Abstract
Human confined placental mosaicism (CPM), where the placental trophoblast is mosaic for a chromosome abnormality but the fetus is chromosomally normal, can cause problems for prenatal diagnosis, but its causes are poorly understood. Tetraploid↔diploid chimeras provide a model for the development of one type of CPM, but animal models for other types of restricted mosaicism are needed. The objective of the present study was to evaluate triploid↔diploid and trisomy-3↔diploid chimeric mouse conceptuses as new models for investigating the development of restricted mosaicism. Novel stocks of mice were generated to produce triploid and trisomy-3 embryos that could be identified by DNA in situ hybridisation to a chromosome 3 transgenic marker. Triploid↔diploid and trisomy-3↔diploid mouse chimeras were produced by embryo aggregation, and the contribution of triploid or trisomy-3 cells was analysed in the fetus and extraembryonic tissues. Only two trisomy-3↔diploid chimeras were analysed but trisomy-3 cells contributed well to all lineages, so these chimeras did not show restricted mosaicism. In contrast, triploid cells usually contributed poorly to all lineages in the ten 3n↔2n chimeras analysed. They contributed more to the primitive endoderm derivatives than other lineages and were present in the primitive endoderm derivatives of all ten chimeras, but excluded from fetuses and trophectoderm derivatives in some cases. This pattern of restricted mosaicism differs from that reported for tetraploid cells in tetraploid↔diploid chimeras, and triploid↔diploid chimeras may provide a useful model for the development of some types of restricted mosaicism in human conceptuses.
Introduction
A high proportion of human preimplantation embryos have chromosomal abnormalities but most are lost either during preimplantation stages or as later spontaneous abortions (Hassold et al. 1980, Hassold & Jacobs 1984). Some survive as mosaics and these include both generalised mosaics, where chromosomally normal and abnormal cells co-exist throughout the conceptus, and restricted (or confined) mosaics, where only some developmental lineages are mosaic. Type-I confined placental mosaicism (CPM) is where the placental trophoblast is mosaic for a chromosome abnormality, but the fetus and other epiblast derivatives are non-mosaic and usually chromosomally normal (Kalousek & Dill 1983, Kalousek 1990, 1994, Wolstenholme et al. 1994, Kalousek & Vekemans 1996, Wolstenholme 1996).
Human CPM usually involves mosaicism for a trisomic (Ts) cell line, but not all trisomies are equally associated with CPM (Wolstenholme 1996) and CPM involving polyploid mosaicism also occurs (Tuerlings et al. 1993, Kalousek 1994, Wolstenholme et al. 1994, Wilkins-Haug et al. 2006). CPM is a clinically significant condition that affects ∼2% of conceptuses. It can produce a false-positive result after prenatal diagnosis by chorionic villus sampling and may be associated with prenatal or perinatal complications (Bennett et al. 1992, Kalousek & Vekemans 1996). There is a need to develop a range of realistic animal models of human CPM and other forms of restricted mosaicism to help understand how different types may arise.
Mouse tetraploid↔diploid (4n↔2n) chimeras provide one useful animal model because tetraploid (4n) cells contribute poorly to derivatives of the fetus and other epiblast derivatives but well to the primitive endoderm and trophectoderm lineages (Tarkowski et al. 1977, 2001, Nagy et al. 1990, James et al. 1995, Goto et al. 2002) (In the mouse, the primitive endoderm tissue produces the endoderm layer of the visceral yolk sac and the parietal endoderm associated with Reichert's membrane. The equivalent lineage in human conceptuses is usually called the hypoblast). The 4n↔2n animal model has been used to investigate the mechanisms responsible for the restricted distribution of chromosomally abnormal cells (James et al. 1995, Everett & West 1996, 1998, Goto & Takagi 1998, Everett et al. 2000, Tang et al. 2000, Goto et al. 2002, Eakin et al. 2005, MacKay & West 2005). However, human 4n/2n CPM is relatively uncommon, so new animal models are required. The aim of this study was to evaluate two other possible models of restricted mosaicism involving, respectively, another class of polyploid cells (triploid) and a class of aneuploid cells (trisomy-3) in mouse chimeras.
The origin of human triploid/diploid mosaics has been discussed by several authors (e.g. Daniel et al. 2003, Golubovsky 2003). Although most die, their fate is variable and may depend on whether triploidy is diandric or digynic, the relative proportions and distributions of triploid and diploid cells, the genetic background and the presence of other chromosomal anomalies. Some human triploid/diploid mosaics show CPM (Sarno et al. 1993, Tuerlings et al. 1993, Daniel et al. 2003) but others survive postnatally as mosaic individuals (van de Laar et al. 2002). The previous reports of mouse triploid↔diploid chimeras showed that 3n cells can contribute to extraembryonic, fetal and adult tissues (Azuma et al. 1991, Suwinska et al. 2005), but it is not yet known whether 3n cells contribute less readily to the fetus (and other epiblast derivatives) than to the primitive endoderm and trophectoderm derivatives.
The ideal model of human CPM would be mosaicism for a mouse trisomy that is equivalent to a human trisomy that is commonly involved in type-I CPM. Spontaneous trisomic/diploid (Ts/2n) mosaicism is very rare in mammals other than humans, so experimental trisomic↔diploid (Ts↔2n) mouse chimeras offer a better chance of developing a realistic model for CPM. Specific trisomic mouse embryos can be produced from mice heterozygous for particular Robertsonian translocations (Gropp et al. 1974, Gropp 1976), but because mouse and human chromosomes are not syntenic, it is difficult to decide which mouse trisomies are likely to be most relevant to the trisomies commonly involved in type-I human CPM.
The production of mouse Ts↔2n chimeras is technically demanding and relatively few have been produced (Magnuson et al. 1982, Cox et al. 1984, Epstein et al. 1984, Fundele et al. 1985, Epstein 1986). In Ts↔2n chimeras studied so far (Ts12, Ts15, Ts16, Ts17 and Ts19), there was little or no selection against Ts cells in the fetus. These previous studies were not intended to model CPM and the extraembryonic tissues were usually ignored or pooled for analysis. For the current study, we investigated the distribution of trisomy-3 cells in mouse Ts3↔2n chimeras. Mouse Ts3 fetuses die at early stage (10–11 days) with hypoplasia and developmental retardation (Gropp et al. 1974, 1983).
For this study, we developed special strains of mice to generate either digynic 3n or Ts3 embryos carrying a transgenic lineage marker on chromosome 3, which is highly reiterated and so readily detectable by DNA in situ hybridisation (Lo 1986, Lo et al. 1987, Katsumata & Lo 1988, Keighren & West 1993, Everett et al. 1994). DNA in situ hybridisation was used to detect chromosome 3, such that two hybridisation signals were produced in interphase nuclei of 3n or Ts3 cells but only one signal was seen in 2n nuclei. We have used this novel approach to assess the contribution of 3n and Ts3 cells to different developmental lineages of postimplantation stage mouse chimeras. Our results suggest that 3n↔2n chimeras may provide a useful new model system for investigating the development of restricted mosaicism.
Results
Frequency and physical characteristics of 3n↔2n chimeras
Three different strain combinations were used to produce 3n↔2n chimeras. The frequency of 3n↔2n chimeras (Table 1) was higher in series TrB and TrC (9/22 chimeras; 40.9%) than in series TrA (2/27 chimeras; 7.4%). Different frequencies were expected because of the nature of the strains used, but the frequency of 3n↔2n chimeras in TrA was lower than that expected. TRIP is an LT/SvKau congenic and so expected to produce about 40% 3n embryos, whereas derived strain PO2 was expected to produce ∼23% 3n embryos (West et al. 1993, Everett et al. 2004). The frequency of non-chimeric conceptuses varied among series and this may also partly reflect differences in strain combination. All were entirely NIH or FVB genotype (embryo 2) and some may have been 3n↔2n chimeras in which 3n cells were depleted to below the detectable level.
Table 1.
The number of chimeric conceptuses recovered from each chimera series analysed (see text for details of mouse strains).
| Number (%) of conceptuses | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Series | Age | Embryo 1 (GPI1-A)↔Embryo 2 (GPI1-B) (female×male)↔(female×male) | Implantations | Empty | Non-chimerica | Unclassified | 2n↔2n | 3n↔2n | Ts3↔2n |
| 3n↔2n chimeras | |||||||||
| TrA | E12.5 | PO2×(BALB/c×A/J)F1↔NIH×NIH | 34 | 0 | 6 (17.6b) | 1c | 25 (92.6d) | 2 (7.4d) | – |
| TrB | E9.5 | TRIP×(BALB/c×A/J)F1↔NIH×NIH | 8 | 1 (12.5) | 1 (8.0) | 0 | 4 (66.7) | 2 (33.3) | – |
| TrC | E9.5 | TRIP×(BALB/c×A/J)F1↔FVB×FVB | 38 | 6 (15.8) | 16 (50.0) | 0 | 9 (56.3) | 7 (43.8) | – |
| Ts3↔2n chimeras | |||||||||
| TS | E9.5 | (BALB/c×A/J)F1×(TGA×RBT)↔FVB×FVB | 51 | 8 (15.7) | 9 (20.9) | 0 | 32 (94.1) | – | 2 (5.9) |
All non-chimeric conceptuses in all four groups were derived from diploid embryo 2 (NIH or FVB).
Percentage of all conceptuses.
Conceptus TrA-6 was chimeric in only the yolk sac mesoderm and was not classified as 2n↔2n or 3n↔2n.
Percentage of chimeric conceptuses (excluding unclassified conceptuses).
Physical parameters of the two E12.5 3n↔2n chimeras are shown in Table 2. Although 3n↔2n chimera TrA-16 had reached the average developmental stage (hind limb morphology index) for its age, it was well below the control average for conceptus weight, fetal weight, placental weight and crown-rump length, and the placental weight was below the minimum for the control group. Triploid↔2n chimera TrA-26 was within the control range for all physical parameters measured.
Table 2.
Physical parameters of E12.5 3n↔2n and 2n↔2n chimeric conceptuses.
| Weight (mg) | |||||
|---|---|---|---|---|---|
| Chimera | Conceptus | Fetus | Placenta | Crown-rump length (mm) | Hind limb morphology indexa |
| 3n↔2n chimeras | |||||
| TrA-16 | 263.2 | 80.3 | 59.1 | 8.3 | 7.5 |
| TrA-26 | 321.6 | 96.4 | 87.4 | 9.3 | 7.0 |
| Mean±s.e.m. | 292.4±29.2 | 88.4±8.1 | 73.3±14.2 | 8.8±0.5 | 7.3±0.3 |
| 2n↔2n chimeras (N=25) | |||||
| Mean±s.e.m. | 338.0±6.4 | 105.3±3.1 | 88.8±1.9 | 9.4±0.1 | 7.3±0.2 |
| Minimum | 261.9 | 61.6 | 71.8 | 8.2 | 5.0 |
| Maximum | 397.3 | 132.4 | 106.1 | 10.2 | 8.0 |
Hind limb morphology index: 7.0=early stage 7; 7.5=late stage 7.
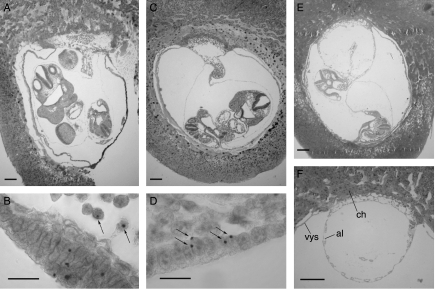
Figure 1 shows examples of sections of E9.5 3n↔2n and 2n↔2n chimeras and the hybridisation signals used to distinguish between chimeras containing Tg/Tg/− 3n cells and those containing Tg/− 2n cells. Abnormalities were seen in two of the E9.5 3n↔2n chimeras. Chimera TrC-26 was an empty gestational sac, without a fetus or amnion, and chimera TrB-3 had an abnormal allantoic vesicle, which had not fused normally with the chorion (Fig. 1F).
Figure 1.
Triploid↔diploid chimeras. Histological sections and DNA in situ hybridisation of (A and B) E9.5 2n↔2n control chimera TrC-21, (C and D) E9.5 3n↔2n chimera TrC-16 and (E and F) E9.5 3n↔2n chimera TrB-3. Arrows show (B) examples of 2n (Tg/−) nuclei with single hybridisation signals and (D) examples of 3n (Tg/Tg/−) nuclei with two hybridisation signals. (F) An abnormal allantoic vesicle in E9.5 3n↔2n chimera TrB-3 (ch, chorion; al, allantoic vesicle; vys, visceral yolk sac). Scale bars=200 μm (A, C, E and F) and 20 μm (B and D).
Contribution of 3n cells to different tissues of 3n↔2n chimeras
The percentages of glucose phosphate isomerase (GPI)1-A in the fetus and different extraembryonic tissues of the E12.5 conceptus are shown in Table 3 for the two 3n↔2n chimeras in series TrA together with their 2n↔2n controls. Overall, the controls showed a balanced chimeric composition with the means for each tissue being ∼50%. Triploid cells made no contribution to the fetus in either of the E12.5 3n↔2n chimeras, but contributed to the primitive endoderm lineage (visceral yolk sac endoderm and parietal endoderm) of both chimeras and also contributed to the extraembryonic epiblast tissues (amnion and visceral yolk sac mesoderm) and the trophectoderm (trophoblast cells overlying Reichert's membrane and placenta) of chimera TrA-16.
Table 3.
Composition (% GPI1-A) of E12.5 3n↔2n and 2n↔2n chimeric conceptuses.
| Epiblast derivatives | Primitive endoderm derivatives | Trophectoderm derivatives | |||||
|---|---|---|---|---|---|---|---|
| Chimera | Fetus | Amnion | YS mes | YS end | P end | Troph (RM) | Placenta |
| 3n↔2n chimeras (uncorrected %GPI1-A)a | |||||||
| TrA-16 | 0.0 | 6.7 | 10.9 | 50.7 | 50.9 | 27.7 | 14.4 |
| TrA-26 | 0.0 | 0.0 | 0.0 | 28.3 | 25.7 | 0.0 | 0.0 |
| Mean±s.e.m. | 0.0±0 | 3.4±3.4 | 5.5±5.5 | 39.5±11.2 | 38.3±12.6 | 13.9±13.9 | 7.2±7.2 |
| 3n↔2n chimeras (corrected %GPI1-A)a,b | |||||||
| TrA-16 | 0.0 | 4.6 | 7.5 | 40.7 | 40.9 | 20.3 | 10.1 |
| TrA-26 | 0.0 | 0.0 | 0.0 | 20.8 | 18.7 | 0.0 | 0.0 |
| Mean±s.e.m. | 0.0±0 | 2.3±2.3 | 3.8±3.8 | 30.8±9.9 | 29.8±11.1 | 10.2±10.2 | 5.0±5.0 |
| 2n↔2n chimeras (N=25)c | |||||||
| Mean±s.e.m. | 47.0±3.6 | 47.1±3.8 | 45.8±3.6 | 56.5±3.3 | 62.2±4.8 | 45.1±7.7 | 42.1±7.1 |
YS mes, visceral yolk sac mesoderm; YS end, visceral yolk sac endoderm; P end, parietal endoderm; Troph (RM), trophoblast overlying Reichert's membrane.
Recipient females were Gpi1c/c, so any maternal contribution (GPI1-C) was excluded from the analysis.
The corrected %GPI1-A=⅔A×100/(⅔A+B), where A=%GPI1-A and B=%GPI1-B. This allows for the expectation that 3n cells would produce 1.5 times the GPI1 activity of 2n cells and probably provides a closer estimate of the percentage 3n cells.
Conceptus TrA-6 was not classified as 3n↔2n or 2n↔2n and is excluded from the table. (It had 15.9% GPI1-A in the yolk sac mesoderm and 0% GPI1-A in the other four tissues analysed.)
Table 4 shows the contribution of 3n cells to the fetus and different extraembryonic tissues of eight E9.5 3n↔2n chimeric conceptuses in series TrB and TrC and five of the E9.5 2n↔2n control chimeras (ordered by contribution to the fetus). 3n↔2n chimera TrC-25 was damaged and so was not analysed and TrC-26 was an incomplete conceptus with no fetus.
Table 4.
Corrected percentage contribution of Tg-positive cells in E9.5 3n↔2n and 2n↔2n chimeras.
| Epiblast derivatives | Primitive endoderm derivatives | Trophectoderm derivatives | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chimera | Fetus | Amnion | YS mes | Allantois | YS end | P end | PT | PTGC | RMGC | Chorion |
| 3n↔2n chimerasa | ||||||||||
| TrC-26 | Absent | Absent | 9.5 | 64.5 | 3.0 | 0.0 | 0.0 | 0.0 | ||
| TrC-11 | 0.0 | 0.0 | 0.0 | 18.8 | 0.8 | 0.0 | 0.0 | 0.0 | ||
| TrC-8 | 0.6 | ND | 0.0 | 11.2 | 13.4 | 0.0 | 0.0 | 0.0 | ||
| TrB-6 | 1.8 | 0.0 | 0.0 | 12.0 | 12.7 | 14.3 | 0.0 | 0.0 | ||
| TrC-16 | 1.8 | 3.4 | 2.5 | 5.8 | 6.7 | 0.3 | 0.0 | 0.0 | ||
| TrC-37 | 10.7 | ND | 6.9 | 0.9 | 1.9 | 17.0 | ND | 3.0 | ||
| TrC-22 | 11.6 | 5.5 | 13.6 | 54.8 | 27.3 | 73.2 | 0.0 | 20.9 | ||
| TrB-3b | 28.2 | 24.1 | 37.0 | 38.0b | 14.9 | 20.2 | ND | ND | 71.2 | 88.0b |
| Mean±s.e.m. | 7.8±3.9 | 6.6±4.5 | 8.7±4.4 | 22.9±8.3 | 10.8±3.3 | 15.0±10.1 | 0.0±0.0 | 11.9±8.8 | ||
| 2n↔2n chimeras | ||||||||||
| TrB-8 | 29.4 | 28.6 | 38.1 | 0.0 | 45.5 | 98.2 | 22.5 | 83.8 | ||
| TrC-5 | 31.0 | 7.5 | 31.9 | 20.6 | 28.7 | 40.0 | 0.0 | 14.4 | ||
| TrC-34 | 38.2 | 42.7 | 33.3 | 85.1 | 78.2 | 16.4 | ND | 30.9 | ||
| TrC-21 | 43.5 | 45.9 | 41.3 | 59.8 | 60.5 | 68.4 | 39.9 | 73.4 | ||
| TrB-9 | 92.0 | 93.4 | 77.1 | 12.8 | 22.2 | 18.0 | 0.0 | 0.0 | ||
| Mean±s.e.m. | 46.8±11.6 | 43.6±14.2 | 44.3±8.4 | 35.7±15.9 | 47.0±10.3 | 48.2±15.7 | 15.6±9.7 | 40.5±16.4 | ||
YS mes, yolk sac mesoderm; YS end, yolk sac endoderm; P end, parietal endoderm; PT, placental trophoblast; PTGC, placental trophoblast giant cells; RMGC, trophoblast giant cells overlying Reichert's membrane; ND, not done.
Triploid↔2n chimera TrC-25 is not included in the table. It was damaged and so not analysed.
Abnormal placental development; an allantoic vesicle was present, which had not fused normally with the chorion. These two tissues were scored separately, but uncorrected values are shown as there were no corresponding tissues in age-matched positive controls.
Overall, the 3n cells made a low contribution to the E9.5 3n↔2n chimeras. Grouping the different tissues into four lineages (excluding the allantois and chorion, which were only scored for TrB-3) showed that 3n cells contributed 7.8±3.9% to the fetus, 7.9±3.1% to the extraembryonic epiblast (amnion plus visceral yolk sac mesoderm), 16.8±4.6% to the primitive endoderm and 9.5±4.7% to the trophectoderm. The contribution of 3n cells varied significantly among these four lineages (P=0.01 by the Kruskal–Wallis test). Although the contribution of 3n cells was lowest in the fetus, Mann–Whitney U-tests revealed no significant differences between the fetus and the extraembryonic epiblast (P=0.75), the primitive endoderm (P=0.09) or the trophectoderm (P=0.09). However, the contribution of 3n cells was significantly lower in the epiblast as a whole (fetus plus extraembryonic epiblast) than the primitive endoderm (P=0.03) but not the trophectoderm lineage (P=0.09).
The contribution of 3n cells to the trophectoderm derivatives was significantly lower than in the primitive endoderm (P=0.002 by Mann–Whitney U-test) but variable. Two of the eight E9.5 3n↔2n chimeras had mean 3n contributions greater than 70% in one of the trophectoderm derivatives sampled but most had low 3n levels, and no 3n cells were detected in any trophectoderm derivatives of three out of the eight chimeras. Chimera TrB-3 had relatively high 3n contributions to all lineages, which might be associated with its apparently abnormal placental development (see above).
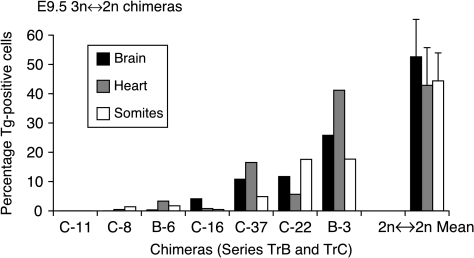
The histograms shown in Fig. 2 indicate that 3n contributions in the fetus did not differ markedly among the three different tissues scored (brain, heart and somites).
Figure 2.
Histogram showing percentage Tg-positive cell contributions to three fetal tissues of seven E9.5 3n↔2n chimeras from series TrB and TrC (identified as B- and C- respectively) and mean contributions (±s.e.m.) for five 2n↔2n control chimeras.
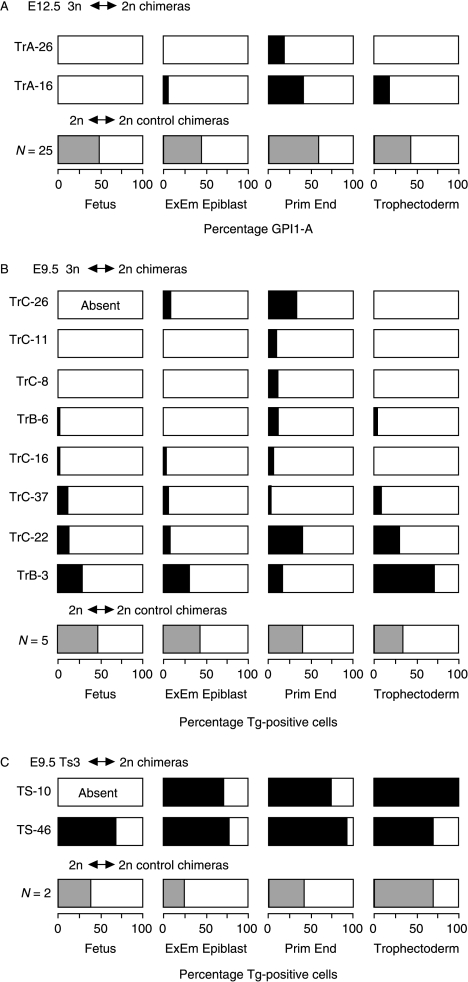
Figure 3A and B summarises the 3n contribution to the fetus and the mean contribution to each of the three extraembryonic lineages for both E12.5 and E9.5 3n↔2n chimeras. It is clear that 3n cells usually contributed poorly to all lineages of these 10 3n↔2n chimeras. Although 3n cells were only excluded completely from three out of the nine E9.5 and E12.5 fetuses in 3n↔2n chimeric conceptuses, only three out of the nine had more than 2% 3n cells in the fetus. This was significantly fewer than that for the combined tissues of the primitive endoderm lineage (9/10; P=0.02) but not significantly different from either the extraembryonic epiblast (4/10; P>0.99) or the trophectoderm lineage (5/10; P=0.65), by Fisher's exact test.
Figure 3.
Diagrammatic summary of the composition of the fetus and three extraembryonic lineages in (A) E12.5 3n↔2n chimeras, (B) E9.5 3n↔2n chimeras and (C) E9.5 Ts3↔2n chimeras. Shaded regions represent the percentage of Tg-positive cells in the different lineages (shading is black for individual 3n↔2n and Ts3↔2n chimeras and grey for means of 2n↔2n control chimeras). The extraembryonic epiblast values are means of amnion and visceral yolk sac mesoderm; primitive endoderm values are means of visceral yolk sac endoderm and parietal endoderm; E12.5 trophectoderm values are means of the placenta (predominantly placental trophoblast) and the trophoblast overlying Reichert's membrane; E9.5 trophectoderm values are means of separate estimates for the placental trophoblast, placental trophoblast giant cells and trophoblast giant cells overlying Reichert's membrane (Tables 3–5). Triploid↔2n chimera TrC-25 is not included because it was damaged and so not analysed. Abbreviation: ExEm, extraembryonic.
There are too few chimeras to evaluate whether 3n cells are depleted between E9.5 and E12.5. Although most 3n↔2n fetuses contained some 3n cells at E9.5 (6/7), none did at E12.5 (0/2), but this difference was not significant by Fisher's exact test (P=0.08). Also, the fetuses of three other E9.5 3n↔2n chimeras contained <2% 3n cells, and this may be below the level detectable by GPI electrophoresis method used to analyse E12.5 chimeras (West & Green 1983). There was less difference in the frequency of fetuses with >2% 3n cells at E9.5 and E12.5 (3/7 vs 0/2; P=0.50).
Both of the E12.5 and one of the E9.5 3n↔2n chimeras showed restricted chimerism where 3n cells were absent from the fetus but present in extraembryonic tissues. In TrA-16, 3n cells were absent from the fetus but present in the extraembryonic epiblast, trophectoderm and primitive endoderm lineages, and in TrA-26 and TrC-11 they were absent from the fetus, extraembryonic epiblast and trophectoderm but present in the hypoblast.
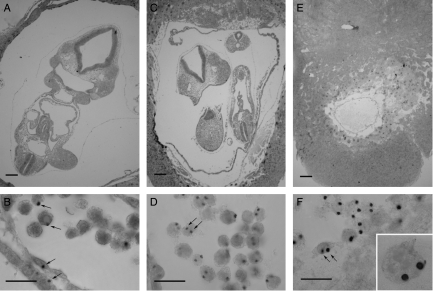
Ts3↔2n chimeras
One series of Ts3↔2n and 2n↔2n control chimeras (TS) were produced and analysed at E9.5 (Table 1). Ts3↔2n chimeras were identified retrospectively by DNA in situ hybridisation to the transgenic sequence (Tg/Tg/− Ts3 cells produce two hybridisation signals per nucleus). Only 2 out of 34 chimeras proved to be Ts3↔diploid (5.9%) but this is close to the expected frequency (see the Discussion section). The composition of the two E9.5 Ts3↔2n chimeras and two of the control 2n↔2n chimeras were analysed by DNA in situ hybridisation (Table 5). Figure 4 shows examples of sections of E9.5 Ts3↔2n and 2n↔2n chimeras and the hybridisation signals used to distinguish between chimeras containing Tg/Tg/− Ts3 cells and those containing Tg/− 2n cells. In contrast to the 3n↔2n chimeras, both Ts3↔2n chimeras had a high Ts3 cell contribution in all lineages (Table 5; Fig. 3C). Ts3↔2n chimera TS-10 was an incomplete conceptus, comprising an empty gestational sac without a fetus or amnion but the trophectoderm lineage and parietal endoderm consisted entirely of Ts3 cells. Ts3↔2n chimera TS-46 had a high contribution of Ts3 cells in all tissues analysed. The overall contribution to the fetus was 74.7% and high levels were seen in all the three fetal tissues scored (76%, 72% and 58% in the brain, heart and somites respectively). The high Ts3 fetal contribution indicated that Ts3↔2n chimeras are unlikely to provide a useful model of CPM.
Table 5.
Corrected percentage contribution of Tg-positive cells in E9.5 Ts3↔2n and 2n↔2n chimeras.
| Epiblast derivatives | Primitive endoderm derivatives | Trophectoderm derivatives | ||||||
|---|---|---|---|---|---|---|---|---|
| Chimera | Fetus | Amnion | YS mes | YS end | P end | PT | PTGC | RMGC |
| Ts3↔2n chimeras | ||||||||
| TS-10 | Absent | Absent | 71.0 | 50.3 | 100a | 100a | 100a | 100a |
| TS-46 | 68.4 | 82.1 | 74.7 | 96.8 | 91.0 | 72.4 | 43.3 | 75.6 |
| 2n↔2n chimeras | ||||||||
| TS-42 | 13.9 | 25.5 | 18.4 | 90.6 | 54.1 | 71.6 | 71.1 | 99.0 |
| TS-6 | 61.8 | 27.8 | 26.8 | 17.4 | 10.8 | 95.1 | 41.6 | 52.1 |
YS mes, yolk sac mesoderm; YS end, yolk sac endoderm; P end, parietal endoderm; PT, placental trophoblast; PTGC, placental trophoblast giant cells; RMGC, trophoblast giant cells on outside of Reichert's membrane.
Corrected values that exceeded 100% are shown as 100%.
Figure 4.
Trisomy-3↔diploid chimeras. Histological sections and DNA in situ hybridisation of (A and B) E9.5 2n↔2n control chimera TS-42, (C and D) E9.5 Ts3↔2n chimera TS-46 and (E and F) E9.5 Ts3↔2n chimera TS-10. Arrows show (B) examples of 2n (Tg/−) nuclei with single hybridisation signals and (D and E) examples of Ts3 (Tg/Tg/−) nuclei with two hybridisation signals. Inset in F shows trophoblast giant cell at the same magnification. Two hybridisation signals are not seen in all sections of Ts3 nuclei. Scale bars=200 μm (A, C and E) and 20 μm (B, D and F).
Discussion
Our new genetic stocks successfully produced trisomy-3 and triploid embryos respectively without the need for experimental manipulations, but yields were low. The reiterated chromosome 3 marker transgene provided a good means of identifying Ts3↔2n and 3n↔2n chimeras and quantifying the Ts3 and 3n contributions in histological sections. The yield of embryos from TRIP strain females was poor and hampered the production of 3n↔2n chimeras. The breeding performance was improved by further genetic crosses but only at the expense of a lower proportion of triploid embryos. Unless higher yields of triploid embryos can be obtained for future experiments (e.g. by selective breeding), it may be worth evaluating the use of cytochalasin B to induce triploidy (Niemierko 1975) in another strain carrying the chromosome 3 transgene.
Only two E9.5 series TS chimeric conceptuses proved to be Ts3↔2n, but this frequency (2/34; 5.9%) is close to that expected if meiotic non-disjunction in Rb1 heterozygotes produces ∼19% aneuploid zygotes (Cattanach & Moseley 1973, Gropp et al. 1974, Everett et al. 1996), comprising about 5% each of trisomy-3, monosomy-3, trisomy-1 and monosomy-1. If monosomic cells contribute infrequently to postimplantation stage chimeras (Magnuson et al. 1982), the expected frequency of Ts3↔2n chimeras is ∼5.6% (five Ts3↔2n chimeras per 90 overt Tg-positive↔−/− chimeras).
Although only one of these two Ts3↔2n chimeric conceptuses had a fetus, this fetus and the extraembryonic tissues of both chimeras had high levels of Ts3 cells, implying that Ts3 cells can survive well in all tissues of chimeric conceptuses. The absence of the fetus in one Ts3↔2n chimera with a very high Ts3 contribution may have been a result of too few diploid cells to correct the abnormal Ts3 phenotype, which is normally lethal around E10–11 days (Gropp et al. 1974, 1983). As Ts3 cells contributed well to the only Ts3↔2n chimeric fetus and to the extraembryonic epiblast lineage in both chimeras, Ts3↔2n chimeras are unlikely to provide a useful model of CPM. This follows the pattern established by earlier studies on other trisomy↔diploid chimeras, involving Ts12, Ts15, Ts16, Ts17 and Ts19, where Ts cells contributed reasonably well to embryos and adults (Magnuson et al. 1982, Cox et al. 1984, Epstein et al. 1984, Fundele et al. 1985, Epstein 1986). None of the trisomy↔diploid chimeras studied so far has been identified as a good model of human CPM.
Some digynic triploid↔diploid chimeras showed morphological abnormalities. E9.5 chimera TrC-26 had no fetus or amnion, but this may not be attributable to the presence of 3n cells as they were not particularly abundant in this chimera. The chimera with the highest contribution of 3n cells (E9.5 chimera TrB-3) had an abnormal allantoic vesicle that was not fused normally with the chorion. This also occurs in some fully digynic triploids (Wróblewska 1971), parthenogenetic↔tetraploid chimeras (Spindle et al. 1996) and several mutants and genetic knockouts (Copp 1995). E12.5 chimera TrA-16 had a small placenta which may reflect poor placental trophoblast growth associated with digynic imprinting abnormalities (Surani et al. 1986).
Unlike Ts3 cells in Ts3↔2n chimeras, digynic triploid cells generally contributed poorly to all lineages of 3n↔2n chimeras. This cannot be attributable to the marker transgene as both Ts3 and 3n cells had an additional copy of chromosome 3 carrying the marker transgene. Only five out of nine (56%) of the 3n↔2n chimeras had any 3n cells in the fetus, and this contribution only exceeded 2% in three out of nine (33%) cases. All of the non-chimeras in the 3n↔2n series were derived from the diploid Gpi1b/b (FVB or NIH strain) embryo, and it seems likely that the 3n cell population was preferentially lost because the 2n↔2n control series showed no such bias in non-chimeric genotype.
Two other studies have demonstrated that 3n cells can contribute to adult tissues of 3n↔2n chimeras. Azuma et al. (1991) identified triploid cells in 24 out of 89 (27%) potential adult 3n↔2n chimeras in two reciprocal series, where digynic triploidy was induced by suppressing extrusion of the second polar body with cytochalasin B. The frequency and genotype of non-chimeras was consistent with loss of 3n cells from the fetal lineage. Suwinska et al. (2005) produced 3n↔2n mouse chimeras by two methods and showed that 3n cells could survive at postimplantation stages and in some adults. The low overall level of 3n contribution estimated by in situ hybridisation in the present study was similar to the cytogenetic results for adult 3n↔2n chimeras reported by Azuma et al. (1991), but rather lower than estimates made by GPI electrophoresis for embryonic, fetal and adult 3n↔2n chimeras reported by Suwinska et al. (2005). Differences in genetic background and methods of production and analysis could each contribute to these differences.
In our 3n↔2n chimeras, 3n cells contributed less to the epiblast (including fetus) and trophectoderm derivatives than to the primitive endoderm derivatives. Triploid cells were present in the primitive endoderm derivatives of all ten 3n↔2n chimeras, but their contribution to the trophectoderm was variable, being absent from three out of ten chimeras but contributing over 70% in one chimera. This variability is consistent with previous analyses of 2n↔2n chimeric placentas (West et al. 1995). Suwinska et al. (2005) also found that 3n cells tended to contribute more to the visceral yolk sac than to the fetus or placenta of 3n↔2n chimeras, but it was unclear whether 3n cells were specifically more abundant in the yolk sac endoderm layer because it was not analysed separately.
In two of our chimeras, 3n cells were absent from the fetus and confined to the primitive endoderm lineage. Although this is not equivalent to CPM, it is not known whether this class of restricted mosaicism occurs spontaneously in humans because the primitive endoderm (hypoblast) lineage is rarely investigated in human conceptuses. While some 3n↔2n chimeras showed some sort of restricted chimerism, 3n cells were not rigorously excluded from the fetus and the extraembryonic epiblast to the extent that tetraploid cells are excluded from these lineages in 4n↔2n chimeras (James et al. 1995). Instead, the low contribution of 3n cells more closely resembles the consistently low contribution of one cell population in genotypically unbalanced 2n↔2n chimeras, where cells of one strain or genotype tend to predominate in all lineages (West & Flockhart 1994). It seems likely that the cases of restricted chimerism, where 3n cells were excluded from the fetus but not from the other tissues, may have arisen partly by chance following a more generalised depletion of 3n cells from all lineages and partly because of a tendency for 3n cells to survive better in the primitive endoderm. It is not yet known how lineage restriction in human CPM occurs, but different mechanisms may apply to different types of restricted mosaicism and different chromosome abnormalities (Wolstenholme 1996). For some types of human restricted mosaicism, 3n↔2n mouse chimeras may provide a more appropriate model than 4n↔2n chimeras.
Materials and Methods
Mice
Four series of chimeras (TrA, TrB, TrC and TS) were made by aggregating embryos from genetically distinct stocks of mice (Table 1). One of each pair of aggregated embryos carried a highly reiterated β-globin transgene (abbreviated to Tg) that is located on chromosome 3 (Lo et al. 1992, Everett et al. 1994) and can be identified readily by DNA in situ hybridisation in histological sections (Lo et al. 1987, Katsumata & Lo 1988, Keighren & West 1993). The pairs of aggregated embryos also differed at the albino locus (Tyr) and Gpi1 (glucose phosphate isomerase). Genetic crosses with specially developed strains of mice were designed to produce some chromosomally abnormal embryos with either a complete extra set of maternal chromosomes (digynic triploid) or an extra paternal copy of chromosome 3 (trisomy-3) carrying the chromosome 3 transgenic marker. All animal work was carried out in accordance with the UK Home Office regulations. Mice were bred and maintained under conventional conditions in the Centre for Reproductive Biology, University of Edinburgh. (For cross-referencing, TrA, TrB, TrC and TS were originally designated XU, C9B, C9H and C9I respectively.) The female is shown first in each genetic cross.
Strains TRIP, PO2, TGA and RBT are all pigmented (Tyr+/+), Gpi1a/a and homozygous for the reiterated β-globin marker transgene, Tg/Tg (Everett et al. 1994, Keighren & West 1994). Inbred strains NIH and FVB are albino (Tyrc/c), Gpi1b/b and Tg negative; (BALB/c × A/J)F1 hybrids are Tyrc/c, Gpi1a/a and Tg negative. Strains TRIP and PO2 were derived from inbred strain LT/SvKau and designed to produce Tg/Tg/− triploid embryos (see below). RBT is homozygous for both the Rb(1.3)1Bnr Robertsonian translocation (abbreviated to Rb1) and the reiterated transgene: Rb1/Rb1, Tg/Tg (Everett et al. 1994). Heterozygous Rb1/+ males undergo meiotic non-disjunction and produce elevated frequencies of sperm with aneuploidy for chromosomes 1 or 3, and so produce some Ts-1 and Ts-3 embryos (Gropp et al. 1974). Hybrid (TGA×RBT)F1 males are Rb1/+, Tg/Tg and their trisomy-3 offspring are Tg/Tg/−.
Production of 3n↔2n chimeras
LT/SvKau females spontaneously ovulate up to 40% of oocytes as diploid primary oocytes which, on fertilisation, produce digynic triploid zygotes (Kaufman & Howlett 1986, Kaufman & Speirs 1987, O'Neill & Kaufman 1987, Speirs & Kaufman 1988). TRIP is an LT/SvKau congenic strain but is homozygous for the chromosome 3 transgenic marker (Tg/Tg) and was developed to generate Tg/Tg/−, digynic 3n embryos for chimera production. However, TRIP females produced a low yield of embryos, so a different strategy was used to produce Tg/Tg/− triploid embryos for the first series of triploid↔diploid chimeras (series TrA). Tg/Tg TRIP males were outcrossed to Tg/Tg TGA females and the Tg/Tg F1 hybrid females (designated PO1: ‘primary oocyte ovulation stock 1’) were backcrossed to Tg/Tg TRIP males to produce Tg/Tg PO2 mice with 75% TRIP (∼75% LT/Kau) genetic background. PO2 females produced a better yield of embryos but a lower proportion were triploid, which proved to be even more of a problem. Thus, Tg/Tg TRIP females were used to produce Tg/Tg/− triploid embryos in series TrB and TrC.
TRIP and PO2 females were superovulated at 3–4 weeks to maximise the yield of digynic, Tg/Tg/− triploid embryos (Speirs & Kaufman 1990) using a standard protocol (West & Flockhart 1994). Other females were superovulated at 6–8 weeks old. Eight-cell stage embryos were collected at E2.5 (vaginal plug date designated E0.5) in M2 handling medium (Quinn et al. 1982). A mixture of Tg/Tg/− 3n and Tg/− 2n embryos were produced from both the PO2×(BALB/c×A/J) and TRIP×(BALB/c×A/J) crosses and aggregated (without identifying the 3n embryos) with either FVB or NIH embryos using methods described previously (James et al. 1993, West & Flockhart 1994), based on the original method of Tarkowski (1961). NIH strain embryos were used as partner 2n embryos for series TrA and TrB, but FVB embryos were used for series TrC because FVB females responded better to our superovulation protocol. Homozygous Gpi1c/c females (hybrid stock ‘CF1’; West & Flockhart 1994) were used as recipients at E2.5 and pregnancies timed according to the recipient female. Series TrA was analysed at E12.5 but series TrB and TrC were analysed at E9.5. Each series produced its own diploid↔diploid control chimeras. DNA in situ hybridisation was used to identify Tg/Tg/− cells in triploid↔2n chimeras and distinguish them retrospectively from control 2n↔2n chimeras.
Production of Ts3↔2n Chimeras
Female (BALB/c×A/J)F1 mice were superovulated and mated to (TGA×RBT)F1 males and embryos were collected and aggregated with FVB embryos as described above. Aggregates were cultured and transferred to pseudopregnant females and conceptuses were analysed at E9.5. Ts3↔2n and 2n↔2n chimeras were distinguished retrospectively by DNA in situ hybridisation to the transgene. Tg/Tg/− Ts3 cells produce two hybridisation signals per nucleus but Tg/− 2n cells produce only one. Ts1 and Ms1 cells that were disomic for chromosome 3 (Tg/−) would also produce single hybridisation signals, whereas Ms3 cells (−) would produce no hybridisation signal.
Analysis of E12.5 chimeras
E12.5 conceptuses were dissected to provide seven tissue samples: placenta, trophoblast overlying Reichert's membrane (plus maternal decidual tissue), parietal endoderm, yolk sac endoderm, yolk sac mesoderm, amnion and fetus (West & Flockhart 1994). The whole conceptus, placenta and fetus were weighed, crown-rump length measured and morphological index assessed by hind limb development (McLaren & Buehr 1990, Palmer & Burgoyne 1991). Tissue preparation, electrophoresis, GPI1 staining and densitometry are described elsewhere (James et al. 1993, West & Flockhart 1994).
The proportions of the two cell populations in the chimeric tissue were estimated from the proportions of GPI1-A and GPI1-B allozymes. The recipient females were Gpi1c/c and produced only GPI1-C enzyme; so any maternal contamination could be excluded from the analysis. The percentage GPI1-A contribution was also corrected to allow for the expectation that triploid cells would produce 1.5 times the activity of diploid cells. The corrected value was calculated as ⅔A×100/(⅔A+B), where A=%GPI1-A and B=%GPI1-B, and this probably provides a closer estimate of the percentage 3n cells. (However, triploid cells do not always produce the predicted 1.5-fold increase in gene product (Epstein 1986), so both uncorrected and corrected GPI1-A levels are presented.) A piece of yolk sac was fixed in acetic alcohol (3 ethanol: 1 acetic acid) and used to distinguish E12.5 3n↔2n and 2n↔2n chimeras by DNA in situ hybridisation on 7 μm histological sections (see below). 3n↔2n chimeras were identified by the presence of Tg/Tg/− triploid cells with two hybridisation signals per nucleus.
Analysis of E9.5 chimeras
E9.5 conceptuses were fixed in acetic alcohol and sections were cut at 7 μm. Probe pMβδ2 was labelled using digoxygenin-labelled deoxyuridine triphosphate (non-radioactive DNA Labelling and Detection Kit; Boehringer Mannheim), DNA–DNA in situ hybridisation was performed and hybridisation signals scored as described previously (Keighren & West 1993). In situ hybridisation of Ts3↔2n and 3n↔2n chimeras produced two hybridisation signals in most Tg-positive nuclei, so they were distinguishable from 2n↔2n control chimeras with one signal per nucleus. Conceptuses with no hybridisation signals were excluded from the analysis, and considered to be non-chimeric, although in series TS some may have been Ms3↔2n chimeras.
Three longitudinal sections of each conceptus were scored (middle plus two flanking sections, approximately midway from the middle to the sides). Fetal brain, heart, somites, amnion, yolk sac mesoderm, yolk sac endoderm, parietal endoderm, placental trophoblast and giant cells were analysed separately and a mean fetal value was calculated from the results for the brain, heart and somites. (The term ‘fetus’ is used at both E9.5 and E12.5 to avoid confusion with the preimplantation embryo, which produces both the fetus and extraembryonic tissues.) The percentage of Tg-positive nuclei was estimated for each tissue by counting positive and negative nuclei in a sample of ∼250–300 cells. Not all 2n nuclei produce the expected (single) hybridisation signal and not all 3n and Ts3 nuclei produce two signals because only part of the nucleus may be included in the section (Keighren & West 1993). Thus, once a chimera had been identified as Ts3↔2n or 3n↔2n, all nuclei with hybridisation signals were scored as Tg positive (and therefore Ts or 3n) irrespective of whether one or two signals were present. The percentages of Tg-positive cells in Tg/Tg/−↔−/− (Ts3↔2n or 3n↔2n) and Tg/−↔−/− (2n↔2n) chimeras were corrected using tissue-specific means from two homozygous Tg/Tg and two hemizygous Tg/− control conceptuses respectively.
Statistical analysis
Statistical tests were performed on an Apple Macintosh computer using StatView software (Abacus Concepts Inc., Berkeley, CA, USA).
Acknowledgements
We dedicate this paper to the memory of Anne McLaren, an outstanding scientist with research interests in reproductive biology, developmental biology and genetics, who introduced J D W to the fascinating world of mouse chimeras when he was her PhD student. We thank Denis Doogan, Maureen Ross and Jim Macdonald for expert mouse husbandry, and Corinne MacLeod, Ronnie Grant and Ted Pinner for assistance with the illustrations. We are grateful to the Wellcome Trust for the financial support (grant 036737 to J D W). The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Footnotes
M A Keighren is now at MRC Human Genetics Unit, Crewe Road, Edinburgh EH4 2XU, Scotland, UK
References
- Azuma S, Fukuda Y, Toyoda Y. Studies on the production of 2n↔3n chimeric mouse embryos. II. Post-implantation development. Japanese Journal of Animal Reproduction. 1991;37:79–87. [Google Scholar]
- Bennett P, Vaughan J, Henderson D, Loughna S, Moore G. Association between confined placental trisomy, fetal uniparental disomy and early uterine growth retardation. Lancet. 1992;340:1284–1285. doi: 10.1016/0140-6736(92)92980-t. [DOI] [PubMed] [Google Scholar]
- Cattanach B, Moseley H. Nondisjunction and reduced fertility caused by tobacco mouse metacentric chromosomes. Cytogenetics and Cell Genetics. 1973;12:264–287. doi: 10.1159/000130462. [DOI] [PubMed] [Google Scholar]
- Copp AJ. Death before birth. Clues from gene knockouts and mutations. Trends in Genetics. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- Cox DR, Smith SA, Epstein LB, Epstein CJ. Mouse trisomy 16 as an animal model for human trisomy 21 (Down syndrome): production of viable trisomy 16↔diploid mouse embryos. Developmental Biology. 1984;101:416–424. doi: 10.1016/0012-1606(84)90156-8. [DOI] [PubMed] [Google Scholar]
- Daniel A, Wu ZH, Darmanian A, Collins F, Jackson J. Three different origins for apparent triploid/diploid mosaics. Prenatal Diagnosis. 2003;23:529–534. doi: 10.1002/pd.634. [DOI] [PubMed] [Google Scholar]
- Eakin GS, Hadjantonakis AK, Papaioannou VE, Behringer RR. Developmental potential and behavior of tetraploid cells in the mouse embryo. Developmental Biology. 2005;288:150–159. doi: 10.1016/j.ydbio.2005.09.028. [DOI] [PubMed] [Google Scholar]
- Epstein CJ. The Consequences of Chromosome Imbalance. Cambridge University Press; New York: 1986. pp 486. [Google Scholar]
- Epstein CJ, Smith SA, Cox DR. Production and properties of mouse trisomy 15↔diploid chimeras. Developmental Genetics. 1984;4:159–165. doi: 10.1016/0012-1606(84)90156-8. [DOI] [PubMed] [Google Scholar]
- Everett CA, West JD. The influence of ploidy on the distribution of cells in chimaeric mouse blastocysts. Zygote. 1996;4:59–66. doi: 10.1017/s0967199400002896. [DOI] [PubMed] [Google Scholar]
- Everett CA, West JD. Evidence for selection against tetraploid cells in tetraploid↔diploid mouse chimaeras before the late blastocyst stage. Genetical Research. 1998;72:225–228. doi: 10.1017/s0016672398003498. [DOI] [PubMed] [Google Scholar]
- Everett CA, Keighren M, West JD. New transgenic Robertsonian strain. Mouse Genome. 1994;92:668. [Google Scholar]
- Everett CA, Searle JB, Wallace BMN. A study of meiotic pairing, nondisjunction and germ-cell death in laboratory mice carrying Robertsonian translocations. Genetical Research. 1996;67:239–247. doi: 10.1017/s0016672300033723. [DOI] [PubMed] [Google Scholar]
- Everett CA, Stark MH, West JD, Davidson D, Baldock RA. Three-dimensional reconstruction of tetraploid↔diploid chimaeric mouse blastocysts. Journal of Anatomy. 2000;196:341–346. doi: 10.1046/j.1469-7580.2000.19630341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett CA, Auchincloss CA, Kaufman MH, Abbott CM, West JD. Genetic influences on ovulation of primary oocytes in LT/Sv strain mice. Reproduction. 2004;128:565–571. doi: 10.1530/rep.00325. [DOI] [PubMed] [Google Scholar]
- Fundele R, Jägerbauer E-M, Kolbus U, Winking H, Gropp A. Viability of trisomy 12 cells in mouse chimaeras. Roux's Archives of Developmental Biology . 1985;194:178–180. [Google Scholar]
- Golubovsky MD. Postzygotic diploidization of triploids as a source of unusual cases of mosaicism, chimerism and twinning. Human Reproduction. 2003;18:236–242. doi: 10.1093/humrep/deg060. [DOI] [PubMed] [Google Scholar]
- Goto Y, Takagi N. Tetraploid embryos rescue embryonic lethality caused by an additional maternally inherited X chromosome in the mouse. Development. 1998;125:3353–3363. doi: 10.1242/dev.125.17.3353. [DOI] [PubMed] [Google Scholar]
- Goto Y, Matsui J, Takagi N. Developmental potential of mouse tetraploid cells in diploid↔tetraploid chimeric embryos. International Journal of Developmental Biology. 2002;46:741–745. [PubMed] [Google Scholar]
- Gropp A 1976 Morphological consequences of trisomy in mammals. In Embryogenesis in Mammals Ciba Symposium 40, pp 155–175, Amsterdam: Elsevier.
- Gropp A, Giers D, Kolbus U. Trisomy in fetal backcross progeny of male and female metacentric heterozygotes of mouse. 1. Cytogenetics and Cell Genetics. 1974;13:511–535. doi: 10.1159/000130304. [DOI] [PubMed] [Google Scholar]
- Gropp A, Winking H, Herbst EW, Claussen CP. Murine trisomy – developmental profiles of the embryo, and isolation of trisomic cellular systems. Journal of Experimental Zoology. 1983;228:253–269. doi: 10.1002/jez.1402280210. [DOI] [PubMed] [Google Scholar]
- Hassold TJ, Jacobs PA. Trisomy in man. Annual Review of Genetics. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- Hassold T, Chen N, Funkhouser J, Jooss T, Manuel B, Matsuura J, Matsuyama A, Wilson C, Yamane JA, Jacobs PA. A cytogenetic study of 1000 spontaneous abortions. Annals of Human Genetics. 1980;44:151–178. doi: 10.1111/j.1469-1809.1980.tb00955.x. [DOI] [PubMed] [Google Scholar]
- James R, Flockhart JH, Keighren M, West JD. Quantitative analysis of mid-gestation mouse aggregation chimaeras: non-random composition of the placenta. Roux's Archives of Developmental Biology . 1993;202:296–305. doi: 10.1007/BF00363218. [DOI] [PubMed] [Google Scholar]
- James RM, Klerkx A, Keighren M, Flockhart JH, West JD. Restricted distribution of tetraploid cells in mouse tetraploid↔diploid chimaeras. Developmental Biology. 1995;167:213–226. doi: 10.1006/dbio.1995.1018. [DOI] [PubMed] [Google Scholar]
- Kalousek DK. Confined placental mosaicism and intrauterine development. Pediatric Pathology . 1990;10:69–77. doi: 10.3109/15513819009067097. [DOI] [PubMed] [Google Scholar]
- Kalousek DK. Current topic: confined placental mosaicism and intrauterine fetal development. Placenta. 1994;15:219–230. doi: 10.1016/0143-4004(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kalousek DK, Dill FJ. Chromosome mosaicism confined to the placenta in human conceptions. Science. 1983;221:665–667. doi: 10.1126/science.6867735. [DOI] [PubMed] [Google Scholar]
- Kalousek DK, Vekemans M. Confined placental mosaicism. Journal of Medical Genetics. 1996;33:529–533. doi: 10.1136/jmg.33.7.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata M, Lo CW. Organization of chromosomes in the mouse nucleus: analysis by in situ hybridization. Journal of Cell Science. 1988;90:193–199. doi: 10.1242/jcs.90.2.193. [DOI] [PubMed] [Google Scholar]
- Kaufman MH, Howlett SK. The ovulation and activation of primary and secondary oocytes in LT/Sv strain mice. Gamete Research. 1986;14:255–264. [Google Scholar]
- Kaufman MH, Speirs S. The postimplantation development of spontaneous digynic triploid embryos in LT/Sv strain mice. Development. 1987;101:383–391. doi: 10.1242/dev.101.2.383. [DOI] [PubMed] [Google Scholar]
- Keighren M, West JD. Analysis of cell ploidy in histological sections of mouse tissues by DNA–DNA in situ hybridization with digoxygenin labelled probes. Histochemical Journal. 1993;25:30–44. doi: 10.1007/BF00161042. [DOI] [PubMed] [Google Scholar]
- Keighren M, West JD. Two new partially congenic transgenic strains. Mouse Genome. 1994;92:666. [Google Scholar]
- van de Laar I, Rabelink G, Hochstenbach R, Tuerlings J, Hoogeboom J, Giltay J. Diploid/triploid mosaicism in dysmorphic patients. Clinical Genetics. 2002;62:376–382. doi: 10.1034/j.1399-0004.2002.620504.x. [DOI] [PubMed] [Google Scholar]
- Lo C. Localization of low abundance DNA sequences in tissue sections by in situ hybridization. Journal of Cell Science. 1986;81:143–162. doi: 10.1242/jcs.81.1.143. [DOI] [PubMed] [Google Scholar]
- Lo CW, Coulling M, Kirby C. Tracking of mouse cell lineage using microinjected DNA sequences: analysis using genomic Southern blotting and tissue-section in situ hybridizations. Differentiation. 1987;35:37–44. doi: 10.1111/j.1432-0436.1987.tb00149.x. [DOI] [PubMed] [Google Scholar]
- Lo CW, Diaz R, Kirby C. Iontophoretic DNA injections and the production of transgenic mice. Mouse Genome. 1992;90:684–686. [Google Scholar]
- MacKay GE, West JD. Fate of tetraploid cells in 4n↔2n chimeric mouse blastocysts. Mechanisms of Development. 2005;122:1266–1281. doi: 10.1016/j.mod.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Magnuson T, Smith S, Epstein CJ. The development of monosomy 19 mouse embryos. Journal of Embryology and Experimental Morphology. 1982;69:223–236. [PubMed] [Google Scholar]
- McLaren A, Buehr M. Development of mouse germ cells in cultures of fetal gonads. Cell Differentiation and Development. 1990;31:185–195. doi: 10.1016/0922-3371(90)90131-f. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Merentes Diaz E, Prideaux V, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Niemierko A. Induction of triploidy in mouse by cytochalasin-B. Journal of Embryology and Experimental Morphology. 1975;34:279–289. [PubMed] [Google Scholar]
- O'Neill GT, Kaufman MH. Ovulation and fertilization of primary and secondary oocytes in LT/Sv strain mice. Gamete Research. 1987;18:27–36. doi: 10.1002/mrd.1120180105. [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Burgoyne PS. The Mus musculus domesticus Tdy allele acts later than the Mus musculus musculus Tdy allele: a basis for XY sex reversal in C57BL/6-YPOS mice. Development. 1991;113:709–714. doi: 10.1242/dev.113.2.709. [DOI] [PubMed] [Google Scholar]
- Quinn P, Barros C, Whittingham DG. Preservation of hamster oocytes to assay the fertilizing capacity of human spermatozoa. Journal of Reproduction and Fertility. 1982;66:161–168. doi: 10.1530/jrf.0.0660161. [DOI] [PubMed] [Google Scholar]
- Sarno AP, Moorman AJ, Kalousek DK. Partial molar pregnancy with fetal survival – an unusual example of confined placental mosaicism. Obstetrics and Gynecology. 1993;82:716–719. [PubMed] [Google Scholar]
- Speirs S, Kaufman MH. Effect of exogenous hormones on the ovulation of primary and secondary oocytes in LT/Sv strain mice. Gamete Research. 1988;21:179–184. doi: 10.1002/mrd.1120210208. [DOI] [PubMed] [Google Scholar]
- Speirs S, Kaufman MH. Effect of maternal age on the incidence of digynic triploidy in LT/Sv strain mice: implications for the ovulation of primary and secondary oocytes in this strain. Journal of Experimental Zoology. 1990;253:83–87. doi: 10.1002/jez.1402530111. [DOI] [PubMed] [Google Scholar]
- Spindle A, Sturm KS, Flannery M, Meneses JJ, Wu K, Pedersen RA. Defective chorioallantoic fusion in mid-gestation lethality of parthenogenone↔tetraploid chimeras. Developmental Biology. 1996;173:447–458. doi: 10.1006/dbio.1996.0039. [DOI] [PubMed] [Google Scholar]
- Surani MAH, Barton SC, Norris ML. Nuclear transplantation in the mouse – heritable differences between parental genomes after activation of the embryonic genome. Cell. 1986;45:127–136. doi: 10.1016/0092-8674(86)90544-1. [DOI] [PubMed] [Google Scholar]
- Suwinska A, Ozdzenski W, Waksmundzka M, Tarkowski AK. Experimentally produced diploid–triploid mouse chimaeras develop up to adulthood. Molecular Reproduction and Development. 2005;72:362–376. doi: 10.1002/mrd.20350. [DOI] [PubMed] [Google Scholar]
- Tang P-C, Ritchie WA, Wilmut I, West JD. The effects of cell size and ploidy on cell allocation in mouse chimaeric blastocysts. Zygote. 2000;8:33–43. doi: 10.1017/s0967199400000800. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK. Mouse chimaeras developed from fused eggs. Nature. 1961;190:857–860. doi: 10.1038/190857a0. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Witkowska A, Opas J. Development of cytochalasin B-induced tetraploid and diploid/tetraploid mosaic mouse embryos. Journal of Embryology and Experimental Morphology. 1977;41:47–64. [PubMed] [Google Scholar]
- Tarkowski AK, Ozdzenski W, Czolowska R. Mouse singletons and twins developed from isolated diploid blastomeres supported with tetraploid blastomeres. International Journal of Developmental Biology. 2001;45:591–596. [PubMed] [Google Scholar]
- Tuerlings JHAM, Breed ASPM, Vosters R, Anders GJPA. Evidence of a second gamete fusion after the 1st cleavage of the zygote in a 47,XX+18/70,XXX+18-mosaic – a remarkable diploid triploid discrepancy after CVS. Prenatal Diagnosis. 1993;13:301–306. doi: 10.1002/pd.1970130409. [DOI] [PubMed] [Google Scholar]
- West JD, Flockhart JH. Genotypically unbalanced diploid↔diploid foetal mouse chimaeras: possible relevance to human confined mosaicism. Genetical Research. 1994;63:87–99. doi: 10.1017/s0016672300032195. [DOI] [PubMed] [Google Scholar]
- West JD, Green JF. The transition from oocyte-coded to embryo-coded glucose phosphate isomerase in the early mouse embryo. Journal of Embryology and Experimental Morphology. 1983;78:127–140. [PubMed] [Google Scholar]
- West JD, Webb S, Kaufman MH. Inheritance of a meiotic abnormality that causes the ovulation of primary oocytes and the production of digynic triploid mice. Genetical Research. 1993;62:183–193. doi: 10.1017/s001667230003189x. [DOI] [PubMed] [Google Scholar]
- West JD, Flockhart JH, Keighren M. Biochemical evidence for cell fusion in placentas of mouse aggregation chimeras. Developmental Biology. 1995;168:76–85. doi: 10.1006/dbio.1995.1062. [DOI] [PubMed] [Google Scholar]
- Wilkins-Haug L, Quade B, Morton CC. Confined placental mosaicism as a risk factor among newborns with fetal growth restriction. Prenatal Diagnosis. 2006;26:428–432. doi: 10.1002/pd.1430. [DOI] [PubMed] [Google Scholar]
- Wolstenholme J. Confined placental mosaicism for trisomies 2, 3, 7, 8, 9, 16, and 22: their incidence, likely origins, and mechanisms for cell lineage compartmentalization. Prenatal Diagnosis. 1996;16:511–524. doi: 10.1002/(SICI)1097-0223(199606)16:6<511::AID-PD904>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Wolstenholme J, Rooney DE, Davison EV. Confined placental mosaicism, IUGR and adverse pregnancy outcome: a controlled retrospective UK collaborative survey. Prenatal Diagnosis. 1994;14:345–361. doi: 10.1002/pd.1970140505. [DOI] [PubMed] [Google Scholar]
- Wróblewska J. Developmental anomaly in the mouse associated with triploidy. Cytogenetics. 1971;10:199–207. doi: 10.1159/000130139. [DOI] [PubMed] [Google Scholar]