Abstract
We have characterized the structure and the function of the 6.6-kDa protein SvtR (formerly called gp08) from the rod-shaped virus SIRV1, which infects the hyperthermophilic archaeon Sulfolobus islandicus that thrives at 85 °C in hot acidic springs. The protein forms a dimer in solution. The NMR solution structure of the protein consists of a ribbon-helix-helix (RHH) fold between residues 13 and 56 and a disordered N-terminal region (residues 1–12). The structure is very similar to that of bacterial RHH proteins despite the low sequence similarity. We demonstrated that the protein binds DNA and uses its β-sheet face for the interaction like bacterial RHH proteins. To detect all the binding sites on the 32.3-kb SIRV1 linear genome, we designed and performed a global genome-wide search of targets based on a simplified electrophoretic mobility shift assay. Four targets were recognized by the protein. The strongest binding was observed with the promoter of the gene coding for a virion structural protein. When assayed in a host reconstituted in vitro transcription system, the protein SvtR (Sulfolobus virus transcription regulator) repressed transcription from the latter promoter, as well as from the promoter of its own gene.
Viruses specifically infecting Archaea, one of the three domains of life, were identified for the first time more than 30 years ago (1). Since then, more than 50 archaeal viruses have been described, all with double-stranded DNA genomes, linear or circular (2). Approximately half of the known species infect hyperthermophilic crenarchaea of the genera Sulfolobus, Acidianus, Stygiolobus, Pyrobaculum, and Thermoproteus (3–5). Based on their morphological and genomic characteristics, the known crenarchaeal viruses were assigned to seven novel families. The Sulfolobus islandicus rod-shaped virus 1, SIRV1 (6), analyzed in this study, belongs, together with SIRV2 (7), Stygiolobus rod-shaped virus, SRV (8), and Acidianus rod-shaped virus 1, ARV1 (9), to the family Rudiviridae.
The study of crenarchaeal viruses is still at an early stage, and the knowledge of their biology and basic molecular processes, including infection, virus-host interactions, DNA replication and packaging, as well as transcription regulation, is limited (10). Viruses belonging to the family Rudiviridae are promising candidates to become a general model for detailed studies of archaeal virus biology. These are indeed easily maintained under laboratory conditions and can be obtained in sufficient yields, which is not the case for many other archaeal viruses. Published data concerning two close representatives of this family of viruses, SIRV1 and SIRV2, brought essential information about three important fields of their biology: viral genes transcription pattern, viral genome replication, and host cell adaptation (6, 7, 11–13).
The transcriptional patterns of the rudiviruses SIRV1 and SIRV2 are relatively simple, with few temporal expression differences (11). Contrastingly, at least ∼10% of SIRV1 genes (5 of 45) were assigned to be putative transcriptional regulators because of the in silico structural prediction of different DNA binding motifs in the proteins they code (5). In particular, the later in silico analysis of available crenarchaeal viruses suggested a high proportion of viral genes coding for DNA binding proteins with the ribbon-helix-helix (RHH)6 motif.
In the Archaea, like in the Bacteria and in contrast to the Eukarya, the RHH fold seems to be a common structural scaffold. Indeed, a large number of RHH proteins have been identified in vitro or in silico both in Bacteria and in Archaea, even though the very low sequence identity (often ≤ 15%) displayed by RHH proteins renders their in silico identification problematic. Their role in the regulation of transcription in Archaea, however, remains poorly studied. It is well established that structurally (14) and functionally (15), the archaeal transcription machinery is similar to the eukaryotic one, but the transcription of archaeal genes is regulated by protein factors similar to those of bacteria (15). The abundance of genes coding for proteins belonging to the RHH superfamily present in the genomes of crenarchaea and their viruses could underline the important role of these proteins in host and viral gene transcription regulation. Even so, no crenarcheal virus protein involved in transcription regulation has so far been studied in detail.
In this work we focused on one of the putative DNA-binding proteins of the rudivirus SIRV1 that infects S. islandicus. The protein, named here SvtR and previously called gp08 or ORF56b (sequence accession numbers AJ414696 and NC_004087) (7), was initially annotated as having a helix-turn-helix motif and was later predicted to show an RHH fold (5). A close homologue of this protein is present in SIRV2 and on the S. islandicus filamentous virus SIFV of the Lipothrixviridae family (16), but these proteins have not been analyzed yet. Here, we present the structural and functional characterization of the protein SvtR (gp08) and the identification of its targets on the SIRV1 genome.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
The gene gp08 of SIRV1 (NP_666596.1, also called ORF56b) was amplified by PCR using primers ORF56F (5′-GGAATTCCATATGCAAACTCAAGAACAGAG-3′) and ORF56R (5′-GGATCCCTCGAGTTAACCGTTTCTCTTTTGCA-3′). The PCR product was digested with NdeI and XhoI and ligated with NdeI- and XhoI-digested pET30a (Novagen, Darmstadt, Germany) plasmid vector. Recombinant SvtR was expressed without any tag or additional residues using Escherichia coli RosettaTM (BL21(DE3)pLysS, Novagen) cells. Cultures were grown at 37 °C in Luria-Bertani (LB) broth or minimal M9 medium, which was supplemented with 1.7 g/liter yeast nitrogen base without amino acids and without ammonium sulfate (Difco). Cells were grown in the presence of kanamycin (100 μg/liter) and chloramphenicol (34 μg/liter). Protein expression was induced during logarithmic growth with 1 mm isopropyl-β-thiogalactopyranoside. Cells were harvested 4 h after induction by centrifugation at 6700 × g and 4 °C, resuspended in buffer A (Tris-HCl, 50 mm, NaCl, 300 mm, pH 8), pelleted by centrifugation, and frozen at −80 °C. For 15N or 15N/13C labeling, the M9 medium was prepared with 15N-labeled ammonium chloride (0.52 g/liter), and uniformly 13C-labeled glucose (2 g/liter) as required.
Frozen cells were thawed, resuspended in buffer A, and lysed by sonication at 4 °C adding phenylmethylsulfonyl fluoride. After centrifugation at 17,000 × g, the supernatant was heated 30 min at 75 °C, allowed to cool down at room temperature, and centrifuged at 17,000 × g at 4 °C. The supernatant was loaded onto a 5-ml HiTrap heparin-Sepharose column (Amersham Biosciences) and eluted with a linear NaCl gradient in buffer A (300–1000 mm NaCl). Fractions were analyzed by SDS-PAGE under reduction conditions. Fractions containing SvtR were pooled, dialyzed against 20 mm ammonium bicarbonate, and lyophilized. Protein preparations were homogeneous as assessed by SDS-PAGE and NMR; protein integrity was verified by electrospray mass spectrometry, which yielded the expected mass for SvtR (6621 Da). The concentration of the protein was determined using a molar extinction coefficient of 4470 m−1.cm−1 calculated from its sequence.
Oligonucleotides
Oligonucleotides were purchased from Proligo (Sigma-Aldrich) or obtained by PCR using Phusion High Fidelity DNA polymerase (Finnzymes) as indicated. Commercial oligonucleotides for fluorescence, NMR, and analytical ultracentrifugation experiments were of reversed-phase high-performance liquid chromatography grade. Double-stranded DNA was obtained by annealing the corresponding single strand nucleotides following standard techniques. For fluorescence measurements, the antisense strand was 5′-labeled with fluorescein. For PAGE experiments, oligonucleotides were 32P radiolabeled using the T4-polynucleotide kinase (New England Biolabs).
DNA-binding Activity of SvtR
A 36-bp double-stranded DNA fragment (36m) corresponding to the promoter region of the gp08 gene (position 5566–5602 in the sequence of SIRV1 var VIII; NP_666596.1) and a 39-bp double-stranded DNA fragment (39m) issued from the promoter region of the gp30 gene (position 16903–16941) were obtained using the corresponding single strand commercial oligonucleotides. Prior to annealing, one strand of each fragment was 32P-radiolabeled. Each double-stranded labeled fragment (1.5 pmol and ∼75 ng) was incubated with 2 μl of 50% (v/v) glycerin, 1 μg of poly[dI,dC] in a total of 20 μl of transcription buffer (50 mm Tris-HCl, pH 8.0, 75 mm KCl, 25 mm MgCl2, and 1 mm dithiothreitol) for 15 min at 48 °C with increasing amounts of SvtR (from 0 to 500 ng). The DNA-protein mixtures were deposited in a non-denaturing 8% 37.5:1 acrylamide/bisacrylamide gel. PAGE was run in TBE buffer (89 mm Tris-borate, 2 mm NaEDTA, pH 8.3). After migration, the gel was vacuum-dried, exposed with Amersham Biosciences HyperfilmTM MP, and developed with a Kodak X-OMAT 2000 processor. This procedure (vacuum drying, exposure, and development) was repeated for all PAGE experiments described below.
Agarose Gel EMSA (AG-EMSA) Target Screening
A library of fragments was constructed by dividing the SIRV1 genome into four large regions of 6956, 8744, 8851, and 7761 bp. Each of these regions was divided into a set of short DNA fragments generated by PCR. These fragments, ranging in size between 2.2 and 0.4 kb, covered the entire SIRV1 genome. The positions of the DNA fragments used for target identification are shown in the supplemental material (Table S1).
The mobility of the generated DNA fragments was analyzed in agarose gels. The DNA fragments of each set were incubated in 20 μl of binding buffer (10 mm Tris-HCl, pH 8.0, 10 mm HEPES, 1 mm EDTA, 1 mm dithiothreitol, 50 mm KCl, 50 μg/ml bovine serum albumin) for 15 min at 48 °C with increasing amounts of SvtR (from 0 to 200 ng). 5 μl of AG-EMSA loading buffer (20 mm Tris-HCl, 10 mm EDTA, 50% glycerol) was added, and 20 μl of the sample was usually analyzed in standard 1.2% agarose gels. Samples were run at 135 V for 10 min, followed by 1 h of migration at 100 V, and visualized under UV light after ethidium bromide staining.
Reducing the Size of the Best Target Candidate, PAGE-EMSA Experiments
A double-stranded DNA fragment of 342 bp identified as the best target in the AG-EMSA experiments was digested with 10 units of AluI for 1 h at 37 °C. The digestion step was followed by 20 min of heat inactivation at 65 °C, dephosphorylation with shrimp alkaline phosphatase (USB Corp.), and 32P-radiolabeled. The generated fragments of 150, 104, and 88 bp were incubated in TBE buffer at 68 °C for 15 min with increasing amounts of SvtR. Five microliters of 5× RB loading dye (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 20% glycerol, 100 μg/ml bovine serum albumin, 1 mg/ml xylene-cyanol) was added, and 20 μl of sample was analyzed in a non-denaturing 12% 37.5:1 acrylamide/bisacrylamide gel in TBE buffer. Samples were run for 2 h at 200 V.
IVT Assays
Sulfolobus solfataricus RNA polymerase, and transcription factors TBP and TFB were obtained as described previously (17). In vitro transcription (IVT) assays were performed to evaluate the influence of the protein SvtR on the transcription from the promoters of genes gp08 and gp30. PCR products of the promoters of genes gp08 (377 bp, position 5398–5774) and gp30 (363 bp, position 16746–17108) were generated from genomic SIRV1 DNA and cloned directly into a pDrive (Qiagen) cloning vector by T/A cloning. A plasmid carrying the SSV T6 promoter, used as a control in IVT experiments, was generated as described in Ref. 18.
In vitro transcription reactions were performed using 100 ng of the corresponding plasmid DNA, 0.2 mm NTPs, 5 ng of TBP, 5 ng of TFB, 125 ng of RNA polymerase, and varying amounts of SvtR as indicated in Fig. 8. The reactions were carried out for 20 min at 70 °C in 50 μl of transcription buffer (50 mm Tris-HCl, pH 8.0, 75 mm KCl, 25 mm MgCl2, and 1 mm dithiothreitol). Reactions were stopped by adding 250 μl of NEW buffer (10 mm Tris-HCl, pH 8.0, 750 mm NaCl, 10 mm EDTA, 0.5% SDS, and 40 mg/ml glycogen). In vitro synthesized RNA was isolated by phenol-chloroform extraction followed by ethanol precipitation. Transcription products were detected by primer extension using sequence-specific primers for the previously described viral promoter templates (11, 18). After addition of 20 μl of 50% formamide loading dye, 20 μl of the denatured sample was analyzed on a 10% denaturing polyacrylamide gel. Gels were fixed in 10% acetic acid-10% ethanol before vacuum drying, exposure, and development.
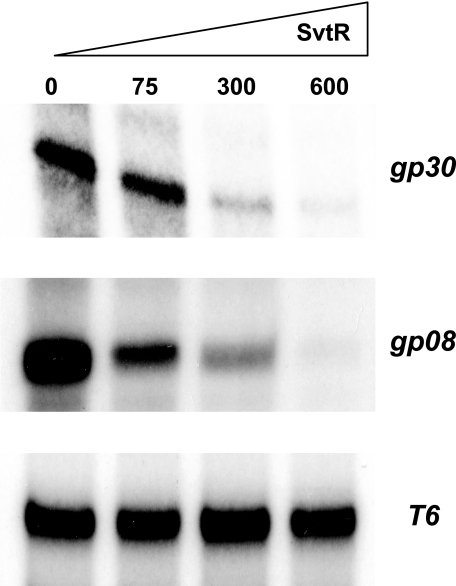
FIGURE 8.
SvtR represses transcription from two SIRV1 promoters. The transcription regulator activity of SvtR was tested using an IVT. All reactions were performed in the presence of limiting amounts of transcription factors TBP and TFB. The added amounts of SvtR are shown in nanograms. Transcription assays from: gp30 promoter (top panel), gp08 promoter (middle panel), and T6 SSV1 promoter (bottom panel). Strong transcription repression was observed for the Pgp30, a weaker repression for the Pgp08, and no effect was seen with the control T6 promoter from an unrelated virus.
NMR Samples
Samples were prepared by dissolving lyophilized SvtR in buffer B (20 mm D4 deuterated sodium acetate 10–15% D2O, pH 5.5). Protein concentration typically ranged between 1.2 and 1.7 mm for unlabeled, 15N-labeled and 13C/15N-labeled samples. For the doubly filtered 13C/15N-13C-edited nuclear Overhauser experiment (NOESY), a 50% doubly labeled/50% unlabeled sample at 2.9 mm concentration was obtained by mixing an unlabeled and a 15N/13C-labeled sample at room temperature.
NMR
Experiments were performed on INOVA 500 or 600 spectrometers (Varian Inc., Palo Alto, CA) with proton-resonating frequencies of 499.8 and 599.4 MHz, respectively. The 600-MHz spectrometer was equipped with a cryo-probe. Experiments were carried out at 25 °C. Spectra were referenced to external sodium 4,4-dimethyl-4-silapentane-1-sulfonate for 1H, and using the indirect chemical shift ratios recommended by the IUPAC for 13C and 15N. Data were collected using Vnmr 6.1C or VnmrJ 2.1B (Varian Inc.), processed with NMRPipe (19) and analyzed with NMRView 5.2.2 (20).
Chemical shifts were assigned to the protein 1H, 13C, and 15N backbone and side-chain nuclei using standard two-dimensional and three-dimensional experiments: 1H homonuclear two-dimensional TOCSY (total correlation spectroscopy (21)) and purged-COSY (correlation spectroscopy (22)), 13C or 15N HSQC (heteronuclear single quantum coherence (23)), HNCO, HNCACB, and CBCA(CO)NH (24), H(CC-TOCSY)NNH and C(CC-TOCSY)NNH (25, 26), 13C-edited HCCH-TOCSY (27), 15N-edited TOCSY-HSQC (28), and two-dimensional (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE (29).
Nitrogen-15 longitudinal (R1) and transverse (R2) relaxation rates and the heteronuclear 1H-15N NOE were determined using pulse sequences described by Kay and co-workers (30). Experiments were performed at 499.8-MHz proton frequency and 25 °C, unless stated otherwise. Ten data points between 20 and 1000 or 10 and 190 ms were collected to calculate R1 and R2, respectively. For the 1H-15N NOE, the 3-s saturation period was preceded by a 2-s recovery delay. Errors were estimated from the noise standard deviation. R1 and R2 data were fitted to single exponential decays without offset. The isotropic rotation correlation time τc of SvtR (1.7 mm) at 25 °C was established from the ratio of R2 and R1 backbone 15N rates of residues selected as described in (31). Relaxation parameters were analyzed using the extended Lipari and Szabo formalism as implemented on the program ModelFree 4.1 (31, 32) and considering isotropic tumbling. Model selection was performed at a confidence level of 90% following the statistical approach proposed in Ref. 31 with some minor modifications.
The hydrodynamic (Stokes) radius of SvtR was determined by NMR using the GCSTECSL (33) pulse sequence with convection compensation (34). The diffusion and the encoding-decoding gradient delays were 100 and 4.0 ms, respectively. The experiment was repeated three times with a protein sample (1.2 mm) prepared in buffer B with 100% D2O and with dioxan (0.005%), which served as an internal hydrodynamic radius standard. The Stokes radius of the protein was determined as described in a previous study (35).
NOE Assignments and Structure Calculations
Five NOE experiments were recorded to obtain distance constraints for structure calculations: 1H-1H two-dimensional NOESY (36) acquired on samples prepared in H2O and in D2O, three-dimensional 13C- and 15N-edited NOESY-HSQC (23, 28), and a 13C/15N-doubly filtered 13C-edited NOESY (37) to distinguish intra- and inter-monomer NOEs. The mixing time was 120 ms for all NOESY spectra, except for the doubly filtered spectrum, in which the mixing time was 150 ms. Proton JHN-HA scalar couplings were determined from an HNHA experiment (38, 39). Coupling constants were transformed into dihedral ϕ angle constraints as follows: −120° ± 25° for 3JHN-Hα ≥ 8.0 Hz, −65° ± 25° for 3JHN-Hα ≤ 5.5 Hz. A backbone hydrogen bond in regions of secondary structure was added as distance constraint if the chemical shift data, the NOE pattern, and the amide hydrogen exchange data were in agreement with a hydrogen bond and if it was present in at least 75% of the structures calculated without any hydrogen bond.
Peaks in NOESY spectra were assigned by means of a β version of ARIA 2.2 (40, 41)7 coupled to CNS 1.1 (42) following ARIA's standard protocols with spin diffusion correction. This version of ARIA handles symmetrical homopolymers. We used a C2 type symmetry for structure calculations. Chemical shift tolerances were set to 0.035 for 1H in two-dimensional experiments, 0.025 for HN protons, and 0.04 ppm for protons in the indirect dimension of three-dimensional experiments, 0.51 ppm for 13C and 0.25 ppm for 15N. Some assignments were obtained manually. To obtain the final structure ensemble, 200 structures were calculated with ARIA 2.2/CNS 1.1 and refined in explicit water using the PARALLHDG 5.3 force field (43). The 10 lowest total energy structures were selected. The quality of the structures was analyzed with PROCHECK 3.5.4 (44), WHAT_CHECK (45), and MOLMOL 2K.2 (46).
SvtR-DNA Interaction Followed by NMR
Proton mono-dimensional spectra and 1H-15N HSQC spectra were recorded to follow the binding of 15N-labeled SvtR to a 17-bp DNA oligomer (17m) issued from the promoter region of the SvtR gene (position 5579–5596, 5′-TAATTCAAAATCTTAAA-3′). Experiments were performed at 25 °C in buffer D: 50 mm D4 deuterated potassium acetate, 100 mm KCl, 1 mm EDTA, 15% D2O, pH 5.5. The protein was added to the 17m sample (100 μm) to obtain the desired protein:DNA ratios.
Fluorescence Experiments
Binding of SvtR to a 36-bp oligonucleotide (36m, position 5566–5602) issued from its own gene promoter or to 39-bp (39m, position 16903–16941), 70-bp (70m, position 16887–16956), and 120-bp (120m, position 16898–17017) oligonucleotides from the promoter region of gene gp30, was followed by competition experiments using the fluorescein-labeled 36m oligonucleotide (flDNA). A 36-bp oligonucleotide composed of CG and CI base pairs (poly[dIG,dC]) was used as nonspecific competitor in independent experiments. The fluorescence of flDNA was attenuated by SvtR, providing a convenient means to monitor protein-DNA binding. Of note, control experiments showed that the quantum yield and emission spectra of the label (fluorescein) at 10 or 50 nm concentration were not significantly affected (1% signal variation) by high concentrations of SvtR (up to 10 μm). These data suggested that the fluorescein label was not directly responsible for the interaction between SvtR and flDNA. However, because the contribution (positive, null, or negative) of the fluorescein moiety of flDNA to binding was not known, we performed competition experiments to analyze the relative apparent binding affinities of the oligonucleotides. We first determined the equilibrium dissociation constant (KD,fl) of the flDNA-SvtR interaction and then performed competition experiments with fixed flDNA and SvtR concentrations and increasing amounts of unlabeled competitor DNA.
Experiments were run at 25 °C on a PerkinElmer Life Sciences LS 50B spectrofluorometer equipped with a thermo-jacket and a stirrer. Samples were excited at 494 nm, and emission was recorded at 519 nm using 0.4 × 1.0 cm Hellma (109.004F QS) quartz cuvettes. Slit-widths depended on the flDNA concentration but were typically 5 and 15 nm for excitation and emission, respectively. Fluorescence was integrated during 1 s and recorded 30 times for each titration point. Samples were prepared in buffer D: 20 mm Tris-HCl, 1 mm Na-EDTA, 1 mm Tris(2-carboxyethyl)phosphine hydrochloride, 0.02% Tween 20, 100 mm KCl, pH 8.0. For flDNA-SvtR experiments, concentrated SvtR was serially added to 5 (or 10) nm flDNA in 1 ml of buffer D. For competition experiments, increasing amounts of unlabeled competitor DNA (36m, 39m, 70m, 120m, or poly[dIG,dC]) were added to a 1-ml sample of 5 nm flDNA and 100 nm SvtR. Sample dilution (<5%) was taken into account to correct fluorescence intensities. Samples were equilibrated at least 5 min before recordings. Experiments were repeated at least twice. We checked by NMR that the 1H-15N HSQC spectra of SvtR, which can be seen as a “fingerprint” of the structure of a protein, were not affected by the addition of KCl, Tween 20, Tris(2-carboxyethyl)phosphine hydrochloride, and Na-EDTA.
Analytical ultracentrifugation indicated that SvtR underwent a monomer-dimer equilibrium with a relatively high dissociation constant. To analyze the DNA-SvtR binding, we assumed that only the SvtR dimers (not the monomers) were able to bind DNA. Furthermore, as stoichiometry measurements (see “Results”) indicated that two dimers bound to one double-stranded flDNA, we analyzed the flDNA-SvtR binding data using a model of a pair of SvtR dimers binding to a single site in double-stranded DNA (or two dimers binding each to a half-site). The equilibrium dissociation constant of the protein monomer-dimer equilibrium was taken into account in the model to calculate the concentration of protein dimers available for the interaction with DNA.
The half-inhibition concentration IC50,u values of unlabeled DNA oligonucleotides were obtained from competition experiments between flDNA and unlabeled DNA oligomers at varying concentrations in the presence of SvtR. The fraction of pairs of protein dimers bound to labeled DNA was described in terms of the dissociation constant of the labeled DNA (KD,fl), the IC50,u of the unlabeled competitor DNA, and the total concentrations of labeled and unlabeled DNA.
The equations used to obtain the equilibrium dissociation constant (KD,fl) of flDNA and SvtR pair of dimers and to fit the competition data with unlabeled DNAs and estimate the IC50,u values are described in the supplemental material.
Analytical Ultracentrifugation
Sedimentation-diffusion equilibrium and sedimentation velocity experiments were performed at 25 °C using a Beckman-Coulter Optima XL-I ultracentrifuge equipped with an AN50-Ti rotor. Experiments were done using buffer E: 20 mm Tris-HCl, pH 8.0, 100 mm KCl, 1 mm Tris(2-carboxyethyl)phosphine hydrochloride. The buffer was supplemented with 1 mm Na-EDTA for experiments involving DNA.
The oligomerization state of SvtR was evaluated by equilibrium experiments run at three protein concentrations (0.5, 5, and 20 μm). Detection of the protein concentration as a function of radial position and time was performed by absorbance measurements at a wavelength of 230 nm. Samples (120 μl) were spun successively at four speeds (26000, 30000, 38000, and 42000 rpm) using a two-channel Epon centerpiece (1.2-mm optic path). Once the equilibrium was reached, radial distributions were analyzed by global fitting of the four speeds to a self-association equilibrium by means of the Ultrascan 9.9 software (47). Ultimately, a monomer-dimer equilibrium model was used. The partial specific volume of the protein (0.731 cm3/g), and the viscosity and density of the buffer at 25 °C were calculated with Sednterp 1.09. The error on the equilibrium constant corresponded to the standard deviation obtained from 10,000 iterations of Monte Carlo simulations of the data.
Binding of SvtR to oligonucleotides 36m, 39m, and 70m was analyzed by sedimentation experiments. Double-stranded DNA and protein concentrations were 2 and 20 μm for experiments with 39m and 70m, or 1 and 8 μm for the 36m samples, respectively. DNA-only and DNA-protein samples (100 μl) were spun at 42,000 rpm using a 3-mm thick two-channel Epon centerpiece. Total intensity at 260 nm was monitored at ∼3-min intervals and converted to pseudo absorbance units with Sedfit 11.7 (48). The partial specific volumes of the DNA-protein complexes were calculated as molecular-mass weighted averages of the partial specific volumes of the DNA (0.57 cm3/g) and the protein. The partial specific volume corresponding to different stoichiometries of the complexes was varied iteratively to obtain the best fit of the data, using the continuous c(s) and c(M) distribution models in Sedfit. Once the stoichiometry was determined by this procedure, the corresponding partial specific volume was used to obtain the sedimentation coefficient of the complexes and their fitted mass.
RESULTS
The Protein SvtR Forms a Dimer in Solution
The protein SvtR was expressed in E. coli and purified to apparent homogeneity as described under “Experimental Procedures.” The hydrodynamic radius Rh of SvtR obtained from self-diffusion experiments at 1.2 mm protein concentration was 20.5 ± 0.8 Å and the isotropic rotation correlation time τc of SvtR (1.7 mm) at 25 °C was 8.2 ± 0.3 ns. Both Rh and τc values determined for SvtR were greater than expected for a compact globular monomer of a 56-residue protein and were more in line with those estimated for a dimer. Sedimentation-diffusion equilibrium experiments run with SvtR indicated a monomer-dimer equilibrium with a molecular mass of 6.6 ± 0.5 kDa for the monomer, which corresponded well to the theoretical mass of SvtR (6621 g/mol). Fitting of the centrifugation profiles (supplemental Fig. S1) yielded a dissociation equilibrium constant KD of 1.0 ± 0.4 μm at 25 °C. Together, hydrodynamics data indicated that SvtR showed a monomer-dimer equilibrium. Because chemical shifts are very sensitive to structure variations, and the 15N-1H HSQC spectra of SvtR were effectively identical between 15 μm and 2.9 mm, we could conclude that SvtR formed a dimer in this concentration range. The fact that 15N-1H HSQC spectra of SvtR showed 54 backbone amide signals out of 55 expected from the sequence, indicated that SvtR formed a symmetrical homodimer.
NMR Spectra Assignments
Sequential assignment of 1H, 13C, and 15N signals was performed following standard procedures. Assignment of backbone and side-chain resonances was almost complete (96%), with only a few atoms missing, mostly on the N-terminal region of SvtR: the N-terminal methionine atoms, the amide group of N2, the CZ carbon of Phe16, the HG12 and CG1 of Ile43 and the side-chain N and H amide atoms of glutamines 4, 6, and 12 for which overlapping resonances were identified but could not be unambiguously assigned to a particular glutamine in the sequence. The assigned 15N-1H HSQC spectrum is shown in Fig. S2 of the supplemental material.
Structure and Dynamics of SvtR
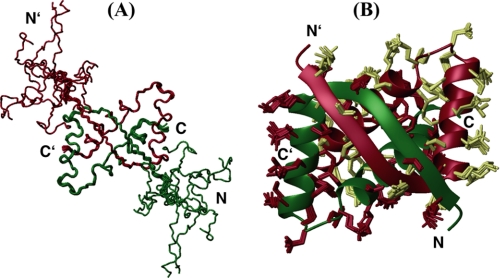
The structural ensemble of SvtR showed a well ordered region between residues 13 and 56 and an N-terminal non-convergent region between residues 1 and 12 (Fig. 1A). Accordingly, most of the relatively few NOEs of the N-terminal region were intra-residue or sequential. Because no medium, long range, or inter-monomer NOEs were observed between the N-terminal tail and the rest of the protein, final structures were calculated using only data from residues 11–56 (Fig. 1B). A total of 2093 restraints per monomer were used in the calculations, which corresponds to ∼46 restraints per residue. Structure statistics are summarized in Table 1.
FIGURE 1.
Structure of SvtR. Main-chain trace representation of the structural ensemble of 10 conformers of the full-length protein (residues 1–56) (A) and ribbon representation of residues 11–56 (B). In B, the heavy side chains of the 10 conformers are shown on the ribbon diagram of the lowest energy structure. Monomers are shown in different colors.
TABLE 1.
Statistics for the ensemble of 10 structures calculated for SvtR
| Constraints per monomer (residues 11–56) | |
| Unambiguous restraints | 1233 |
| Ambiguous distance restraints | 803 |
| Total number of distance restraintsa | 2036 |
| Intra-residue |j-i| = 0 | 537 |
| Sequential |j-i| = 1 | 404 |
| Medium range 2 ≤ |j-i| < 4 | 370 |
| Long range |j-i| ≥ 4 | 227 |
| Inter-monomer | 498 |
| Backbone dihedral ϕ angle restraints | 30 |
| Total number of hydrogen bonds | 27 |
| Inter-monomer hydrogen bonds | 4 |
| Residual distance constraint violations | |
| Number ≥ 0.3 Å | 0 |
| Number ≥ 0.1 Å | 50 ± 2 |
| Root mean square deviation from NOEs (Å) | 0.0205 ± 0.0001 |
| Residual dihedral angle constraint violations | |
| Number ≥ 5.0° | 4 |
| Root mean square deviation from dihedrals (°) | 10.3 ± 0.0001 |
| Energies (kcal/mol) | |
| Total | −2870 ± 53 |
| Van der Waals | −276 ± 9 |
| Electrostatic | −3572 ± 87 |
| Mean of pairwise root mean square deviation (Å) (13–56)b | |
| Backbone atoms N, Ca, and C′ | 0.24 ± 0.06 |
| Heavy atoms | 0.72 ± 0.07 |
| Ensemble Ramachandran plot (13–56)b | |
| Residues in most favored regions | 86.3% |
| Additionally allowed | 12.4% |
| Generously allowed | 1.2% |
| Structure Z scores (13–56)b | |
| Second generation packing quality | 3.9 ± 0.2 |
| Ramachandran plot appearance | −4.4 ± 0.4 |
| Chi1/Chi2 rotamer normality | −2.3 ± 0.2 |
| Backbone conformation | −5.5 ± 1.2 |
| Unsatisfied H-bond donors per dimerb | 1.2 |
| Unsatisfied H-bond acceptors per dimerb | 0 |
| Bumps per dimer (13–56)b | 12.1 |
a Distance constraints used for structure calculations, which excluded fixed intra-residue distances.
b Values for the structured region (between residues 13 and 56).
The structure of SvtR displayed an RHH fold, which consisted of an antiparallel inter-monomer β-sheet between residues 13 and 21, followed by two long α-helices of 13 (H1:22–34) and 16 (H2:39–54) residues separated by a 4-residue loop. Helices H1 and H2 within a monomer formed an angle of 111.0 ± 0.8°. The structure showed extensive inter-monomer contacts and a large interface with a buried surface of 2047 Å per monomer. The protein SvtR showed a larger number of inter-monomer NOEs (498) than of intra-monomer long range NOEs (227). Accordingly, most of the inter-residue contacts between atoms in elements of secondary structure were intermolecular. Residues in strand B1 of monomer A made numerous van der Waals contacts with residues in B1 and in H2, as well as some with the N-terminal part of H1 of monomer B; residues in H1 made intermolecular contacts with residues in H2, and residues in H2 made numerous contacts with residues in H2 of the other monomer as well. Only α-helix H1 did not participate in intermolecular contacts with its equivalent counterpart. The C2 axis of symmetry, which was roughly perpendicular to the β-sheet plane, crossed through the middle of the β-sheet and between α-helices H2 at the level of residues Gly17 and Ala46, respectively.
The core of the dimer included several fully buried hydrophobic residues belonging to the three secondary structure elements: Phe16, Ile18, Met20, Leu24, Leu28, Tyr31, Cys32, Ala42, Ile43, Ala46, Ile47, Tyr50, and Leu51. All these residues were located at the inter-monomer interface, and, except for Cys32, made numerous inter-monomer contacts. In addition to the secondary structure-related backbone-backbone hydrogen bonds, SvtR structures displayed only a few backbone-side-chain or side-chain-side-chain intramolecular (Tyr31–Asn35, Leu51–Asn55, and Asn55–Gly56) or intermolecular (Asp21–Glu44, Arg27–Gly56, Tyr31–Arg54, and Glu45–Arg54) hydrogen bonds. Finally, the highly rich in positive residues SvtR dimer showed a network of intermolecular salt bridges Glu45 ↔ Arg54 ↔ Glu49 ↔ Lys53, and Lys48 ↔ Asp23, with Asp23 also forming an intra-monomer salt bridge with Arg27.
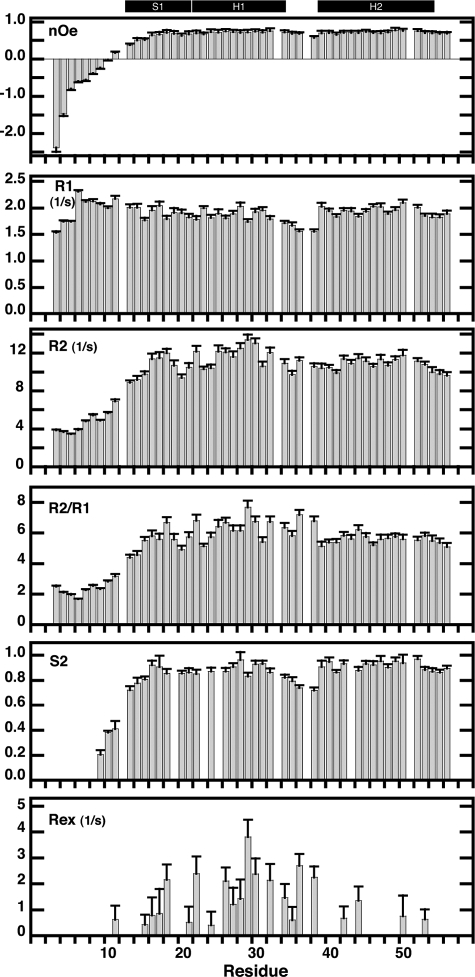
The backbone dynamics of SvtR were monitored through the longitudinal (R1) and transverse (R2) relaxation rates and the heteronuclear 15N-1H Overhauser effect (NOE) of the amide nitrogens (Fig. 2). The heteronuclear NOE showed negative values or very low positive values for the N-terminal residues (3–12), as well as slow transverse relaxation rates (R2) relative to the rest of the protein, which indicated that the N-terminal tail was disordered and that rapid internal fluctuations on the picoseconds-nanoseconds time-scale were responsible for the lack of convergence in structure calculations for the N-terminal region. In agreement with this, the values of the chemical shift of the backbone atoms were very close to the values expected for disordered polypeptides, as evidenced by the high values of the “random coil index” empirical correlation (49) observed for the N-terminal residues of the protein (supplemental Fig. S3). The rest of the backbone nitrogens showed relatively homogeneous relaxation values typical of folded proteins.
FIGURE 2.
15N relaxation rates (R2, R1 and R2/R1), heteronuclear 15N-1H NOE, and order (S2) and conformational exchange (Rex) Lipari-Szabo parameters of SvtR plotted as a function of residue number. Experiments were performed at 25 °C and a proton frequency of 499.8 MHz as described under “Experimental Procedures.” The secondary structure of SvtR is indicated on the top with black boxes (S = strand, H = helix). Relaxation data for residues 12, 33, 37, and 51 are not shown because of partial overlap of their corresponding signals. Data for residues 9 (overlap with 12) and 10 (overlap with 33) are included, however, because the amide group of these residues, which are located in the unfolded N-terminal region, gave strong signals that were less influenced by the overlapping signals. Order parameters are shown only for residues that were fit at a confidence level ≥ 0.9.
The relaxation parameters of SvtR were further analyzed using the extended Lipari and Szabo formalism to describe the internal dynamics of the protein on the nanosecond to picosecond and microsecond to millisecond time scales. The tumbling of the molecule was assumed to be isotropic. The order parameter (S2) reflects the amplitude of the internal motions on the picosecond to nanosecond time scale, that is motions faster than the tumbling of the molecule (τc = 8.2 ns for SvtR at 25 °C). A value close to 1 would correspond to small amplitude motions, whereas a value of 0 would indicate completely unrestricted motions. The mean of the S2 values obtained for SvtR between residues 13 and 56 was 0.88 ± 0.07, a value in line with those observed for compact proteins (Fig. 2). Residues Lys13, Ala14 and to a lesser extend Val15, showed somewhat lower order parameter values, indicating some fraying of the N-terminal end of the inter-monomer β-sheet. Residues (35–38) in the loop between α-helices H1 and H2 also showed higher amplitude motions than the rest of the backbone nitrogens. The S2 values were well correlated with the random coil index values, the backbone mean pair-wise root mean square deviations of the structures, and the number of NOEs used to calculate the structures. Finally, only Ile18 (β-sheet) and some residues located in H1 (Lys22, Thr26, Lys29, Val30, and Cys32) and the H1–H2 loop (Asn36 and Gln38) displayed high values of the Rex parameter (≥1.5 s−1), which indicated significant conformational exchange on the microsecond to millisecond time scale for these residues (Fig. 2).
Comparison of the Structure of SvtR with That of Other RHH Proteins
We searched the DALI data base (50) for structural homologues of SvtR and superposed its structure with the best hits using the Combinatorial Extension software (51). We found that the structure of SvtR was remarkably similar to that of bacterial/viral RHH proteins despite the low sequence similarity with these proteins. In particular, the structure of SvtR resembled the most, in decreasing order, to the structure of the proteins CopG (copy plasmid regulator, PDB code 1B01 (52)), Arc (1BDV (53)), NickR (nickel-responsive element, 1Q5V (54)), ParG (1P94 (55)), Omega (1IRQ (56)), Helicobacter pylori hypothetical protein HP0222 (1X93 (57)), MetJ (methionine repressor, 1MJ2 (58)), trafficking protein A (FitA, 2BSQ (59)), and PutA (proline utilization A, 2GPE (60)). Except for HP0222 of unknown function, all these proteins are transcription repressors. The Z-scores, CA root mean square deviations over 44–39 residues and sequence identities ranged from 4.4 to 4.2 and 1.19 to 1.92 Å and 22 to 7%, respectively. The small differences in the backbone structure between SvtR and the latter proteins were mainly located in the loop between α-helices H1 and H2 and in the β-strand B1, with small deviations in the angle between α-helices. The sequence similarity observed between SvtR and bacterial/viral RHHs of known structure was limited. Nevertheless, the sequence alignment obtained from the structure superpositions indicated that the hydrophobic character of the residues at positions occupied by residues in secondary structures at the interface between monomers of SvtR and corresponding to the hydrophobic core described above, was conserved (supplemental Fig. S4). Importantly, these positions showed also hydrophobic residues and some degree of conservation in the alignment of putative RHH proteins of crenarchaeal viruses described in the literature (5), again, despite the limited sequence similarity, supporting the correctness of the prediction.
The SvtR Protein Binds Specifically Its Own Gene Promoter
Because many of the characterized RHH proteins regulate the expression of their own gene and bind to the corresponding promoter, we tested whether the recombinant protein SvtR expressed in E. coli could bind in vitro to its own gene promoter region. The results of PAGE-EMSA experiments with SvtR and a labeled 36-bp oligonucleotide (36m) situated just upstream of the transcription initiation site of its own gene indicated that the protein could bind specifically this fragment (data not shown; this result was reproduced in another experiment displayed below in Fig. 5 comparing two SvtR targets). Indeed, SvtR was able to bind to the promoter region of its own gene in the presence of an excess of unlabeled nonspecific DNA, producing a strong retardation of the 36m DNA fragment migration. The absence of retardation of DNA fragments of similar size corresponding to the promoters of genes gp26 and gp35 under the same conditions pointed toward a specificity of binding (data not shown). Noteworthy, the 36m oligonucleotide used in the EMSA experiments contained a short imperfect inverted repeat (see Fig. 4C), and this type of repeat is often found in the binding sites of characterized RHH proteins. To summarize, these results confirmed the conclusions of the structural analysis and directly demonstrated the capacity of the protein to bind DNA. One of its putative targets in the SIRV1 genome is the promoter of its own gene. It was interesting to identify other genes of the viral genome regulated by SvtR and to characterize the role of the protein in transcription regulation.
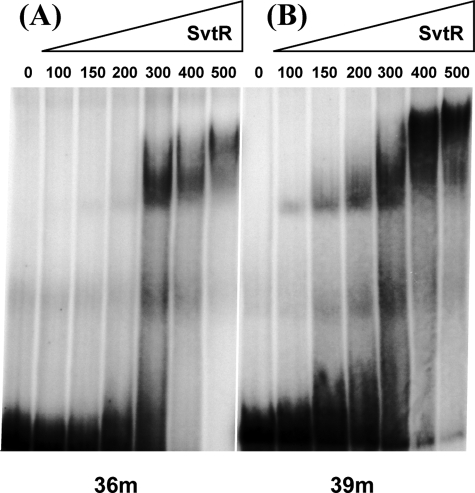
FIGURE 5.
The binding of SvtR to the 36m (A) and 39m (B) oligonucleotides is specific. The PAGE-EMSA were performed in the presence of a strong excess (13-fold) of competitor nonspecific DNA, poly[dI,dC]. The amounts of SvtR used are indicated in nanograms. Binding is observed from 300 ng of SvtR for the 36m and from 100 ng for the 39m.
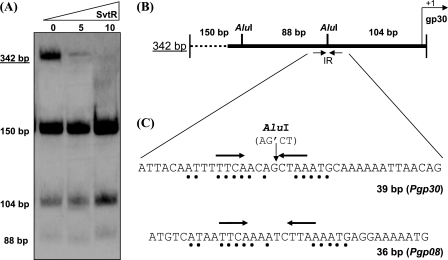
FIGURE 4.
Target identification within the promoter of gp30. A, PAGE-EMSA analysis of the radioactively labeled 342-bp DNA fragment and of its derivatives obtained by digestion with the AluI restriction enzyme. Protein amounts are shown in nanograms. B, scheme presenting a physical map of the 342-bp fragment. The positions of the AluI restriction sites are indicated. The black arrows show the position of the inverted repeat (IR) upstream of the gp30 gene transcription start site. C, sequence comparison of two identified SvtR short targets, 36m (Pgp08) and 39m (Pgp30). Black dots indicate the identical parts present in the inverted repeat region of both sequences. Black arrows show the position of the inverted repeats identified in both promoter regions.
Global SIRV1 Viral Genome Target Screening: System Design
A common approach used to validate the role of a hypothetical transcriptional regulator is to use its own gene promoter for target identification and functional studies. This normally allows one to identify a binding site for the regulator or a minimal DNA fragment sufficient for its specific binding. For SvtR, however, this direct approach was a particularly difficult task. Indeed, footprint experiments, which could lead to the identification of the protein-protected nucleotides at the binding site, were not successful (data not shown). The same difficulty has been described by other authors (61) and might be explained by a high general affinity of these proteins to DNA.
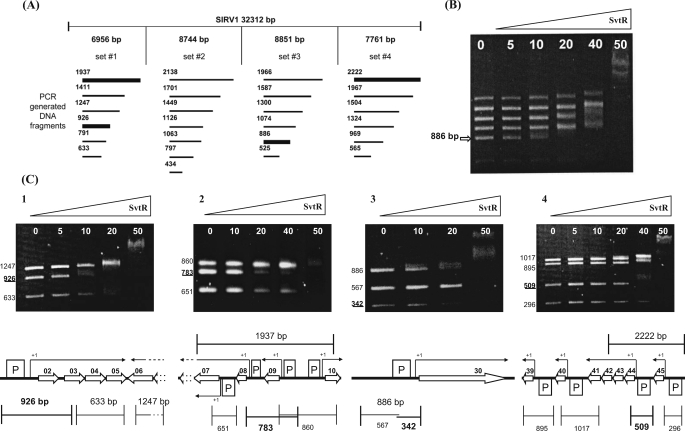
To overcome this problem, we designed a global screening strategy applicable to the analysis of small genomes (viruses, plasmids, etc.) to identify, in a one-step experiment, all the targets recognized by DNA-binding proteins. The proposed simplified electrophoretic mobility shift assay (AG-EMSA) is based on the fast analysis of the migration pattern of unlabeled DNA fragments separated in an agarose gel in the presence and in the absence of the studied protein. This approach can be successful if the affinity of the studied DNA-binding protein to its targets is strong enough to produce clearly identifiable migration retardation. In the case of the SvtR protein and the SIRV1 genome, sets of short DNA fragments between 0.5 and 3 kb were created covering all of the viral genome. The end points of the generated fragments were chosen to be located inside the coding part of the genes to avoid an eventual inactivation of the promoters (Fig. 3). After incubation with SvtR, the targets bound to the protein appeared in the gel as an upward shifted diffuse smear at low protein concentrations, instead of the normal sharp DNA band.
FIGURE 3.
Identification of the SvtR binding sites by the AG-EMSA approach. A, AG-EMSA strategy of the global protein-DNA interaction study for SIRV1. The SIRV1 genome was divided into four sets incubated separately with the SvtR protein in varying amounts (indicated in B and C in nanograms) and migrated on 1.2% agarose gels. The bold lines represent DNA fragments whose mobility changed specifically, i.e. at low concentrations of SvtR. B, typical results obtained with initial sets of DNA fragments (illustrated with set 3). The arrow indicates the position of the specifically responding fragment of 886 bp. C, AG-EMSA results that led to the identification of the SvtR targets and their positions on the genetic map of SIRV1. The strongest binding was observed in C3 with the gp30 promoter region. C1, a fragment of 926 bp was identified as specifically responding to SvtR in the first round of analysis. The scheme indicates this fragment's position that covered the promoter region for the operon gp02/03/04/05. C2, a 1937-bp fragment initially identified was divided into three subfragments. The 783-bp subfragment showed the strongest response to SvtR. This fragment covers the gp08 and gp09 promoters. because the gp09 promoter region was also included in a 860-bp fragment that did not specifically respond in the assay, it was concluded that the gp08 promoter region was involved in the specific binding of SvtR. C3, the initially identified 886-bp fragment was divided into two subfragments. In this analysis, the 342-bp fragment responded specifically. As indicated in the scheme, this fragment covers the gp30 promoter region. C4, a 2222-bp fragment covering two promoter regions was identified in the initial round of AG-EMSA. A 509-bp fragment was identified when subfragments were analyzed; its position covering the promoter region for the operon gp44-43-42-41 is indicated in the corresponding scheme.
Protein SvtR Recognized Specifically Four Targets in the SIRV1 Genome
The described strategy was successful to identify all the targets of the protein SvtR on the SIRV1 genome. The viral genome was divided into four regions. In each of these regions a set of 6 to 7 subfragments, all inside a requested size range, were amplified by PCR (25 fragments) as described under “Experimental Procedures” and Fig. 3A. The mobility of the fragments included in each generated set was analyzed in the presence of increasing amount of the purified protein.
Four targets were identified by these experiments (Fig. 3). The strongest binding occurred with an 886-bp fragment present in set number 3. This DNA fragment included the promoter region for gene gp30, which codes for a SIRV1 structural protein (8). The same AG-EMSA strategy was used to analyze the subfragments generated from the strongest binding region (886 bp). This allowed us to identify a 342-bp DNA fragment as the smallest region interacting with SvtR.
Three other fragments, a 926-bp and a 1937-bp fragment from set 1 and a 2222-bp fragment from set 4 showed slightly weaker binding when compared with the 886-bp region. The 926-bp region included the promoter region for an operon gp02-03-04-05 (Fig. 3C). The 2222-bp fragment overlapped two promoter regions, one corresponding to the operon gp44-43-42-41 and the second one to gene gp45. The later fragment was divided into smaller subfragments, which were re-analyzed by AG-EMSA. A 509-bp fragment was identified as being clearly specifically retarded, thus confirming that protein SvtR specifically binds to the promoter of the operon gp44-43-42-41 (unknown function). Both the 926- and the 509-bp fragments contained promoter regions that displayed the same sequence and were located within the long internal terminal repeats (ITRs) present at both ends of the viral genome. The 1937-bp fragment initially identified included the promoter region for an operon composed of three genes, gp08, 07, and 06, an internal gp07 promoter active 2 h after infection (11) and the promoter regions for genes gp09 and gp10. Genes gp07 and gp06 are predicted to code for a multitransmembrane permease similar to the FrlA putative fructoselysine transporter from E. coli and for a queuine-archaeosine tRNA ribosyltransferase, respectively (5), whereas genes gp09 and gp10 code for proteins of unknown function. The 1937-bp fragment was divided into three subfragments. The 783-bp subfragment, which covers the gp08 and gp09 gene promoters, was best recognized by SvtR. Because the gp09 promoter region was also included in the 860-bp fragment that did not respond specifically in the assay, we concluded that the gp08 promoter region was involved in the specific binding of SvtR.
Minimal Target Identification within the gp30 Promoter Region
The strongest binding of SvtR was observed with the fragment containing the gp30 gene promoter. This could indicate that this region contained a binding site that could be the main target of the protein in vivo. We thus decided to better define the region recognized by SvtR in this fragment. Because the AG-EMSA approach did not allow the analysis of fragments smaller than 250 bp, we switched to classic PAGE-EMSA experiments with radioactively labeled candidate DNA fragments.
The 342-bp fragment identified as the best target was digested with the AluI restriction enzyme generating three sub fragments, 150, 104, and 88 bp long. When the mobility of these fragments was analyzed in PAGE-EMSA experiments, none of them was shifted, whereas the 342-bp initial fragment was clearly retarded by the SvtR protein (Fig. 4A). This observation suggested that the digestion by AluI inactivates the SvtR binding site. One of the two AluI sites is situated too far from the TATA box of the gp30 promoter to be taken in consideration while the second one is situated only 103 bp upstream of the gp30 transcription initiation site, and is thus well located to include the SvtR recognition site. Interestingly, this restriction site is part of an inverted repeat very similar to that found in the already described 36m fragment specifically recognized by the SvtR protein (Fig. 4B).
To test whether the SvtR binding site was located in the inverted repeat region containing the AluI site, we used a 39-bp double-stranded oligonucleotide covering this region (Fig. 5) This allowed us to compare the affinity of SvtR to two different short targets, 36m (Pgp08) and 39m (Pgp30) (Fig. 5).
The binding was considered to be specific for both oligonucleotides, because it was observed in the presence of a strong excess of unspecific DNA (dI,dC polymer). Twice less of the SvtR protein was required to shift the 39m fragment migration compared with that of 36m. A longer fragment of 70 bp (70m) centered at the middle of the 39m target was slightly better recognized by SvtR. There was effectively no difference in the retardation efficiency between the 70m fragment and a 120-bp fragment (120m) that covers the gp30 promoter region upstream of the transcription +1 point (not shown; the relative positions of the fragments from the gp30 promoter region are displayed in supplemental Fig. S5).
Interaction of SvtR with Short DNA Oligomers Followed by Analytical Ultracentrifugation and Fluorescence
We performed velocity experiments with the 39m and 70m oligonucleotides from the gp30 promoter region and with the 36m from the SvtR gene promoter to determine the number of species involved in the interaction and the stoichiometry of the complexes for each of the oligonucleotides. The molecular weight c(M) profiles of the DNA samples without protein showed a single peak with a molecular mass in agreement with double-stranded DNA (supplemental Fig. S6): 36m: 22.4 ± 0.8 kDa (theoretical mass = 22.1 kDa), 39m: 23.7 ± 0.7 kDa (24.0 kDa), and 70m: 44.2 ± 1.6 kDa (43.1 kDa). In the presence of protein, the three samples showed a minor peak with a molecular mass corresponding to free double-stranded DNA and major peaks with higher molecular mass arising from protein-DNA complexes: 36m.SvtR: 56.9 ± 3.0 kDa, 39m.SvtR: 52.0 ± 4.1 kDa and 68.1 ± 4.5 kDa, and 70m.SvtR: 92.3 ± 6.1 kDa. The molecular mass of these peaks was in agreement with two SvtR dimers bound per DNA for the 36m and the 39m oligonucleotides (theoretical mass = 48.6 and 50.5 kDa, respectively), as well as with four protein dimers per DNA for a second, less populated peak, detected for the 39m oligonucleotide (68.1 ± 4.5 kDa, theoretical mass = 76.9 kDa). Finally, the data revealed that the 70m oligomer could bind four dimers per DNA (theoretical mass = 96.1 kDa).
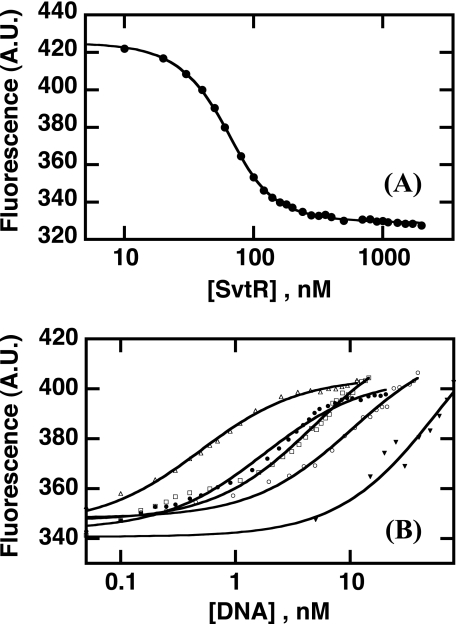
To establish the relative binding strengths of the different oligonucleotides issued from promoters Pgp30 (39m, 70m, and 120m) and Pgp08 (36m), we studied their binding to SvtR by fluorescence. We also characterized the binding of SvtR to nonspecific DNA, namely a 36-bp poly[dIG,dC] oligonucleotide. We first studied the binding of SvtR to the fluorescein labeled 36m oligonucleotide (flDNA) and determined the dissociation constant of the complex as explained under “Experimental Procedures.” As shown in Fig. 6A, binding of SvtR quenched the fluorescence of the fluorescein-labeled DNA. To fit the experimental data, we considered a simple binding mechanism of protein pairs of dimers binding to the fluorescein-labeled 36m, and took into account the monomer-dimer equilibrium of SvtR. The fluorescence data were well described by this model of protein binding non-cooperatively to flDNA and yielded a dissociation constant of the flDNA-SvtR pair of dimers complex of 1.6 ± 0.2 nm. The apparent half-dose effect (∼25 nm protein expressed as dimer concentration or 50 nm total protein) was of course higher due to the relatively high dissociation constant of the monomer-dimer equilibrium.
FIGURE 6.
Interaction of SvtR with oligonucleotides 36m, 39m, 70m, 120m, and nonspecific DNA (poly[dIG,dC]) followed by fluorescence. A, binding of SvtR to fluorescein-labeled 36m oligonucleotide (flDNA). Fluorescence intensity (arbitrary units) as a function of total protein concentration. The solid line corresponds to the fit of the data (Equation 1 in the supplemental material) The dissociation constant (KD,fl) of the SvtR pair of dimers-flDNA complex obtained by non-linear least squares fitting was 1.6 ± 0.2 nm. B, competition of unlabeled DNA with fluorescein-labeled flDNA. Unlabeled oligonucleotides were added to 5 nm flDNA-100 nm protein samples. Poly[dIG,dC] was used as a nonspecific competitor in an independent assay. Solid lines correspond to fits of the data to Equation 5 in the supplemental material. The IC50,u values in nm units were, in decreasing affinity order 0.3 ± 0.2 (120m, ▵), 0.7 ± 0.2 (70m, ●), 1.3 ± 0.1 (39m, □), 1.6 ± 0.3 (36m, ○), and 15.2 ± 1.7 (poly[dIG,dC], ▾).
To compare the binding of different DNA oligomers to SvtR, we performed displacement experiments in which we added increasing amounts of unlabeled DNA competitor to an flDNA-SvtR sample (Fig. 6B). The unlabeled DNA-SvtR dimer IC50,u values computed from the fit of the data (see “Experimental Procedures” and supplemental material for details) revealed that SvtR dimers bound all the tested DNAs, including poly[dIG,dC] DNA, with high affinity (∼nanomolar IC50,u values or lower) but with different strength. The protein SvtR recognized with higher affinity the oligonucleotides from the Pgp30 promoter than the 36m DNA issued from the Pgp08 promoter, which was better recognized than the nonspecific poly[dIG,dC] DNA.
Mapping the DNA Interaction Surface of SvtR by NMR
To monitor the interaction of SvtR with DNA from its promoter region, we used 15N-labeled protein and unlabeled DNA. 1H-15N HSQC spectra were recorded to follow the protein. We first studied the interaction of SvtR with the 36m oligomer (mixture of one SvtR dimer per DNA). Compared with the spectrum of the protein alone, only a handful of signals could be detected in the spectrum of the 1:1 mixture (excess DNA relative to the expected stoichiometry of two dimers per DNA). Here, “isolated” stands for the protein in the absence of DNA and “free” for the free protein in the presence of DNA. None of these signals corresponded to the isolated-protein chemical shifts of residues in the structured region of SvtR (13–56). Given that the line width depends on the rate of molecular tumbling, this observation suggested that the protein was bound to the DNA and formed a high molecular weight complex with a slow rotational correlation rate that resulted in very broad, and hence invisible, signals. Except for two relatively narrow signals and some very broad signals, the observed signals in the spectrum of the SvtR:36m mixture corresponded to the N-terminal unfolded region of the protein without significant changes in chemical shifts (residues 3–10 and 12). This clearly indicated that most of the N-terminal region remained flexible in the complex and, hence, that the residues mentioned above were not involved in the interaction with the 36m oligomer.
Because the sensitivity of NMR is highly dependent on molecular size, we aimed at reducing the size of the protein-DNA complex. We thus studied a 17-bp oligomer (17m) issued from the 36-bp oligomer. The 17m DNA contained a short imperfect inverted repeat. Fluorescence experiments with fluorescein-labeled DNA indicated that the labeled 17m and 36m DNA molecules bound SvtR with similar binding isotherms (not shown). The imino region (12–14.5 ppm) of 1H one-dimensional spectra was used to follow the DNA (no protein signals resonate in this region of the spectrum), and, as for the 36m, two-dimensional 1H-15N HSQC served to monitor the protein (Fig. 7). The HSQC spectrum of the protein at a 1:1 ratio showed the same signals than the SvtR:36m mixture, indicating again that the N-terminal residues mentioned above were not involved in the complex, and that complex formation caused severe line broadening of signals of the structured region of SvtR. The DNA imino signal envelope also became broader. In addition to the broad signals at the frequencies corresponding to the free DNA signals, some broad new signals, indicative of complex formation, were apparent. At a ratio of 2 dimers:1 DNA, which corresponded to the stoichiometry observed by fluorescence experiments, the spectrum of the protein was effectively identical to that at a 1 dimer:1 DNA ratio (not shown). The DNA imino signals, however, completely disappeared from the spectrum. This severe line broadening could be due to the formation of a high molecular weight complex and/or to conformational exchange on the microsecond to millisecond time scale. Line broadening was also observed in the rest of the one-dimensional spectrum, and in particular in the region close to 6 ppm, where the ribose anomeric protons resonate. When the protein was added in excess (3:1), together with the broad signals observed for bound SvtR, the HSQC spectrum of the protein showed all the signals corresponding to the chemical shifts of the free protein, without any significant difference. No shift of the signals was observed when increasing the protein:DNA ratio up to six dimers per DNA. Thus, the free and bound proteins seemed to be in slow exchange on the chemical shift time scale. However, the free-protein signals were very broad compared with that of the isolated protein, and their relative intensities did differ. To understand the different dynamical behavior of the free protein in the presence or absence of DNA, we determined the 15N transverse (T2) and longitudinal (T1) relaxation times of both states under the same conditions (25 °C, 600-MHz proton field-strength, 100 mm KCl). Although the means of the T1 values of residues 14–56 were rather similar (free: 0.73 ± 0.05 s, isolated: 0.67 ± 0.03 s) the T2 values, which are influenced by conformational exchange processes on the microsecond to millisecond time scale, were shorter by factor of 2.2 for the free protein (free: 0.039 ± 0.008 s, isolated: 0.087 ± 0.004 s). Hence, broadening of free SvtR signals was due to the exchange between different conformations, presumably between free and invisible bound states.
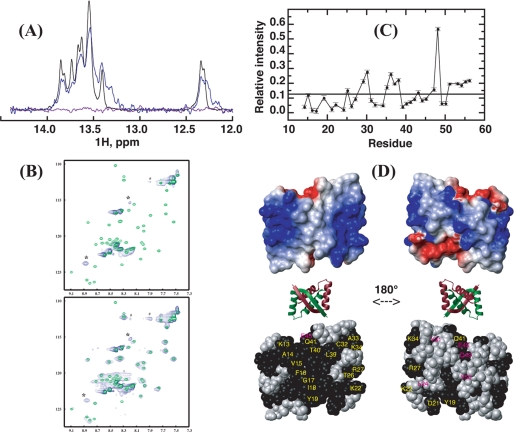
FIGURE 7.
Interaction of SvtR and a 17-bp oligomer (17m) derived from the promoter region of the gp08 gene followed by NMR. Spectra were recorded at 25 °C with buffer D. A, imino region of the one-dimensional 1H spectrum of 17m DNA alone (black), and in the presence of SvtR dimer at a molar ratio of 1:1 (blue) or 1:2 (violet). B, superpositions of the 1H-15N HSQC spectra of SvtR dimer alone (green) and in the presence of the 17m DNA (blue). The molar ratio of protein dimer:DNA was 1:1 (top spectrum) or 6:1 (bottom). *, resonances that do not match the frequencies of isolated SvtR are indicated; #, noise ridge spurious signal; +, folded arginine side-chain signal, their 15N positions in the spectra are different because isolated protein and SvtR:17m spectra were acquired with different spectral widths; C, relative intensity (RI) of the 1H-15N HSQC signals of free SvtR signals in the presence of 17m (3 dimers per DNA molar ratio) and of isolated SvtR as a function of residue number. A threshold value was arbitrarily drawn at 0.12 RI. RI values are given in arbitrary units. Signals of residues 3–12, which showed high apparent RI values (1.7–7.2) are not displayed, because their intensities in the protein-DNA mixture were due to both free and bound protein. In D: top, surface electrostatic potential of SvtR best structure (blue = positive charge, red = negative charge); middle, ribbon diagram of the structure of SvtR to show the orientation of the surface representations; bottom, CPK representation of SvtR best structure. All the atoms of the residues with amide relative intensities lower than 0.12 are shown in black. Labels are shown only for one monomer in yellow for residues on the β-sheet face or in magenta for residues that are not exposed on the β-sheet face.
We calculated the ratio of intensities (RIs) of the signals of the free (3 SvtR dimers per 17m) and isolated protein signals to map the interaction site on SvtR, under the rationale that the residues that were most affected by the exchange process should be involved (or close to residues involved) in protein-DNA (or protein-protein) interactions within the complex. It should be mentioned that this assumption neglects possible rearrangements of the structure of the protein bound to DNA. The data are summarized in Fig. 7C for residues 13–56. Clearly, the exchange process influenced the amide signals differently. The most affected amide groups showed broader lines and thereby lower intensity signals in the free state and concomitant lower RI values. Taking an arbitrary cut-off value for visualization purposes, we plotted the RI values on a CPK representation of the protein structure (Fig. 7D). A residue was colored according to its amide group RI value. Strikingly, most of the residues with lower RI amide values (black spheres) were located on the β-sheet face. These were residues 13–19 on the β-sheet, Lys22, Thr26, Arg27, Cys32, Ala33, and Lys34 on α-helix H1 and Leu39, Thr40, and Gln41 on α-helix H2. Only six residues with low amide RI values were not exposed on the β-sheet face of the protein. Four of those (Leu24, Glu45, Gln49, and Tyr50) were located on the opposite face of the molecule, Tyr31 was buried in the core of the protein and the side chain of Asp21, the C-terminal residue of the β-sheet, was at the border between both faces, and its amide group was buried. In summary, even though the nature of the conformational exchange displayed by SvtR in the presence of DNA cannot be determined, the data presented here indicated that the β-sheet side of the molecule was implicated almost exclusively. Interestingly, this face of the SvtR molecule shows a highly positive electrostatic potential (Fig. 7D) that may be important to attract the protein to the negatively charged DNA polyanion and to stabilize the protein-DNA complex. In this sense, it is noteworthy that four residues that show low RI values on this face are positively charged (Lys13, Lys22, Arg27, and Lys34) and could be directly involved in the interaction with the negatively charged DNA.
Protein SvtR Is a Transcriptional Repressor
The structure, the DNA-binding capability, the specificity of binding to certain gene promoter regions, and the interface used by SvtR to interact with DNA showed that the protein was indeed an RHH protein with a similar DNA-binding mechanism to that of bacterial RHH proteins. As this structural protein superfamily includes many characterized transcriptional regulators, we tested SvtR for this property. Its biological role was thus studied using the Sulfolobus in vitro transcription system as described under “Experimental Procedures.” The activity of SvtR was tested with two identified targets, promoters Pgp08 and Pgp30. The in vitro transcription reactions were performed varying the quantity of SvtR in the presence of limiting amounts of TBP and TFB as described previously (13). In these conditions, transcription from the gp30 promoter was strongly repressed (Fig. 8). The SvtR protein also repressed transcription from promoter Pgp08, but not as strongly as for Pgp30. In contrast, SvtR didn't show any effect on the transcription efficiency of the heterologous promoter PT6 from the SSV1 (18) virus used as a control.
These results clearly indicated that the RHH protein coded by the gp08 gene of virus SIRV1 was a transcriptional repressor. This repressor activity motivated the name SvtR (Sulfolobus viral transcriptional regulator) that we propose for the protein.
DISCUSSION
The genes coding for putative DNA-binding proteins bearing the RHH motif are well represented in archaeal genomes and in the genomes of the viruses that infect them. In bacteria, many representatives of this structural superfamily, such as CopG, MetJ, NikR, and others, are well characterized structurally and functionally (62). They code for transcriptional regulators (mainly repressors) involved in the regulation of many distinct biological processes, such as control of plasmid copy number and partitioning, conjugation, amino acid biosynthesis, metal uptake, antitoxin activity, and motility. The main characteristics of the interaction of the RHH proteins with their cognate DNAs were initially unraveled by the structure of the complex of MetJ from E. coli with its operator DNA (63) and have been extended by the structural and functional studies of several RHH proteins (see Ref. 62 for a recent review). In addition to their common fold and despite their generally low sequence similarity, the RHH proteins share several features in their mode of binding to their targets: these proteins bind to DNA in a sequence-specific manner by inserting the antiparallel β-sheet in the major groove of the DNA and making specific contacts between the side chains of the residues in the β-sheet and the DNA bases; the N terminus of the second α-helix of the RHH motif, makes nonspecific helix-dipole-DNA backbone phosphate interactions; these proteins recognize repeat sequences in DNA, which are most often inverted repeats but can also be direct repeats; the RHH proteins exist usually as dimers in solution and bind to DNA as dimers that recognize two or more repeats and oligomerize into higher order oligomers; finally, in those cases in which the structures of the free and bound proteins are available, the structures of the bound and free dimer units show only minor rearrangements.
The presumed RHH proteins from the Archaea, despite their abundance in this domain of life, have not been thoroughly characterized. The best studied protein of this family in Archaea is probably the protein ORF56, coded by the S. islandicus plasmid pRN1 (61, 64–66). This protein, which contains a predicted RHH fold, is involved in the regulation of the number of copies of the PRN1 plasmid. The protein exists as a dimer in solution and binds to its cognate DNA as tetramers without any apparent cooperativity.
In this study, we performed a detailed analysis of the protein gp08 coded by the S. islandicus virus SIRV1, now called SvtR. Identified in the virus genome by in silico analysis, the protein was predicted to present a RHH motif, suggesting that it could be an important transcriptional regulator of viral functions. The protein gene was cloned and expressed in E. coli. Using a gene candidate approach we showed that, on the one hand, SvtR was indeed a DNA-binding protein, and, on the other hand, it was able to bind to a 36-bp nucleotide (36m) issued from its own gene promoter in the presence of excess of competing nonspecific DNA.
We solved the solution structure of SvtR by NMR and found that the protein contained a folded domain between residues 13–56 and an unfolded N terminus (residues 1–12). The folded region of the protein displayed a RHH structure that was remarkably similar to that of other RHH proteins and in particular to the structure (CA root mean square deviation of 1.2 Å over 44 residues) of the distantly related (16% sequence identity) bacterial plasmid copy regulator CopG (52).
We monitored by NMR the interaction of SvtR with two oligonucleotides (17m and 36m) corresponding to the region of the promoter of its own gene. When the protein was in excess relative to the 17m DNA (≥ 3 dimers per DNA), the HSQC spectra showed broad signals corresponding to the free protein. The signal broadening was due to a conformational exchange phenomenon on the microsecond to millisecond time scale, which was mandatory between free and bound forms of the protein because the experiments in the absence or presence of DNA were performed under the same experimental conditions. We defined the RI ratio as a measure of the broadening of each residue backbone amide signal and used this ratio to map the region of interaction of SvtR with DNA (and most likely with bound protein) on the structure of the free protein. We compared the residues highlighted by the RI analysis with the residues that contact DNA or/and make dimer-dimer contacts in the available crystal structures of RHH proteins in complex with DNA, i.e. CopG, Arc, Omega, FitA, MetJ, and NikR (52, 53, 58, 59, 67, 68), we identified the DNA contacting or dimer-dimer contacting residues and color-coded the structurally equivalent residues of SvtR accordingly (supplemental Fig. S7). Although the details differed from one protein to the other, analysis of these data suggested that many of the low RI residues were located at similar positions than those implicated in protein-DNA interactions for other RHH domains, mainly on the β-sheet side of the corresponding structures. Interestingly, the N terminus of helix H2 of SvtR (residues Leu39 and Thr40), which in the complexes of other RHH proteins forms a helix-dipole-DNA phosphate backbone interaction, was also significantly affected by the exchange process. Finally, the stoichiometry of binding of SvtR to the 36m, 39m, and 70m oligonucleotides was in agreement with SvtR binding to DNA as pairs of dimers (two or four dimers per DNA). Although this observation strongly suggests that SvtR may bind to DNA as dimers of dimers like other RHH proteins, we cannot rule out the possibility that each dimer may bind to DNA independently of each other. Nevertheless, some of the residues that showed low RI values could eventually be implicated in dimer-dimer interactions in the complex with DNA. Unfortunately, we cannot identify these residues, in part because the RHH proteins with known protein-DNA complex structures show different dimer-dimer interfaces and similar, but not identical, protein-DNA interfaces. To summarize, these data suggested that the mode of binding to DNA of SvtR, a protein from a crenarchaeal virus, was similar to that of bacterial/bacteriophage RHH domains; SvtR interacts with DNA through its β-sheet face, and most likely, via insertion of its β-strands on the major groove of the DNA and making dimers of dimers.
The specificity of RHH proteins for DNA is coded in part by the exposed side chains of the residues in the β-sheet, which establish specific interactions with DNA bases (62). These proteins typically show hydrophilic residues at positions 3, 5, and 7 of the β-sheet (numbering relative to the SvtR β-sheet starting at residue Lys13) that contact DNA nucleotide bases, although some proteins present an exposed hydrophobic residue at any of these positions (supplemental Fig. S4). Usually, a basic residue (Lys or Arg) is found either at position 3 or 7. The latter makes several DNA contacts and is important for the specificity. In the case of SvtR, positions 3, 5, or 7 are occupied by a valine (Val15), a glycine (Gly17), and a tyrosine (Tyr19), three residues that are seldom found in known RHH proteins. The exposed valine side-chain methyls of SvtR could in principle make specific van der Waals contacts with DNA thymine methyls. The glycine, which has no side chain, is at the middle of the β-sheet at a position that is often occupied by a serine or a threonine that establish hydrogen bonds with the DNA. Glycines are also found in the PutA family of RHH proteins at this position. Finally, the exposed side chain of Tyr19, which is also present in the predicted RHH proteins of some crenarchaeal viruses, could potentially hydrogen bond or stack with DNA bases.
Sedimentation-equilibrium experiments ran at 25 °C indicated that SvtR showed a monomer-dimer equilibrium with a relatively high equilibrium dissociation constant. Interestingly, the dimer stability was adversely affected by the ionic strength of the solution. Indeed, in analytical ultracentrifugation experiments, SvtR was unstable in 200 mm KCl at low concentrations (0.5–50 μm, data not shown), whereas at 100 mm KCl the protein was stable at these concentrations and showed a KD of dimerization of 1.0 ± 0.4 μm. This observation pointed out the importance of the network of intermolecular salt bridges (intermolecular: Glu45 ↔ Arg54 ↔ Glu49 ↔ Lys53, and Lys48 ↔ Asp23, intra-monomer: Asp23 ↔ Arg27) displayed by the structure of SvtR in stabilizing the dimer. The protein SvtR must accomplish its biological function at temperatures above 80 °C, and we cannot extrapolate the dissociation constant obtained at 25 °C to these temperatures, nor assess quantitatively the effect of molecular crowding in vivo on the monomer and dimer populations. The same limitations apply to the apparent affinities (IC50,u) of SvtR to its cognate DNAs. However, we speculate that the network of inter-monomer salt bridges (69, 70), together with the compact network of intermolecular hydrophobic interactions might be important in stabilizing the dimer at higher temperatures. If this were not the case, the monomer-dimer equilibrium could play a role as a switch between inactive (monomer) and active (dimer) conformations. Nevertheless, given the high affinity of the dimer for its target DNAs, but also for DNA in general, the high concentration of DNA within the host cells will favor the dimer.
It is well documented in the literature that usually the RHH transcriptional regulators control the expression of their own genes. Using a gene candidate approach, the binding of SvtR to its own promoter was demonstrated, and a short 36-bp target was identified. Taking advantage of the fact that the SIRV1 virus is one of the best studied crenarchaeal virus with well established genetic and transcriptional maps, a global target search system was established and used to find all the SvtR targets on the SIRV1 genome. We focused on the most efficiently recognized target, which was situated in the promoter of the gp30 gene, and we identified a minimal fragment of 39 bp specifically recognized by SvtR. An imperfect inverted repeat is present in this sequence and its integrity is absolutely essential for the binding of SvtR, because the binding is abolished after AluI digestion of this site. The two identified fragments, 36m and 39m, share some regions, especially those included in the inverted repeats (Fig. 4). Longer DNA fragments that included the 39-bp sequence bound SvtR with ∼3 (70m)- or 5 (120m)-fold higher apparent affinities (IC50,u) in competition experiments, most probably due to the capacity of this longer fragments to bind more protein dimers per molecule and/or to possible cooperative effects.
It should be mentioned that SvtR showed a rather low difference in apparent affinities to specific and nonspecific DNA (e.g. ∼10-fold between 39m and poly[dIG,dC]) in both fluorescence competition assays and in AG-EMSA experiments. Although usually transcription regulators display affinities >1000-fold higher for specific DNA than for nonspecific DNA, some transcription regulators such as Lrp show discrimination factors that can be of the order of 20-fold (71) as observed in this work for SvtR.
To understand the biological role of SvtR we undertook its functional analysis by following its effect on the in vitro transcription activity from two SIRV1 promoters: Pgp08 and Pgp30. The results of these assays indicated that the protein SvtR acts as a transcriptional repressor.
The negative control of these promoters could probably be due to the competition of SvtR with the transcription initiation factors, because the inhibitory effect could only be observed when these factors were present in limiting amounts. The position of the 36m fragment on the gp08 promoter fits well with this hypothesis. As for the gp30 promoter, the identified fragment of 39 bp starts 83 nucleotides upstream of the TATA box and thus cannot overlap it. It is known that many of the RHH proteins described in bacteria regulate their targets by cooperatively occupying adjacent DNA operator sub sites (62). An eventual formation of higher order oligomers of SvtR could explain its negative effect on the distantly situated gp30 promoter. That the 70m fragment is bound by four dimers as shown by the ultracentrifugation experiments lends support to this hypothesis. In this respect, it should be mentioned that the present data do not allow us to state on the cooperativity of binding of SvtR to the fragments of the gp30 promoter.
The AG-EMSA experiments, the comparison of the relative apparent affinities of SvtR to different DNA fragments containing a binding site for this regulator, as well as the IVT assays indicated that the gp30 promoter is the preferential target of SvtR. The gp30 gene codes for a structural protein involved in the formation of the three tail fibers of the virion (72). The protein that this gene codes for should only be required during the later stages of the virus development, which fits well with the presence of a strong negative transcriptional regulation by SvtR. This protein also regulates its own synthesis, repressing the promoter of the gp08 gene. This strategy allows the cell to avoid overproducing the repressor protein and to keep its functional concentration.
The transcriptional regulator targets in the Archaea are usually identified by the gene candidate approach (15) leaving other potential targets uncharacterized. This limitation slows down the complete characterization and full understanding of the functional role and biological importance of the regulators and the regulation pathways. In the present report, for the first time, a RHH regulator SvtR from an archaeal virus was characterized in details, structurally and functionally, and all its targets in the genome of SIRV1 were identified. The results presented in this report will help to better understand the regulation of the archaeal virus cycle, an interesting but poorly explored domain of archaeal virology.
Supplementary Material
Acknowledgments
We thank Thierry Rose for initial sedimentation/diffusion equilibrium experiments of SvtR, Georg Lipps for initial discussions on protein function, and the Plate-forme de Biophysique of the Institut Pasteur for providing access to the fluorometer.
The work was supported in part by the Institut Pasteur and the CNRS (CNRS URA 2185). The 600-MHz spectrometer was funded by the Région Ile de France and the Institut Pasteur.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
The atomic coordinates and structure factors (code 2KEL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
B. Bardiaux, T. Malliavin, and M. Nilges, personal communication.
- RHH
- ribbon-helix-helix
- AG
- agarose gel
- EMSA
- electromobility shift assay
- IR
- inverted repeat
- IVT
- in vitro transcription assay
- NOE
- nuclear Overhauser effect
- TBP
- Sulfolobus Tata-Box binding protein
- TFB
- Sulfolobus transcription factor B
- NOESY
- nuclear Overhauser effect spectroscopy
- TOCSY
- total correlation spectroscopy
- HSQC
- heteronuclear single quantum coherence
- flDNA
- fluorescein-labeled 36m oligonucleotide
- ITR
- internal terminal repeat.
REFERENCES
- 1.Torsvik T., Dundas I. D. (1974) Nature 248, 680–681 [DOI] [PubMed] [Google Scholar]
- 2.Prangishvili D., Stedman K., Zillig W. (2001) Trends Microbiol. 9, 39–43 [DOI] [PubMed] [Google Scholar]
- 3.Prangishvili D., Garrett R. A. (2004) Biochem. Soc. Trans. 32, 204–208 [DOI] [PubMed] [Google Scholar]
- 4.Prangishvili D., Forterre P., Garrett R. A. (2006) Nat. Rev. Microbiol. 4, 837–848 [DOI] [PubMed] [Google Scholar]
- 5.Prangishvili D., Garrett R. A., Koonin E. V. (2006) Virus Res. 117, 52–67 [DOI] [PubMed] [Google Scholar]
- 6.Blum H., Zillig W., Mallok S., Domdey H., Prangishvili D. (2001) Virology 281, 6–9 [DOI] [PubMed] [Google Scholar]
- 7.Peng X., Blum H., She Q., Mallok S., Brügger K., Garrett R. A., Zillig W., Prangishvili D. (2001) Virology 291, 226–234 [DOI] [PubMed] [Google Scholar]
- 8.Vestergaard G., Shah S. A., Bize A., Reitberger W., Reuter M., Phan H., Briegel A., Rachel R., Garrett R. A., Prangishvili D. (2008) J. Bacteriol. 190, 6837–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vestergaard G., Häring M., Peng X., Rachel R., Garrett R. A., Prangishvili D. (2005) Virology 336, 83–92 [DOI] [PubMed] [Google Scholar]
- 10.Lawrence C. M., Menon S., Eilers B. J., Bothner B., Khayat R., Douglas T., Young M. J. (2009) J. Biol. Chem. 284, 12599–12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessler A., Brinkman A. B., van der Oost J., Prangishvili D. (2004) J. Bacteriol. 186, 7745–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng X., Kessler A., Phan H., Garrett R. A., Prangishvili D. (2004) Mol. Microbiol. 54, 366–375 [DOI] [PubMed] [Google Scholar]
- 13.Kessler A., Sezonov G., Guijarro J. I., Desnoues N., Rose T., Delepierre M., Bell S. D., Prangishvili D. (2006) Nucleic Acids Res. 34, 4837–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata A., Klein B. J., Murakami K. S. (2008) Nature 451, 851–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell S. D. (2005) Trends Microbiol. 13, 262–265 [DOI] [PubMed] [Google Scholar]
- 16.Arnold H. P., Zillig W., Ziese U., Holz I., Crosby M., Utterback T., Weidmann J. F., Kristjanson J. K., Klenk H. P., Nelson K. E., Fraser C. M. (2000) Virology 267, 252–266 [DOI] [PubMed] [Google Scholar]
- 17.Abella M., Rodríguez S., Paytubi S., Campoy S., White M. F., Barbé J. (2007) Nucleic Acids Res. 35, 6788–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qureshi S. A., Bell S. D., Jackson S. P. (1997) EMBO J. 16, 2927–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 20.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 21.Griesinger C., Otting G., Wüthrich K., Ernst R. R. (1988) J. Am. Chem. Soc. 110, 7870–7872 [Google Scholar]
- 22.Marion D., Bax A. (1988) J. Magn. Reson. 80, 528–533 [Google Scholar]
- 23.Kay L. E., Keifer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 24.Muhandiram D. R., Kay L. E. (1994) J. Magn. Reson. 103, 203–216 [Google Scholar]
- 25.Grzesiek S., Anglister J., Bax A. (1993) J. Magn. Reson. 101, 114–119 [Google Scholar]
- 26.Logan T. M., Olejniczak E. T., Xu R. X., Fesik S. W. (1993) J. Biomol. NMR 3, 225–231 [DOI] [PubMed] [Google Scholar]
- 27.Kay L. E., Xu G., Singer A. U., Muhandiram D. R., Forman-Kay J. D. (1993) J. Magn. Reson. 101, 333–337 [Google Scholar]
- 28.Zhang O., Kay L. E., Olivier J. P., Forman-Kay J. D. (1994) J. Biomol. NMR 4, 845–858 [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki T., Forman-Kay J. D., Kay L. E. (1993) J. Am. Chem. Soc. 115, 11054–11055 [Google Scholar]
- 30.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 31.Mandel A. M., Akke M., Palmer A. G., 3rd. (1995) J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 32.Palmer A. G., Rance M., Wright P. E. (1991) J. Am. Chem. Soc. 113, 4371–4380 [Google Scholar]
- 33.Pelta M. D., Barjat H., Morris G. A., Davis A. L. (1998) Magn. Reson. Chem. 36, 706–714 [Google Scholar]
- 34.Jerschow A., Müller N. (1997) J. Magn. Reson. 125, 372–375 [Google Scholar]
- 35.Wilkins D. K., Grimshaw S. B., Receveur V., Dobson C. M., Jones J. A., Smith L. J. (1999) Biochemistry 38, 16424–16431 [DOI] [PubMed] [Google Scholar]
- 36.States D. J., Haberkorn R. A., Ruben D. J. (1982) J. Magn. Reson. 48, 286–292 [Google Scholar]
- 37.Zwahlen C., Legault P., Vincent S. J. F., Greenblatt J., Konrat R., Kay L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 38.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 39.Grzesiek S., Kuboniwa H., Hinck A. P., Bax A. (1995) J. Am. Chem. Soc. 117, 5312–5315 [Google Scholar]
- 40.Nilges M., Macias M. J., O'Donoghue S. I., Oschkinat H. (1997) J. Mol. Biol. 269, 408–422 [DOI] [PubMed] [Google Scholar]
- 41.Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., Nilges M. (2007) Bioinformatics 23, 381–382 [DOI] [PubMed] [Google Scholar]
- 42.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 43.Linge J. P., Habeck M., Rieping M., Nilges M. (2003) Prot. Struct. Funct. Genet. 50, 496–506 [Google Scholar]
- 44.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 45.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graphics 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 47.Demeler B. (2005) in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., Rowe A. J. eds) pp. 210–229, Royal Society of Chemistry, UK [Google Scholar]
- 48.Brown P. H., Schuck P. (2006) Biophys. J. 90, 4651–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berjanskii M. V., Wishart D. S. (2005) J. Am. Chem. Soc. 127, 14970–14971 [DOI] [PubMed] [Google Scholar]
- 50.Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 51.Shindyalov I. N., Bourne P. E. (1998) Protein Eng. 11, 739–747 [DOI] [PubMed] [Google Scholar]
- 52.Gomis-Rüth F. X., Solá M., Acebo P., Párraga A., Guasch A., Eritja R., González A., Espinosa M., del Solar G., Coll M. (1998) EMBO J. 17, 7404–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schildbach J. F., Karzai A. W., Raumann B. E., Sauer R. T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreiter E. R., Sintchak M. D., Guo Y., Chivers P. T., Sauer R. T., Drennan C. L. (2003) Nat. Struct. Biol. 10, 794–799 [DOI] [PubMed] [Google Scholar]
- 55.Golovanov A. P., Barillà D., Golovanova M., Hayes F., Lian L. Y. (2003) Mol. Microbiol. 50, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 56.Murayama K., Orth P., de la Hoz A. B., Alonso J. C., Saenger W. (2001) J. Mol. Biol. 314, 789–796 [DOI] [PubMed] [Google Scholar]
- 57.Popescu A., Karpay A., Israel D. A., Peek R. M., Jr., Krezel A. M. (2005) Proteins 59, 303–311 [DOI] [PubMed] [Google Scholar]
- 58.Garvie C. W., Phillips S. E. (2000) Structure 8, 905–914 [DOI] [PubMed] [Google Scholar]
- 59.Mattison K., Wilbur J. S., So M., Brennan R. G. (2006) J. Biol. Chem. 281, 37942–37951 [DOI] [PubMed] [Google Scholar]
- 60.Larson J. D., Jenkins J. L., Schuermann J. P., Zhou Y., Becker D. F., Tanner J. J. (2006) Protein Sci. 15, 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lipps G., Stegert M., Krauss G. (2001) Nucleic Acids Res. 29, 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schreiter E. R., Drennan C. L. (2007) Nat. Rev. Microbiol. 5, 710–720 [DOI] [PubMed] [Google Scholar]
- 63.Somers W. S., Phillips S. E. (1992) Nature 359, 387–393 [DOI] [PubMed] [Google Scholar]
- 64.Zeeb M., Lipps G., Lilie H., Balbach J. (2004) J. Mol. Biol. 336, 227–240 [DOI] [PubMed] [Google Scholar]
- 65.Lipps G. (2004) Biochem. Soc. Trans. 32, 240–244 [DOI] [PubMed] [Google Scholar]
- 66.Berkner S., Lipps G. (2007) J. Bacteriol. 189, 1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weihofen W. A., Cicek A., Pratto F., Alonso J. C., Saenger W. (2006) Biochem. Soc. Trans. 34, 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schreiter E. R., Wang S. C., Zamble D. B., Drennan C. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13676–13681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elcock A. H. (1998) J. Mol. Biol. 284, 489–502 [DOI] [PubMed] [Google Scholar]
- 70.Robinson-Rechavi M., Alibés A., Godzik A. (2006) J. Mol. Biol. 356, 547–557 [DOI] [PubMed] [Google Scholar]
- 71.Peterson S. N., Dahlquist F. W., Reich N. O. (2007) J. Mol. Biol. 369, 1307–1317 [DOI] [PubMed] [Google Scholar]
- 72.Steinmetz N. F., Bize A., Findlay K. C., Lomonossoff G. P., Manchester M., Evans D. J., Prangishvili D. (2008) Adv. Funct. Materials 18, 3478–3486 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.