Abstract
Density-enhanced phosphatase-1 (DEP-1) is a trans-membrane receptor protein-tyrosine phosphatase that plays a recognized prominent role as a tumor suppressor. However, the mechanistic details underlying its function are poorly understood because its primary physiological substrate(s) have not been firmly established. To shed light on the mechanisms underlying the anti-proliferative role of this phosphatase, we set out to identify new DEP-1 substrates by a novel approach based on screening of high density peptide arrays. The results of the array experiment were combined with a bioinformatics filter to identify eight potential DEP-1 targets among the proteins annotated in the MAPK pathway. In this study we show that one of these potential targets, the ERK1/2, is indeed a direct DEP-1 substrate in vivo. Pulldown and in vitro dephosphorylation assays confirmed our prediction and demonstrated an overall specificity of DEP-1 in targeting the phosphorylated tyrosine 204 of ERK1/2. After epidermal growth factor stimulation, the phosphorylation of the activation loop of ERK1/2 can be modulated by changing the concentration of DEP-1, without affecting the activity of the upstream kinase MEK. In addition, we show that DEP-1 contains a KIM-like motif to recruit ERK1/2 proteins by a docking mechanism mediated by the common docking domain in ERK1/2. ERK proteins that are mutated in the conserved docking domain become insensitive to DEP-1 de-phosphorylation. Overall this study provides novel insights into the anti-proliferative role of this phosphatase and proposes a new mechanism that may also be relevant for the regulation of density-dependent growth inhibition.
DEP-14 (also known as CD148, HPTPη, and PTPRJ) is a class III receptor protein-tyrosine phosphatase, characterized by eight fibronectin type III repeats within the extracellular domain, a trans-membrane region, and a single cytosolic catalytic domain (1, 2). DEP-1 is expressed in all human hematopoietic cell lineages and was shown to negatively regulate T cell activation. In addition, several epithelial cell types display DEP-1 on their cell membranes (3). Homozygous DEP-1 mutant mice die before embryonic day 11.5, displaying severe defects in vascular organization (4). Interestingly, DEP-1 expression levels were found to augment with increased cell density (2), suggesting a role for this tyrosine phosphatase in sensing cell-cell contacts and in density-dependent growth inhibition (5). Moreover, accumulating evidence supports a prominent role for DEP-1 as a tumor suppressor as it negatively regulates cell proliferation and is poorly expressed in many cancer cell lines (6–10). The observed anti-proliferative effect may be accounted for by the ability of DEP-1 to down-regulate growth factor signaling through the dephosphorylation of various receptor tyrosine kinases, such as PDGFR, VEGFR2, and MET (11–13), resulting in quenching of the downstream RAS-MAPK pathway. However, given the complex pleiotropic functions of DEP-1, it is also possible that additional regulatory circuits mediated by yet unknown DEP-1 substrates may play a functional role in contact inhibition and control of cell proliferation.
A variety of in vivo and in vitro approaches has led us to propose a number of DEP-1 substrates as mediators of its function. These include PDGFR, p120 catenin (CTND1), hepatocyte growth factor receptor, SRC kinase, VEGFR2, phosphatidylinositol 3-kinase regulatory subunit α (P85A), and RET receptor kinase (5, 11–16).
Here we report a novel, unbiased strategy based on the screening of high density phosphopeptide arrays for their ability to bind phosphatase trapping mutants. A large portion of the phosphoproteome could be explored by this approach, thus unveiling a long list of potential substrates. A selected list of potentially relevant substrates has been obtained by applying a bioinformatics context filter. In this study we report the detailed characterization of one of these substrates, and we propose that DEP-1 modulates the RAS pathway by directly dephosphorylating Tyr-204 of ERK1/2. In addition, we show that the efficient removal of the phosphate group from Tyr-204 requires the integrity of a docking site on the ERK1/2 proteins.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies
Anti-hemagglutinin (HA) and anti-FLAG were from Sigma; anti-DEP-1, anti-SRC, and anti-tubulin were from Santa Cruz Biotechnology; anti-ERK1/2(P), anti-ERK1/2, anti-MEK(P), anti-MEK, anti-p38(P) and anti-p38 were from Cell Signaling, and anti-4G10 was from Upstate Biotechnology, Inc. Peroxidase-conjugated anti-rabbit, anti-mouse secondary antibodies were from Jackson ImmunoResearch, and anti-mouse TRICT was purchased from Molecular Probes.
Plasmids
pRSα DEP-1WT/HA, pRSα DEP-1CS/HA, pRSα DEP-1DA/HA, and pRSα DEP-1ΔCy/HA were kindly provided by Dr T. Takahashi. The pGEX-4TK expression plasmids coding for the fusion proteins GST-PTPs (PTP1B, TC-PTP, MEG-2, MEG-1, FAP-1, LYP-1, PTPH1, PEST, DEP-1, SAP-1, LAR, PTPβ, PTPα, SHP1, and SHP2) were kindly provided by Rob Hooft van Huijsduijnen. PMV rat HA-ERK2 plasmid was donated by Dr. Tartaglia (17); cDNA encoding SRC Y527F was cloned in pSGT (18). The full-length TC-PTP was cloned in p3XFLAG-CMV14 vector (NotI/BamHI). The two shRNA constructs directed against DEP-1 transcript were from OriGene Technologies (TI340203-4). The expression plasmid p3XFLAG-CMV7.1 coding for rat FLAG-ERK2 was kindly donated by Prof. Cobb (19). The FLAG-ERK2 D319A and HA-DEP-1 K1016A mutants were generated by site-directed mutagenesis using the QuikChange kit (Stratagene).
GST Fusion Proteins
After transformation, colonies of Escherichia coli BL21 Rosetta containing the indicated GST fusion proteins were used to inoculate LB cultures with 100 μg/ml ampicillin and chloramphenicol. Cultures were grown at 37 °C until an A600 of 0.5 was reached. Protein expression was at 30 °C for 4 h after the addition of isopropyl 1-thio-β-d-galactopyranoside at a final concentration of 100 μm. Pelleted bacteria were resuspended in lysis buffer (50 mm Tris, pH 8.0, 5 mm EDTA, 0.1% Triton X-100, 150 mm NaCl) plus a 250 mm proteinase inhibitor mixture (Roche Applied Science) and lysed by treatment with lysozyme (200 μg/ml final) for 1 h on ice followed by three rounds of sonication. The lysates were clarified by spinning at 14,000 rpm for 20 min and bound to glutathione-Sepharose beads (Amersham Biosciences) at 4 °C for 1 h. Finally, beads were extensively washed in phosphate-buffered saline (PBS) with 100 mm dithiothreitol, and the PTPs were eluted in 50 mm Tris, pH 8.0, with 10 mm glutathione. Glycerol was added to a final concentration of 20%; the amount of proteins produced was determined with a Bio-Rad protein assay, and aliquots were stocked at −80 °C until use.
Peptide Arrays
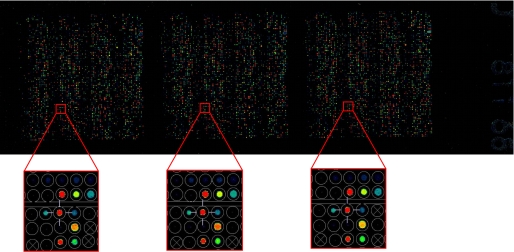
A chip containing ∼6000 phosphopeptides in three repeated subarrays was used.5 To choose the peptides to be synthesized and printed, we first downloaded from Phospho-ELM (20) 1604 phosphorylated peptides that have been identified in high and low throughput experiments with the aim to characterize the phospho-proteome in Homo sapiens. To this list we added 4453 peptides that had a high score according to the NetPhos predictive algorithm (21). Each chip contains three identical replicated arrays (subarray), and each subarray contains the phosphorylated peptides and several control spots arranged in a grid of 6400 positions. As shown in the Fig. 1, each peptide maintains the same grid position in the three subarrays. The peptides were synthesized and arrayed by Jerini Peptide Technology according to their proprietary technology. After 1 h of blocking in 5% PBS/bovine serum albumin, the phosphopeptide array was incubated with 1 μg/ml GST-DEP-1. The chip was washed in PBS and incubated at 1:1000 with anti-GST Cy-5-conjugated antibody (Amersham Biosciences) avoiding light exposure. The chip was extensively washed in PBS, and fluorescence intensity was revealed with ScanArray Gx Plus (PerkinElmer Life Science).
FIGURE 1.
Strategy to identify putative candidate DEP-1 substrates involved in the MAPK pathway. The trapping mutant D1205A of the catalytic domain of DEP-1 was challenged with a library of 6400 tyrosine-phosphorylated peptides. The chip regions from the three array repeats containing the phosphorylated (Tyr-204) 13-mer in the ERK1/2 activating loop (spot circled in yellow) were enlarged to demonstrate experimental reproducibility.
Cell Culture, Transfection, and EGF Stimulation
Human embryonic kidney (HEK)293 cells and human epithelial carcinoma (HeLa) cells were purchased from the ATCC. Cells were maintained in a humidified atmosphere at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum (Sigma) and 0.1% penicillin/streptomycin (Invitrogen). HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. After 5 h of starvation in fetal calf serum-free media, cell were stimulated with 100 μg/ml EGF (Invitrogen) for the indicated time.
Pulldown Assay
After EGF stimulation, confluent HeLa cells were washed with ice-cold PBS and lysed in RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, 1% Nonidet P-40, 0.25% sodium deoxycholate) supplemented with 1 mm pervanadate, 1 mm NaF, protease inhibitor mixture 200× (Sigma), inhibitor phosphatase mixture I and II 100× (Sigma). The samples were kept on ice for 30 min and centrifuged at 15,000 rpm at 4 °C for 30 min. The supernatant was collected, and the total amount of protein was determined by Bradford colorimetric assay (Bio-Rad). The whole cell lysates were incubated with 50 μg of the indicated GST fusion protein at 4 °C for 1 h. Thus glutathione-Sepharose 4B beads were blocked by incubating with 3% bovine serum albumin with rocking at 4 °C for 1 h, and then after centrifugation for 3 min at 4000 × g, at 4 °C, the dry beads were bound to lysates mixed with GST fusion proteins at 4 °C for 1 h. The supernatant was discarded by centrifugation, and the beads were washed six times with lysis buffer for 3 min at 4000 × g, at 4 °C, and then the dry beads were resuspended in 50 μl of 3× SDS sample buffer, boiled for 10 min, and analyzed by SDS-PAGE and Western blotting on nitrocellulose membrane.
In Vitro Dephosphorylation Assay
HEK293 cells were transfected with SRC and HA-ERK2 expression plasmids. Cells were collected and lysed in RIPA buffer containing 1 mm pervanadate, 1 mm NaF, protease inhibitor mixture 200× (Sigma), inhibitor phosphatases mixture I and II 100×, and supernatants were clarified by spinning at 15,000 rpm, at 4 °C, for 30 min. Lysates were immunoprecipitated with antibody anti-HA overnight at 4 °C, and then protein A-Sepharose beads were added and incubated for 1 h at 4 °C. After washing with lysis buffer, the dry beads were incubated with 1 μg of the indicated GST fusion protein in reaction buffer for 15 or 30 min. For the vanadate competition assay, 1 mm Na3VO4 was added to PTPase assay buffer. The immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted with antibodies. The catalytic activity of the fusion proteins was checked using p-nitrophenyl phosphate assay. The reaction was stopped by adding 2 m NaOH, and the enzymatic activity was monitored by measuring the absorbance at 405. The enzyme activity (μmol/min/μg) was calculated from Beer-Lambert law.
Immunoprecipitation and Immunoblot Analysis
HEK293 cells were lysed as described previously. The whole cell lysates were incubated with antibodies overnight at 4 °C, and then protein A-Sepharose beads were added and incubated for 1 h at 4 °C. The beads were washed with lysis buffer, and the immunoprecipitated proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and immunoblotted with antibodies. The immunoreactions were visualized using ECL detection system (Amersham Biosciences).
Fluorescence Microscopy
HEK293 cells plated onto glass coverslips at ∼50% confluency were cotransfected using Lipofectamine with pRSα DEP-1WT/HA and pEGFP-C1 (Clontech). 24 h following transfection cells were fixed for 10 min in 4% paraformaldehyde (EM Sciences), washed with PBS, 0.5% Triton X-100, and permeabilized with the blocking solution (0.1% Triton 10% fetal calf serum) for 30 min. Coverslips were incubated in humidified chambers, in blocking solution, with anti-DEP-1 antibody for 1 h at room temperature. Cells were rinsed in PBS and stained with anti-mouse TRITC antibodies for 30 min at room temperature. Cells were stained with 4′,6-diamidino-2-phenylindole in PBS, 0.1% Triton for 5 min at room temperature. Coverslips were mounted on slides as described and analyzed by indirect immunofluorescence microscopy Leica DMR.
Data Base Submission
The interactions described in this study have been submitted to the MINT data base (22) following the MIMIX recommendations (23).
RESULTS
Identification of Tyr-204 of ERK1/2 Proteins as Potential Substrate of DEP-1 Phosphatase
To investigate the enzymatic specificity of DEP-1 and to define its substrate landscape, we purified the trapping mutant phosphatase enzymatic domain as a GST fusion, and we probed it with a library of ∼6,000 tyrosine-phosphorylated peptides arrayed on a glass surface. After measuring phosphatase binding with an anti-GST antibody conjugated to a fluorophore, we defined “DEP-1 putative targets” as the 215 peptides with a signal intensity larger than the median signal plus two standard deviations (Fig. 1). This relatively large list of putative DEP-1 substrates was mapped onto the MAPK pathway, as annotated in the KEGG data base (24), and only peptides whose in vivo phosphorylation could be supported by experimental evidence in the Phosphosite data base (25) were considered. Using this filtering procedure, we re-discovered PDGFR, a known DEP-1 substrate (11), and we inferred seven new putative substrates (EGF receptor, GRB2, RAF-1, ERK1/2, AKT, FLNA, and JIP3).
In this study we focused on the 199TGFLTEpYVATRWY210 peptide shared by ERK1/2 proteins, as previous experimental evidence had already revealed a negative correlation between DEP-1 expression and ERK1/2 activation in tumor cell lines and in response to different growth stimuli (11–13). Validation of the remaining inferred substrates is hampered by the presence of multiple phosphorylated tyrosine residues and requires the development of reagents to specifically monitor their phosphorylation levels.
DEP-1 Trapping Mutant Specifically Binds Phosphorylated ERK1/2 Proteins
The results of the array experiment suggested that the DEP-1 trapping mutant binds the ERK1/2 peptide containing Tyr-204 in its phosphorylated form. To confirm this interaction in a full-length protein and cellular context and to assess its specificity, we performed a pulldown assay with 15 trapping mutants of different PTPs. As shown in Fig. 2A, DEP-1 can efficiently bind ERK1/2, whereas other phosphatases bind to different phosphorylated proteins but cannot affinity purify ERK1/2. Interestingly SAP-1, whose amino acid sequence is related to DEP-1, also binds ERK1/2.
FIGURE 2.
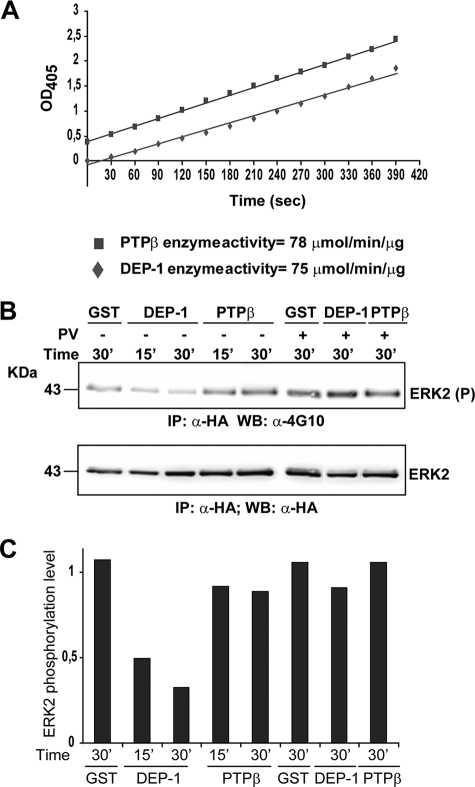
DEP-1 specifically binds and dephosphorylates ERK1/2 proteins. A, GST fusion proteins of several tyrosine phosphatase trapping mutants were incubated with a protein extract of HeLa cells induced with EGF for 5 min. Affinity-purified phosphatase ligands were separated by SDS-PAGE and transferred onto cellulose membranes. The blots were revealed with anti-phosphotyrosine antibody (WB: α-4G10). The cell lysate (input) and the sample affinity-purified with the GST protein (GST) were used as controls. The same samples were revealed with the anti-ERK1/2 (WB: α-ERK1/2) and the anti-phospho-ERK1/2 (WB: α-ERK1/2(P)) antibodies. B, HeLa cells were starved and treated with EGF for 5 min. After cell lysis, the protein extract was incubated with GST and with GST fused the to the wild type (WT) catalytic domain of DEP-1 (DEP-1 WT) or to a mutant variant carrying the D1205A mutation in the catalytic pocket (DEP-1 D/A). The interacting proteins were affinity-purified over glutathione-agarose beads, separated by SDS-PAGE, and visualized by immunoblotting with anti-phosphotyrosine (α-4G10), anti-ERK1/2 (α-ERK1/2), and anti-phosphorylated ERK1/2 (α-ERK1/2(P)) antibodies recognizing phospho-Tyr-204. C, after starvation (0 min), HeLa cells were incubated for 5, 10, and 30 min with EGF. Protein extracts were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. The blot was incubated with anti-phospho-Tyr-204-ERK1/2 (WB: α-ERK1/2(P)) and anti ERK1/2 (WB: α-ERK1/2) antibodies. Equal gel loading was assessed by re-probing with anti-tubulin (WB: α-tubulin). D, protein extracts were incubated with the DEP-1 trapping mutant expressed as a GST fusion (DEP-1 D/A). Affinity-purified proteins were analyzed by SDS-PAGE and revealed with anti-phospho-ERK1/2 (WB: α-ERK1/2(P)) and anti ERK1/2 (WB: α-ERK1/2).
Next, to exclude that the observed interaction could occur outside the PTP catalytic pocket, we compared two pulldown experiments using as a bait the wild type or the trapping mutant version of the catalytic domain of DEP-1 (Fig. 2B). Only the trapping mutant could be shown to bind to ERK1/2 proteins providing further support to the notion of a direct interaction of phosphorylated ERK1/2 and the catalytic site of the DEP-1 phosphatase. To demonstrate that the observed binding is dependent on the phosphorylation of Tyr-204, we performed the pulldown experiment at different times after EGF induction. The observation that the amount of affinity-purified ERK1/2 is proportional to Tyr-204 phosphorylation levels, as probed with a specific antibody recognizing the phosphorylated Tyr-204, indicates that the interaction occurs only when ERK1/2 is phosphorylated in the activation loop (Fig. 2, C and D). The association of phospho-ERK1/2 proteins with the DEP-1 trapping mutant but not with the wild type catalytic domain implies that ERK1/2 is a substrate of DEP-1 enzymatic activity. To investigate whether purified ERK1/2 can be dephosphorylated in vitro by this phosphatase, the same amount of purified DEP-1 and PTPβ wild type catalytic domains were incubated with immunoprecipitated ERK2, after co-expression with a constitutively active SRC kinase in HEK293 cells. Although the two enzymes have the same activity on a generic synthetic substrate (Fig. 3A), only DEP-1 dephosphorylated ERK2 in a time-dependent manner (Fig. 3, B and C). These results taken together are consistent with DEP-1 phosphatase directly and specifically targeting the phosphorylated Tyr-204 in ERK1/2.
FIGURE 3.
DEP-1 directly dephosphorylates ERK1/2 in vitro. A, DEP-1 and PTPβ wild type catalytic domains were incubated with p-nitrophenyl phosphate substrate in assay buffer. The increase in the absorbance at 405 was plotted, and the specific activity of the two PTPs were calculated from the slopes of the straight lines and the Beer Lambert law. B, HEK293 cells were transiently co-transfected with 5 μg of DNA of an expression plasmid directing the synthesis of the Y527F constitutively active mutant of the SRC kinase and with a second expression plasmid for HA-ERK1/2. After cell lysis, the protein extract was immunoprecipitated with the anti-HA antibody. The beads were washed with lysis buffer and treated (+) or not (−) with 1 mm sodium pervanadate (PV) to inhibit phosphatase activity. The anti-HA immunoprecipitate (IP) was incubated with 1 μg of GST, GST-DEP-1 wild type, and GST-PTPβ wild type for the indicated times at 37 °C in assay buffer. The tyrosine phosphorylation level of ERK2 was analyzed by immunoblotting using anti-phosphotyrosine antibody (α-4G10) and anti-HA antibody (α-HA). C, relative quantitations of the phospho-ERK1/2 signal versus total ERK1/2 were plotted and represented as bar graph.
ERK1/2 Proteins Are Directly Dephosphorylated by DEP-1 in Intact Cells
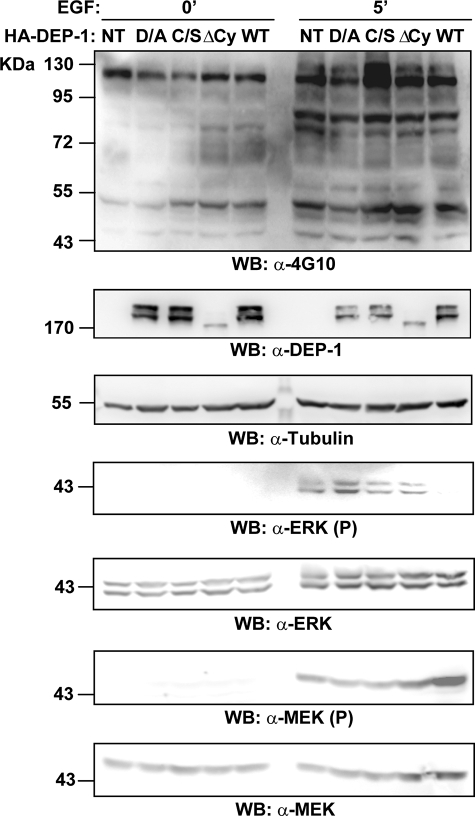
Next we asked whether ERK1/2 dephosphorylation by DEP-1 could also occur in vivo. To this end, HEK293 cells were transiently transfected with an HA-tagged construct directing the expression of full-length DEP-1 phosphatase. Similar transfections were carried out with mutant versions of DEP-1 carrying single substitutions in the catalytic site (D1205A or C1239S trapping mutants) or a deletion of the entire cytoplasmic domain (ΔCy). The transfected cell cultures were stimulated for 5 min with EGF to induce ERK1/2 protein phosphorylation. As shown in Fig. 4, wild type DEP-1 overexpression remarkably reduced Tyr-204 phosphorylation levels without affecting the activation of the upstream kinase MEK, as measured with antibody specific for the phosphorylation of the MEK activation loop. By contrast, the DEP-1 C1239S and ΔCy mutants did not affect ERK1/2 or MEK phosphorylation levels. Moreover, the overexpression of a third DEP-1 mutant carrying an Asp-1205 to Ala substitution (D/A trapping mutant) increased ERK1/2 kinase phosphorylation acting as a dominant negative effector. Presumably, the stable association between trapping mutant phosphatase and its substrate prevents ERK1/2 dephosphorylation by cellular wild type DEP-1 or other phosphatases. These results suggest that DEP-1 directly inactivates ERK1/2 in a RAS-MEK independent manner.
FIGURE 4.
DEP-1 directly dephosphorylates ERK1/2 in vivo. HEK293 cells were transiently transfected with full-length wild type (WT) DEP-1 or with its catalytic mutants (D/A; C/S; ΔCy). The transfected cultures were incubated with EGF, and aliquots were sampled at time 0 and after 5 min. After cell lysis, whole protein extracts were analyzed by SDS-PAGE and probed by Western blotting with an anti-phosphotyrosine antibody (WB: α-4G10). Transfection efficiency was assessed by revealing the blot with antibodies anti-DEP-1 (α-DEP-1) and anti-tubulin (α-tubulin). The phosphorylation levels of the ERK1/2 proteins were monitored by probing with anti-phospho-ERK1/2 (α-ERK1/2(P)) and anti-ERK1/2 (α-ERK1/2) antibodies. The activation levels of the MEK1/2 kinases were observed by immunoblotting with anti-phospho-MEK (α-MEK(P)) and anti-MEK (α-MEK) antibodies.
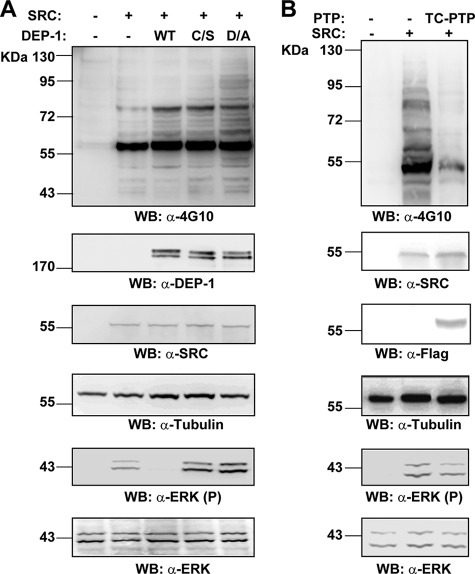
Because DEP-1 is known to suppress the activity of various growth factor receptors by direct de-phosphorylation (11, 13), we also analyzed the DEP-1/ERK1/2 interaction in a growth factor independent experimental system. To enhance ERK1/2 protein phosphorylation, HEK293 cells were transiently co-transfected with the constitutively active mutant of SRC tyrosine kinase (Y527F) and with expression plasmids coding for wild type or substrate trapping mutants (C/S or D/A) of the DEP-1 phosphatase. As shown in Fig. 5A, DEP-1 overexpression resulted in a marked reduction of the phosphorylation levels of ERK1/2 proteins, whereas ERK1/2 phosphorylation was not affected by DEP-1 C/S, and it was remarkably increased by the DEP-1 D/A trapping mutant. The inactivation of ERK1/2 proteins in c-SRC-transfected cells was not observed upon TC-PTP overexpression (Fig. 5B) confirming the selective dephosphorylation of ERK1/2 kinases by DEP-1.
FIGURE 5.
DEP-1 dephosphorylates ERK1/2 in SRC-activated cell lines. A, HEK293 cells were transiently co-transfected with full-length HA-DEP-1 or with its inactive mutants (D/A or C/S) and with 5 μg of Y527F SRC kinase (constitutively activated) expression plasmid. Tyrosine phosphorylation levels in whole protein extracts were analyzed by immunoblotting with anti-4G10 antibody (α-4G10). Transfection efficiency was assessed with anti-DEP-1 and anti-SRC (α-DEP-1; α-SRC) antibodies. Equal gel loading was assessed by re-probing with anti-tubulin antibody (α-tubulin). The phosphorylation profile of ERK1/2 proteins was analyzed by immunoblotting with anti-phospho-ERK1/2 (α-ERK1/2(P)) and anti-ERK1/2 (α-ERK1/2) antibodies. B, HEK293 cells were transiently co-transfected with 5 μg of Y527F SRC kinase and with expression plasmids coding for full-length wild type (WT) TC-PTP. The tyrosine phosphorylation profiles of whole cell lysates were analyzed by immunoblotting with anti-phosphotyrosine antibody (WB: α-4G10). The transfection efficiency of SRC and TC-PTP was verified by revealing the lysates with the correspondent antibodies (WB: α-SRC; α-TC-PTP). Equal gel loading was assessed with anti-tubulin (WB: α-tubulin). The phosphorylation level of ERK1/2 proteins was analyzed with anti-phospho-ERK1/2 (α-ERK1/2(P)) and anti-ERK1/2 (α-ERK1/2) antibodies.
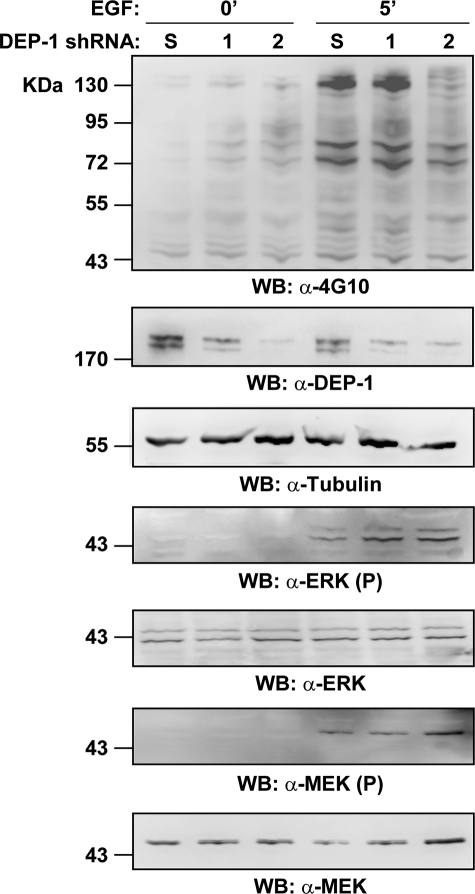
To assess whether down-regulation of endogenous DEP-1 levels would have any effect on ERK1/2 activation, we transiently transfected HeLa cells with two interfering shRNA constructs targeting different sites on the phosphatase transcript. After 5 min of EGF induction, the decrease in DEP-1 expression was assessed by immunoblot (Fig. 6). Consistent with the observed decrease in activated ERK under conditions of DEP-1 overexpression, cells transfected with DEP-1 shRNA constructs showed a sustained ERK1/2 phosphorylation when compared with cells transfected with a shRNA plasmid targeting an unrelated sequence. The fraction of activated MEK was comparable in the two conditions (Fig. 6). These results are consistent with the DEP-1 phosphatase directly targeting the ERK1/2 kinases.
FIGURE 6.
DEP-1 depletion increases ERK1/2 activation. HeLa cells were transiently transfected with two different shRNA plasmids targeting the DEP-1 transcript (lanes 1 and 2) or with a scramble shRNA (S) as a negative control. After incubation with EGF, lysates were analyzed by immunoblot with anti-4G10 antibody (α-4G10). Knockdown expression of DEP-1 was assessed with anti-DEP-1 and anti-tubulin antibodies (α-DEP-1; α-tubulin). The phosphorylation levels of the ERK1/2 proteins were monitored by probing with anti-phospho-Tyr-204-ERK1/2 (α-ERK1/2(P)) and anti-ERK1/2 (α-ERK1/2) antibodies. The activation levels of the MEK1/2 kinases were observed by immunoblotting with anti-phospho-MEK (α-MEK(P)) and anti-MEK (α-MEK) antibodies.
DEP-1 Interacts with ERK1/2 via a Conserved Docking Site
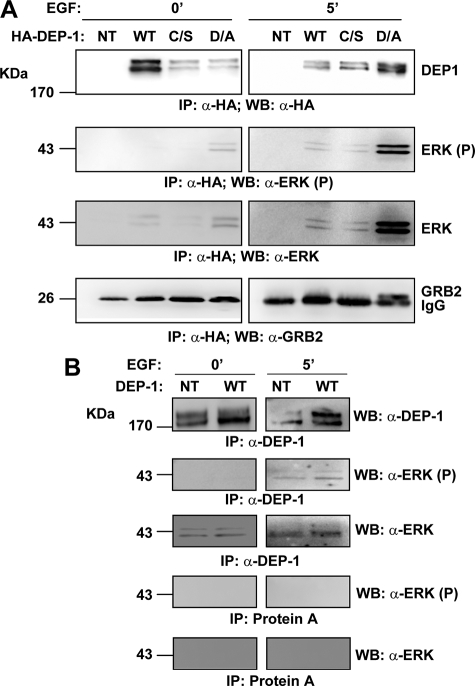
To explore whether ERK1/2 form a physical complex with DEP-1, HEK293 were transfected with HA-tagged DEP-1. After lysis and immunoprecipitation with anti-HA antibody, phosphorylated ERK1/2 was found to associate strongly with the DEP-1 D/A trapping mutant and, to a much lesser extent, with the wild type and the C/S phosphatase mutant (Fig. 7A). Interestingly, some significant association was also observed with the wild type and D/A trapping mutant in conditions where ERK1/2 is not phosphorylated. By contrast GRB2, a second DEP-1 target predicted by our approach, only formed a complex with the D/A trapping mutant when the cells were stimulated with EGF. As shown in Fig. 7B, the interaction between DEP-1 and ERK1/2 can be detected at endogenous expression levels in HeLa cells, in the absence of EGF. Taken together these results are consistent with the presence of an ERK1/2 docking site in the intracellular domain of DEP-1. In the D/A trapping mutant this complex was stabilized by the synergic interaction of the inactive “trapping” catalytic domain with the target phosphopeptide in the ERK1/2 proteins.
FIGURE 7.
DEP-1 interacts with ERK1/2 in mammalian cells. A, HEK293 cells were transiently transfected with full-length DEP-1 phosphatase or with its catalytic mutants (D/A; C/S) and were stimulated with EGF for 5 min. Aliquots were sampled at times 0 and 5 min, and after cell lysis, whole protein extracts were immunoprecipitated with anti-HA antibody. The beads were washed with lysis buffer, and the immunoprecipitation (IP) was revealed with anti-HA (WB: α-HA), anti-phospho-ERK1/2 (WB: α-ERK1/2 (P)), anti-ERK1/2 (WB: α-ERK1/2), and anti-GRB2 antibodies. B, lysates of HeLa cells transiently transfected with full-length DEP-1 wild type (WT) and not transfected (NT) were induced with EGF for 5 min and immunoprecipitated with anti-DEP-1 antibody. The immunoprecipitation (IP) was revealed with anti-DEP-1 (WB: α-DEP-1), anti-phospho ERK1/2 (WB: α-ERK1/2 (P)), and anti-ERK1/2 (WB: α-ERK1/2).
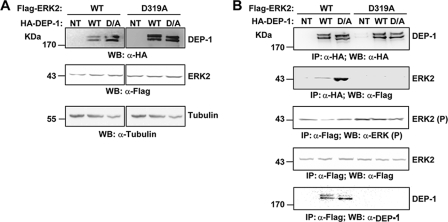
As reported in the literature, the common docking (CD) domain of ERK1/2 proteins directly interacts with the KIM motif of several phosphatases (HE-PTP, STEP, PTP-SL, MKP3, and MKP4), kinases (MEK1 and MEK2), and substrates (SAP1, ELK1, RSK, and LIN) (26, 27). To test whether such a docking site was acting as a receptor for DEP-1, HEK293 cells were transiently co-transfected with plasmids expressing DEP-1 and rat ERK2. In parallel, a transfection was carried out with an ERK2 mutant carrying the D319A substitution, a mutation in the CD domain that impairs binding to the KIM domain of the MKP3 phosphatase (28). After cell lysis, whole protein extracts were immunoprecipitated with anti-FLAG or anti-HA antibodies to recover ERK2 and DEP-1 together with the associated proteins, respectively (Fig. 8A). As shown in Fig. 8B, differently from wild type, the D319A ERK2 mutant is not dephosphorylated by DEP-1. Moreover the interaction between DEP-1 and the ERK2 protein is impaired by the D319A mutation, suggesting that this binding is mediated by a putative KIM motif contained in the cytoplasmic domain region of the DEP-1 phosphatase.
FIGURE 8.
DEP-1 forms a complex with ERK1/2 by binding its CD. A, HEK293 cells were transiently transfected with full-length DEP-1 phosphatase or with its catalytic mutants D/A and were stimulated with EGF for 5 min. After cell lysis, transfection was assessed by immunoblotting with anti-HA (WB: α-HA), anti-FLAG (WB: α-FLAG), and anti-tubulin (WB: α-Tubulin) antibodies. NT, not transfected. B, whole protein extracts were immunoprecipitated with anti-FLAG antibody to purify the wild type (WT) or the Asp-319 mutant of ERK2 and with anti-HA antibody to recover DEP-1 phosphatase. The beads were washed with lysis buffer, and the immunoprecipitate (IP) was revealed with anti-FLAG (WB: α-FLAG), anti-HA (WB: α-HA), anti-phospho ERK1/2 (WB: α-ERK1/2 (P)), and anti-DEP-1 (WB: α-DEP-1).
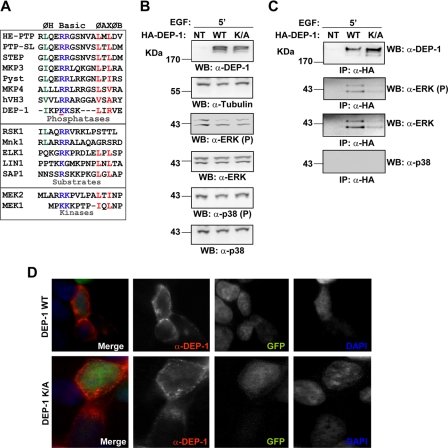
As shown in Fig. 9A, the DEP-1 positively charged peptide 1013IKPKKSKLIRVE1024 in the juxtamembrane region has some resemblance to the KIM domain sequence motif. To investigate the involvement of this region in ERK1/2 recruitment, HEK293 cells were transiently transfected with a DEP-1 mutant carrying a substitution in one of the residues of the putative KIM domain (K1016A). Because the mutation is close to the trans-membrane region, we first checked that the membrane localization is not affected by the mutation (Fig. 9D). We next tested the interaction with ERK1/2, and we observed a reduction of ∼80% as a consequence of the K1016A substitution, suggesting a prominent role of this residue in the recruitment of ERK1/2 (Fig. 9C). We observed, however, that the mutated DEP-1 phosphatase maintains its ability to dephosphorylate ERK1/2. This result suggests that the residual interaction between DEP-1 K/A and ERK1/2, observed in Fig. 9B, is sufficient to mediate an efficient dephosphorylation of the ERK1/2 substrates.
FIGURE 9.
K1016A substitution in the DEP-1 KIM-like motif strongly impairs ERK1/2 recruitment. A, alignment of ERK1/2 KIM domains. These sites share an N-terminal hydrophobic residue (ØH), one or two positively charged residues (basic), and the ØAXØB motif. We identified in DEP-1 phosphatase a similar sequence (modified from Ref. 27). B, HEK293 cells were transiently transfected with full-length K1016A DEP-1 mutant and with the wild type (WT) phosphatase. After 5 min of EGF induction, cells were lysed and the transfection efficiency monitored by immunoblotting with anti-DEP-1 and anti-tubulin antibodies. The phosphorylation levels of the ERK and p38 MAPKs were monitored by probing the protein extracts with anti-phospho-ERK1/2 (WB: α-ERK1/2 (P)), anti-ERK1/2 (WB: α-ERK1/2), anti-phospho-p38 (WB: α-p38 (P)), and anti-p38 (WB: α-p38) antibodies. NT, not transfected. C, whole protein extracts were immunoprecipitated with anti-HA antibody to recover DEP-1 protein. The immunoprecipitated sample (IP) was probed with anti-HA (WB: α-HA), anti-phospho-ERK1/2 (WB: α-ERK1/2 (P)), anti-ERK1/2 (WB: α-ERK1/2), and anti-p38 (WB: α-p38). D, HEK293 cells were transiently co-transfected with full-length wild type phosphatase or its mutant K1016A and 3 μg of green fluorescent protein expression plasmid. After fixation, cells were stained with anti-DEP-1 antibody and analyzed by indirect immunofluorescence microscopy.
We finally asked whether the KIM motif identified in DEP-1 is specific for the ERK1/2 kinases. To this end we tested whether the p38 MAPK would form a complex with DEP-1. The results in Fig. 9C demonstrate that neither wild type DEP-1 nor its mutants co-immunoprecipitate p38. Accordingly, the overexpression of DEP-1 WT or its K1016A mutant did not affect the phosphorylation level of Tyr-182 in the activation loop of the p38 kinase (Fig. 9B).
DISCUSSION
Over the past decade evidence has been accumulating that DEP-1 is a tumor suppressor that potently inhibits cell proliferation (6–8, 10, 29). The anti-proliferative role of this phosphatase has been correlated with its ability to dephosphorylate receptor tyrosine kinases, including PDGFR, MET, and VEGFR2 (11–13). Nevertheless, it could not be excluded that the inactivation of different DEP-1 substrates, more downstream in the MAPK pathway, could play an equivalent or even more prominent role in regulating cell proliferation. Our phosphoproteome-wide screening of DEP-1 targets uncovered seven new potential substrates, functionally associated with the RAS-MAPK pathway. Among the putative new substrates, ERK1/2 attracted our attention because of its central role in propagating receptor kinase proliferation signals. Here we have provided compelling evidence that ERK1/2 proteins are modulated by the activity of the DEP-1 protein in vivo after stimulation with EGF.
Until now the role of DEP-1 in the EGF pathway has not received much attention, although DEP-1 is expressed in epithelial cells and may play a role, for instance, in gastrointestinal epithelial cells, where EGF stimulates cell migration, proliferation, and wound closure through the activation of MAPK signaling (30). In addition, DEP-1 overexpression or knockdown was found to affect cell proliferation and migration in colon epithelial cells, suggesting a possible role of this phosphatase in the modulation of the EGF pathway (6). Accordingly, we have found that upon EGF induction, DEP-1 markedly dephosphorylates and inactivates ERK1/2 proteins. This activity was not observed when a phosphatase-defective C/S mutant was overexpressed, whereas the D/A trapping mutant displayed a dominant negative effect resulting in ERK1/2 hyper-phosphorylation. Finally, knocking down the expression of endogenous DEP-1 by shRNA also resulted in increased phosphorylation of ERK1/2.
Although DEP-1-mediated negative regulation of the RAS pathway has been attributed to its ability to dephosphorylate receptor tyrosine kinases such as platelet-derived growth factor, MET, or vascular endothelial growth factor, several lines of evidence presented here suggest that the observed negative regulation of ERK1/2 activity, in our experimental system, can be accounted for by a direct dephosphorylation of Tyr-204 in the activation loop of ERK1/2.
First, we have shown that the DEP-1 D/A trapping mutant can bind a synthetic 13-mer peptide centered on phosphorylated Tyr-204 and specifically pulls down ERK1/2 from an EGF-activated cell extract. In the same experiment, other phosphatase domains, although similar in primary sequence, do not bind ERK1/2, thereby attesting to the specificity of the interaction (Fig. 2A). Next, purified activated ERK1/2 proteins were dephosphorylated in vitro by the purified DEP-1 phosphatase domain (Fig. 3B). These results are consistent with a specific interaction between the phosphatase domain of DEP-1 and the Tyr-204 of ERK1/2, which, as a result, is dephosphorylated in vitro and in vivo (Fig. 4). In addition, we show that the upstream kinase MEK is not affected by DEP-1 overexpression and depletion, suggesting that the phosphatase activity acts downstream of MEK (Fig. 6). Consistently, we observe that in an EGF-independent experimental system, DEP-1 is able to dephosphorylate ERK1/2 proteins when they are activated by a constitutively active SRC kinase (Fig. 5).
Although the level of selectivity of protein-tyrosine phosphatases is still being debated, all the experimental systems that we have used to characterize the interaction between DEP-1 and ERK1/2 have demonstrated a certain level of specificity. Among the 15 phosphatases that we have analyzed, only the trapping mutant catalytic domains of DEP-1 and SAP-1 bind ERK1/2 proteins. Similarly in the in vitro dephosphorylation test the catalytic region of PTPβ, which shares the highest sequence homology with DEP-1 (31), was not able to dephosphorylate ERK1/2 proteins in vitro. In addition, overexpression of TC-PTP did not down-regulate ERK1/2 phosphorylation in our system. These results, supporting a substantial level of selectivity, are in accord with the report by Zhang and co-workers (32) showing that among 11 phosphatases tested, only three enzymes (HE-PTP, PP2A, and MKP3) were able to dephosphorylate ERK1/2 proteins in vitro.
Taken together these results suggest that DEP-1 down-regulates the EGF-activated MAPK pathway through a direct and specific dephosphorylation of Tyr-204 in the activation loop of the ERK1/2 kinases. Nevertheless, we cannot rule out the possibility that DEP-1 phosphatase also acts on the EGF receptor because the peptide centered on Tyr-998 gave a signal in our peptide array screening. EGF receptor has multiple phosphorylation sites, and we have not been able to detect a decrease of receptor tyrosine phosphorylation upon DEP-1 phosphorylation by probing with a generic anti-phosphotyrosine antibody (supplemental Fig. 1). We also attempted to monitor the formation of complexes with SHC1 and SRC. Both are predicted to bind to phosphorylated Tyr-998 via the Src homology 2 or phosphotyrosine binding domains. No difference in the formation of these complexes with or without DEP-1 expression was observed (supplemental Fig. 1). However, it must be borne in mind that both proteins have the potential to bind different phosphotyrosine peptides in the cytoplasmic tail of the EGF receptor.
ERK1/2 are key members of MAPK signaling and are activated by the phosphorylation on threonine and tyrosine regulatory residues in the kinase domain activation loop (33). Protein serine/threonine phosphatases, as well as dual specificity phosphatases, are known to be involved in the inactivation of MAPKs by dephosphorylating their phosphothreonine and phosphotyrosine regulatory residues (34, 35). However, because dephosphorylation of the regulatory tyrosine residue is sufficient to inactivate ERK1/2, a crucial role for protein-tyrosine phosphatases has been proposed (36, 37). For instance ERK1/2 proteins have been shown to be inactivated by HE-PTP in hematopoietic cell lines (32, 38), whereas in neuronal cells they are inhibited by PTP-SL and STEP (39). Our results add DEP-1 as a physiological negative modulator of ERK1/2 activity in epithelial cells (Fig. 6 and Fig. 7), which is consistent with the established role of DEP-1 as a tumor suppressor.
A variety of reports have shown that phosphatases have an intrinsic enzymatic preference and that, when challenged with different peptide substrates, they display a distinct bias that is often associated with specific sequence contexts flanking the phosphotyrosine (40–43). Sequence preference, however, cannot fully explain the specificity of in vivo substrate selection because considerable flexibility exists for PTPs to recognize phosphopeptides whose sequence deviates from their “ideal” in vitro substrate. Thus, although enzymatic specificity plays a role in determining substrate choice, it is by now clear that the phosphatase-substrate vicinity, mediated by docking domains, contributes to a large extent to target selection.
The physiological relevance of our findings is strengthened by the observation that DEP-1 forms a complex with ERK1/2. ERK1/2 has evolved a strategy to recruit many of their substrates or effectors via the CD, the common docking domain, that acts as a receptor for a positively charged motif, which has been named KIM (kinase interaction motif) (44). We have demonstrated that such a docking mechanism is responsible for the formation of a complex between DEP-1 and ERK1/2 irrespective of the ERK phosphorylation state. ERK mutated in the conserved aspartate 319 in the CD domain cannot any longer act as a DEP-1 substrate. Interestingly, a glutamic acid-to-lysine substitution at codon 322 (E322K) in the CD domain of ERK2 has been reported to result in constitutively phosphorylated ERK2 in an oral squamous cell carcinoma cell line (45). We have identified a positively charged peptide that loosely matches the KIM consensus motif in the juxta-membrane region of DEP-1. Its involvement in ERK1/2 recruitment was proven by mutating Lys-1016 into an Ala and by showing that its interaction with ERK is decreased by ∼80%. This decrease, however, is not sufficient to abolish the dephosphorylation of ERK following DEP-1 overexpression.
The newly identified KIM motif is specific for the ERK CD domain because the p38 MAPK does not form a complex with DEP-1 in the same conditions. This observation is consistent with the report that other PTPs (HE-PTP, PTP-SL, and STEP) show some level of specificity in their interaction with ERK and p38 MAPK (44, 46).
It has been suggested that ERK1/2 proteins are activated in colon cancer cells via recruitment to the plasma membrane by binding to integrin β6, thus promoting cell proliferation (47). Because loss or heterozygosity for DEP-1 has been described previously in about 71% of the cases of colon carcinoma (48), we propose that in normal cells DEP-1 counterbalances the activating role of integrins.
Finally, DEP-1 expression is enhanced at high cell density (2). It has been suggested that DEP-1 plays an important role in cell-cell contact sensing by forming a complex with p120 (ctn) at the adherent junctions. Our data point to a possible role for DEP-1/ERK1/2 interaction in the regulation of density-dependent growth inhibition (Fig. 10). Growth inhibition by cell-cell contact is a complex process that requires the coordinated action of several signals mediated by diverse molecules on the cell surface (49). Heterotropic interaction between up-regulated gangliosides and up-regulated receptor protein-tyrosine phosphatase-sigma on adjacent cells has been proposed as a key mechanism for growth arrest (50). Because cellular confluence increases DEP-1 expression, our observation that DEP-1 can down-regulate the RAS pathway by directly dephosphorylating ERK1/2 provides a second putative mechanism underlying high density growth arrest.
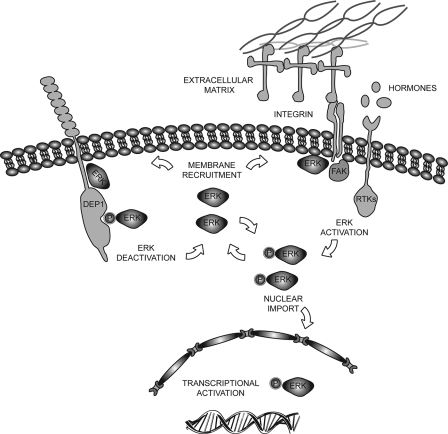
FIGURE 10.
Growth contact inhibition mediated by the direct interaction of ERK1/2 and the DEP-1 phosphatase. According to the illustrated model DEP-1 has two anti-proliferative activities. By binding to inactive ERK1/2, it sequesters them and limits their activation by integrins and growth factors. In addition it can shut down ERK activity by directly dephosphorylating it. Cellular confluence increases DEP-1 expression that in turns inhibits cells proliferation by inactivating ERK1/2.
Supplementary Material
Acknowledgments
F. Bohmer provided plasmid pNRTIS33 for DEP-1 expression. S. Gonfloni donated pSGT cDNA encoding SRC Y527F. The PMV rat HA-ERK2 plasmid was provided by M. Tartaglia. M. Cobb provided the expression plasmid p3XFLAG-CMV7.1 coding for rat FLAG-ERK2.
This work was supported by a grant from the Italian Association for Cancer Research, by the ENFIN Network of Excellence, and by the Interaction Proteome Integrated Project of the European Union FP6.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
M. Tinti, S. Costa, L. Kiemer, M. Miller, F. Sacco, J. Olsen, M. Carducci, S. Paoluzi, C. Workman, N. Blom, K. Machida, C. Thomson, M. Schutkowski, S. Brunak, M. Mann, B. Mayer, L. Castagnoli, and G. Cesareni, manuscript in preparation.
- DEP-1
- density-enhanced phosphatase-1
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- EGF
- epidermal growth factor
- VEGFR2
- vascular endothelial growth factor receptor 2
- PDGFR
- platelet-derived growth factor receptor
- PBS
- phosphate-buffered saline
- HEK
- human embryonic kidney
- GST
- glutathione S-transferase
- PTP
- protein-tyrosine phosphatase
- TRITC
- tetramethylrhodamine isothiocyanate
- KIM
- kinase interaction motif
- shRNA
- short hairpin RNA
- CD
- common docking.
REFERENCES
- 1.Jallal B., Mossie K., Vasiloudis G., Knyazev P., Zachwieja J., Clairvoyant F., Schilling J., Ullrich A. (1997) J. Biol. Chem. 272, 12158–12163 [DOI] [PubMed] [Google Scholar]
- 2.Ostman A., Yang Q., Tonks N. K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9680–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autschbach F., Palou E., Mechtersheimer G., Rohr C., Pirotto F., Gassler N., Otto H. F., Schraven B., Gaya A. (1999) Tissue Antigens 54, 485–498 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T., Takahashi K., St. John P. L., Fleming P. A., Tomemori T., Watanabe T., Abrahamson D. R., Drake C. J., Shirasawa T., Daniel T. O. (2003) Mol. Cell. Biol. 23, 1817–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holsinger L. J., Ward K., Duffield B., Zachwieja J., Jallal B. (2002) Oncogene 21, 7067–7076 [DOI] [PubMed] [Google Scholar]
- 6.Balavenkatraman K. K., Jandt E., Friedrich K., Kautenburger T., Pool-Zobel B. L., Ostman A., Böhmer F. D. (2006) Oncogene 25, 6319–6324 [DOI] [PubMed] [Google Scholar]
- 7.Keane M. M., Lowrey G. A., Ettenberg S. A., Dayton M. A., Lipkowitz S. (1996) Cancer Res. 56, 4236–4243 [PubMed] [Google Scholar]
- 8.Trapasso F., Iuliano R., Boccia A., Stella A., Visconti R., Bruni P., Baldassarre G., Santoro M., Viglietto G., Fusco A. (2000) Mol. Cell. Biol. 20, 9236–9246 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Trapasso F., Yendamuri S., Dumon K. R., Iuliano R., Cesari R., Feig B., Seto R., Infante L., Ishii H., Vecchione A., During M. J., Croce C. M., Fusco A. (2004) Carcinogenesis 25, 2107–2114 [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Martelli M. L., Battaglia C., Trapasso F., Tramontano D., Viglietto G., Porcellini A., Santoro M., Fusco A. (1997) Exp. Cell Res. 235, 62–70 [DOI] [PubMed] [Google Scholar]
- 11.Kovalenko M., Denner K., Sandström J., Persson C., Gross S., Jandt E., Vilella R., Böhmer F., Ostman A. (2000) J. Biol. Chem. 275, 16219–16226 [DOI] [PubMed] [Google Scholar]
- 12.Lampugnani M. G., Orsenigo F., Gagliani M. C., Tacchetti C., Dejana E. (2006) J. Cell Biol. 174, 593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palka H. L., Park M., Tonks N. K. (2003) J. Biol. Chem. 278, 5728–5735 [DOI] [PubMed] [Google Scholar]
- 14.Pera I. L., Iuliano R., Florio T., Susini C., Trapasso F., Santoro M., Chiariotti L., Schettini G., Viglietto G., Fusco A. (2005) Oncogene 24, 3187–3195 [DOI] [PubMed] [Google Scholar]
- 15.Tsuboi N., Utsunomiya T., Roberts R. L., Ito H., Takahashi K., Noda M., Takahashi T. (2008) Biochem. J. 413, 193–200 [DOI] [PubMed] [Google Scholar]
- 16.Iervolino A., Iuliano R., Trapasso F., Viglietto G., Melillo R. M., Carlomagno F., Santoro M., Fusco A. (2006) Cancer Res. 66, 6280–6287 [DOI] [PubMed] [Google Scholar]
- 17.Fragale A., Tartaglia M., Wu J., Gelb B. D. (2004) Hum. Mutat. 23, 267–277 [DOI] [PubMed] [Google Scholar]
- 18.Gonfloni S., Williams J. C., Hattula K., Weijland A., Wierenga R. K., Superti-Furga G. (1997) EMBO J. 16, 7261–7271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson F. L., Whitehurst A. W., Raman M., Cobb M. H. (2002) J. Biol. Chem. 277, 14844–14852 [DOI] [PubMed] [Google Scholar]
- 20.Diella F., Cameron S., Gemünd C., Linding R., Via A., Kuster B., Sicheritz-Pontén T., Blom N., Gibson T. J. (2004) BMC Bioinformatics 5, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blom N., Gammeltoft S., Brunak S. (1999) J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 22.Chatr-aryamontri A., Ceol A., Palazzi L. M., Nardelli G., Schneider M. V., Castagnoli L., Cesareni G. (2007) Nucleic Acids Res. 35, D572–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orchard S., Salwinski L., Kerrien S., Montecchi-Palazzi L., Oesterheld M., Stümpflen V., Ceol A., Chatr-aryamontri A., Armstrong J., Woollard P., Salama J. J., Moore S., Wojcik J., Bader G. D., Vidal M., Cusick M. E., Gerstein M., Gavin A. C., Superti-Furga G., Greenblatt J., Bader J., Uetz P., Tyers M., Legrain P., Fields S., Mulder N., Gilson M., Niepmann M., Burgoon L., De Las Rivas J., Prieto C., Perreau V. M., Hogue C., Mewes H. W., Apweiler R., Xenarios I., Eisenberg D., Cesareni G., Hermjakob H. (2007) Nat. Biotechnol. 25, 894–898 [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M. (2002) Novartis Found. Symp. 247, 91–101, 119,–128, 244–252 [PubMed] [Google Scholar]
- 25.Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 26.Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 27.Zhou T., Sun L., Humphreys J., Goldsmith E. J. (2006) Structure 14, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., Zhou B., Zheng C. F., Zhang Z. Y. (2003) J. Biol. Chem. 278, 29901–29912 [DOI] [PubMed] [Google Scholar]
- 29.Jandt E., Denner K., Kovalenko M., Ostman A., Böhmer F. D. (2003) Oncogene 22, 4175–4185 [DOI] [PubMed] [Google Scholar]
- 30.Frey M. R., Golovin A., Polk D. B. (2004) J. Biol. Chem. 279, 44513–44521 [DOI] [PubMed] [Google Scholar]
- 31.Tonks N. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 32.Zhou B., Wang Z. X., Zhao Y., Brautigan D. L., Zhang Z. Y. (2002) J. Biol. Chem. 277, 31818–31825 [DOI] [PubMed] [Google Scholar]
- 33.Farooq A., Zhou M. M. (2004) Cell. Signal. 16, 769–779 [DOI] [PubMed] [Google Scholar]
- 34.Sun H., Charles C. H., Lau L. F., Tonks N. K. (1993) Cell 75, 487–493 [DOI] [PubMed] [Google Scholar]
- 35.Anderson N. G., Maller J. L., Tonks N. K., Sturgill T. W. (1990) Nature 343, 651–653 [DOI] [PubMed] [Google Scholar]
- 36.Payne D. M., Rossomando A. J., Martino P., Erickson A. K., Her J. H., Shabanowitz J., Hunt D. F., Weber M. J., Sturgill T. W. (1991) EMBO J. 10, 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarcevic B., Erikson E., Maller J. L. (1993) J. Biol. Chem. 268, 25075–25083 [PubMed] [Google Scholar]
- 38.Pettiford S. M., Herbst R. (2000) Oncogene 19, 858–869 [DOI] [PubMed] [Google Scholar]
- 39.Pulido R., Zúñiga A., Ullrich A. (1998) EMBO J. 17, 7337–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wälchli S., Espanel X., Harrenga A., Rossi M., Cesareni G., Hooft van Huijsduijnen R. (2004) J. Biol. Chem. 279, 311–318 [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z. Y., Thieme-Sefler A. M., Maclean D., McNamara D. J., Dobrusin E. M., Sawyer T. K., Dixon J. E. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4446–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z. Y., Maclean D., McNamara D. J., Sawyer T. K., Dixon J. E. (1994) Biochemistry 33, 2285–2290 [DOI] [PubMed] [Google Scholar]
- 43.Barr A. J., Ugochukwu E., Lee W. H., King O. N., Filippakopoulos P., Alfano I., Savitsky P., Burgess-Brown N. A., Müller S., Knapp S. (2009) Cell 136, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharrocks A. D., Yang S. H., Galanis A. (2000) Trends Biochem. Sci. 25, 448–453 [DOI] [PubMed] [Google Scholar]
- 45.Arvind R., Shimamoto H., Momose F., Amagasa T., Omura K., Tsuchida N. (2005) Int. J. Oncol. 27, 1499–1504 [PubMed] [Google Scholar]
- 46.Muñoz J. J., Tárrega C., Blanco-Aparicio C., Pulido R. (2003) Biochem. J. 372, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed N., Niu J., Dorahy D. J., Gu X., Andrews S., Meldrum C. J., Scott R. J., Baker M. S., Macreadie I. G., Agrez M. V. (2002) Oncogene 21, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 48.Ruivenkamp C. A., van Wezel T., Zanon C., Stassen A. P., Vlcek C., Csikós T., Klous A. M., Tripodis N., Perrakis A., Boerrigter L., Groot P. C., Lindeman J., Mooi W. J., Meijjer G. A., Scholten G., Dauwerse H., Paces V., van Zandwijk N., van Ommen G. J., Demant P. (2002) Nat. Genet. 31, 295–300 [DOI] [PubMed] [Google Scholar]
- 49.Nelson P. J., Daniel T. O. (2002) Kidney Int. 61, S99–105 [DOI] [PubMed] [Google Scholar]
- 50.Pestana M., Oliveira G., Xavier P., Mendes A., Guerra L. E. (1999) Transplant. Proc. 31, 306–307 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.