Abstract
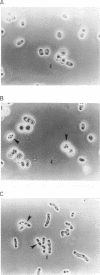
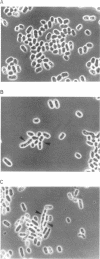
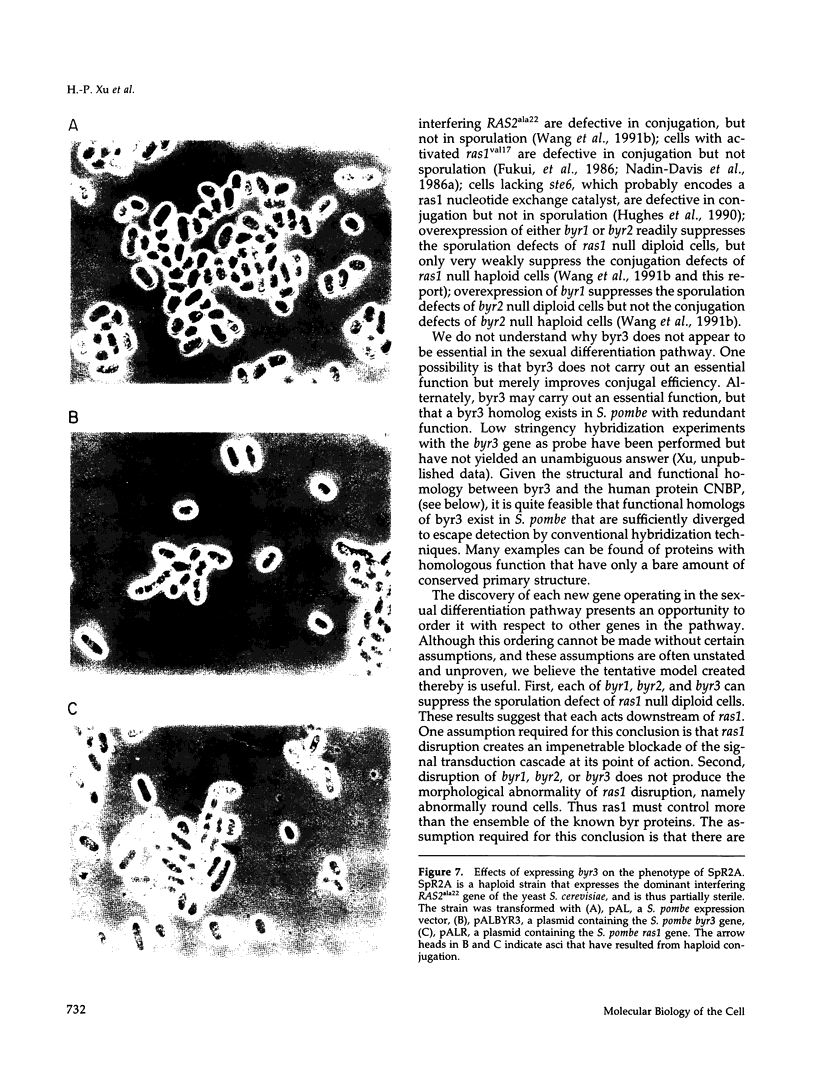
Byr3 was selected as a multicopy suppressor of the sporulation defects of diploid Schizosaccharomyces pombe cells that lack ras1. Like cells mutant at byr1 and byr2, two genes that encode putative protein kinases and that in multiple copies are also suppressors of the sporulation defects of ras1 null diploid cells, cells mutant at byr3 are viable but defective in conjugation. Nucleic acid sequence indicates byr3 has the capacity to encode a protein with seven zinc finger binding domains, similar in structure to the cellular nucleic acid binding protein (CNBP), a human protein that was identified on the basis of its ability to bind DNA. Expression of CNBP in yeast can partially suppress conjugation defects of cells lacking byr3.
Full text
PDF


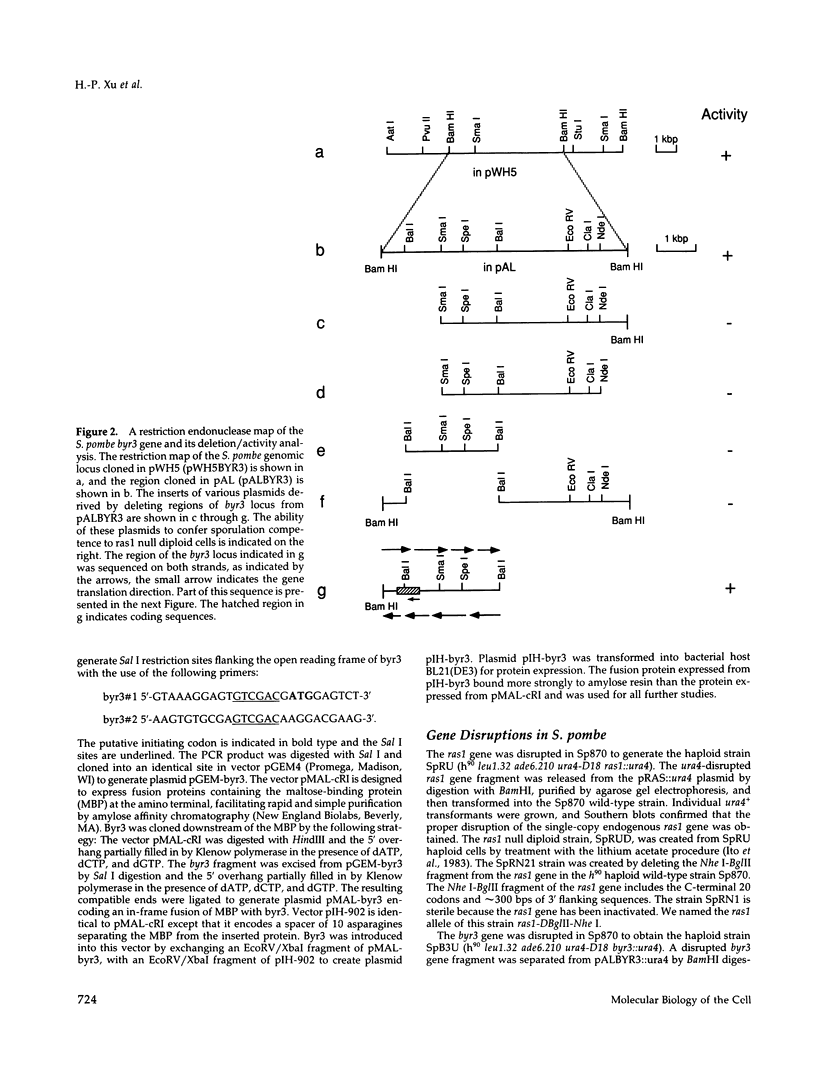






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Beckner S. K., Hattori S., Shih T. Y. The ras oncogene product p21 is not a regulatory component of adenylate cyclase. Nature. 1985 Sep 5;317(6032):71–72. doi: 10.1038/317071a0. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C., Broek D., Wigler M. ras proteins can induce meiosis in Xenopus oocytes. Cell. 1985 Dec;43(3 Pt 2):615–621. doi: 10.1016/0092-8674(85)90233-8. [DOI] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Covey S. N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986 Jan 24;14(2):623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Kozasa T., Kaziro Y., Takeda T., Yamamoto M. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 1986 Jan 31;44(2):329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hughes D. A., Fukui Y., Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990 Mar 22;344(6264):355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K. W., Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990 Feb;10(2):634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K. W., Vogelstein B. Whole genome PCR: application to the identification of sequences bound by gene regulatory proteins. Nucleic Acids Res. 1989 May 25;17(10):3645–3653. doi: 10.1093/nar/17.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Hui H. X., Kappes J. C., Haggarty B. S., Hoxie J. A., Arya S. K., Shaw G. M., Hahn B. H. Molecular characterization of an attenuated human immunodeficiency virus type 2 isolate. J Virol. 1990 Feb;64(2):890–901. doi: 10.1128/jvi.64.2.890-901.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Plasmids carrying the yeast OMP decarboxylase structural and regulatory genes: transcription regulation in a foreign environment. Cell. 1983 Feb;32(2):371–377. doi: 10.1016/0092-8674(83)90456-7. [DOI] [PubMed] [Google Scholar]
- McLeod M., Stein M., Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987 Mar;6(3):729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. T., Hill K. A., Schimmel P. Evidence for a "cysteine-histidine box" metal-binding site in an Escherichia coli aminoacyl-tRNA synthetase. Biochemistry. 1991 Jul 16;30(28):6970–6976. doi: 10.1021/bi00242a023. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis S. A., Nasim A. A gene which encodes a predicted protein kinase can restore some functions of the ras gene in fission yeast. EMBO J. 1988 Apr;7(4):985–993. doi: 10.1002/j.1460-2075.1988.tb02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis S. A., Nasim A., Beach D. Involvement of ras in sexual differentiation but not in growth control in fission yeast. EMBO J. 1986 Nov;5(11):2963–2971. doi: 10.1002/j.1460-2075.1986.tb04593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis S. A., Nasim A. Schizosaccharomyces pombe ras1 and byr1 are functionally related genes of the ste family that affect starvation-induced transcription of mating-type genes. Mol Cell Biol. 1990 Feb;10(2):549–560. doi: 10.1128/mcb.10.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis S. A., Yang R. C., Narang S. A., Nasim A. The cloning and characterization of a RAS gene from Schizosaccharomyces pombe. J Mol Evol. 1986;23(1):41–51. doi: 10.1007/BF02100997. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavashisth T. B., Taylor A. K., Andalibi A., Svenson K. L., Lusis A. J. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989 Aug 11;245(4918):640–643. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Wang Y., Boguski M., Riggs M., Rodgers L., Wigler M. sar1, a gene from Schizosaccharomyces pombe encoding a protein that regulates ras1. Cell Regul. 1991 Jun;2(6):453–465. doi: 10.1091/mbc.2.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu H. P., Riggs M., Rodgers L., Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991 Jul;11(7):3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A., Maundrell K., Heyer W. D., Beach D., Nurse P. Vectors for the construction of gene banks and the integration of cloned genes in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Plasmid. 1986 Mar;15(2):156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]