selection with ricin.
EXPERIMENTAL PROCEDURES
Chemical Mutagenesis of HEK 293T Cells and Isolation of Mutants Resistant to Ricin
HEK 293T cells (ATCC CRL-1573) were cultured adherently in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; Sigma), l-glutamine (Invitrogen), and penicillin-streptomycin-neomycin (Sigma). Cells were plated out at a density of 4 × 106 cells/75-cm2 T-flasks. After incubation at 37 °C for 20 h (5% CO2), the medium was replaced with fresh medium (20 ml) containing 200 μg/ml ethyl methanesulfonate (EMS; Sigma). Cells were grown in the presence of EMS for 20 h, washed twice with phosphate-buffered saline (PBS), trypsinized. The survival rate after EMS treatment was ∼50%. One aliquot of frozen mutagenized cells was thawed and plated out at 2 × 105 cells/75-cm2 T-flask and left to expand for 5 days (∼80% confluence). These cells were then split 1:10 and grown for a further 2 days. The medium was replaced with fresh medium containing either 1 ng/ml or 10 ng/ml ricin (laboratory stocks, J. Michael Lord, University of Warwick). Cells were refed with this medium every 4 days (six treatments in total) until colonies formed. No cells survived multiple treatments with 10 ng/ml ricin. Seventy colonies surviving treatment with 1 ng/ml ricin were expanded to confluence in 6-well plates in medium containing ricin. The colonies were then expanded and maintained in medium with no ricin, and two vials of each of these colonies were stored in FBS, 10% DMSO in liquid nitrogen.
Cytotoxicity Assays
Cytotoxicity was measured as the ability of cells to incorporate [35S]methionine into acid-precipitable material after ricin or Pseudomonas exotoxin A (PEx) treatment. Ricin-resistant cell lines were seeded at 2 × 104 cells/well in 96-well plates. For initial screens, after overnight growth and washing with PBS, cells were incubated for 4 h with 100 μl of fresh DMEM/FBS (control, untreated cells) or with 100 μl of DMEM/FBS containing ricin graded in 2-fold dilutions from 10.0 to 0.156 ng/ml. Subsequently, cells were washed twice with PBS and incubated in PBS containing 10 μCi/ml [35S]methionine (Amersham Biosciences) for 1 h. After the cells were washed twice with PBS, labeled proteins were precipitated with three washes in 5% (w/v) trichloroacetic acid (Sigma). Then the wells were washed twice with PBS, and the amount of radiolabel incorporated was determined after the addition of 200 μl of Optiphase SuperMix scintillation fluid (PerkinElmer Life Sciences) by scintillation counting in a Wallac Micro-Beta 1450 Trilux counter (PerkinElmer Life Sciences). Sensitivity to ricin is defined as the concentration (IC50) of ricin required to reduce protein synthesis to 50% of that in untreated cells. With approximate IC50 values established, assays on selected cell lines were repeated using 2-fold dilutions of ricin from 640 to 0.16 ng/ml. Accurate sensitivity values (IC50) were determined using a median effect plot (35). Cell lines were also screened for nonselected sensitivity to PEx by incubating them for 4 h with PEx in DMEM/FBS, graded in 2-fold dilutions from 5000 to 2.44 ng/ml.
Binding and Flow Cytometry Analysis
Ricin B chain (RTB) was purchased from Vector Laboratories (Burlingame, CA) and labeled with Alexa Fluor-647 dye using an Alexa Fluor protein labeling kit (Molecular Probes, Eugene, OR). The labeled RTB (RTB-Alexa 647) was divided into small aliquots, protected from light, and stored at −20 °C. 3 × 105 cells were washed twice with PBS and incubated in 100 μl of PBS/2 mm NaN3 with a final concentration of 10 μg/ml RTB-Alexa 647 on ice for 30 min. After being washed twice with 2 ml of PBS/2 mm NaN3, cells were resuspended in PBS/2 mm NaN3 and kept on ice until being subjected to flow cytometry using a BD Biosciences immunocytometry system.
Preparation of cDNA from HEK 293T Parental Cells and HEK 293T Lec36 Cells
2 × 106 cells were resuspended in residual medium after centrifugation. TRIzol reagent (Invitrogen) was added (1 ml), and cells were immediately lysed by repetitive pipetting. The homogenized samples were incubated at room temperature for 5 min. After extraction with 0.2 ml of chloroform and precipitation with 0.5 ml of isopropanol, the pellet was washed in 75% ethanol and dissolved in 100 μl of RNase-free water (Sigma). For the generation of cDNA by reverse transcription-polymerase chain reaction (RT-PCR), 10 μl of RNA (500 ng/μl) was transferred into an RNase-free 0.6-ml Microfuge tube with 0.5 ng of Oligo(dT)12–18 primer (Invitrogen). The mixture was heated at 70 °C for 10 min. An RT-PCR master mix was generated using SuperScriptTM II reverse transcriptase kit (Invitrogen) and RNasin® RNase inhibitor (Promega, Madison, WI). The RNA/Oligo(dT) mix was cooled at room temperature for 5 min before the addition of 9 μl of RT-PCR master mix and incubation at 42 °C for 40 min followed by 10 min at 90 °C and 5 min on ice. The cDNA product was diluted 1 in 5 using deionized water and stored at −20 °C.
Sequence Analysis
For a comparison of the sequences of the MAN2A1 (Golgi α-mannosidase II) and MAN2A2 (Golgi α-mannosidase IIx) genes of HEK 293T and HEK 293T Lec36 cells, the following primers were used to amplify the coding regions of the two genes: MAN2A1, forward (5′-ATGAAGTTAAGCCGCCAGTTCACCG-3′; nucleotides 1–25 of Swiss-Prot entry Q16706) and reverse (5′-TCACCTCAACTGGATTCGGAATGTGC-3′; nucleotides 3435-3410 of Q16706); and MAN2A2, forward (5′-ATGAAGCTGAAAAAGCAGGTGACAGTGTGTG-3′; nucleotides 1–31 of Swiss-Prot entry P49641) and reverse (5′-CTAACCCAAGCGGAGGCGAAAGGTAG-3′; nucleotides 3420-3395 of P49641). The PCR product pools were then sequenced directly, and the experiment was repeated to confirm the sequence traces. MAN2A1 from the cDNA of Lec36 cells was also cloned into pCR®4-TOPO vector using the TOPO TA Cloning Kit for Sequencing (Invitrogen) to obtain the sequences of the two MAN2A1 alleles separately. Thirty clones were picked and sequenced to confirm the mutations found in each allele.
Assessment of the mRNA Expression of MAN2A1 and MAN2A2
To compare the relative mRNA expression levels of MAN2A1 and MAN2A2 in HEK 293T cells, PCR analysis was performed using serial dilutions (neat, 1:4, 1:16, 1:64, and 1:256) of cDNA prepared from HEK 293T cells. The coding regions of the two genes were amplified using the primers described above.
Cloning of cDNA Fragments for Expression
A cDNA fragment encoding the human soluble (s) 19A extracellular region (residues 1–222) was amplified by PCR and inserted into the pEE14 vector (36, 37) in-frame with a C-terminal LysHis6 tag. For the complementation experiments involving the co-expression of wild-type or mutant Golgi α-mannosidase II with s19A, the wild-type MAN2A1 sequence from HEK 293T cells, and the mutant alleles from HEK 293T Lec36 cells cloned into the TOPO TA vector as described above were each transferred to the pLEXm expression vector (38). In the process, a Kozak sequence (gccacc) was inserted at the 5′- end of the cDNA adjacent to the start codon (atg) of the Golgi α-mannosidase II gene. The sequences were confirmed by dideoxy sequencing.
Large-scale Expression of s19A in HEK 293T and HEK 293T Lec36 Cells
HEK 293T and HEK 293T Lec36 cells were grown to 90% confluence and transiently transfected with pEE14/s19A using polyethyleneimine (39), and the medium was replaced immediately with DMEM, 2% FBS. When used, swainsonine (Toronto Research Chemicals, North York, Ontario, Canada) was added immediately after transfection to a final concentration of 20 μm. Conditioned media, containing the secreted fusion protein, were collected 4 days post-transfection, centrifuged, sterile-filtered, and diluted with two volumes of PBS. The pH was adjusted to 8.0 with 1 m Tris-HCl. The His-tagged protein was incubated with Ni2+-charged chelating Sepharose (Amersham Biosciences) at 16 °C for 4 h. The beads were then packed in Bio-Rad Econo columns and washed with 20 bed volumes of PBS/10 mm Tris-HCl, pH 8, supplemented with 0.05% Tween-20 followed by 20 bed volumes of PBS/10 mm Tris-HCl, pH 8. Bound proteins were eluted in 500 mm imidazole/PBS, pH 8, and concentrated to ∼300 μl for Sephacryl S-200 gel filtration in 10 mm HEPES, pH 7.4, 150 mm NaCl. Fractions containing the target protein at >95% purity (assessed by SDS-PAGE) were pooled, quantified, and stored at 4 °C.
Complementation Experiments in HEK 293T Lec36 Cells
HEK 293T Lec36 cells were co-transfected at about 40% confluence using Novagen GeneJuice® (Merck Chemicals Ltd.) with an 8:1 molar ratio of s19A and either the wild type, allele 1, or allele 2 of mutant Golgi α-mannosidase II. Twelve hours post-transfection the medium was replenished, and 2 days post-transfection the conditioned growth medium was harvested by centrifugation (6300 × g, 15 min). s19A was purified from the supernatant using Ni2+-charged chelating Sepharose as described above. The activities of the wild-type and mutant forms of Golgi α-mannosidase II were evaluated by testing the endoglycosidase H (Endo H) sensitivity of purified, co-expressed s19A and by MALDI-TOF MS analysis of the sugars released from s19A.
Endo H Sensitivity
The purified proteins were treated with Endo H (New England Biolabs). Seven micrograms of each protein was digested at 37 °C with 1000 units of Endo H in a final volume of 25 μl of 0.1 m NaOAc at pH 5.2. For comparison, proteins were also treated with protein N-glycanase F (PNGase F; New England Biolabs). Five micrograms of proteins was digested at 37 °C with 500 units of PNGase F, pH 7.4. The extent of digestion was monitored by SDS-PAGE analysis. Sampling after 1.5 and 3 h of digestion confirmed that it had gone to completion within 1.5 h.
Enzymatic Release of N-Linked Glycans
Oligosaccharides were released from bands containing ∼10 μg of s19A excised from Coomassie Blue-stained reducing SDS-polyacrylamide gels, which were washed with 20 mm NaHCO3, pH 7.0, and dried in a vacuum centrifuge before rehydration with 30 μl of 30 mm NaHCO3, pH 7.0, containing 100 units/ml PNGase F (Prozyme, San Leandro, CA) and incubation for 12 h at 37 °C (40). The enzymatically released N-linked glycans were eluted with water. Salts were removed by incubation at ∼24 °C (5 min) with 200 μl of acid-activated AG-50W (200–400 mesh) slurry (Bio-Rad), which was removed by filtration with a 0.45-μl pore size filter (Millex-LH, hydrophobic polytetrafluoroethylene).
Matrix-assisted Laser Desorption/Ionization (MALDI) Time-of-flight (TOF) Mass Spectrometry
Aqueous solutions of the glycans were cleaned with a Nafion 117 membrane (41). Positive ion MALDI-TOF mass spectra were recorded with a Waters-Micromass TofSpec reflectron-TOF mass spectrometer (Waters-Micromass MS Technologies Ltd., Manchester, UK) fitted with delayed extraction and a nitrogen laser (337 nm). The acceleration voltage was 20 kV, the pulse voltage was 3200 V, and the delay for the delayed extraction ion source was 500 ns. The spectra in Fig. 7 were recorded with a Waters MALDI Micro MS mass spectrometer with an extraction voltage of 12 kV. Samples were prepared by adding 0.5 μl of an aqueous solution of the sample to the matrix solution (0.3 μl of a saturated solution of 2,5-dihydroxybenzoic acid in acetonitrile) on the stainless steel target plate and allowing it to dry at room temperature. The sample/matrix mixture was then recrystallized from ethanol.
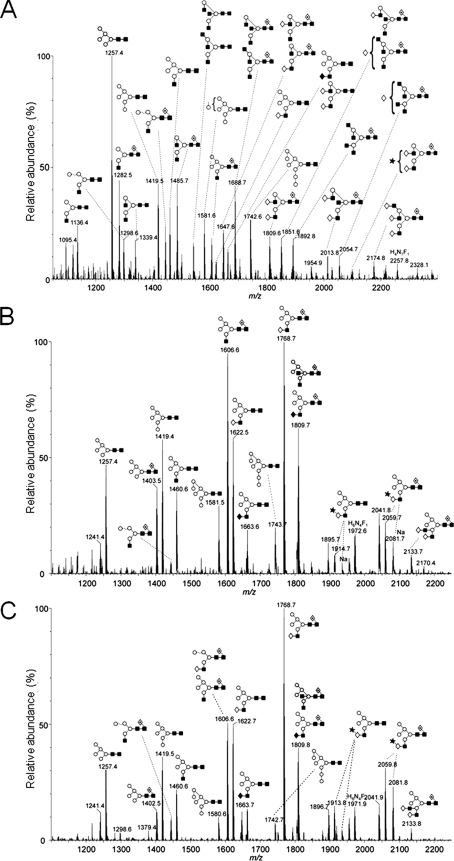
FIGURE 7.
MALDI-TOF MS of N-linked glycans ([M+Na]+ ions) from s19A co-expressed in HEK 293T Lec36 cells with wild-type Golgi α-mannosidase II (A), the G574E substituted form of Golgi α-mannosidase II encoded by MA1 (B), and the ΔILVQ-deleted form of Golgi α-mannosidase II encoded by MA2 (C). Symbols are same as described in the legend for Fig. 2. Spectra were processed with the MaxEnt 2 function of MassLynx 4.1.
Electrospray Ionization (ESI) Mass Spectrometry
Electrospray mass spectrometry was performed with a Waters-Micromass quadrupole time-of-flight (Q-Tof) Ultima Global instrument in negative ion mode. Samples in 1:1 (v:v) methanol:water were infused through Protana (Odense, Denmark) borosilicate nanospray capillaries. Ion source conditions were the following: temperature 120 °C, nitrogen flow 50 liters/h, infusion needle potential 1.2 kV, cone voltage 100 V, RF-1 voltage 150 V. Spectra (2-s scans) were acquired with a digitization rate of 4 GHz and accumulated until a satisfactory signal:noise ratio had been obtained. The ions were observed as [M+H2PO4]− adducts, the phosphate arising from residual phosphate in the solution. For MS/MS data acquisition, the parent ion was selected at low resolution (∼5 m/z mass window) to allow transmission of isotope peaks and fragmented with argon at a pressure (recorded on the instrument's pressure gauge) of 0.5 mbar. The voltage on the collision cell was adjusted with mass and charge to give an even distribution of fragment ions across the mass scale (typically 80–120 V). Other voltages were as recommended by the manufacturer. Instrument control and data acquisition and processing were performed with MassLynx software, version 4.0 (Waters-Micromass MS Technologies). Negative ion collision-induced dissociation (CID) spectra provide highly diagnostic fragmentation spectra that enable structural assignment of the glycans. For a more extensive discussion of these techniques, see Harvey et al. (42–45).
RESULTS AND DISCUSSION
Selection Strategy
HEK 293T cells were chosen for selection with ricin because of their recent emergence as a host for large-scale production of human glycoproteins for structural and functional analysis. Ricin is a heterodimeric toxin, comprising a toxic A chain (RTA), which depurinates a specific base on the 28S rRNA and is disulfide-linked to a cell-binding B chain (RTB) with specificity for terminal β1→4-linked galactose residues (46). Following cell binding and endocytosis, the holotoxin traffics in a retrograde manner via early endosomes and the trans-Golgi network to the ER (Ref. 47 and references therein). In the ER, the toxin is processed, and reductive separation of RTA and RTB occurs (48). RTA then retrotranslocates (dislocates) to the cytosol, where scrutiny by the Hsc70 and Hsp90 chaperones leads to recovery of catalytically active toxin that depurinates rRNA, blocking protein synthesis and, ultimately, inducing cell death (49).
Ricin resistance could, in principle, result from mutations of genes controlling retrograde trafficking, ER processing, dislocation, and cytosolic processing of the toxin or of genes that alter the glycosylation of proteins functioning as ricin receptors. Following selection, therefore, we screened our ricin-resistant HEK 293T cells for nonselected resistance to PEx, which also traffics in a retrograde manner to the ER and in which the A chain also dislocates to the cytosol (Ref. 47 and references therein) to eliminate mutations generally affecting trafficking, toxin processing, and dislocation. Screens of ricin binding site expression, ability to support high level expression of transfected genes, and electrophoretic analysis of the glycoprotein products of the transfected genes were then used to refine our search for a stable, viable, Golgi α-mannosidase II-deficient cell line.
Isolation of Ricin-resistant HEK 293T Mutants
Lectin-resistant HEK 293T cells were generated using the method of Reeves et al. (29), who derived the HEK 293S Lec1 cell line. Briefly, EMS-mutagenized HEK 293T cells were grown in the presence of 1 ng/ml or 10 ng/ml ricin, and single cell-derived colonies were obtained after 3 weeks at a ricin concentration of 1 ng/ml. Although Reeves et al. (29) selected 293S cell-derived clones deficient in GnT I activity in medium containing 10 ng/ml ricin, under the conditions of our experiments, no EMS-mutagenized cells survived such a challenge. To examine this further, we grew the GnT I-deficient HEK 293S Lec1 cells in the presence of two rounds of 15.6, 7.8, 3.9, 1.9, and 0.9 ng/ml ricin. At 15.6 ng/ml and 7.8 ng/ml ricin, all Lec1 cells died after one round of treatment. At 3.9 ng/ml ricin, only a few cells survived after the two rounds of ricin treatment, and at the end of the experiment, there was only one surviving colony comprising ∼100 vacuolated cells that were not growing. At concentrations of 1.9 and 0.9 ng/ml, ∼70 and >200 viable colonies grew, respectively. We are therefore confident that under our conditions, even 293S Lec1 cells cannot survive challenge with 10 ng/ml ricin. The difference between the apparent sensitivities of Lec1 cells in the two laboratories may simply reflect differences in the specific activity of the ricin used.
Screening of Ricin-resistant HEK 293T Mutant Cell Lines
Screen 1: Ricin Sensitivity
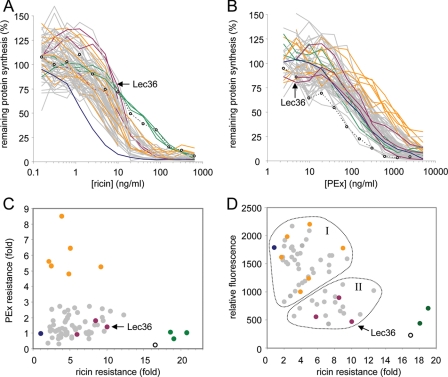
From the original set of EMS-mutagenized HEK 293T-derived clones, we collected 70 that survived six rounds of challenge with 1 ng/ml ricin and expanded these clones into lines in the absence of ricin. The cell lines were initially screened in a cytotoxicity-based, ricin-sensitivity assay (Fig. 1A, Table 1). Three cell lines (coded green in Fig. 1A) were ∼20-fold more resistant to ricin than were the parental HEK 293T cells (coded dark blue) and thus had cytotoxicity resistance profiles matching that of the 293S Lec1 cells (open circles and dotted line). The remainder of the clones exhibited a 2–12-fold variation in resistance.
FIGURE 1.
Screening of ricin-resistant clones generated by EMS mutagenesis. A, ricin resistance of ricin-selected cell lines. Individual cell lines were treated with graded doses of ricin for 4 h, and their subsequent ability to synthesize proteins was determined. Blue, parental HEK 293T (n = 24); open circles and dotted lines, HEK 293S Lec1 (n = 4); gray (n = 1–4), orange (n = 1, 2, 3, or 6), violet (n = 1–3), and green (n = 6 or 7) indicate cell lines belonging to various selection screen-defined classes. Error bars have been omitted for clarity. B, nonselected sensitivity of all of these cell lines to PEx. Cell lines were treated with graded doses of PEx for 4 h, and their subsequent ability to synthesize proteins was determined. Blue (n = 10), open circles (n = 2), gray (n = 1–4), violet (n = 1 or 3), and green (n = 3) are same as in A; orange (n = 1–5) indicates cells with ∼5-fold or greater resistance to PEx. Error bars have been omitted for clarity. C, scatter plot of PEx resistance versus ricin resistance for all of the cell lines generated. Sensitivity of the cell lines to toxin was determined as described in A by measuring the concentration (IC50, ng/ml) of toxin required to reduce protein synthesis to 50% of that of untreated cells, and resistance (-fold) was calculated from the ratio of the IC50 for the cell line versus the IC50 for HEK 293T cells. PEx-resistant cells (orange) cluster as a group with 2–9-fold ricin resistance; green, a cluster of cell lines with very high ricin resistance. D, scatter plot of relative number of ricin B chain-binding sites versus ricin resistance for 62 of the cell lines generated (7 “gray” and 1 “green” cell line were not tested). The relative number of ricin-binding sites on the cell lines was determined by flow cytometry using Alexa 647-labeled ricin B chain. Apart from the highly ricin-resistant lines (green), the cell lines cluster into two broad groups, with either medium to high (group I) or low to medium (group II) numbers of ricin-binding sites. Violet-colored symbols, candidate HEK 293T Lec36 cells.
TABLE 1.
Screening ricin-resistant cell lines
| Ricin resistancea | PEx resistancea | Color codeb | No. ricin binding sitesc | No. cell linesd | Candidate loss of function |
|---|---|---|---|---|---|
| -Fold | -Fold | ||||
| 18–21 | ∼1 | Green | Very low | 3 | Loss of ricin receptors |
| 2–9 | 5–9 | Orange | Medium-normal | 6 | Toxin trafficking, processing, or dislocation |
| 2–13 | 0.3–3 | Gray | Medium-normal (group I) | ∼38 | Toxin trafficking, processing, or dislocation |
| 2–13 | 0.3–3 | Gray | Low-medium (group II) | ∼20 | Loss of ricin receptors |
| 6–10 | 1–2 | Violet | Low | 3 | Loss of ricin receptors; Golgi α-mannosidase II |
a -Fold resistance over parental (IC50 clone: IC50 HEK 293T).
b Color code is defined in Fig. 1.
c Determined by flow cytometry after binding Alexa Fluor 647-labeled ricin B chain.
d Eight cell lines (seven “gray” and one “green”) were not tested for Alexa Fluor 647-labeled ricin B chain binding.
Screen 2: PEx Sensitivity
All of the cell lines were screened for sensitivity to PEx (Fig. 1B, Table 1). Six of these (coded orange) showed ∼5–9-fold nonselected resistance to PEx and 2–9-fold ricin resistance versus HEK 293T cells (Fig. 1, B and C; Table 1), suggesting that these cell lines carry mutations in gene(s) having products that limit the toxicity of both toxins. Candidate disruptions could involve (a) retrograde trafficking machineries, as both ricin and PEx traffic from the cell surface via the trans-Golgi network to the ER; (b) ER processing, as the A chain of each toxin is separated from its cell-binding B partner in the ER; (c) dislocation systems that permit the retrotranslocation of the A chain into the cytosol; and (d) cytosolic processing events.
Screen 3: Ricin B Chain Binding
Ricin binds mammalian cells primarily through interactions of its B chain with exposed terminal β1→4-linked galactose residues. To estimate the relative number of ricin-binding sites, most of the cell lines were tested by flow cytometry analysis using a fluorescent ricin B chain probe (Fig. 1D, Table 1). Highly ricin-resistant cells (Fig. 1D, green) display the much reduced RTB binding characteristic of 293S Lec1 cells and were not characterized further. The remaining clones cluster into two broad groups, one (I) with close to parental (Fig. 1D, blue) numbers of RTB-binding sites and the other (II) with reduced RTB binding. Most of the cell lines with high PEx resistance (Fig. 1D, orange) cluster in the former group, with close to normal numbers of ricin-binding sites, consistent with the notion that these do not carry mutations in ricin receptors. Group I cell lines were not examined any further. Group II cell lines have reduced numbers of ricin-binding sites, and it was from within this set that we expected to identify novel glycosylation mutations.
Screen 4: High Level Expression of Transfected Cell-surface Molecules
Group II cell lines were transiently transfected with a pEE14 plasmid encoding the soluble extracellular region of stem cell marker 19A. Supernatants from these cultures were then screened by Western blot for high level expression of s19A (data not shown). Cells capable of high level expression of s19A were selected, and s19A was purified. Three cell lines (Fig. 1, violet) expressed s19A with an apparent molecular weight 5–10 kDa larger than that of the s19A produced in 293S Lec1 cells. Of these, one was selected for further study and, following the nomenclature of Patnaik and Stanley (15), is designated HEK 293T Lec36 hereafter.
Characterization of Lec36 Cells
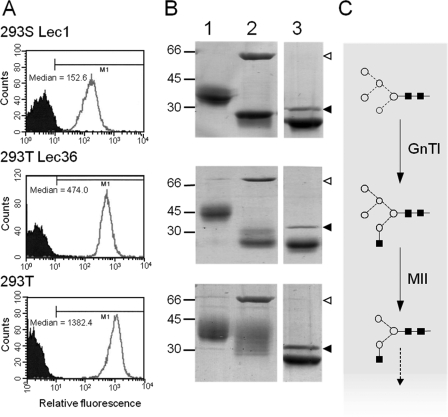
Parental HEK 293T cells, GnT I-deficient 293S Lec1 cells, and HEK 293T Lec36 cells were retested for ricin sensitivity. Parental HEK 293T cells are very sensitive to ricin with an IC50 (the ricin concentration that inhibits protein synthesis by 50% versus untreated cells) of 1.69 ± 0.46 ng/ml (1 S.D., n = 7), whereas HEK 293T Lec36 cells were highly resistant (IC50 = 13.02 ± 0.77 ng/ml (1 S.D., n = 3)). This is about half the resistance of GnT I-deficient 293S cells (IC50 of 28.21 ± 5.39 ng/ml (1 S.D., n = 3)). Because ricin binds terminal β1→ 4-linked galactose residues, this intermediate sensitivity suggests that these cells have a disruption in the glycosylation pathway differing from that of Lec1 cells. The levels of ricin resistance were mirrored in the flow cytometric analysis where ricin B chain binding for HEK 293T Lec36 cells fell between that of the parental HEK 293T cells and GnT I-deficient (Lec1) 293S cells (Fig. 2A).
FIGURE 2.
Identification of Lec36 cells. A, flow cytometric analysis of HEK 293S Lec1, Lec36, and parental HEK 293T cells using Alexa 647-labeled ricin B. B, Endo H sensitivities of s19A expressed in HEK 293S Lec1, HEK 293T Lec36, and HEK 293T cells analyzed by SDS-PAGE (undigested, and Endo H-treated s19A in lanes 1 and 2, respectively). For comparison, the lane containing PNGase F-treated protein from the same gel is also shown (lane 3). Open arrowheads, Endo H; filled arrowheads, PNGase F. C, schematic view of the activities of GnT I and Golgi α-mannosidase II (MII). Processing of the Glc3Man9GlcNAc2 precursor to Man5GlcNAc2 by α-glucosidase I and II and the class I α-mannosidases has been omitted for clarity as have the potential elaborations of the basic hybrid-type glycan. Symbols used to represent monosaccharide constituents in this and subsequent figures: ◇, Gal; ♦, GalNAc; ■, GlcNAc; ○, Man; ★, sialic acid;  , Fuc. The linkage position is shown by the angle of the lines linking the sugar residues: vertical line, 2-link; forward slash, 3-link; horizontal line, 4-link; back slash, 6-link. Anomericity is indicated by solid lines for β-bonds and broken lines for α-bonds.
, Fuc. The linkage position is shown by the angle of the lines linking the sugar residues: vertical line, 2-link; forward slash, 3-link; horizontal line, 4-link; back slash, 6-link. Anomericity is indicated by solid lines for β-bonds and broken lines for α-bonds.
So that putative changes in glycan processing could be examined directly, HEK 293T Lec36 cells were again transiently transfected with a pEE14 plasmid encoding the s19A glycoprotein. The s19A glycoprotein was selected as a reporter molecule for glycosylation because it contains six N-linked glycosylation sequons distributed over two immunoglobulin domains, and consequently changes in glycan processing can be readily assessed by Endo H digestion and SDS-PAGE (23). In addition to HEK 293T Lec36, s19A was also expressed in 293S Lec1 cells, parental HEK 293T cells, and in HEK 293T cells in the presence of 20 μm swainsonine, yielding 1.6, 2.2, 5.4, and 6.0 mg/liter, respectively. Variation in the apparent molecular weight according to SDS-PAGE analysis (Fig. 2B) can be attributed to the N-linked glycans, as all variation between the samples is eliminated by digestion with PNGase F (data for swainsonine-treated s19A are published elsewhere (21)). GnT I deficiency in 293S cells results in the accumulation of Man5GlcNAc2 N-glycans (Fig. 2C (29, 50)). s19A from HEK 293T Lec36 cells migrated at ∼45 kDa, whereas s19A from the GnT I-deficient 293S cells migrated as 35–40 kDa, indicating that Lec36 glycans are larger than Man5GlcNAc2. The presence of hybrid- or oligomannose-type glycans on s19A from HEK 293T Lec36 cells was confirmed by Endo H digestion (Fig. 2B).
Identification of N-Linked Glycans from s19A
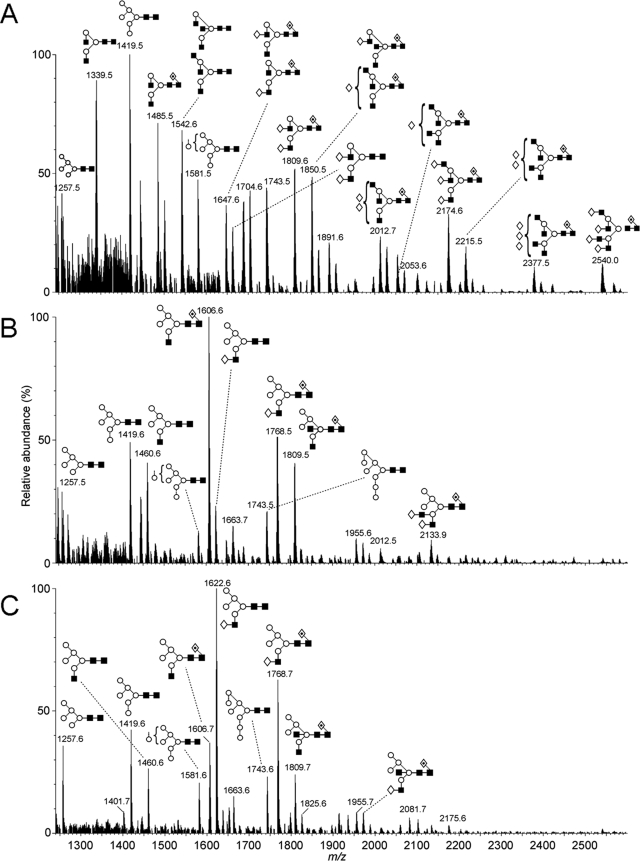
The Lec36 N-glycans were further characterized by MALDI-TOF (Fig. 3) and nano-ESI mass spectrometry (Fig. 4) following release from s19A with PNGase F. Importantly, glycans were identified not only by composition but also by negative ion MS/MS (42–45, 51). MALDI rather than ESI spectra are shown owing to the greater clarity of the spectra resulting from the ionization of the glycans as single sodium adducts. HEK 293T cells are capable of sialylation as observed in the hybrid structures of Fig. 3, B and C. The ESI spectra revealed a slightly higher abundance of sialic acid compared with the MALDI spectra because of the differing ionization efficiencies. The N-linked glycans from s19A expressed in HEK 293T cells were predominantly of the complex- and oligomannose-type with a low abundance of hybrid-type glycans (Fig. 3A). The most abundant glycan was Man5GlcNAc2 ([M+Na]+ m/z = 1257.4). A series of other oligomannose-type glycans, Man6–8GlcNAc2 at m/z = 1419.5, 1581.6, and 1742.6, were also identified. The remaining glycans were complex-type, revealing significant Golgi α-mannosidase II activity, and exhibited a range of elaborations from, for example, a monoantennary structure (m/z = 1136.4) to a core fucosylated triantennary glycan (m/z = 2174.8).
FIGURE 3.
MALDI-TOF MS of N-linked glycans ([M+Na]+ ions) from s19A expressed in HEK 293T cells (A), HEK 293T Lec36 cells (B), and HEK 293T cells (C) in the presence of 20 μm swainsonine. Spectra were processed with the MaxEnt 2 function of MassLynx 4.1. Symbols are same as described in the legend for Fig. 2.
FIGURE 4.
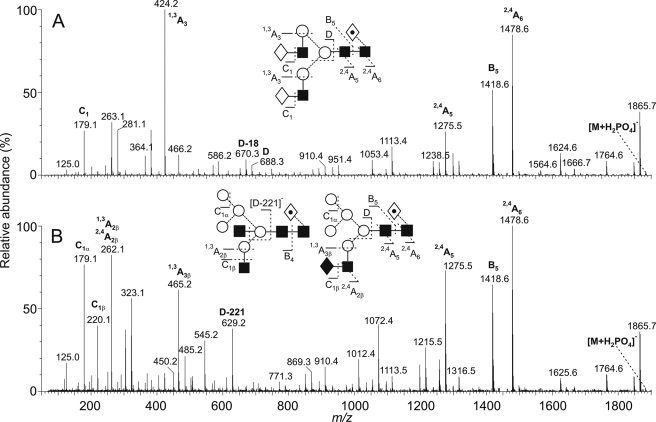
Negative ion ESI-CID spectrum of isobaric complex-type (A) and hybrid-type (B) N-linked glycans ([M+H2PO4]− ions; m/z 1883.7) from s19A. The complex- and hybrid-type glycans were from s19A expressed in HEK 293T and HEK 293T Lec36 cells, respectively. Symbols are the same as described in the legend for Fig. 2. Ion nomenclature follows the method of Domon and Costello (54).
In contrast to parental HEK 293T cell-derived s19A, the mass spectra for s19A expressed in the HEK 293T Lec36 cells or in HEK 293T cells in the presence of swainsonine (Fig. 3, B and C, respectively) were completely free of complex-type N-glycans. All of the major glycans were of the classical Man5 hybrid type. The striking similarity of the two spectra strongly implies disruption of Golgi α-mannosidase II activity in the HEK 293T Lec36 cells. Negative ion fragmentation allowed assignment of all the peaks to oligomannose-type or hybrid-type structures.
We also noted that a small population of Man4 hybrid-type glycans was released from s19A produced under both sets of conditions, for example at m/z = 1298.6 (Fig. 3). The presence of α-mannosidase activity of unknown origin had been reported previously in lectin-resistant cell lines; CHO Lec1 displays an as yet undetermined α-mannosidase activity that digests the dominant Man5GlcNAc2 to Man3–4GlcNAc2 (22, 50). In addition, Golgi α-mannosidase IIx is inhibited by swainsonine (52). Given that trace amounts of Man4-based hybrids were detected in s19A expressed in HEK 293T Lec36 cells and in swainsonine-treated 293T cells (Fig. 3, B and C, respectively), this activity is unlikely to be attributable to a class II α-mannosidase.
Structural Assignment of the N-Linked Glycans
The structural assignment of the glycans in Fig. 3 is based on highly diagnostic fragmentation in the negative ion CID spectra (42–45, 53). An example of the assignment of ions to hybrid-type structures as opposed to isobaric complex-type glycans is illustrated in Fig. 4 for the glycan of composition Hex5HexNAc4dHex1 (m/z 1883.7). The CID spectrum of the ion from s19A expressed in the parental cell line contains features characteristic of a core fucosylated bi-galactosyl biantennary complex type glycan (Fig. 4A). The complex nature of the 6-antenna was revealed by the presence of D (containing the 6-antenna and core-branching mannose) and [D-18]− ions at m/z 688.3 and 670.3, respectively, and the ion at m/z 424, which is a 1,3A cleavage of the antenna mannose residues confirming the composition of the antennae as Hex-HexNAc (ion nomenclature is as described by Domon and Costello (54)). The C1 ion at m/z 179 confirmed hexose (galactose) at the nonreducing terminus and the 2,4A6, B6, and 2,4A5 ions at m/z 1478.6, 1418.6, and 1275.5, respectively, located the fucose to the 6-position of the asparagine-linked GlcNAc residue. 2,4A and B ions from the core GlcNAc residues appeared at the same m/z values in the spectrum of the hybrid glycans shown in Fig. 4B, again indicating the presence of a core fucose residue. However, the remainder of the spectrum was very different to that shown in Fig. 4A. The ion at m/z 424 was of very low abundance but was replaced by two ions at m/z 262 and 465 with compositions of HexNAc+59 and HexNAc2+59, respectively. The C1 ion at m/z 220 confirmed HexNAc at the nonreducing terminus in addition to hexose, which gave the corresponding ion at m/z 179. The D and [D-18]− ions appeared at m/z 647 and 629, and these ions, together with the cross-ring products at m/z 575 and 545, defined a 6-antenna containing three hexose (mannose) residues. In summary, the spectrum appeared to be that of a mixture of two hybrid compounds: the high abundance of m/z 629 relative to m/z 670 was characteristic of a glycan containing a bisecting GlcNAc and was consistent with the presence of the HexNAc+59 (1,3A2β) ion at m/z 262; the second hybrid appeared to contain a HexNAc2 chain (probably GalNAc-GlcNAc) and accounted for the HexNAc2+59 ion at m/z 465.
Molecular Basis of the Lec36 Mutation
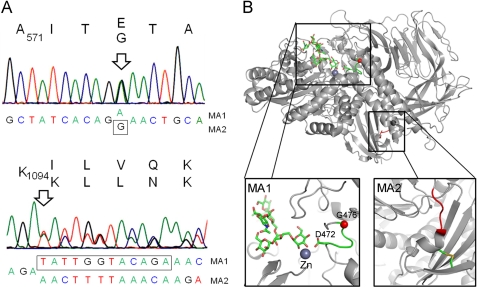
The observed differences in glycosylation of wild type- and HEK 293T Lec36 cell-produced s19A were consistent with there being null mutations of the Golgi α-mannosidase II gene in the HEK 293T Lec36 cells. We used RT-PCR to obtain cDNAs from both the cell lines and sequenced the PCR products of MAN2A1 and MAN2A2 (encoding Golgi α-mannosidase II and IIx) from the two cDNAs. The sequencing results show that there are two mutations in MAN2A1 of the HEK 293T Lec36 cells (Fig. 5A). One is a single nucleotide mutation, with superimposable traces for G and A, resulting in a highly nonconservative Gly → Glu mutation at position 574 of the mature sequence (using the UniProtKB/Swiss-Prot numbering). The second is a deletion of 12 nucleotides leading to an expected four amino acid deletion (1095ILVQ1098; ΔILVQ) in the predicted translation product. The sequence is otherwise identical to that reported previously (55) except that we observed the same polymorphism at residue 608 as referred to in UniProtKB/Swiss-Prot entry Q16706, wherein the codon for this residue in both our sequences (parental wild-type HEK 293T and HEK 293T Lec36 cells) is TTG (Leu) rather than GGG (Gly). We proceeded to clone the PCR products from MAN2A1 of the HEK 293T Lec36 cells to determine whether the mutations affected each allele. The sequencing of 30 clones revealed that the mutations are heterozygous. Sixteen sequences yielded mutant allele 1 (MA1) bearing the point mutation (GGA→GAA; G574E), and 14 sequences yielded the second mutation (mutant allele 2 (MA2); ΔILVQ), wherein 12 nucleotides were deleted. Beyond the mutated regions, the sequences of the two alleles are identical to that of the parental wild-type HEK 293T cells. The ease with which the bi-allelic null Lec36 mutant was isolated (i.e. from 2 × 105 EMS-mutated cells) is somewhat surprising given that EMS mutation of the single functional MgatI allele in Pro−5 CHO cells to give Lec1 cells only occurs at a frequency of 10−3 (55).
FIGURE 5.
Molecular basis of the Lec36 mutation. A, sequence of Golgi α-mannosidase II (MAN2A1) from HEK 293T Lec36 cells indicating the positions of the point mutation in MA1 encoding G574E and the in-frame deletion in MA2 resulting in the ΔILVQ deletion. B, mapping of the Lec36 mutations onto the crystal structure of D. melanogaster Golgi α-mannosidase II (Protein Data Bank ID code 3CZN) complexed with zinc and oligomannose-type substrate (56). The box on the left shows the area of the active site containing the residue analogous to the Lec36 G574E substitution encoded by MA1. The active site comprises a loop, depicted in green, containing both the ligand-binding Asp472 and the buried Gly476 (equivalent to Asp570 and Gly574, respectively, of the human enzyme). The Gly476 Cα carbon is depicted as a red sphere. The box on the right highlights in red the region analogous to the ΔILVQ (995QKLD998) deletion encoded by MA2. The molecular graphics images were generated using PyMOL (DeLano Scientific, San Carlos, CA).
In contrast to MAN2A1, no mutations were detected in MAN2A2 of the Lec36 cell line, indicating that catalysis by Golgi α-mannosidase IIx does not contribute significantly to the formation of complex-type N-glycans in HEK 293T cells. One possible explanation is that Golgi α-mannosidase IIx is poorly expressed in HEK 293T cells. Consistent with this, MAN2A2 transcripts are substantially less abundant (by at least 64-fold) in cDNA isolated from wild-type HEK 293T cells than are MAN2A1 transcripts (supplemental Fig. S1).
Wild-type Golgi α-Mannosidase II Restores Complex-type Glycosylation in HEK 293T Lec36 Cells
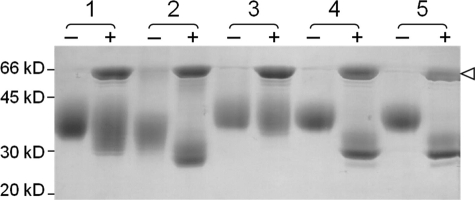
To demonstrate that the Lec36 mutation disrupts Golgi α-mannosidase II activity and to show that the Golgi α-mannosidase II deficiency is not due to a mutation in another gene that regulates its folding or activity, we performed complementation experiments by coexpressing s19A as a secreted marker of glycosylation alongside the mutated and wild-type Golgi α-mannosidases in Lec36 cells. s19A expressed alone in HEK 293T cells exhibits some Endo H sensitivity (Fig. 6, sample 1) consistent with the presence of some oligomannose- and hybrid-type glycans (Fig. 3A). In contrast, s19A expressed alone in HEK 293T Lec36 cells is largely Endo H-sensitive (Fig. 6, sample 2; also as seen in Fig. 2B). As no complex-type glycosylation was detected (Fig. 3B), the incomplete digestion can be attributed to the presence of core fucosylation, which limits cleavage by Endo H. In complementation experiments using HEK 293T Lec36 cells, co-transfection of the wild type, but not the mutant MAN2A1 alleles, restored glycan processing to complex-type forms as assessed by monitoring Endo H sensitivity (Fig. 6, samples 3, 4, and 5). Expression of the wild-type Golgi α-mannosidase II resulted in glycosylation that was more resistant to Endo H than s19A expressed in HEK 293T cells. This is consistent with a reduction in the hybrid- or oligomannose-type glycans and suggests that in HEK 293T cells the activity of Golgi α-mannosidase II is limiting for the generation of complex glycans. Consistent with this finding, overexpression of wild-type Golgi α-mannosidase II in HEK 293T cells also reduced the Endo H sensitivity of co-expressed s19A (data not shown). We note that the Endo H sensitivity of s19A expressed in the presence of the mutated Golgi α-mannosidase II (either mutant allele 1 or mutant allele 2) in HEK 293T Lec36 cells is slightly enhanced as compared with that observed in its absence (Fig. 6, sample 2 versus samples 4 and 5). s19A is more weakly expressed in the presence of the Golgi α-mannosidases (either wild-type or mutant alleles; data not shown), and the relative abundance of the hybrid-type glycans changes slightly (Figs. 3B and 7A). Subtle changes in processing, e.g. in the levels or distribution of fucose addition, may account for the variation in Endo H sensitivity observed at these different levels of protein expression. The effects of the HEK 293T Lec36 mutations on the activity of Golgi α-mannosidase II were confirmed with a second reporter protein, sPD-L1 (soluble programmed cell death 1-ligand 1; data not shown).
FIGURE 6.
Complementation of Golgi α-mannosidase II deficiency. Coomassie-stained SDS-polyacrylamide gel run under reducing conditions showing undigested (−) and Endo H-digested (+) s19A expressed alone in HEK 293T (sample 1) and Lec36 cells (sample 2) and of s19A co-expressed in HEK 293T Lec36 cells with wild-type Golgi α-mannosidase II (sample 3), the G574E substituted form of Golgi α-mannosidase II encoded by MA1 (sample 4), and the ΔILVQ-deleted form of Golgi α-mannosidase II encoded by MA2 (sample 5). Open arrowhead, Endo H.
The effects were further investigated using MALDI MS. The MALDI mass spectrum of the s19A N-linked glycans reveals that the co-expression of wild-type Golgi α-mannosidase II results in predominantly complex-type glycosylation (Fig. 7A). The low abundance of hybrid-type glycans at m/z 1460.6, 1606.6, and 1622.6, observed in the HEK 293T spectrum (Fig. 3A), demonstrates that the overexpression of wild-type Golgi α-mannosidase II does indeed drive the processing of these substrates to completion. In contrast, the oligomannose-type glycans in the HEK 293T spectrum (Fig. 3A) were observed at m/z 1257.5 and 1419.6, demonstrating that processing upstream of Golgi α-mannosidase II was not significantly affected. The presence of a greater abundance of tri- and tetra-antennary glycans indicates that the processing downstream of Golgi α-mannosidase II has been stimulated, consistent with a change in the processing kinetics. In contrast to the complementation observed with wild-type enzyme, the two mutated forms of Golgi α-mannosidase II of Lec36 cells failed to restore any complex-type glycosylation (Fig. 7, B and C), demonstrating that the G574E and ΔILVQ mutations each disrupt catalysis.
Potential Structural Effects of the HEK 293T Lec36 Mutations
To consider the possible structural effects of the HEK 293T Lec36 mutations, we mapped the changes onto the structure of Drosophila melanogaster Golgi α-mannosidase II, which shares ∼40% sequence homology with human Golgi α-mannosidase II. For this we used both the Drosophila crystal structure and sequence alignment of Shah et al. (56). The sequence alignment suggests that the residue mutated in HEK 293T Lec36 mutant allele 1 (MA1), i.e. Gly574, is equivalent to a highly conserved residue, Gly476, which is close to the active site of Drosophila Golgi α-mannosidase II (Fig. 5B). In the insect structure (56), Gly476 forms part of a loop containing Asp472, which forms critical hydrogen bonds with the substrate, in this case the hybrid-type glycan GlcNAcβ1→2-Man5GlcNAc2. It can readily be envisaged that the G574E mutation would alter the conformation of the equivalent loop in human Golgi α-mannosidase II, destabilizing substrate binding. The deletion in HEK 293T Lec36 allele 2 (ΔILVQ, MA2), on the other hand, maps to the C-terminal region of the α-mannosidase (Fig. 5B). The corresponding tetrapepetide in the D. melanogaster structure, 995QKLD998, is adjacent to an α-helix containing a cysteine that forms a stabilizing disulfide with a β-strand cysteine nine residues toward the C terminus. Sequence conservation in this region is poor, and the cysteines are not present, but it is possible that the ILVQ deletion disrupts analogous stabilizing interactions in the human protein.
Conclusions
The loss of Golgi α-mannosidase II activity in Lec36 cells mimics that observed in some patients with HEMPAS (10, 11). We anticipate that Lec36 cells could find application both as a sensitive host for detecting mutations in Golgi α-mannosidase II and to further investigate the interaction of the altered cell surface with the mannose-specific lectins of the innate immune system. Although the precise molecular basis of HEMPAS remains to be determined, it is characterized by either a reduction in β1→4-galactosyltransferase, GlcNAcTII, or Golgi α-mannosidase II activity (11, 12). In some cases, the alteration in oligosaccharide processing may be a secondary effect of, for example, changes in enzyme localization or membrane trafficking (57). An increase in hybrid-type glycosylation has been reported in a CDG patient with apparently unmutated MAN2A1 and MAN2A2 (58), perhaps reflecting a defect in the localization of Golgi a-mannosidase II. Indeed, the requirement for precise localization of processing enzymes became apparent in the reconstruction of human-like glycosylation in yeast (26, 59) and is also revealed by the effects of defects in intra-Golgi trafficking on glycosylation in cells and patients (60, 61).
Despite the existence of numerous lectin-resistant cell lines, the HEK 293T Lec36 cell line reported herein represents the first confirmed Golgi α-mannosidase II-deficient cell line. Among several types of applications, lectin-resistant cell lines have provided powerful tools for the analysis of CDG pathology (15–17) and for the manipulation of the oligosaccharide chains of glycoprotein therapeutics. The Golgi α-mannosidase II -deficient Lec36 cells described herein extend the utility of this important resource.
Supplementary Material
Acknowledgments
We thank Drs. H. G. Khorana and P. J. Reeves (Massachusetts Institute of Technology) for the gift of GnT I-deficient HEK 293S cells and Dr. T. A. Bowden for helpful discussions. We thank the Wellcome Trust for an equipment grant to purchase the Q-Tof mass spectrometer and the Biotechnology and Biological Sciences Research Council for a grant to purchase the MALDI-TOF mass spectrometer.
This work was supported in part, by National Institutes of Health Grant 5 U01AI 65869 (to J. M. L. and R. A. S.). This work was also supported by the Wellcome Trust, Cancer Research UK, the United Kingdom Medical Research Council, and the Oxford Glycobiology Institute Endowment and by Structural Proteomics in Europe (SPINE) Contract QLG2-CT-2002-00988 from the European Commission under the Integrated Programme “Quality of Life and Management of Living Resources.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ER
- endoplasmic reticulum
- CDG
- congenital disorders of glycosylation
- CHO
- Chinese hamster ovary
- CID
- collision-induced dissociation
- DMEM
- Dulbecco's modified Eagle's medium
- EMS
- ethyl methanesulfonate
- Endo H
- endoglycosidase H
- ESI
- electrospray ionization
- FBS
- fetal bovine serum
- GnT
- N-acetylglucosaminyltransferase
- HEK
- human embryonic kidney
- HEMPAS
- hereditary erythroblastic multinuclearity with positive acidified serum lysis test
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- Lec
- lectin
- MALDI
- matrix-assisted laser desorption/ionization
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- PBS
- phosphate-buffered saline
- PEx
- Pseudomonas exotoxin A
- PNGase F
- peptide N-glycosidase F
- Q
- quadrupole
- RT
- reverse transcription
- RTA
- ricin A chain
- RTB
- ricin B chain
- s
- soluble
- TOF
- time-of-flight.
REFERENCES
- 1.Kornfeld R., Kornfeld S. (1985) Annu. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 2.Balaguer E., Demelbauer U., Pelzing M., Sanz-Nebot V., Barbosa J., Neusüss C. (2006) Electrophoresis 27, 2638–2650 [DOI] [PubMed] [Google Scholar]
- 3.Neusüss C., Demelbauer U., Pelzing M. (2005) Electrophoresis 26, 1442–1450 [DOI] [PubMed] [Google Scholar]
- 4.Parodi A. J. (2000) Annu. Rev. Biochem. 69, 69–93 [DOI] [PubMed] [Google Scholar]
- 5.Herscovics A. (2001) Biochimie 83, 757–762 [DOI] [PubMed] [Google Scholar]
- 6.Tremblay L. O., Herscovics A. (2000) J. Biol. Chem. 275, 31655–31660 [DOI] [PubMed] [Google Scholar]
- 7.Lal A., Pang P., Kalelkar S., Romero P. A., Herscovics A., Moremen K. W. (1998) Glycobiology 8, 981–995 [DOI] [PubMed] [Google Scholar]
- 8.Schachter H. (1991) Glycobiology 1, 453–461 [DOI] [PubMed] [Google Scholar]
- 9.Jaeken J., Matthijs G. (2007) Annu. Rev. Genomics Hum. Genet. 8, 261–278 [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M. N., Masri K. A., Dell A., Luzzatto L., Moremen K. W. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7443–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda M. N., Gaetani G. F., Izzo P., Scartezzini P., Dell A. (1992) Br. J. Haematol. 82, 745–752 [DOI] [PubMed] [Google Scholar]
- 12.Lowe J. B., Marth J. D. (2003) Annu. Rev. Biochem. 72, 643–691 [DOI] [PubMed] [Google Scholar]
- 13.Green R. S., Stone E. L., Tenno M., Lehtonen E., Farquhar M. G., Marth J. D. (2007) Immunity 27, 308–320 [DOI] [PubMed] [Google Scholar]
- 14.Paulson J. C. (2007) Cell 130, 589–591 [DOI] [PubMed] [Google Scholar]
- 15.Patnaik S. K., Stanley P. (2006) Methods Enzymol. 416, 159–182 [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Duncker I., Dupré T., Piller V., Piller F., Candelier J. J., Trichet C., Tchernia G., Oriol R., Mollicone R. (2005) Blood 105, 2671–2676 [DOI] [PubMed] [Google Scholar]
- 17.Hong Y., Sundaram S., Shin D. J., Stanley P. (2004) J. Biol. Chem. 279, 49894–49901 [DOI] [PubMed] [Google Scholar]
- 18.Sethuraman N., Stadheim T. A. (2006) Curr. Opin. Biotechnol. 17, 341–346 [DOI] [PubMed] [Google Scholar]
- 19.Hoppe H. (2000) J. Biotechnol. 76, 259–261 [DOI] [PubMed] [Google Scholar]
- 20.Davis S. J., Puklavec M. J., Ashford D. A., Harlos K., Jones E. Y., Stuart D. I., Williams A. F. (1993) Protein Eng. 6, 229–232 [DOI] [PubMed] [Google Scholar]
- 21.Davis S. J., Davies E. A., Barclay A. N., Daenke S., Bodian D. L., Jones E. Y., Stuart D. I., Butters T. D., Dwek R. A., van der Merwe P. A. (1995) J. Biol. Chem. 270, 369–375 [DOI] [PubMed] [Google Scholar]
- 22.Butters T. D., Sparks L. M., Harlos K., Ikemizu S., Stuart D. I., Jones E. Y., Davis S. J. (1999) Protein Sci. 8, 1696–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang V. T., Crispin M., Aricescu A. R., Harvey D. J., Nettleship J. E., Fennelly J. A., Yu C., Boles K. S., Evans E. J., Stuart D. I., Dwek R. A., Jones E. Y., Owens R. J., Davis S. J. (2007) Structure 15, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley P. (1992) Glycobiology 2, 99–107 [DOI] [PubMed] [Google Scholar]
- 25.Dwek R. A. (1998) Dev. Biol. Stand. 96, 43–47 [PubMed] [Google Scholar]
- 26.Hamilton S. R., Bobrowicz P., Bobrowicz B., Davidson R. C., Li H., Mitchell T., Nett J. H., Rausch S., Stadheim T. A., Wischnewski H., Wildt S., Gerngross T. U. (2003) Science 301, 1244–1246 [DOI] [PubMed] [Google Scholar]
- 27.Li H., Sethuraman N., Stadheim T. A., Zha D., Prinz B., Ballew N., Bobrowicz P., Choi B. K., Cook W. J., Cukan M., Houston-Cummings N. R., Davidson R., Gong B., Hamilton S. R., Hoopes J. P., Jiang Y., Kim N., Mansfield R., Nett J. H., Rios S., Strawbridge R., Wildt S., Gerngross T. U. (2006) Nat. Biotechnol. 24, 210–215 [DOI] [PubMed] [Google Scholar]
- 28.Hamilton S. R., Davidson R. C., Sethuraman N., Nett J. H., Jiang Y., Rios S., Bobrowicz P., Stadheim T. A., Li H., Choi B. K., Hopkins D., Wischnewski H., Roser J., Mitchell T., Strawbridge R. R., Hoopes J., Wildt S., Gerngross T. U. (2006) Science 313, 1441–1443 [DOI] [PubMed] [Google Scholar]
- 29.Reeves P. J., Callewaert N., Contreras R., Khorana H. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gleeson P. A., Feeney J., Hughes R. C. (1985) Biochemistry 24, 493–503 [DOI] [PubMed] [Google Scholar]
- 31.Hughes R. C., Feeney J. (1986) Eur. J. Biochem. 158, 227–237 [DOI] [PubMed] [Google Scholar]
- 32.Hughes R. C., Mills G., Stojanovic D. (1983) Carbohydr. Res. 120, 215–234 [DOI] [PubMed] [Google Scholar]
- 33.Foddy L., Feeney J., Hughes R. C. (1986) Biochem. J. 233, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paschinger K., Hackl M., Gutternigg M., Kretschmer-Lubich D., Stemmer U., Jantsch V., Lochnit G., Wilson I. B. (2006) J. Biol. Chem. 281, 28265–28277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aghi M., Kramm C. M., Chou T. C., Breakefield X. O., Chiocca E. A. (1998) J. Natl. Cancer Inst. 90, 370–380 [DOI] [PubMed] [Google Scholar]
- 36.Bebbington C., Hentschell C. (1987) in DNA Cloning III: A Practical Approach (Glover D. M. ed) pp. 168–188, IRL Press, Oxford [Google Scholar]
- 37.Davis S. J., Ward H. A., Puklavec M. J., Willis A. C., Williams A. F., Barclay A. N. (1990) J. Biol. Chem. 265, 10410–10418 [PubMed] [Google Scholar]
- 38.Aricescu A. R., Lu W., Jones E. Y. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 39.Durocher Y., Perret S., Kamen A. (2002) Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Küster B., Wheeler S. F., Hunter A. P., Dwek R. A., Harvey D. J. (1997) Anal. Biochem. 250, 82–101 [DOI] [PubMed] [Google Scholar]
- 41.Börnsen K. O., Mohr M. D., Widmer H. M. (1995) Rapid Commun. Mass Spectrom. 9, 1031–1034 [DOI] [PubMed] [Google Scholar]
- 42.Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 622–630 [DOI] [PubMed] [Google Scholar]
- 43.Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 631–646 [DOI] [PubMed] [Google Scholar]
- 44.Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 647–659 [DOI] [PubMed] [Google Scholar]
- 45.Harvey D. J., Royle L., Radcliffe C. M., Rudd P. M., Dwek R. A. (2008) Anal. Biochem. 376, 44–60 [DOI] [PubMed] [Google Scholar]
- 46.Olsnes S., Pihl A. (1982) in Molecular Action of Toxins and Viruses (Cohen P., van Heyningen S. eds) pp. 51–105, Elsevier, Amsterdam [Google Scholar]
- 47.Watson P., Spooner R. A. (2006) Adv. Drug Deliv. Rev. 58, 1581–1596 [DOI] [PubMed] [Google Scholar]
- 48.Spooner R. A., Watson P. D., Marsden C. J., Smith D. C., Moore K. A., Cook J. P., Lord J. M., Roberts L. M. (2004) Biochem. J. 383, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spooner R. A., Hart P. J., Cook J. P., Pietroni P., Rogon C., Höhfeld J., Roberts L. M., Lord J. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17408–17413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crispin M., Harvey D. J., Chang V. T., Yu C., Aricescu A. R., Jones E. Y., Davis S. J., Dwek R. A., Rudd P. M. (2006) Glycobiology 16, 748–756 [DOI] [PubMed] [Google Scholar]
- 51.Crispin M., Aricescu A. R., Chang V. T., Jones E. Y., Stuart D. I., Dwek R. A., Davis S. J., Harvey D. J. (2007) FEBS Lett. 581, 1963–1968 [DOI] [PubMed] [Google Scholar]
- 52.Oh-Eda M., Nakagawa H., Akama T. O., Lowitz K., Misago M., Moremen K. W., Fukuda M. N. (2001) Eur. J. Biochem. 268, 1280–1288 [DOI] [PubMed] [Google Scholar]
- 53.Harvey D. J., Crispin M., Scanlan C., Singer B. B., Lucka L., Chang V. T., Radcliffe C. M., Thobhani S., Yuen C. T., Rudd P. M. (2008) Rapid Commun. Mass Spectrom. 22, 1047–1052 [DOI] [PubMed] [Google Scholar]
- 54.Domon B., Costello C. E. (1988) Glycoconj. J. 5, 397–409 [Google Scholar]
- 55.Chen W., Stanley P. (2003) Glycobiology 13, 43–50 [DOI] [PubMed] [Google Scholar]
- 56.Shah N., Kuntz D. A., Rose D. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9570–9575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moremen K. W. (2002) Biochim. Biophys. Acta 1573, 225–235 [DOI] [PubMed] [Google Scholar]
- 58.Butler M., Quelhas D., Critchley A. J., Carchon H., Hebestreit H. F., Hibbert R. G., Vilarinho L., Teles E., Matthijs G., Schollen E., Argibay P., Harvey D. J., Dwek R. A., Jaeken J., Rudd P. M. (2003) Glycobiology 13, 601–622 [DOI] [PubMed] [Google Scholar]
- 59.Choi B. K., Bobrowicz P., Davidson R. C., Hamilton S. R., Kung D. H., Li H., Miele R. G., Nett J. H., Wildt S., Gerngross T. U. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5022–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X., Steet R. A., Bohorov O., Bakker J., Newell J., Krieger M., Spaapen L., Kornfeld S., Freeze H. H. (2004) Nat. Med. 10, 518–523 [DOI] [PubMed] [Google Scholar]
- 61.Foulquier F., Vasile E., Schollen E., Callewaert N., Raemaekers T., Quelhas D., Jaeken J., Mills P., Winchester B., Krieger M., Annaert W., Matthijs G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3764–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.