Abstract
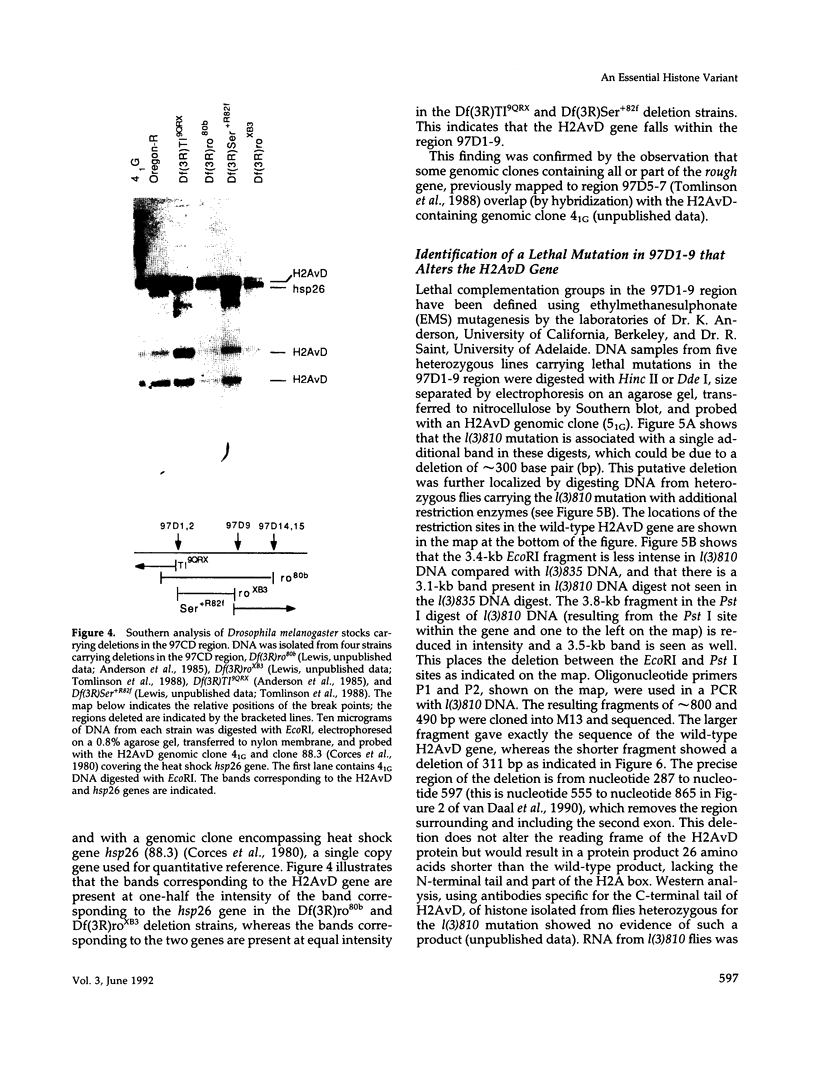
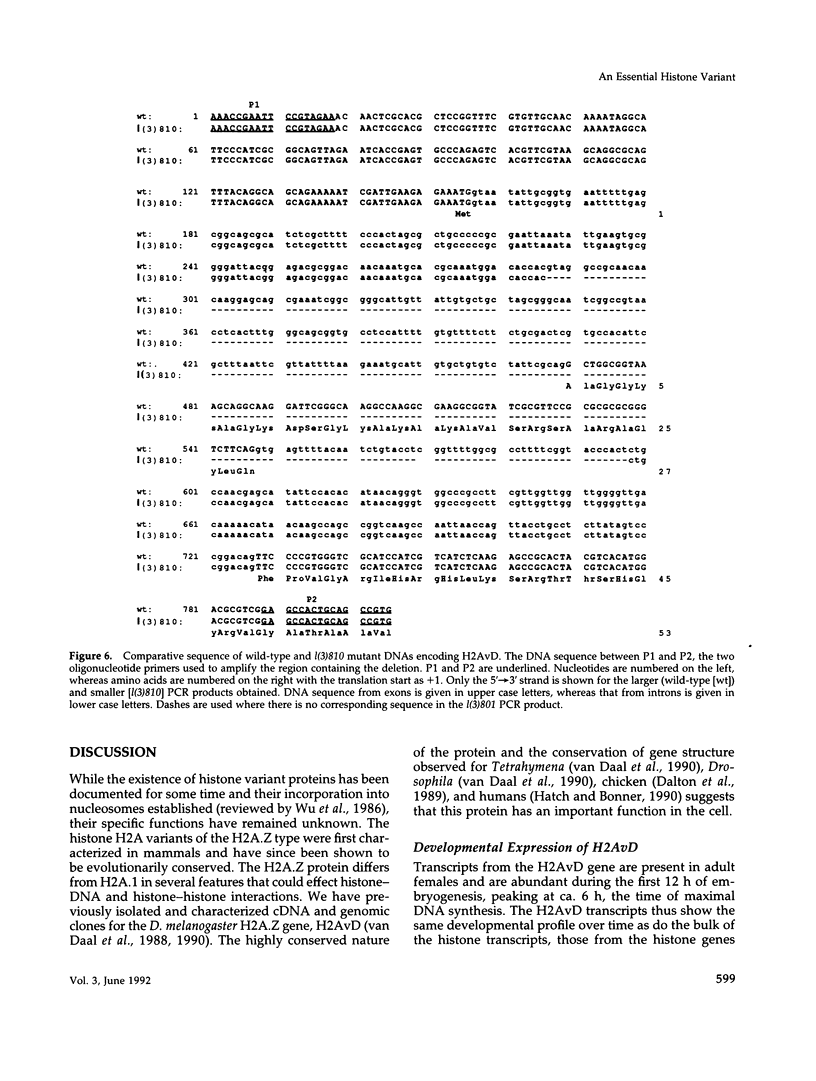
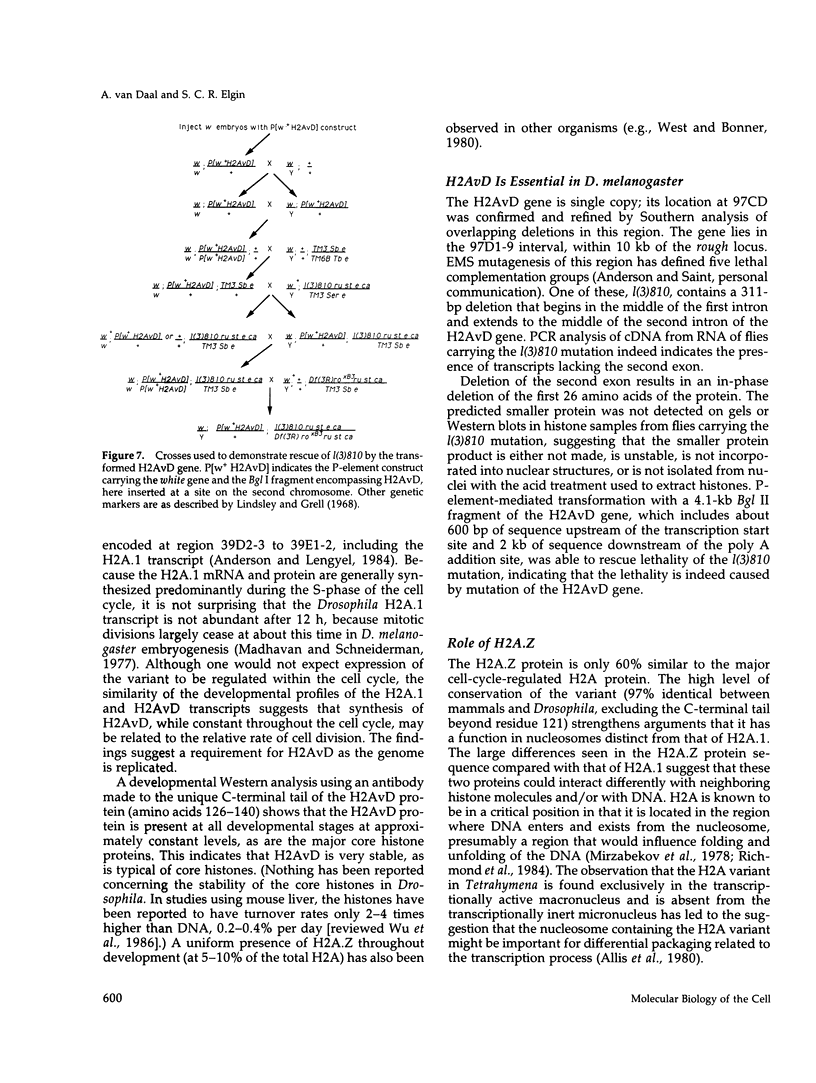
H2AvD, a Drosophila melanogaster histone variant of the H2A.Z class, is encoded by a single copy gene in the 97CD region of the polytene chromosomes. Northern analysis shows that the transcript is expressed in adult females and is abundant throughout the first 12 h of embryogenesis but then decreases. The H2AvD protein is present at essentially constant levels in all developmental stages. Using D. melanogaster stocks with deletions in the 97CD region, we have localized the H2AvD gene to the 97D1-9 interval. A lethal mutation in this interval, l(3)810, exhibits a 311-base pair deletion in the H2AvD gene, which removes the second exon. P-element mediated transformation using a 4.1-kilobase fragment containing the H2AvD gene rescues the lethal phenotype. H2AvD is therefore both essential and continuously present, suggesting a requirement for its utilization, either to provide an alternative capability for nucleosome assembly or to generate an alternative nucleosome structure.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Anderson K. V., Jürgens G., Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. 1985 Oct;42(3):779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Kolodrubetz D., Grunstein M. The two yeast histone H2A genes encode similar protein subtypes. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1484–1487. doi: 10.1073/pnas.79.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces V., Holmgren R., Freund R., Morimoto R., Meselson M. Four heat shock proteins of Drosophila melanogaster coded within a 12-kilobase region in chromosome subdivision 67B. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5390–5393. doi: 10.1073/pnas.77.9.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Robins A. J., Harvey R. P., Wells J. R. Transcription from the intron-containing chicken histone H2A.F gene is not S-phase regulated. Nucleic Acids Res. 1989 Feb 25;17(4):1745–1756. doi: 10.1093/nar/17.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C., Hood L. E. Chromosomal proteins of Drosophila embryos. Biochemistry. 1973 Nov 20;12(24):4984–4991. doi: 10.1021/bi00748a026. [DOI] [PubMed] [Google Scholar]
- Ernst S. G., Miller H., Brenner C. A., Nocente-McGrath C., Francis S., McIsaac R. Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res. 1987 Jun 11;15(11):4629–4644. doi: 10.1093/nar/15.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Green N., Alexander H., Olson A., Alexander S., Shinnick T. M., Sutcliffe J. G., Lerner R. A. Immunogenic structure of the influenza virus hemagglutinin. Cell. 1982 Mar;28(3):477–487. doi: 10.1016/0092-8674(82)90202-1. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Whiting J. A., Coles L. S., Krieg P. A., Wells J. R. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc Natl Acad Sci U S A. 1983 May;80(10):2819–2823. doi: 10.1073/pnas.80.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M. Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res. 1988 Feb 11;16(3):1113–1124. doi: 10.1093/nar/16.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M. The human histone H2A.Z gene. Sequence and regulation. J Biol Chem. 1990 Sep 5;265(25):15211–15218. [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Shick V. V., Belyavsky A. V., Bavykin S. G. Primary organization of nucleosome core particle of chromatin: sequence of histone arrangement along DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4184–4188. doi: 10.1073/pnas.75.9.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEELIN J. M., CALLAHAN P. X., LAMB D. C., MURRAY K. THE HISTONES OF CHICKEN ERYTHROCYTE NUCLEI. Can J Biochem. 1964 Dec;42:1743–1752. doi: 10.1139/o64-185. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Alfageme C. R., Nardi R. V., Cohen L. H. Histone changes during chromatin remodeling in embryogenesis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):421–431. doi: 10.1101/sqb.1978.042.01.045. [DOI] [PubMed] [Google Scholar]
- Norris D., Osley M. A. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol Cell Biol. 1987 Oct;7(10):3473–3481. doi: 10.1128/mcb.7.10.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R. Histone genes: not so simple after all. Cell. 1984 Oct;38(3):624–626. doi: 10.1016/0092-8674(84)90256-3. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Kedes L. H., Weinberg E. S., Birnstiel M. L. Localization of sequences coding for histone messenger RNA in the chromosomes of Drosophila melanogaster. Chromosoma. 1977 Aug 25;63(2):135–151. doi: 10.1007/BF00292726. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Wangh L. J., Littau V. C., Allfrey V. G. Changes in histone acetyl content and in nuclear non-histone protein composition of avian erythroid cells at different stages of maturation. J Biol Chem. 1974 Nov 25;249(22):7358–7368. [PubMed] [Google Scholar]
- Schümperli D. Multilevel regulation of replication-dependent histone genes. Trends Genet. 1988 Jul;4(7):187–191. doi: 10.1016/0168-9525(88)90074-1. [DOI] [PubMed] [Google Scholar]
- Shimamura A., Worcel A. The assembly of regularly spaced nucleosomes in the Xenopus oocyte S-150 extract is accompanied by deacetylation of histone H4. J Biol Chem. 1989 Aug 25;264(24):14524–14530. [PubMed] [Google Scholar]
- Silver L. M., Elgin S. C. Distribution patterns of three subfractions of drosophila nonhistone chromosomal proteins: possible correlations with gene activity. Cell. 1977 Aug;11(4):971–983. doi: 10.1016/0092-8674(77)90308-7. [DOI] [PubMed] [Google Scholar]
- Spiker S., Isenberg I. Evolutionary conservation of histone-histone binding sites: evidence from interkingdom complex formation. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):157–163. doi: 10.1101/sqb.1978.042.01.017. [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Kimmel B. E., Rubin G. M. rough, a Drosophila homeobox gene required in photoreceptors R2 and R5 for inductive interactions in the developing eye. Cell. 1988 Dec 2;55(5):771–784. doi: 10.1016/0092-8674(88)90133-x. [DOI] [PubMed] [Google Scholar]
- West M. H., Bonner W. M. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980 Jul 8;19(14):3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Panusz H. T., Hatch C. L., Bonner W. M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20(2):201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- van Daal A., White E. M., Elgin S. C., Gorovsky M. A. Conservation of intron position indicates separation of major and variant H2As is an early event in the evolution of eukaryotes. J Mol Evol. 1990 May;30(5):449–455. doi: 10.1007/BF02101116. [DOI] [PubMed] [Google Scholar]
- van Daal A., White E. M., Gorovsky M. A., Elgin S. C. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res. 1988 Aug 11;16(15):7487–7497. doi: 10.1093/nar/16.15.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]