Abstract
Little is known about the regulation of the innate host defense peptide cathelicidin at the mucosal surfaces. Expression is believed to be transcriptionally regulated, and several cis-acting elements have been identified in the cathelicidin putative promoter. However, the trans-acting factors have not been clearly defined. We have recently reported that bacterial exotoxins suppress cathelicidin expression in sodium butyrate-differentiated intestinal epithelial cells (ECs), and this may be mediated through inducible cAMP early repressor. Here we have shown that cAMP-signaling pathways transcriptionally regulate cathelicidin expression in various ECs. cAMP-response element-binding protein (CREB) and AP-1 (activator protein-1) bind to the cathelicidin putative promoter in vitro. Additionally, transcriptional complexes containing CREB, AP-1, and cathelicidin upstream regulatory sequences are formed within ECs. We have also shown that these complexes may activate cathelicidin promoter and are required for its inducible expression in ECs. This is underscored by the fact that silencing of CREB and AP-1 results in failure of ECs to up-regulate cathelicidin, and hepatitis B virus X protein may use CREB to induce cathelicidin. On the other hand, inducible cAMP early repressor competes with CREB and AP-1 for binding to the cathelicidin promoter and represses transcription, thus functioning as a counter-regulatory mechanism. Finally, both CREB and AP-1 were shown to play major roles in the regulation of cathelicidin in sodium butyrate-differentiated HT-29 cells. This is the first report of a detailed mechanistic study of inducible cathelicidin expression in the mucosal ECs. At the same time, it describes a novel immunomodulatory function of cAMP.
hCAP-18 is the only known member of the cathelicidin family of small cationic antimicrobial peptides in humans that constitutes an essential component of the host innate immune system (1–4). It has direct microbicidal activities against a wide range of bacteria, viruses, fungi, and protozoa in vitro (5–10). In addition, its antimicrobial functions in vivo are significantly contributed by multiple immunomodulatory properties (11–13). The physiological importance of cathelicidin in host defense is underscored by the increased susceptibility of the knock-out animals, whereas the transgenics develop resistance to various infections (14–18).
Cathelicidin is widely expressed by many cells and tissues of the body (11, 19). Epithelial surfaces, such as the skin and the mucosal and squamous epithelia of the gastrointestinal, respiratory, and genitourinary tracts, are the major sites of cathelicidin function that includes homeostasis as well as immune responses. Accumulating evidence suggests that the expression, which is either constitutive or modulated by external stimuli, as well as the regulatory mechanisms, may be stimulus- and tissue-specific (20–22). Thus, normal skin and the colonic epithelium express very low levels of cathelicidin, although high basal expression is found in bone marrow, thymus, and several other tissues (19, 22, 23). Inflammatory lesions have been reported to increase its expression in the airway and cervical epithelium as well as in the keratinocytes (19, 23, 24). In addition, cytokines and growth factors may regulate cathelicidin expression in the skin epithelial cells (25–27). However, the underlying mechanisms of regulation remain poorly understood. On the other hand, pro- or anti-inflammatory cytokines play no role in the regulation of cathelicidin expression in the colonic epithelium (22).
It is generally believed that cathelicidin expression in different tissues is transcriptionally regulated (1, 11, 28, 29). Researchers have suggested complex regulation by both transcriptional activators and repressors (30). Vitamin D3 has been extensively studied for its role in the regulation of cathelicidin in the keratinocytes and monocytes (31–33). It functions through vitamin D receptor (VDR),2 a member of the nuclear hormone receptor superfamily that binds to the consensus VDR element repeats over the cathelicidin promoter (34, 35). Elegant studies published recently have demonstrated VDR-mediated induction of cathelicidin in response to TLR2 activation (36, 37). On the other hand, sodium butyrate (NaB) so far remains the most potent inducer of cathelicidin in the colon epithelial cells (22, 38). Although researchers have described several NaB-responsive elements in the cathelicidin upstream regulatory region, efforts to identify specific trans-acting factors remain largely elusive (39). Recent reports have suggested a role for intracellular signaling molecules like ERK1/2, p38 MAPK, and transforming growth factor-β1 kinase in NaB-mediated up-regulation of cathelicidin expression (38, 40), and it is believed that histone deacetylase inhibition by NaB may also contribute to this effect (21, 41). Several pathogenic microorganisms have been demonstrated to either up- or down-regulate cathelicidin in the mucosal ECs (5, 22, 42), and we have recently reported that bacterial exotoxins markedly suppress cathelicidin expression in the differentiated intestinal ECs in vitro and in vivo in a cAMP-dependent mechanism (43).
Activation of cAMP-signaling pathways involves accumulation of cAMP second messenger inside the cells and subsequent phosphorylation of the cellular kinases (44). Protein kinase A (PKA) is the best known cAMP effector that regulates transcription mainly through direct phosphorylation and activation of the bZip family members CREB, CREM τ, and ATF1. Activated bZip family transcription factors bind the consensus cAMP-response element (CRE) sequences over the promoters of the cAMP-responsive genes (45, 46). A similar sequence called AP-1-response element/TPA-response element (ARE/TRE) is occupied by activated AP-1 family proteins c-Fos and c-Jun. These molecules may be transcriptionally induced by CREB (47) or regulated post-translationally through phosphorylation by MAPKs (48), which extensively cross-talk with the cAMP-PKA pathway. Although p38 MAPK is usually activated by cAMP, JNK and ERK may be either positively or negatively regulated (49, 50). Activated ERK and p38 MAPK in turn may phosphorylate CREB. CRE- and ARE/TRE-binding factors also include several transcriptional repressors. Although the majority of them (CREMα, -β, and -γ, E4BP4, and CREB2) are regulated via phosphorylation by PKA, a group of CREM family members known as ICER is stringently controlled at the transcriptional level by CREB and an autoregulatory feedback mechanism (45, 46). Our previous studies have suggested that ICER may be involved in the transcriptional repression of cathelicidin (43).
Here, we have studied transcriptional regulation of cathelicidin by cAMP signal transduction pathways. We have extensively investigated the role of CREB, AP-1, and ICER in cAMP-induced regulation of cathelicidin expression in the mucosal epithelial cells. In addition, we have also studied their role in NaB-regulated cathelicidin expression.
EXPERIMENTAL PROCEDURES
Cathelicidin
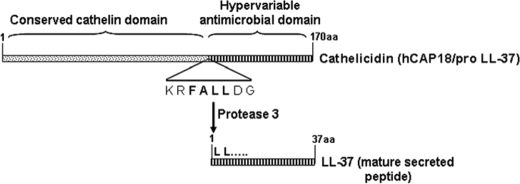
Cathelicidin is a family of small cationic antimicrobial peptides, the sole member of which in humans is called hCAP-18 (human cathelicidin antimicrobial peptide 18) (Scheme 1). The full-length molecule is synthesized in the cells and stored within vesicles. It is cleaved off by protease 3, and the mature 37-amino acid-long peptide (LL-37) with antimicrobial functions is secreted outside the cells.
SCHEME 1.
Cells and Reagents
HT-29 (HTB-38), Caco-2 (HBT-37), INT-407 (CCL6), and A459 (CCL-185) cell lines were purchased from the American Type Culture Collection (ATCC). Sodium butyrate (NaB), purified cholera toxin, 8-bromo-cAMP, N6,2′-O-dibutyryladenosine-cAMP, forskolin (FSK), 2′,5′-dideoxyadenosine (DDA), prostaglandin E2, actinomycin D (ActD), and (Rp)-cAMP were purchased from Sigma. Heat-labile toxin (LT) of enterotoxigenic E. coli was a generous gift from Dr. T. Hamabata of the International Medical Center of Japan, Tokyo. Cathelicidin antibody was procured from Santa Cruz Biotechnology, and CREB, c-Jun, and c-Fos antibodies and their respective phosphoantibodies were purchased from Cell Signaling Technology. Synthetic inhibitors (U0126, SB203580, MG132, and JNK inhibitor II) were procured from Calbiochem. Oligonucleotides for the cloning of c-jun (FP, 5′-GAATTCATGACTGCAAAGATGGAAACG-3′; RP, 5′-CTCGAGGACGGTCTCTCTTCTAAATG-3′), c-fos (FP, 5′-GGAATTCATGATGTTCTCGGGCTTCAACGC-3′; RP, 5′-CTCGAGTCACAGGGCCAGCAGCGTGGGTG-3′), and cathelicidin upstream regulatory sequences (FP1, 5′-TAGATGGAGCAGAGCCTTCG-3′; FP2, 5′-CTGTTACCCAGGCTGGAGTG-3′; FP3, 5′-CTGCTTCCCGGGTTCAATG-3′; FP4, 5′-TACAGGTGTGAGCCATCATG-3′; FP5, 5′-CTTGAGCACCCCTGGCTATGAC-3′, and RP, 5′-GGCCCAGCAGCAGGAGCACCA-3′) were custom-synthesized from IDT. Primers for qPCR amplification of cathelicidin (FP, 5′-CCAAGCCTGTGAGCTTCACAG-3′; RP, 5′-GGACTCTGTCCTGGGTACAAG-3′), CREB (FP, 5′-ATGACCATGGAATCTGGAGC-3′; RP, 5′-TTAATCGGATTTGTGGCAG-3′), c-jun (FP, 5′-ACAGAGCATGACCCTGAACC-3′; RP, 5′-CCGTTGCTGGACTGGATTAT-3′), c-fos (FP, 5′-GAATCCGAAGGGAAAGGAA-3′; RP, 5′-CTTCTCCTTCAGCAGGTTGG-3′), and ICER (FP, 5′-TGGAGATGAAACAGATGAGGAA-3′; RP, 5′-TCTCTGAGGGCCTTGAGTTC-3′) were also received from IDT.
Cell Culture and Stimulation
All the cell lines were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 2 mmol/liter l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). For experimental purposes, 2 × 105 cells were seeded in each well of a 24-well plate and allowed to grow. Cells near confluence were cultured overnight in serum-free DMEM before the experiment except for NaB treatment where 1-day post-confluent cells were cultured in serum-free DMEM for 48 h in the presence of NaB (4 mmol/liter).
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted with TRIzolTM reagent (Invitrogen) following the manufacturer's instructions. cDNA was prepared from 2 μg of extracted RNA using Superscript II reverse transcriptase (Invitrogen) following the standard protocol.
SYBR-Green® Real Time PCR
Real time quantitative PCR was performed using ABI7300 (Applied Biosystems). Relative quantitation was done by the comparative CT method. PCR was performed with SYBR Green® Mastermix (Applied Biosystems), where SYBR green is the fluorescent reporter. The internal control gene GAPDH was amplified simultaneously in separate reaction tubes. To eliminate primer dimers, the fluorescent signal was collected at a temperature of 82 °C, where the primer dimers melted, but the PCR products were still in their double-stranded form, thereby emitting fluorescence. The reaction conditions were set as follows: initial heating at 95 °C for 5 min, followed by 40 cycles of reactions at 95 °C (1 min), followed by 62 °C (2.5 min), and finally 82 °C (45 s). Final extension was carried out at 62 °C for 8 min. Threshold cycle number (CT) of triplicate reactions was determined using the ABI-SDS software, and the mean CT of triplicate reactions was determined. The levels of expression of the genes of interest were normalized against GAPDH using the formula 2−ΔΔCT, where −ΔΔCT = ΔCT (sample) − ΔCT (calibrator) and ΔCT is the CT of the target gene subtracted from the CT of the housekeeping gene (GAPDH). The calibrator used in our experiments was the unstimulated HT-29 and A549 cells.
Cloning and Expression
CREB and dominant negative CREB (dnCREB) constructs were the generous gifts from Marc Montiminy (Salk Institute), and the ICER expression construct was kindly provided by P. Sassone-Corsi (University of California, Irvine). c-jun and c-fos were PCR-amplified from cDNAs derived from HT-29 cells using Pfu TurboTM (Stratagene). Cathelicidin putative promoter sequences and the deletion mutants were also amplified by PCR. Amplified DNAs were cloned into pBluescriptSK+ (Novagen). Positive clones were selected by blue-white screening and confirmed by sequencing in an ABI automated sequencer. Overexpression constructs of all the above genes were generated by subcloning the DNAs into the pIRES-hrGFP vector (Stratagene), and the reporter constructs of cathelicidin promoter sequences were generated by subcloning the latter into the pGL3-Basic vector (Promega) upstream of the luciferase gene. Expression construct of hepatitis B “X” gene (HBX) was generated by PCR amplification of the gene using a clone of hepatitis B virus whole genome (kindly provided by Dr. Hidenori Shiraha, Okayama University, Japan) as template, followed by cloning into pBluescriptSK+ and finally subcloning into pcDNA3.1 vector (Invitrogen).
Transient and Stable Transfection
Transfection of HT-29 cells was done using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Briefly, cells (1 × 105/well) were seeded in 24-well plates 18 h prior to transfection and cultured in DMEM supplemented with 10% fetal bovine serum but no antibiotics. Cells in each well were incubated with transfection mix containing 0.75 μg of total DNA for 6 h followed by addition of complete DMEM. Transient transfection was checked after 48 h. HepG2 cells stably expressing HBX were generated by transfecting the cells as described above followed by selection with G418 (600 μg/ml) for 4 weeks.
Reporter Assay
Reporter assay was done 48 h post-transfection using dual luciferase assay kit (Promega) following the manufacturer's protocol. Briefly, cells were washed with PBS followed by lysis with 1× passive lysis buffer (Promega). The lysate was cleared by brief centrifugation, and firefly luciferase reporter activity in the clear supernatant was measured with a luminometer (Berthold). Renilla luciferase activity in the same lysate was used as transfection control.
ELISA
LL-37 ELISA was performed by coating the ELISA plates (Nunc) with the secreted LL-37 protein present in the culture supernatants of unstimulated or FSK-stimulated HT-29 cells. 50 μl of the culture supernatant containing 50 μg of total protein was added to each well and incubated overnight at 4 °C. Excess supernatants were withdrawn from the wells, which were washed twice with PBS. 200 μl of blocking buffer containing 1% bovine serum albumin in PBS (1×) was added to each well and incubated at room temperature for 1 h. The wells were washed three times with the wash buffer (0.05% Tween 20 in 1× PBS) followed by the addition of 100 μl of LL-37 primary antibody (Santa Cruz Biotechnology) diluted (1:100) in blocking buffer. After incubation for 1 h at room temperature, wells were washed four times with the wash buffer and further incubated at room temperature for 1 h with 100 μl of HRP-conjugated secondary antibody (Pierce) diluted (1:1000) in the blocking buffer. Wells were washed again four times and incubated with 50 μl (1 mg/ml) of o-phenylenediamine (Sigma) diluted in phosphate/citrate buffer, pH 5.0. After 30 min of incubation at room temperature, the reaction was stopped by adding 50 μl of 1 n H2SO4, and absorbance was taken at 590 nm.
Western Blot Analysis
Cell lysates were prepared by harvesting cells in Triton X-100 lysis buffer (50 mm HEPES, pH 7.4, 250 mm NaCl, 2 mm EDTA, 1% Triton X-100, 10% glycerol, 50 mm β-glycerophosphate, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 1 mm NaF, 10 μg/ml leupeptin, and 5 μg/ml aprotinin). After clearing the lysate by centrifugation at 14,000 rpm for 10 min, supernatants were collected, and total protein content was measured by the Bradford assay. 30 μg of total protein per well was resolved in a 12% SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane. The blot was probed with the specific primary antibody followed by HRP-conjugated secondary antibody (Pierce) and developed using SuperSignal West Pico chemiluminescence substrate (Pierce).
Nuclear Run-on Assay
A nonradioactive nuclear run-on assay (51) was used in conjunction with qRT-PCR to measure the transcription rate of cathelicidin in the HT-29 cell line. Briefly, 107 numbers of cells were trypsinized and washed twice with Ca2+- and Mg2+-free PBS. The cell pellet was resuspended in lysis buffer containing 10 mm Tris-HCl, pH 7.4, 3 mm MgCl2, 10 mm NaCl, 150 mm sucrose, and 0.5% Nonidet P-40 and passed through a 21-gauge needle five times, which ensures lysis of the cell membrane. Nuclei were collected by centrifugation at 14,000 rpm for 10 min at 4 °C. The run-on reaction was carried out in a 2× transcription buffer containing 200 mm KCl, 20 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 5 mm dithiothreitol, 4 mm each of ATP, GTP, and CTP, 200 mm sucrose, and 20% glycerol. The reaction was initiated by adding 4 mm biotin-16-UTP (Ambion) and continued for 1 h at 29 °C. uMACS column (Miltenyi Biotec) packed with magnetic beads covalently linked to streptavidin was used to isolate the biotin-labeled run-on RNA. The beads were resuspended in RNase-free water and used for reverse transcription and real time PCR. Total cellular RNAs from parallel experiments were isolated using TRIzol reagent (Invitrogen) and subjected to qRT-PCR.
Preparation of Nuclear and Cytoplasmic Extracts
Nuclear and cytoplasmic fractions were prepared as described previously (52). Briefly, 5 × 106 HT-29 or A549 cells seeded 24 h prior to the experiment were harvested by scraping. Cells were resuspended in low salt buffer A (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl) and incubated on ice for 15 min. After the addition of Nonidet P-40 (0.5%, final concentration), the cell suspensions were passed through a 21-gauge needle 5–6 times. Cytoplasmic and nuclear fractions were separated by centrifugation at 14,000 rpm for 10 min at 4 °C. Nuclear proteins were extracted by incubating the pellet in high salt buffer C (20 mm HEPES, pH 7.9, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 25% glycerol) for 30 min on ice with vigorous shaking (250 rpm).
Electrophoretic Mobility Shift Assays (EMSAs)
CREB and AP-1 EMSAs were carried out using standard protocols. Briefly, nuclear extracts of HT-29 and A549 cells were incubated with biotin-labeled CRE (5′-CATGATCTCA-3′ in three tandem repeats) and ARE (5′-TGCCTCATTC-3′ in three tandem repeats) consensus oligonucleotides in a binding buffer (50 mm HEPES, 250 mm KCl, 0.5 mm EDTA, 50% glycerol, 1 mm dithiothreitol, 5 mm MgCl2, 0.4 μg/ml single-stranded DNA) at room temperature for 30 min. For oligonucleotide competition assays, nuclear extracts were preincubated with a 100-fold excess of the mutated (CRE, 5′-CATGATTGTC-3′; ARE, 5′-TGTTTCGCGC-3′) or unlabeled oligonucleotides prior to incubation with the labeled oligonucleotides, while for the supershift assays, the DNA-protein complexes were incubated with antibodies against the protein(s) in the complexes. The reaction mixture was resolved in a 6% nondenaturing polyacrylamide gel at 200 V for 2 h followed by transfer to a charged nylon membrane. The charged nylon membrane was developed using streptavidin-HRP followed by chemiluminescence.
DNA Pulldown Assay
DNA pulldown assay was performed as described previously with minor modifications (53). Briefly, CRE or ARE oligonucleotides used in EMSA were labeled with biotinylated dUTP by random labeling kit (Pierce) following the manufacturer's protocol. 0.3 μg of labeled oligonucleotides and nuclear extracts (500 μg of total protein) of untreated or FSK-treated HT-29 or A549 cells was incubated together in the binding buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 5% glycerol, 1 mm dithiothreitol, and 0.1% Triton X-100) at 4 °C for 90 min. After the reaction was completed, 100 μl of streptavidin-coated magnetic beads (Miltenyi Biotec) were added to the reaction mix and loaded onto a pre-equilibrated column placed in a high magnetic field. The column was washed three times with appropriate wash buffer, and the bound proteins were eluted with the elution buffer containing 1 m NaCl. Eluted proteins were resolved in 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Transferred proteins were probed with CREB, c-Jun, and c-Fos antibodies (phospho and total), and the blot was developed using SuperSignal West Pico chemiluminescence substrate (Pierce).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using EZChIPTM chromatin immunoprecipitation kit (Upstate) following the manufacturer's instructions. Briefly, HT-29 and A549 cells treated with or without FSK were cross-linked using formaldehyde and harvested by scraping. The cell pellet was resuspended in the lysis buffer provided with the kit, and chromatin was immunoprecipitated using CREB, c-Jun, or c-Fos antibodies as well as anti-RNA polymerase II antibody. Immunoprecipitated genomic DNA of cathelicidin promoter was amplified by qPCR using specific primers flanking the CRE (FP, 5′-TTGGGGGTGGCTACTGTCTT-3′; RP, 5′-AGCTGAGATCATGCCACTGC-3′) and ARE sequences (FP, 5′-TGCTGGGATTATAGGCGTGA-3′; RP, 5′-ATAGCCAGGGGTGCTCAAGA-3′). The specific region of the GAPDH promoter was also amplified (FP, 5′-TACTAGCGGTTTTACGGGC-3′; RP, 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′) from anti-RNA polymerase II-precipitated genomic DNA and was used to normalize cathelicidin promoter usage in the unstimulated and stimulated cells.
Gene Silencing by siRNA
siRNAs for CREB, c-jun, and c-fos as well as the transfection kit were purchased from Santa Cruz Biotechnology. Growing HT-29 and A549 cells at 70–80% confluence were transiently transfected following the manufacturer's protocol. siRNA duplex and siRNA transfection reagent were diluted into the siRNA transfection medium and incubated together at room temperature for 30 min. Cells were overlaid with the transfection mixture and cultured for 5 h at 37 °C in the presence of 5% CO2. The efficiency of gene silencing was verified by qRT-PCR 48 h post-transfection. siRNAs for ICER were designed using the siRNA data base (Ambion). Briefly, four predicted target sequences of the ICER gene were selected and corresponding oligonucleotides were synthesized from Sigma. Oligonucleotides were cloned into the pSilencer-puro 4.1 vector (Ambion). Clones were transfected into HT-29 and A549 cells using siPortTM transfection reagent (Ambion) following the manufacturer's protocol. Transfection efficiency in the form of silencing of ICER was verified 48 h post-transfection by qRT-PCR.
Statistical Analysis
Statistical significance was analyzed by the Student's t test. The results were considered less significant at p ≤ 0.01 and highly significant at p ≤ 0.001.
RESULTS
cAMP Regulates Cathelicidin Expression in the Epithelial Cells of Diverse Origins
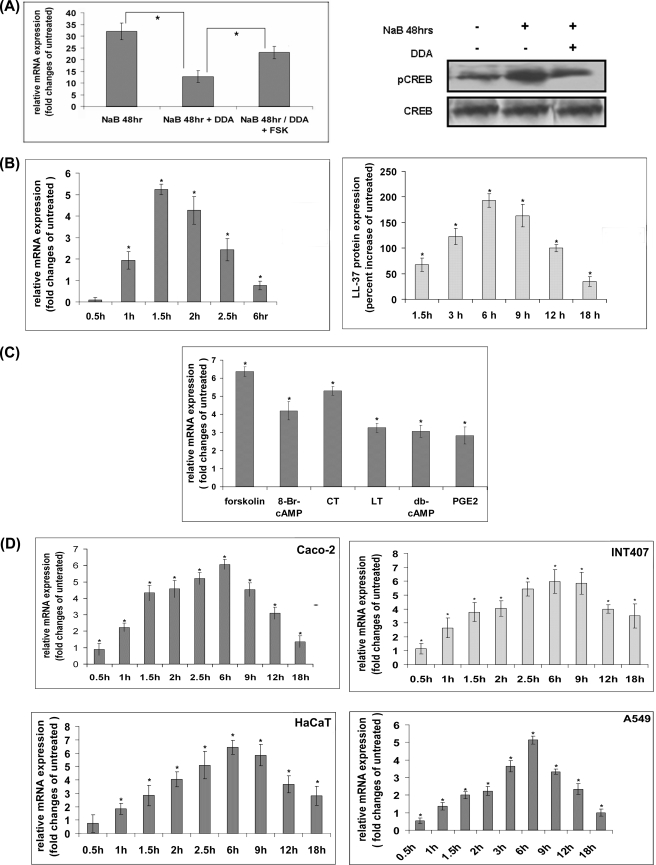
We have recently reported that cholera toxin (CT) and LT of enterotoxigenic Escherichia coli down-regulate cathelicidin expression in NaB-differentiated intestinal ECs, possibly through ICER (43). Although the latter is known to repress transcription of genes induced by cAMP, we failed to detect induction of cathelicidin in the differentiated intestinal ECs upon stimulation of the cAMP signal transduction pathways by CT or LT. We postulated that these pathways might already be activated by NaB treatment, and this resulted in elevated cathelicidin expression in the differentiated cells. Cathelicidin levels failed to increase upon further stimulation; instead, such stimulation induced repressor gene ICER, which suppressed cathelicidin expression. To prove this hypothesis, we studied cathelicidin levels in the colon epithelial cells HT-29, treated for 48 h with NaB with or without adenylate cyclase inhibitor 2′,5′-DDA that interfered with cAMP accumulation inside the cells. The results showed marked induction of cathelicidin in the NaB-treated cells, which was significantly obliterated by concomitant treatment with the adenylate cyclase inhibitor. Stimulation of the inhibitor-treated cells with the cAMP agonist FSK augmented cathelicidin expression, further supporting the hypothesis (Fig. 1A, left panel). Western blot analysis showed that CREB was activated in NaB-differentiated HT-29 cells, but significantly less in the 2′,5′DDA-treated cells (Fig. 1A, right panel). The above results together suggest that NaB activates cAMP signal transduction pathways and induces cathelicidin expression in the HT-29 cells. Moreover, cathelicidin is significantly more elevated in the NaB-treated cells that have activated cAMP-signaling pathways compared with similarly treated cells where the signaling pathways are inhibited. To investigate if cAMP regulates cathelicidin expression in the epithelial cells, undifferentiated HT-29 cells were stimulated with FSK for various durations. The highest levels of cathelicidin mRNA and LL-37 protein were induced after 90 min and 6 h, respectively, of stimulation that returned to the original levels over next several hours (Fig. 1B). Simultaneous stimulation of NaB-differentiated cells with FSK showed no up-regulation of cathelicidin; instead, its expression was reduced by severalfold compared with the cells treated with NaB alone (supplemental Fig. S1). Role of cAMP in cathelicidin regulation was further supported by stimulating the HT-29 cells with other cAMP agonists. These included bacterial ADP-ribosylating toxins CT and LT as well as the cell-permeable cAMP analogs (8-bromo-cAMP and dibutyryl cyclic AMP) and an endogenous agonist of cAMP (prostaglandin E2). All these agents significantly induced cathelicidin expression, albeit to a lesser extent compared with FSK (Fig. 1C). To rule out the possibility that cAMP effects were cell- or tissue-restricted, we stimulated other intestinal epithelial cell lines (Caco-2 and INT407) as well as keratinocyte (HaCaT) and lung ECs (A549) with FSK. Despite different kinetics, cathelicidin expression essentially followed a similar pattern as in the HT-29 cells (Fig. 1D). The above results together suggest that cAMP-signaling pathways may regulate cathelicidin expression in multiple ECs of human origin.
FIGURE 1.
Induction of cathelicidin expression by cAMP. A, 1-day post-confluent HT-29 cells were treated with NaB with or without 2′,5′-DDA for 48 h. A separate set of cells treated with both the agents was subsequently stimulated with FSK for 1.5 h. Left panel, cathelicidin mRNA expression was analyzed in triplicate samples by qRT-PCR and data presented as mean (±S.D.) relative (GAPDH normalized) expression in the treated cells compared with that in the untreated cells. Right panel, Western blot analysis with the cells treated as above using phospho-CREB (pCREB) and CREB (total) antibodies. B, HT-29 cells were stimulated with FSK for different durations. Left panel, cathelicidin mRNA expression in triplicate samples was analyzed by qRT-PCR, and data are presented as in A in the stimulated compared with the unstimulated cells. Right panel, ELISA showing LL-37 protein expression (mean ±S.D.) in the FSK-stimulated compared with the unstimulated quadruplicate samples. C, qRT-PCR analysis of cathelicidin expression in the HT-29 cells stimulated with different cAMP analogs or exogenous or endogenous activators of the cAMP signal transduction pathways for 1.5 h. D, cathelicidin mRNA expression in the epithelial cells of diverse origins stimulated with FSK for various durations was analyzed by qRT-PCR. The experiment was done in triplicate and data presented as in B. Each of the above data is representative of three independent experiments.
Cathelicidin Regulation by cAMP Is Transcriptionally Mediated and Involves Multiple Signal Transduction Pathways
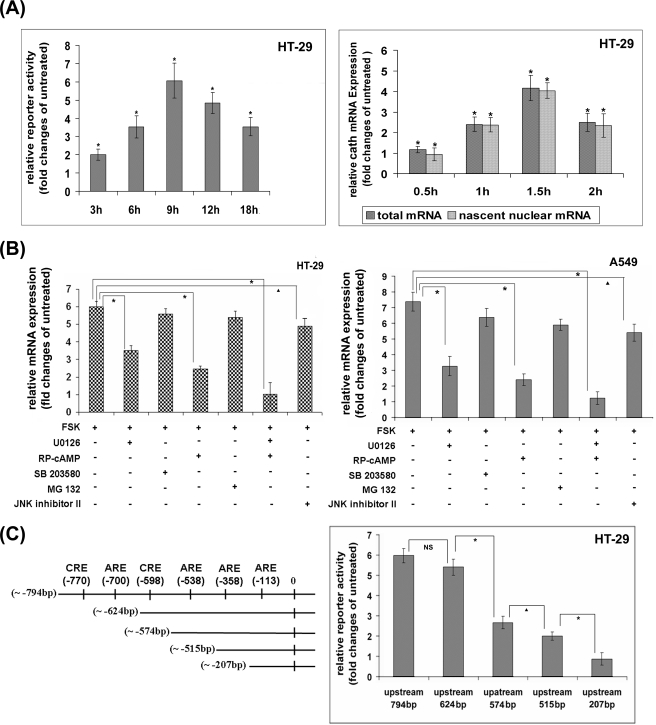
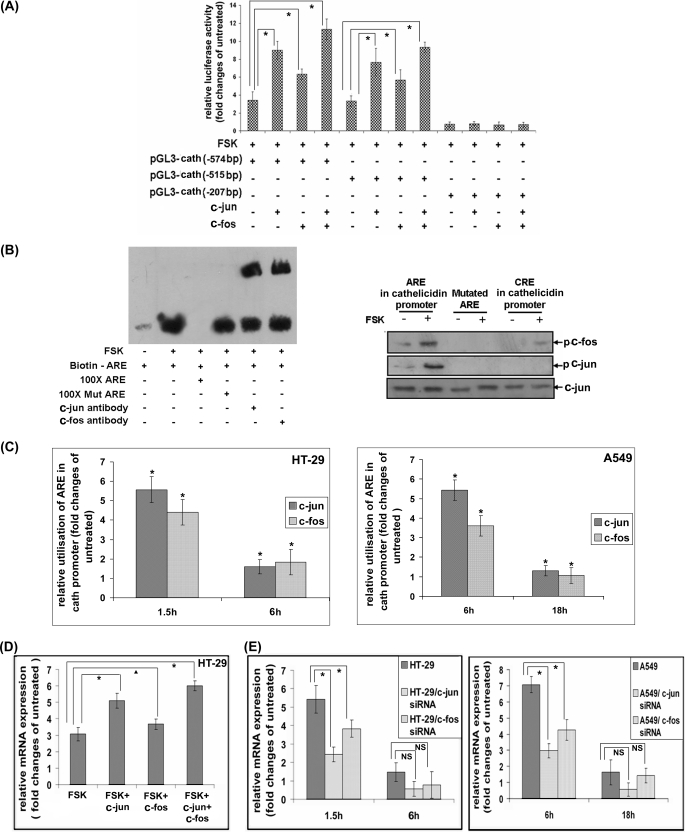
Although several published studies have suggested that the regulatory mechanisms for cathelicidin expression in different tissues mainly operate at the transcriptional level (11, 19), it has never been experimentally proved for mucosal expression. On the contrary, NaB has been suggested to post-transcriptionally regulate cathelicidin in the colonic epithelial cells (20). To investigate whether elevated cathelicidin mRNA expression upon activation of the cAMP-signaling pathways is caused by increased transcriptional activity or a post-transcriptional mechanism like mRNA stability, we transiently overexpressed into the HT-29 cells a reporter plasmid containing the cathelicidin putative regulatory sequences (−794 bp upstream to +200 bp downstream of the transcription start site) cloned upstream of a luciferase gene. Stimulation of the cells with FSK increased the reporter activity, suggesting transcriptional activation (Fig. 2A, left panel). This was further confirmed by nuclear run-on assays with unstimulated and FSK-stimulated cells. Induction of nascent cathelicidin mRNAs incorporating biotinylated 16-UTP and the corresponding total mRNAs followed the same kinetics, confirming that increased transcriptional rate rather than post-transcriptional mechanisms led to the elevated cathelicidin mRNA and protein levels inside the cells (Fig. 2A, right panel). To study the pathways activated downstream of cAMP that may regulate cathelicidin expression, HT-29 and A549 cells were treated with synthetic inhibitors of the signaling pathways followed by FSK or FSK alone. The results showed significantly reduced induction of cathelicidin when the cells were pretreated with the PKA or the MEK inhibitor (Fig. 2B). Concomitant use of both the inhibitors almost completely obliterated cathelicidin induction, although the inhibitors of the NF-κB pathway, p38 MAPK, and JNK had little effect. The above results suggest essential and cooperative roles played by the ERK MAPK and PKA in the regulation of cathelicidin expression. PKA and ERK regulate gene expression mainly through the activation of transcription factors CREB and AP-1, respectively, and the cathelicidin putative promoter has several consensus CRE and ARE sites (43). We therefore studied the role of the above transcription factors in cathelicidin regulation by luciferase reporter assays. To this end, reporter constructs of the cathelicidin putative promoter having progressive deletions of the CRE and the ARE sequences were transiently overexpressed into the HT-29 cells followed by activation of CREB and AP-1 by FSK. The results showed significant loss of reporter activity when the CRE and the ARE sites at −598 and −358 bp, respectively were deleted, suggesting that CREB and AP-1 binding to these sites may play critical roles in the transactivation of cathelicidin gene by cAMP (Fig. 2C, right panel).
FIGURE 2.
Transcriptional regulation of cathelicidin by cAMP. A, left panel, luciferase reporter construct of cathelicidin putative promoter (−794 to +200 bp) was transiently overexpressed into the HT-29 cells followed by stimulation of the cells with FSK for various durations. Data are presented as the ratio of the mean (±S.D.) reporter (firefly luciferase) activities, normalized against the Renilla luciferase reporter activities, of the stimulated and the unstimulated cells. Right panel, qRT-PCR analysis of cathelicidin expression was performed with RNAs derived from nuclear run-on assays. Biotin-16-UTP-incorporated nascent RNAs were extracted from the nuclei of unstimulated and FSK-stimulated HT-29 cells. In parallel experiments, total cellular RNAs were extracted from similarly treated cells. Ratio of the mean (±S.D.) relative cathelicidin expression of the stimulated and unstimulated cells in the nuclear as well as the total lysates is presented. B, HT-29 (left) and A549 (right) cells were treated with synthetic signaling pathway inhibitors, either alone or in different combinations, followed by FSK for 1.5 and 6 h, respectively. Cathelicidin mRNA expression was analyzed by qRT-PCR, and data are presented as in Fig. 1B. C, left, selective cis-acting elements over the cathelicidin putative promoter, as retrieved from CSHL promoter data base and analyzed by TRANSFAC are graphically represented. Right, luciferase reporter activities were analyzed with the lysates of HT-29 cells transiently transfected with the reporter constructs of the cathelicidin putative promoter or one of its deletion mutants. All the experiments under A–C were performed with triplicate samples and repeated three times, and one representative for each experiment is shown. Cath, cathelicidin; U0126, MEK1 and 2 inhibitor; RP-cAMP, PKA inhibitor; SB 203580, p38 MAPK inhibitor; MG132, proteasome inhibitor; JNK inhibitor II (SP 600125), JNK inhibitor.
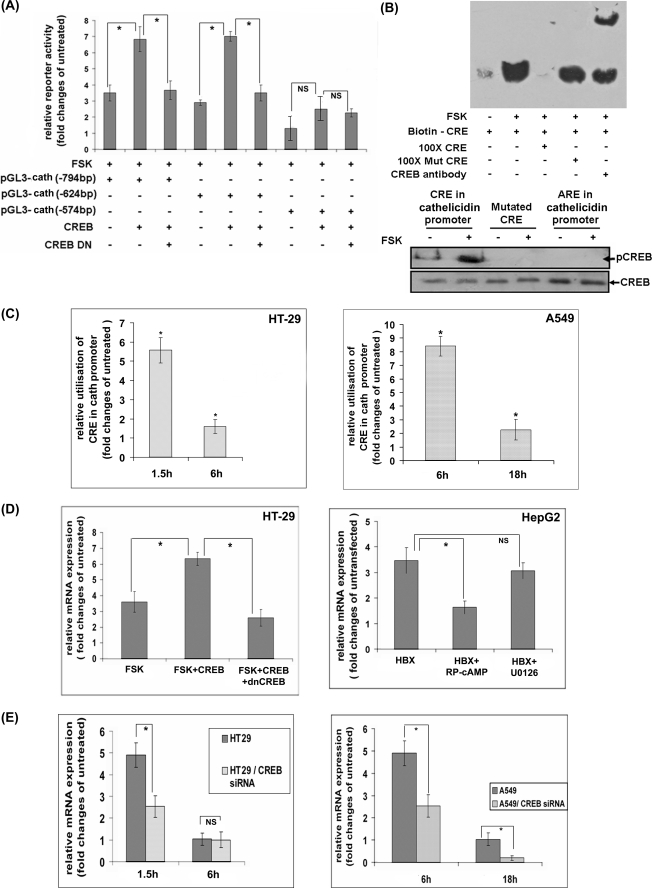
CREB Is Required for cAMP-induced Transcriptional Activation of Cathelicidin
The above results suggest that CREB may be required for cathelicidin promoter activity. To investigate whether CREB can drive transcription by this promoter, we analyzed luciferase reporter activity following co-expression of CREB and a reporter construct of the cathelicidin putative promoter. The results showed ∼100% increase in the basal luciferase activity when CREB was expressed along with the reporter. This rise was completely abolished by simultaneous expression of the dnCREB, indicating that it was indeed due to CREB. Deletion of the CRE site at −598 bp, but not at −770 bp, completely abolished CREB-induced reporter activation. This suggests that CREB may preferentially bind to the former site to transcribe cathelicidin (Fig. 3A). To investigate CREB binding to the cathelicidin promoter, we first carried out in vitro DNA/protein binding assays with the biotinylated oligonucleotides containing the CRE consensus sequence at −598 bp of the cathelicidin promoter (the “probe”) and the nuclear extracts of the HT-29 cells. When the reaction mixture was resolved in EMSA, the probe that was incubated with the nuclear extracts of the unstimulated cells migrated with the free probe (data not shown). On the other hand, the probe incubated with the FSK-stimulated nuclear extracts migrated considerably slower (the “shift”), suggesting formation of DNA-protein complex (Fig. 3B, upper panel). Although the excess of unlabeled wild type probe competed out the labeled probe from the complex, 100-fold excess of the mutant probe failed to do so. This suggests specificity of binding of the nuclear proteins(s) of FSK-stimulated cells to the CRE sequence of the cathelicidin promoter. The identity of the protein present in this complex was confirmed by incubating the DNA-protein complex with the CREB antibody that resulted in further delayed migration of the probe (supershift assay), indicating that CREB was indeed present there. In a separate assay (pulldown assay), the existence of CREB in the in vitro-generated DNA-protein complex was confirmed by Western blot analysis. Although significant amounts of CREB were detected in the complex containing the wild type CRE oligonucleotides, no CREB was bound to the mutated CRE or ARE oligonucleotides (Fig. 3B, lower panel). The above results together suggest that activated CREB of FSK-stimulated cells may bind to the cathelicidin promoter. To further address this issue, ChIP assays were performed where transcriptional complexes formed inside the unstimulated and FSK-stimulated HT-29 and A549 cells were immunoprecipitated using CREB antibody. qPCR amplification with specific primers using the immunoprecipitate as the template resulted in a single amplified product that matched the cathelicidin promoter sequences. The amounts of PCR-amplified products were significantly higher with the stimulated as opposed to the unstimulated cells, suggesting higher utilization of the promoter by CREB in the stimulated cells (Fig. 3C). In addition, cells having maximum cathelicidin expression (1.5 h post-stimulation) utilized CRE to a significantly larger extent compared with the cells with basal cathelicidin levels (6 h post-stimulation). These results strongly suggest that the complex described above may transactivate cathelicidin. To further study if CREB can mediate up-regulation of cathelicidin expression in the ECs, we transiently overexpressed CREB in the HT-29 cells or engineered liver epithelial HepG2 cells to stably express HBX protein that is known to enhance CREB-mediated transcription in a PKA-dependent manner (54). Cathelicidin expression was significantly augmented upon stimulation of the transfected HT-29 cells with FSK compared with the stimulated untransfected cells (Fig. 3D, left panel). On the other hand, HBX-expressing HepG2 cells constitutively expressed severalfold more cathelicidin than the parental HepG2 cells (Fig. 3D, right panel). These results indicate that CREB plays an important role in cAMP-induced cathelicidin expression. This is further supported by abolition of cathelicidin up-regulation by dnCREB in the HT-29 cells or treatment of the HepG2 cells expressing HBX with the PKA inhibitor. To finally prove that induction of cathelicidin by cAMP requires CREB, we silenced the CREB gene by siRNAs in the HT-29 and A549 cells and studied cathelicidin expression in response to the stimulus (FSK) that increases cAMP. FSK was remarkably less efficient in inducing cathelicidin following silencing of the CREB gene, confirming the essential role the latter plays in this regulation (Fig. 3E).
FIGURE 3.
Regulation of cathelicidin by CREB. A, HT-29 cells were transiently transfected with the luciferase reporter constructs of the cathelicidin putative promoter sequence (pGL3-cath), either alone or in combination with CREB or CREB and dnCREB constructs. Mean (±S.D.) reporter activities of the unstimulated and FSK-stimulated triplicate cell samples were normalized and presented as in Fig. 2A. B, upper panel, EMSAs performed with the biotinylated oligonucleotides of the CRE consensus sequence at −598 bp over the cathelicidin promoter, incubated with the nuclear extracts of the unstimulated and FSK-stimulated (1.5 h) HT-29 cells with or without 100-fold excess of unlabeled or mutated oligonucleotides or CREB antibody. DNA-protein complexes were resolved in 6% PAGE, transferred to a charged nylon membrane, and developed with streptavidin-HRP. Lower panel, DNA-protein complexes as generated above were pulled down using streptavidin-coated magnetic beads. The complexes were studied by Western blot analysis using phospho-CREB and CREB antibodies. C, ChIP assays. DNA-protein complexes from the unstimulated and FSK-stimulated HT-29 (left) and A549 (right) cells were immunoprecipitated with CREB (for cathelicidin promoter) and RNA polymerase II (for GAPDH promoter) antibodies. DNAs in the immunoprecipitates were amplified by qPCR using specific primers corresponding to the flanking sequences of the CRE sites at −598 bp of the cathelicidin promoter and to the GAPDH promoter, respectively. The ratio of GAPDH-normalized amplification of the CRE sequences of the stimulated and unstimulated cells is presented here. D, left, qRT-PCR analysis of cathelicidin expression in FSK-stimulated (1.5 h) untransfected HT-29 cells or in cells transfected with CREB or CREB along with dnCREB constructs. The results of the experiments done with triplicate samples are presented as in Fig. 1B. Right, cathelicidin and GAPDH mRNAs were amplified by qRT-PCR using total RNAs extracted from the untransfected HepG2 cells and cells stably expressing HBX protein. Cells in triplicate were either left untreated or treated with PKA or MEK inhibitor before RNAs were extracted. Data presented as above as relative cathelicidin expression of the engineered and parental HepG2 cells. E, CREB gene was silenced in the HT-29 (left) and A549 (right) cells using siRNAs. Cathelicidin was amplified by qRT-PCR from the triplicate parental and CREB-silenced cells stimulated with FSK for different durations. Data are presented as mean relative cathelicidin expression of the stimulated and unstimulated parental and silenced cells. All the experiments under A–E were performed three times, and the results of a representative one in each case is presented here. Cath, cathelicidin.
cAMP-induced Cathelicidin Expression Requires AP-1
In our studies, ARE/TRE sequences were found to be necessary for cathelicidin promoter activity (Fig. 2C). Hence, we investigated if AP-1 can augment cathelicidin promoter-driven transcription. To this end, HT-29 cells were transfected with the luciferase reporter construct of the cathelicidin promoter or one of its deletion mutants along with c-jun and/or c-fos. The results showed significant increase of reporter activity in the dually transfected cells as opposed to the cells that were transfected with the reporter alone when they were stimulated with FSK (Fig. 4A). Although c-Jun was transcriptionally more active than c-Fos, they acted cooperatively as their combined expression enhanced cathelicidin reporter activity more than when either one was expressed. Deletion of the ARE sequence at −358 bp resulted in complete loss of AP-1-induced reporter activity, suggesting that AP-1 predominantly uses this sequence over the cathelicidin promoter for transactivation. To experimentally prove the binding of AP-1 to the cathelicidin promoter, we carried out in vitro DNA/protein binding assays by incubating ARE oligonucleotides with the nuclear extracts of the HT-29 cells. The reaction mixture as resolved in EMSAs showed slower migration of the probe that was incubated with the nuclear extracts of FSK-stimulated cells as compared with that incubated with the unstimulated nuclear extracts (Fig. 4B, left panel). This suggests formation of the DNA-protein complex in the former case. Binding specificity of the nuclear protein(s) to the ARE sequence was ascertained by competition assays as described above. The mutated oligonucleotides failed to replace the labeled probe, which was efficiently competed out by the unlabeled wild type oligonucleotides. The presence of AP-1 in the DNA-protein complex was confirmed by supershift assays following incubation of the DNA-protein complex with c-Jun or c-Fos antibodies. Supershift was observed with both antibodies, indicating the presence of c-Jun as well as c-Fos in the complex. The identity of the proteins was also studied by Western blot analysis where the DNA-protein complex pulled down with streptavidin-coated magnetic beads was probed with c-Jun or c-Fos antibodies. AP-1 subunits significantly bound to the ARE sequence of the cathelicidin promoter but not to the CRE or mutated ARE (Fig. 4B, right panel). To investigate if endogenous AP-1 binds to the cathelicidin promoter, we performed ChIP assays as described above with unstimulated and FSK-stimulated HT-29 and A549 cells. Bound DNA was amplified in a qPCR after the DNA-protein complex was immunoprecipitated with c-Jun or c-Fos antibodies. Significantly higher amounts of amplified PCR product corresponding to the ARE sequence when the immunoprecipitate was generated from the FSK-treated as compared with the untreated cells suggest greater utilization of the above sequence in the former cells (Fig. 4C). As expected, c-Jun binding to the cathelicidin promoter was more efficient than c-Fos binding. In addition, the amounts of the PCR product were considerably higher with the HT-29 and A549 cells stimulated for 1.5 and 6 h, respectively. Given that the cells expressed highest levels of cathelicidin at those time points, these results strongly suggest that AP-1 may be involved in the transcriptional induction of cathelicidin. Next, we studied the functional significance of AP-1 binding to the cathelicidin promoter. To this end, we first investigated if AP-1 can augment cathelicidin expression in the ECs. c-Jun and c-Fos, both individually and cooperatively, significantly enhanced cathelicidin expression in the HT-29 cells upon stimulation with FSK, suggesting a role for AP-1 in cAMP-induced cathelicidin regulation (Fig. 4D). Finally, we confirmed the essential role played by c-Jun and c-Fos in this regulation by silencing these genes in the HT-29 and A549 cells. Cathelicidin induction was significantly abrogated in the FSK-stimulated cells that lacked either AP-1 subunit, more in the absence of c-Jun as expected (Fig. 4E).
FIGURE 4.
AP-1-induced regulation of cathelicidin. A, activity of the pGL3-cath reporter constructs transiently overexpressed in the HT-29 cells, along with c-fos or c-jun or both followed by stimulation of the cells with FSK for 1.5 h. The relative increase of reporter activity in FSK-stimulated versus unstimulated cells is presented as in Fig. 3A. B, left, EMSAs performed as in Fig. 3B with the biotinylated oligonucleotides of the ARE sequences at −358 bp over the cathelicidin promoter. Competition and supershift assays were done by incubating the above complexes with 100-fold excess of unlabeled or mutated ARE and c-Jun or c-Fos antibodies, respectively. The complexes were subsequently analyzed as in Fig. 3B. Right, DNA-protein complexes as generated above were pulled down using streptavidin-coated magnetic beads followed by Western blot analysis with phospho-c-Jun, phospho-c-Fos, and c-Jun (total) antibodies. C, ChIP assays. DNA-protein complexes immunoprecipitated with c-Jun or c-Fos antibodies from the unstimulated and FSK-stimulated HT-29 (left) and A549 (right) cells were subjected to qPCR analysis using primers flanking the ARE sequence as above. Data are normalized and presented as in Fig. 3C. D, cathelicidin gene amplified by qRT-PCR from the untransfected or c-jun/c-fos/c-jun plus c-fos transfected HT-29 cells stimulated with FSK. The experiment was done in triplicate, and data are presented as in Fig. 3D. E, FSK-induced cathelicidin expression in the parental and c-fos or c-jun gene-silenced HT-29 (left) and A549 (right) cells. Mean (±S.D.) relative (GAPDH-normalized) expression of the triplicate samples is presented as in Fig. 3E. All the above experiments were repeated three times and data from a representative one are shown here. Cath, cathelicidin.
Transcriptional Induction of AP-1 as Well as Phosphorylation of CREB and AP-1 by Upstream Kinases Regulate Cathelicidin Expression
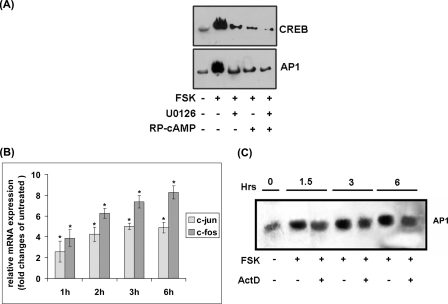
PKA and ERK are known to be the major cAMP-induced kinases that post-translationally regulate the activation of CREB and AP-1, respectively, by phosphorylation (50, 55). ERK has also been reported to independently phosphorylate CREB, whereas the latter molecule may transactivate both c-jun and c-fos (47, 56). To investigate if phosphorylation of CREB and AP-1 by the upstream kinases regulates cathelicidin transcription, we carried out DNA/protein binding assays using oligonucleotide probes of the CRE and ARE sequences as described above and the nuclear extracts of A549 cells treated with PKA and/or MEK inhibitor(s) followed by FSK. The results showed marked inhibition of transcriptional complex formation with the CRE oligonucleotides by both the kinase inhibitors, PKA inhibitor being more efficient (Fig. 5A). In addition, the inhibitors worked cooperatively to reduce DNA binding, suggesting that the above kinases acted independently as well as in collaboration to regulate cathelicidin. Similarly, PKA and ERK-MAPK also cooperated in the formation of transcriptional complexes with the ARE oligonucleotides. As AP-1 may be transcriptionally induced by the cAMP signal transduction pathways, we investigated whether such regulation exists in the A549 cells. To this end, we studied c-jun and c-fos mRNA expression in these cells variously stimulated with FSK. The results showed significant induction of c-fos and less of c-jun expression upon cAMP activation (Fig. 5B). To study if transcriptional induction of AP-1 contributes to cathelicidin expression, we carried out EMSAs with the transcriptional complexes formed by incubating ARE oligonucleotides and the nuclear extracts of cells treated with or without ActD followed by FSK. DNA binding with the nuclear extracts of dually treated cells at 6 h post-stimulation when cathelicidin was maximally induced was somewhat less efficient as compared with the cells treated with FSK alone for the same duration, suggesting that transcriptional induction of AP-1 by cAMP may also contribute to cathelicidin transactivation (Fig. 5C).
FIGURE 5.
Transcriptional and post-translational regulation of CREB and AP-1. A, EMSAs performed with the nuclear extracts of unstimulated HT-29 cells as well as cells stimulated with FSK alone or PKA or MEK inhibitor followed by FSK. Each nuclear extract was incubated with the biotinylated CRE or ARE oligonucleotides before being resolved in EMSA. DNA-protein complexes were analyzed as in Fig. 3B. B, qRT-PCR analysis of c-jun and c-fos expression in the HT-29 cells stimulated with FSK for various durations. The results from triplicate samples are presented as mean (±S.D.) relative expression in the stimulated versus unstimulated cells. C, EMSAs carried out with the FSK or FSK and ActD-treated nuclear extracts of HT-29 cells incubated with the biotinylated ARE oligonucleotides. The DNA-protein complexes were blotted to a membrane that was developed as described under “Experimental Procedures.” Each of the above data is representative of three independent experiments.
ICER Represses CREB and AP-1-regulated Transactivation of Cathelicidin
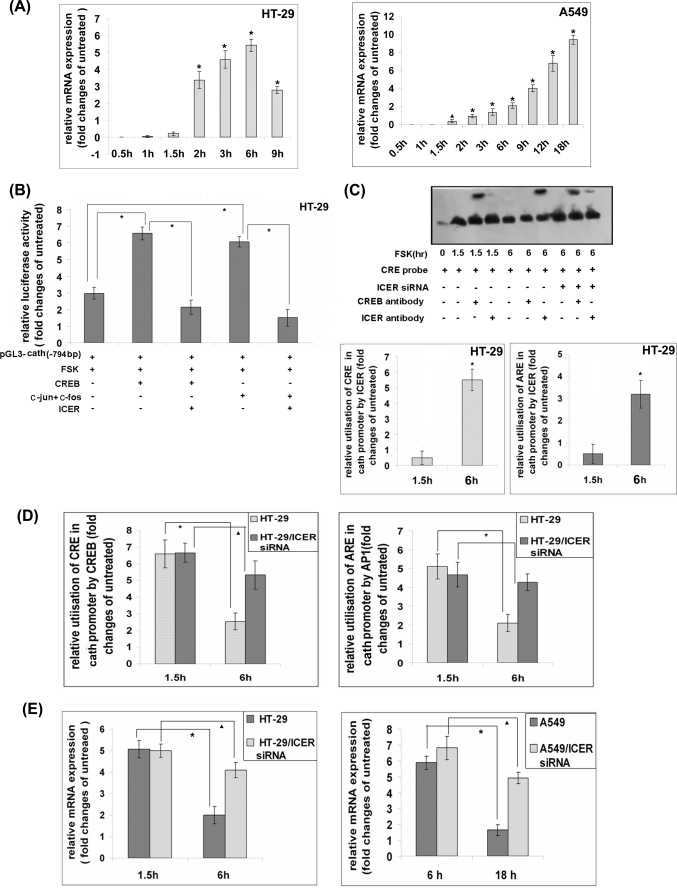
CREB and AP-1 are mainly activated by phosphorylation, whereas dephosphorylation remains the major counter-regulatory mechanism that controls expression of genes induced by them (46, 57). However, considerable CREB phosphorylation persisted after 6 h of stimulation of the HT-29 cells with FSK, whereas basal cathelicidin expression level was restored (supplemental Fig. S2). Some of the CREB and AP-1 regulated genes are known to be counter-regulated by an inducible repressor called ICER (45, 46, 58). We had earlier reported that cathelicidin and ICER expression in the NaB-differentiated HT-29 cells bears an inverse relation (43). To investigate if ICER may act as a transcriptional repressor of cathelicidin, we first studied the temporal profile of ICER expression in FSK-stimulated cells. ICER appeared after cathelicidin expression reached the maximum levels and gradually increased as the cathelicidin came down. Maximum ICER mRNA expression was observed 6 and 18 h post-stimulation in the HT-29 and A549 cells, respectively, when cathelicidin levels were minimal. The above results suggest that ICER may counter-regulate cathelicidin. Next, we studied if ICER may suppress cathelicidin promoter activity induced by CREB and AP-1. To this end, HT-29 cells were transfected with a luciferase reporter construct of cathelicidin promoter (−794 bp upstream) with or without CREB/AP-1. Reporter activity was significantly more elevated in the dually transfected cells compared with the ones expressing the reporter only. Concomitant expression of ICER resulted in complete abrogation of both CREB- and AP-1-induced reporter activities, suggesting that ICER may repress CREB/AP-1-induced transactivation of cathelicidin. It was earlier reported that ICER transrepresses genes either by binding to ARE/TRE sequences as homodimers and competing CREB/AP-1 out or by forming a heterodimer with the latter and interfering with their binding to the above sequences (45). To investigate the mechanism behind ICER-mediated repression of cathelicidin transcription, we carried out EMSAs with the nuclear extracts of untreated HT-29 cells as well as cells treated with FSK for two different durations. The results showed generation of large DNA-protein complex with both the FSK-stimulated extracts and retarded migration of the CRE oligonucleotide probe. The complex generated with the cells stimulated for 1.5 h, when cathelicidin expression reached the peak, contained CREB but not ICER as suggested by the supershift assays (Fig. 6C, 3rd and 4th lanes). In contrast, relatively less amounts of DNA-protein complex were formed with the nuclear extracts of 6-h stimulated cells that had significantly reduced cathelicidin expression, and it was composed of ICER rather than CREB (Fig. 6C, 5th to 7th lanes). This suggests that ICER, which had the highest levels of expression at the latter time point, replaced CREB and preferentially bound to the CRE sequence. To further prove that ICER competitively replaced CREB from binding to the cathelicidin promoter, EMSAs were carried out with the cells knocked down of the ICER gene. CREB remained bound to the oligonucleotide probe at 6 h post-stimulation in the absence of ICER (Fig. 6C, 8th to 10th lanes). ICER binding to the cathelicidin URR was also studied by ChIP assays. To this end, transcriptional complexes formed inside the FSK-stimulated HT-29 cells were immunoprecipitated with the CREM/ICER antibody. qPCR amplification showed that significantly higher amounts of DNA corresponding to the CRE and ARE sequences of the cathelicidin promoter were present in the complexes immunoprecipitated from the cells stimulated for 6 h as compared with 1.5-h stimulated cells (Fig. 6C, lower panel). These results suggest greater utilization of the above sequences by ICER in the FSK-stimulated cells where cathelicidin expression returned back to the original levels. To investigate if endogenous ICER and CREB/AP-1 compete for binding to the cathelicidin promoter, transcriptional complexes were immunoprecipitated from the FSK-stimulated wild type and ICER knocked down HT-29 cells using CREB and AP-1 antibodies. qPCR with the immunoprecipitated DNAs resulted in almost identical amplification of the CRE and ARE sequences with the cells stimulated for 1.5 h. In contrast, the amounts of the amplified products were much smaller with the wild type HT-29 cells stimulated for 6 h compared with the similarly treated cells where ICER gene was silenced (Fig. 6D). This suggests that although ICER replaced CREB and AP-1 from the cathelicidin promoter, significantly larger amounts were retained in the absence of ICER. Given that cathelicidin expression was the least at the 6-h time point, this result suggests that ICER may transcriptionally repress cathelicidin. Similar results were observed with A549 cells where the amounts amplified at the earlier time points (6 h post-stimulation) were comparable between the parental and ICER knocked down cells but were significantly larger with the latter cells 18 h after stimulation (supplemental Fig. S3). That ICER is required for the counter-regulation of cAMP-induced cathelicidin expression was further studied by analyzing cathelicidin in the parental and ICER knocked down HT-29 and A549 cells variously stimulated with FSK. The results showed significantly less suppression of cathelicidin expression at the later time points in the cells silenced for ICER expression (Fig. 6E), strongly suggesting a mandatory role for ICER in this down-regulation.
FIGURE 6.
Transrepression of CREB and AP-1-regulated cathelicidin expression by ICER. A, qRT-PCR analysis of ICER expression in HT-29 (left) and A549 (right) cells stimulated for various durations with FSK. Data presented as in Fig. 1B. B, reporter construct of cathelicidin promoter (−794 bp) was transfected into HT-29 cells, either alone or in combination with CREB or AP-1 expression constructs with or without the ICER construct. Triplicate cells were stimulated with FSK for 1.5 h, and the relative (Renilla luciferase activity normalized) reporter activities compared with that in the unstimulated cells are shown. C, upper panel, EMSAs with the nuclear extracts of the unstimulated and FSK-stimulated HT-29 cells incubated with biotin-labeled CRE with or without CREB or ICER antibody. Nuclear extracts were also prepared from the ICER gene-silenced cells that were stimulated with FSK for 6 h. DNA-protein complexes were analyzed as in Fig. 3B and Fig. 4B. Lower panel, ChIP assays done with the nuclear extracts of unstimulated and FSK-stimulated HT-29 cells immunoprecipitated with CREM/ICER antibody. The immunoprecipitate was used as a template to amplify CRE (left) or ARE (right) sequences as mentioned above by qPCR using flanking sequence-specific primers. GAPDH promoter was also amplified from the same samples, and the data are presented as in Fig. 3C and Fig. 4C. D, ChIP assays were performed with the nuclear extracts of parental or ICER gene-silenced HT-29 cells that were either left unstimulated or stimulated with FSK for two different durations. DNA-protein complexes were immunoprecipitated with CREB (left) or AP-1 (c-Jun plus c-Fos) (right) antibodies along with RNA polymerase II antibodies in both cases, and the protein-bound DNAs were amplified by qPCR using CRE, ARE, and GAPDH sequence-specific primers. Data are presented as above. E, cathelicidin mRNAs were amplified from the parental and ICER gene-silenced HT-29 (left) and A549 (right) cells, which were either left unstimulated or stimulated with FSK. Data were analyzed and presented as in Fig. 3E and Fig. 4E. All the above experiments were carried out at least three times, and data from one representative experiment is shown here. Cath, cathelicidin.
NaB-induced Up-regulation of Cathelicidin Is Mediated through CREB and AP-1
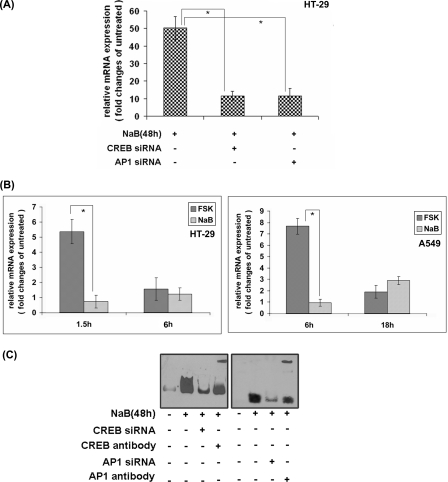
NaB markedly up-regulates cathelicidin expression in several epithelial cells (22, 38), but the molecular mechanism behind this regulation remains poorly understood. We investigated if CREB and AP-1 may also regulate NaB-induced cathelicidin expression. To this end, parental as well as CREB or AP-1 gene-silenced HT-29 cells were treated with NaB for 48 h. Around 50-fold induction of cathelicidin was found in the parental HT-29 cells. In contrast, the siRNA-transfected cells exhibited markedly reduced cathelicidin induction, suggesting that both CREB and AP-1 play significant roles in cathelicidin regulation by NaB (Fig. 7A). However, the gene was not induced at the time points when FSK induced it, indicating that NaB exerted delayed effects (Fig. 7B). The role of CREB and AP-1 was further addressed by EMSAs (Fig. 7C). Large DNA-protein complexes were generated when the nuclear extracts of NaB-treated cells were incubated with the CRE or ARE oligonucleotides. Complex formation was markedly abrogated in CREB or AP-1 knocked down cells, suggesting critical roles played by these molecules in the formation of transcriptional complexes within NaB-treated HT-29 cells. Their presence in these complexes was further confirmed by incubating the complexes with CREB or AP-1 (both c-Jun and c-Fos) antibodies that resulted in a “supershift” of the bound probe.
FIGURE 7.
CREB and AP-1-mediated regulation of NaB-induced cathelicidin expression. A, cathelicidin expression as analyzed by qRT-PCR in the parental and CREB or AP-1 gene-silenced HT-29 cells treated with NaB for 48 h. Data from triplicate samples are presented as the ratio of expression of NaB-treated and untreated cells. B, qRT-PCR analysis of cathelicidin expression in the HT-29 (left) or A549 (right) cells stimulated in triplicate with FSK or NaB. C, EMSAs with the nuclear extracts of untreated and NaB-treated parental or CREB/AP-1 gene-silenced HT-29 cells. Nuclear extracts from each cell population were incubated with the CRE or ARE oligonucleotides, and supershift assays were done by incubating DNA-protein complexes with CREB or AP-1 (c-Jun plus c-Fos) antibodies, respectively. The blot containing the DNA-protein complexes was prepared and developed as described under “Experimental Procedures.”
DISCUSSION
In this report, we have undertaken studies on the regulation of cathelicidin expression in the mucosal epithelial cells. Although previous reports had shown tissue-specific regulation by multiple stimuli (19–22), we observed a similar pattern of expression, albeit with different kinetics, in the epithelial cells of diverse origins upon activation of the cAMP signal transduction pathways (Fig. 1). Mechanisms operating at the transcriptional level may predominantly regulate cathelicidin, as mRNA and protein expression in different tissues closely follows each other (22, 23, 43). Reporter assays carried out previously (30, 39) as well as here with cathelicidin promoter constructs have lent further support to this idea. However, a recent study found discordance between cathelicidin promoter activity and gene expression. In the HT-29 cells, vitamin D3 was able to drive the cathelicidin promoter, but the gene expression remained unaltered. In contrast, butyrate augmented cathelicidin expression but not the promoter activity (20). This suggests post-transcriptional rather than transcriptional regulation, although we had reported activation of the cathelicidin promoter by NaB (43). This difference in observation may have resulted from the different time points when the assays were performed after NaB treatment. Noticeable impact of NaB on cathelicidin is seen only at later time points, and the optimum effect takes as long as 48 h to develop (38). Nuclear run-on assay is currently the best method to study transcriptional versus post-transcriptional regulations, as it directly measures the amounts of newly generated (nascent) mRNAs (51). Using this assay, we have convincingly proved for the first time that cathelicidin may indeed be transcriptionally regulated.
Analysis of the cathelicidin putative promoter by several independent groups has reported consensus binding sequences for many transcription factors (1, 28, 29, 43). However, the evidence to date for such interactions is, at best, indirect except for VDR in the keratinocytes and monocytes (31–33). Although transcriptional regulation of cathelicidin by NF-IL6 in several squamous epithelia has been suggested (23), it has never been experimentally demonstrated. Instead, interleukin-6 failed to induce cathelicidin in the colon epithelial cells (22). The role of VDR, however, in the mucosal cathelicidin expression remains unclear despite ubiquitous distribution of the former. As stated above, vitamin D3 fails to induce cathelicidin in the colonic epithelial cell line HT-29 (20), but it augments its promoter activity in the presence of NaB, and either of them may induce binding of the Ets family transcription factor PU.1 to the cathelicidin promoter (39). On the other hand, NaB-induced expression of cathelicidin in the Caco-2 cells may require VDR (40). The latter may result from differentiation of the colon epithelial cells by NaB treatment, which leads to major re-programming of gene expression, including significantly elevated cathelicidin levels (22, 38, 59, 60). This is supported by the fact that induction of cathelicidin is a relatively late event that is apparent between 24 and 48 h after NaB treatment when the cells may be already differentiated (30, 38, 39). On the other hand, we have investigated early regulatory events in the undifferentiated mucosal epithelial cells.
cAMP may regulate gene expression by activating multiple intracellular signaling pathways (50, 61). However, cathelicidin in our studies was regulated by PKA and ERK-MAPK only. Whereas PKA is directly activated by cAMP because of the removal of the inhibitory subunit (44), ERK may be either activated or inhibited (50, 55). In most cells, PKA inhibits ERK, but it may activate ERK through small GTPase Rap-1 in the B-Raf-expressing cells. In addition, cAMP may function in a PKA-independent way through Epacs to induce ERK activation (50, 62). The ERK, in our studies, was activated in both a PKA-dependent and -independent manner, as it cooperated with PKA to regulate CREB activation and cathelicidin transcription (Fig. 2B and Fig. 5A). However, we did not address the specific mechanisms of ERK activation by cAMP in the HT-29 and A549 cells. Our results are in agreement with the earlier studies that indicated critical involvement of ERK-MAPK in NaB-induced cathelicidin expression in the colon epithelial cells (38). p38 MAPK, on the other hand, has a rather controversial role, and both positive and negative regulation of cathelicidin by this molecule has been reported (38, 40). This suggests cell- and stimulus-specific regulation of cathelicidin in response to NaB. However, we found that p38 MAPK, JNK, and NF-κB play no significant role in the regulation of cathelicidin expression by cAMP in the colon or lung epithelial cells (Fig. 2B).
PKA and ERK-MAPK activate a large number of transcription factors, mainly through phosphorylation (50, 61). Computational analysis showed several consensus CRE and ARE sequences in the cathelicidin promoter (43). These sequences are bound by CREB and AP-1, two major transcription factors activated by PKA and ERK kinases, respectively. Our studies here have shown that cathelicidin promoter activity is predominantly regulated by the CRE and ARE sequences at −598 and −358 bp, respectively.
CREB primarily binds to the CRE sites, although it may also occupy ARE because of the high degree of similarity between the two sequences. Binding of CREB to ARE usually represses gene expression by interfering with the transcriptional activities of AP-1 (63, 64). In our studies, CREB strongly and specifically bound to the CRE sequence but not to the ARE (Fig. 3B). This interaction is critical for the induction of cathelicidin transcription by cAMP, as significantly larger amounts of endogenous CREB bound to the CRE sequence when cathelicidin expression level was the highest than when the expression was the least. In addition, silencing of the CREB gene resulted in marked suppression of cathelicidin induction (Fig. 3). HBX has been reported to directly interact with CBP/p300 and to enhance PKA-induced phosphorylation and activation of CREB (54). Augmented cathelicidin expression in the cells expressing HBX protein further indicates the role of CREB in cathelicidin transactivation. Considering the critical role played by cathelicidin in innate immunity, the above result suggests that cAMP-induced cathelicidin may contribute to anti-viral immune responses.
Transactivator complex of AP-1 usually consists of c-Jun/c-Fos heterodimers or c-Jun homodimers that bind the ARE/TRE sequences over the promoter (45, 65). Our studies have suggested that c-Jun and c-Fos work cooperatively to regulate cAMP-induced cathelicidin transactivation, although c-Jun plays a greater role compared with c-Fos (Fig. 4). c-Jun may also heterodimerize with ATF2 to bind the CRE sequences and activate transcription (46). In our studies, however, it did not bind CRE over the cathelicidin promoter.
AP-1 activities are mainly regulated through phosphorylation by the MAPKs (48, 66). However, cAMP may regulate their amounts within the cells by acting through CRE-binding factors, especially CREB (47, 56). We observed induction of c-jun and c-fos mRNA expression by cAMP in A549 cells, and this corroborated with increased cathelicidin expression (Fig. 5). Pretreatment of the cells with transcriptional inhibitor ActD resulted in decreased binding of the cathelicidin promoter sequence by AP-1. However, ActD had much less effect compared with that of kinase inhibitors, which suppressed both transcriptional and post-translational regulations of AP-1. This suggests that post-translational regulation of AP-1 predominantly contributes to its role in cathelicidin transactivation.
Negative regulation of cAMP-induced genes operates through multiple mechanisms. Whereas dephosphorylation of the cellular kinases and activated transcription factors by various phosphatases remains a key mechanism (46, 57), CRE-binding specific repressors also exist. The latter include the CREM family members that are either constitutively active (CREM-α, -β, and -γ) or induced (ICER) by signals that activate cAMP pathways. ICER, which has multiple isoforms expressed in tissue-dependent and physiological condition-dependent manner, is transcriptionally induced by CRE sequence-binding bZip transcription factors from an alternative, intronic promoter within the CREM gene (45, 58, 67). The gene products contain only the DNA-binding domain, but lack any transactivation domain, and function as transcriptional repressors when bound to the CRE/ARE sequences (58, 68). The characteristic kinetics of ICER mRNA expression that usually reaches the peak within first 6 h suggests that it belongs to the class of “early response genes” (58). In our studies, peak ICER expression in the HT-29 cells was found at 6 h post-stimulation. However, the kinetics may vary between cells and tissues (69), and we observed the highest mRNA expression in the A549 cells as late as 18 h. ICER imposes the most stringent control over cAMP-induced genes, such as MIP-1β, IL-2, GABA receptor, and tyrosine hydrolase (70–73). We have shown here that ICER mediates the major counter-regulatory mechanisms for cathelicidin expression (Fig. 6). Strict regulation by ICER is physiologically important, as persistently high cathelicidin levels may be detrimental to the host cells (1, 74). It would be interesting to study the specific ICER isoform(s) that regulate cathelicidin at different mucosal sites. The constitutive CREM repressors, however, played only a minor role, if any, in our studies as very little CRE- and ARE-binding transcriptional complexes exist in the unstimulated HT-29 and A549 cells.
Several studies have described NaB as a potent inducer of cathelicidin in various epithelial cells (21, 38). We, however, observed little effect of NaB on cathelicidin expression levels in the HT-29 and A549 cells at the time points when they exhibited the highest induction by cAMP. This reflects the fact that NaB probably functions indirectly through its role in cellular differentiation and expression of other genes, rather than directly, to regulate cathelicidin levels. Our data indicating major roles for CREB and AP-1 in NaB-induced cathelicidin expression are in agreement with the previous studies, which suggested that cAMP-mediated transactivation of the target genes in response to histone deacetylase inhibitors is a late event (75). In addition, AP-1 was earlier reported to be involved in cathelicidin expression in the lung epithelial cells EBC-1 (41). However, the induction was significantly poorer compared with that in the HT-29 cells and AP-1 bound to a single site between −109 and −68 bp of the cathelicidin promoter in EBC-1. On the other hand, we observed roles for multiple CRE and ARE sequences in the HT-29 cells. Multiple cis-acting regulatory elements have also been reported by other investigators (30, 39). Interestingly, one group of researchers found an NaB-responsive PU.1-binding sequence in the cathelicidin promoter (39). As the consensus PU.1 site reported by the authors is significantly homologous to the ARE sequences, it would be interesting to investigate whether AP-1 may bind to this site to regulate cathelicidin expression.
cAMP regulates a wide range of cellular processes, including differentiation, secretion, gene transcription, cytoskeletal remodeling, cell proliferation, apoptosis, leukocyte chemotaxis, and angiogenesis (61, 76). The physiological stimuli as well as the agonists that trigger cAMP production may exert both pro- and anti-inflammatory effects. Thus, Cox-2 and p38 MAPK are the major pro-inflammatory targets of cAMP-signaling pathways (77–79). On the other hand, prostaglandin E2 may function as both agonist and antagonist of cAMP and suppresses several pro-inflammatory cytokines and chemokines, while increasing the others (80). Dual pro- and anti-inflammatory activities are also exerted by PKA (80). Although the determinants of the pro- and anti-inflammatory role of cAMP are not clearly known, it may depend on the type or the strength of the stimulus as well as the specific cellular compartments (44). In addition, this may regulate the immunomodulatory functions that are long appreciated for cAMP and its agonists (81–83). CT has been recognized as a potent immunomodulator, a property that largely depends on its ability to induce cAMP production, and has been extensively studied as a vaccine adjuvant (84). CT and FSK were found to induce several cytokines and chemokines, pro- as well as anti-inflammatory, in the peripheral blood mononuclear cells (85, 86). Given that cathelicidin expressed at the mucosal surfaces shows multiple immunological functions (87), many of which, such as chemotaxis, angiogenesis, cytokine, and chemokine production, overlap with those of cAMP, it would be important to investigate if it may mediate immunomodulatory roles of cAMP at different mucosal sites.
Supplementary Material
This work was supported in part by Program of Funding Research Center for Emerging and Reemerging Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan and extramural funds from Indian Council of Medical Research Grant IRIS ID 2006-05630.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- VDR
- vitamin D receptor
- CREB
- cAMP-response element-binding protein
- dnCREB
- dominant negative CREB
- PKA
- protein kinase-A
- ICER
- inducible cAMP early repressor
- EMSA
- electrophoretic mobility shift assay
- ChIP
- chromatin immunoprecipitation
- NaB
- sodium butyrate
- FSK
- forskolin
- CRE
- cAMP-response element
- ARE
- AP-1-response element
- qRT
- quantitative reverse transcription
- ELISA
- enzyme-linked immunosorbent assay
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- HRP
- horseradish peroxidase
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK kinase
- DDA
- 2′,5′-dideoxyadenosine
- ActD
- actinomycin D
- JNK
- c-Jun NH2-terminal kinase
- HBX
- hepatitis B virus X
- siRNA
- small interfering RNA
- TRE
- 12-O-tetradecanoylphorbol-13-acetate-response element
- CREM
- cAMP-response element modulator
- FP
- forward primer
- RP
- reverse primer
- LT
- heat-labile toxin.
REFERENCES
- 1.Bals R., Wilson J. M. (2003) Cell. Mol. Life Sci. 60, 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dürr U. H., Sudheendra U. S., Ramamoorthy A. (2006) Biochim. Biophys. Acta 1758, 1408–1425 [DOI] [PubMed] [Google Scholar]
- 3.Zanetti M. (2004) J. Leukocyte Biol. 75, 39–48 [DOI] [PubMed] [Google Scholar]
- 4.Lehrer R. I., Ganz T. (2002) Curr. Opin. Hematol. 9, 18–22 [DOI] [PubMed] [Google Scholar]
- 5.Islam D., Bandholtz L., Nilsson J., Wigzell H., Christensson B., Agerberth B., Gudmundsson G. (2001) Nat. Med. 7, 180–185 [DOI] [PubMed] [Google Scholar]
- 6.Bowdish D. M., Davidson D. J., Lau Y. E., Lee K., Scott M. G., Hancock R. E. (2005) J. Leukocyte Biol. 77, 451–459 [DOI] [PubMed] [Google Scholar]
- 7.Gordon Y. J., Huang L. C., Romanowski E. G., Yates K. A., Proske R. J., McDermott A. M. (2005) Curr. Eye Res. 30, 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G., Watson K. M., Buckheit R. W., Jr. (2008) Antimicrob. Agents Chemother. 52, 3438–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg A., Krisanaprakornkit S., Dale B. A. (1998) Crit. Rev. Oral Biol. Med. 9, 399–414 [DOI] [PubMed] [Google Scholar]
- 10.Niyonsaba F., Ogawa H. (2005) J. Dermatol. Sci. 40, 157–168 [DOI] [PubMed] [Google Scholar]
- 11.Dommett R., Zilbauer M., George J. T., Bajaj-Elliott M. (2005) Mol. Immunol. 42, 903–912 [DOI] [PubMed] [Google Scholar]
- 12.Braff M. H., Hawkins M. A., Di Nardo A., Lopez-Garcia B., Howell M. D., Wong C., Lin K., Streib J. E., Dorschner R., Leung D. Y., Gallo R. L. (2005) J. Immunol. 174, 4271–4278 [DOI] [PubMed] [Google Scholar]
- 13.Mookherjee N., Brown K. L., Bowdish D. M., Doria S., Falsafi R., Hokamp K., Roche F. M., Mu R., Doho G. H., Pistolic J., Powers J. P., Bryan J., Brinkman F. S., Hancock R. E. (2006) J. Immunol. 176, 2455–2464 [DOI] [PubMed] [Google Scholar]
- 14.Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R. A., Pestonjamasp V., Piraino J., Huttner K., Gallo R. L. (2001) Nature 414, 454–457 [DOI] [PubMed] [Google Scholar]
- 15.Morrison G., Kilanowski F., Davidson D., Dorin J. (2002) Infect. Immun. 70, 3053–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberger C. M., Gallo R. L., Finlay B. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chromek M., Slamová Z., Bergman P., Kovács L., Podracká L., Ehrén I., Hökfelt T., Gudmundsson G. H., Gallo R. L., Agerberth B., Brauner A. (2006) Nat. Med. 12, 636–641 [DOI] [PubMed] [Google Scholar]
- 18.Bals R., Wang X., Meegalla R. L., Wattler S., Weiner D. J., Nehls M. C., Wilson J. M. (1999) Infect. Immun. 67, 3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bals R., Wang X., Zasloff M., Wilson J. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9541–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schauber J., Dorschner R. A., Yamasaki K., Brouha B., Gallo R. L. (2006) Immunology 118, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schauber J., Iffland K., Frisch S., Kudlich T., Schmausser B., Eck M., Menzel T., Gostner A., Lührs H., Scheppach W. (2004) Mol. Immunol. 41, 847–854 [DOI] [PubMed] [Google Scholar]
- 22.Hase K., Eckmann L., Leopard J. D., Varki N., Kagnoff M. F. (2002) Infect. Immun. 70, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frohm M., Agerberth B., Ahangari G., Stâhle-Bäckdahl M., Lidén S., Wigzell H., Gudmundsson G. H. (1997) J. Biol. Chem. 272, 15258–15263 [DOI] [PubMed] [Google Scholar]
- 24.Frohm Nilsson M., Sandstedt B., Sørensen O., Weber G., Borregaard N., Ståhle-Bäckdahl M. (1999) Infect. Immun. 67, 2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erdag G., Morgan J. R. (2002) Ann. Surg. 235, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sørensen O. E., Cowland J. B., Theilgaard-Mönch K., Liu L., Ganz T., Borregaard N. (2003) J. Immunol. 170, 5583–5589 [DOI] [PubMed] [Google Scholar]
- 27.Howell M. D., Novak N., Bieber T., Pastore S., Girolomoni G., Boguniewicz M., Streib J., Wong C., Gallo R. L., Leung D. Y. (2005) J. Invest. Dermatol. 125, 738–745 [DOI] [PubMed] [Google Scholar]
- 28.Larrick J. W., Lee J., Ma S., Li X., Francke U., Wright S. C., Balint R. F. (1996) FEBS Lett. 398, 74–80 [DOI] [PubMed] [Google Scholar]
- 29.Gudmundsson G. H., Agerberth B., Odeberg J., Bergman T., Olsson B., Salcedo R. (1996) Eur. J. Biochem. 238, 325–332 [DOI] [PubMed] [Google Scholar]
- 30.Elloumi H. Z., Holland S. M. (2008) Mol. Immunol. 45, 204–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson J., Carlsson G., Larne O., Andersson M., Pütsep K. (2008) J. Leukocyte Biol. 84, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 32.Martineau A. R., Wilkinson K. A., Newton S. M., Floto R. A., Norman A. W., Skolimowska K., Davidson R. N., Sørensen O. E., Kampmann B., Griffiths C. J., Wilkinson R. J. (2007) J. Immunol. 178, 7190–7198 [DOI] [PubMed] [Google Scholar]
- 33.Gombart A. F., O'Kelly J., Saito T., Koeffler H. P. (2007) J. Steroid Biochem. Mol. Biol. 103, 552–557 [DOI] [PubMed] [Google Scholar]
- 34.Wang T. T., Nestel F. P., Bourdeau V., Nagai Y., Wang Q., Liao J., Tavera-Mendoza L., Lin R., Hanrahan J. W., Mader S., White J. H., Hanrahan J. H. (2004) J. Immunol. 173, 2909–2912 [DOI] [PubMed] [Google Scholar]
- 35.Gombart A. F., Borregaard N., Koeffler H. P. (2005) FASEB J. 19, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 36.Liu P. T., Stenger S., Li H., Wenzel L., Tan B. H., Krutzik S. R., Ochoa M. T., Schauber J., Wu K., Meinken C., Kamen D. L., Wagner M., Bals R., Steinmeyer A., Zügel U., Gallo R. L., Eisenberg D., Hewison M., Hollis B. W., Adams J. S., Bloom B. R., Modlin R. L. (2006) Science 311, 1770–1773 [DOI] [PubMed] [Google Scholar]
- 37.Schauber J., Dorschner R. A., Coda A. B., Büchau A. S., Liu P. T., Kiken D., Helfrich Y. R., Kang S., Elalieh H. Z., Steinmeyer A., Zügel U., Bikle D. D., Modlin R. L., Gallo R. L. (2007) J. Clin. Invest. 117, 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schauber J., Svanholm C., Termén S., Iffland K., Menzel T., Scheppach W., Melcher R., Agerberth B., Lührs H., Gudmundsson G. H. (2003) Gut 52, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Termén S., Tollin M., Rodriguez E., Sveinsdóttir S. H., Jóhannesson B., Cederlund A., Sjövall J., Agerberth B., Gudmundsson G. H. (2008) Mol. Immunol. 45, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 40.Schwab M., Reynders V., Shastri Y., Loitsch S., Stein J., Schröder O. (2007) Mol. Immunol. 44, 2107–2114 [DOI] [PubMed] [Google Scholar]
- 41.Kida Y., Shimizu T., Kuwano K. (2006) Mol. Immunol. 43, 1972–1981 [DOI] [PubMed] [Google Scholar]
- 42.Bergman P., Johansson L., Asp V., Plant L., Gudmundsson G. H., Jonsson A. B., Agerberth B. (2005) Cell. Microbiol. 7, 1009–1017 [DOI] [PubMed] [Google Scholar]
- 43.Chakraborty K., Ghosh S., Koley H., Mukhopadhyay A. K., Ramamurthy T., Saha D. R., Mukhopadhyay D., Roychowdhury S., Hamabata T., Takeda Y., Das S. (2008) Cell. Microbiol. 10, 2520–2537 [DOI] [PubMed] [Google Scholar]
- 44.Schwartz J. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13482–13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sassone-Corsi P. (1994) EMBO J. 13, 4717–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalli E., Sassone-Corsi P. (1994) J. Biol. Chem. 269, 17359–17362 [PubMed] [Google Scholar]
- 47.Nakamura T., Datta R., Sherman M. L., Kufe D. (1990) J. Biol. Chem. 265, 22011–22015 [PubMed] [Google Scholar]
- 48.Angel P., Karin M. (1991) Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 49.Schulte G., Fredholm B. B. (2003) Cell. Signal. 15, 813–827 [DOI] [PubMed] [Google Scholar]
- 50.Stork P. J., Schmitt J. M. (2002) Trends Cell Biol. 12, 258–266 [DOI] [PubMed] [Google Scholar]
- 51.Kwei K. A., Finch J. S., Thompson E. J., Bowden G. T. (2004) Neoplasia 6, 440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das S., Cho J., Lambertz I., Kelliher M. A., Eliopoulos A. G., Du K., Tsichlis P. N. (2005) J. Biol. Chem. 280, 23748–23757 [DOI] [PubMed] [Google Scholar]
- 53.Portis T., Longnecker R. (2003) J. Virol. 77, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cougot D., Wu Y., Cairo S., Caramel J., Renard C. A., Lévy L., Buendia M. A., Neuveut C. (2007) J. Biol. Chem. 282, 4277–4287 [DOI] [PubMed] [Google Scholar]
- 55.Dumaz N., Marais R. (2005) FEBS J. 272, 3491–3504 [DOI] [PubMed] [Google Scholar]
- 56.Kacich R. L., Williams L. T., Coughlin S. R. (1988) Mol. Endocrinol. 2, 73–77 [DOI] [PubMed] [Google Scholar]
- 57.Hagiwara M., Alberts A., Brindle P., Meinkoth J., Feramisco J., Deng T., Karin M., Shenolikar S., Montminy M. (1992) Cell 70, 105–113 [DOI] [PubMed] [Google Scholar]
- 58.Sassone-Corsi P. (1998) Int. J. Biochem. Cell Biol. 30, 27–38 [DOI] [PubMed] [Google Scholar]
- 59.Getzenberg R. H., Light B. W., Lapco P. E., Konety B. R., Nangia A. K., Acierno J. S., Dhir R., Shurin Z., Day R. S., Trump D. L., Johnson C. S. (1997) Urology 50, 999–1006 [DOI] [PubMed] [Google Scholar]
- 60.Kállay E., Bises G., Bajna E., Bieglmayer C., Gerdenitsch W., Steffan I., Kato S., Armbrecht H. J., Cross H. S. (2005) Carcinogenesis 26, 1581–1589 [DOI] [PubMed] [Google Scholar]
- 61.Sands W. A., Palmer T. M. (2008) Cell. Signal. 20, 460–466 [DOI] [PubMed] [Google Scholar]
- 62.Gerdin M. J., Eiden L. E. (2007) Sci. STKE 2007 382, pe15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rutberg S. E., Adams T. L., Olive M., Alexander N., Vinson C., Yuspa S. H. (1999) Oncogene 18, 1569–1579 [DOI] [PubMed] [Google Scholar]
- 64.Masquilier D., Sassone-Corsi P. (1992) J. Biol. Chem. 267, 22460–22466 [PubMed] [Google Scholar]
- 65.Verma I. M., Ransone L. J., Visvader J., Sassone-Corsi P., Lamph W. W. (1990) CIBA Found. Symp. 150, 128–146 [DOI] [PubMed] [Google Scholar]
- 66.Barber J. R., Verma I. M. (1987) Mol. Cell. Biol. 7, 2201–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Servillo G., Della Fazia M. A., Sassone-Corsi P. (2002) Exp. Cell Res. 275, 143–154 [DOI] [PubMed] [Google Scholar]
- 68.Bodor J., Habener J. F. (1998) J. Biol. Chem. 273, 9544–9551 [DOI] [PubMed] [Google Scholar]
- 69.Tamai K. T., Monaco L., Nantel F., Zazopoulos E., Sassone-Corsi P. (1997) Recent Prog. Horm. Res. 52, 121–140 [PubMed] [Google Scholar]
- 70.Barabitskaja O., Foulke J. S., Jr., Pati S., Bodor J., Reitz M. S., Jr. (2006) J. Leukocyte Biol. 79, 378–387 [DOI] [PubMed] [Google Scholar]
- 71.Bodor J., Fehervari Z., Diamond B., Sakaguchi S. (2007) Eur. J. Immunol. 37, 884–895 [DOI] [PubMed] [Google Scholar]
- 72.Hu Y., Lund I. V., Gravielle M. C., Farb D. H., Brooks-Kayal A. R., Russek S. J. (2008) J. Biol. Chem. 283, 9328–9340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trocmé C., Ravassard P., Sassone-Corsi P., Mallet J., Biguet N. F. (2001) J. Neurosci. Res. 65, 91–99 [DOI] [PubMed] [Google Scholar]
- 74.Lau Y. E., Bowdish D. M., Cosseau C., Hancock R. E., Davidson D. J. (2006) Am. J. Respir. Cell Mol. Biol. 34, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Michael L. F., Asahara H., Shulman A. I., Kraus W. L., Montminy M. (2000) Mol. Cell. Biol. 20, 1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beavo J. A., Brunton L. L. (2002) Nat. Rev. Mol. Cell Biol. 3, 710–718 [DOI] [PubMed] [Google Scholar]
- 77.Howe L. R. (2007) Breast Cancer Res. 9, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bergamaschi A., Corsi M., Garnier M. J. (2006) Cell. Signal. 18, 1679–1684 [DOI] [PubMed] [Google Scholar]
- 79.Harfi I., D'Hondt S., Corazza F., Sariban E. (2004) J. Immunol. 173, 4154–4163 [DOI] [PubMed] [Google Scholar]
- 80.Lorenowicz M. J., Fernandez-Borja M., Hordijk P. L. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1014–1022 [DOI] [PubMed] [Google Scholar]
- 81.Horrigan L. A., Kelly J. P., Connor T. J. (2006) Pharmacol. Ther. 111, 877–892 [DOI] [PubMed] [Google Scholar]
- 82.Taskén K., Stokka A. J. (2006) Biochem. Soc. Trans. 34, 476–479 [DOI] [PubMed] [Google Scholar]
- 83.Iwahashi H., Takeshita A., Hanazawa S. (2000) J. Immunol. 164, 5403–5408 [DOI] [PubMed] [Google Scholar]
- 84.Lycke N. (1997) Res. Immunol. 148, 504–520 [DOI] [PubMed] [Google Scholar]
- 85.Royaee A. R., Jong L., Mendis C., Das R., Jett M., Yang D. C. (2006) Mol. Immunol. 43, 1267–1274 [DOI] [PubMed] [Google Scholar]
- 86.Royaee A. R., Mendis C., Das R., Jett M., Yang D. C. (2006) Mol. Immunol. 43, 702–709 [DOI] [PubMed] [Google Scholar]
- 87.Bowdish D. M., Davidson D. J., Hancock R. E. (2006) Curr. Top. Microbiol. Immunol. 306, 27–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.