Abstract
The human cancer susceptibility gene, BRCA2, functions in double-strand break repair by homologous recombination, and it appears to function via interaction of a repetitive region (“BRC repeats”) with RAD-51. A putatively simpler homolog, dmbrca2, was identified in Drosophila melanogaster recently and also affects mitotic and meiotic double-strand break repair. In this study, we examined patterns of repeat variation both within Drosophila pseudoobscura and among available Drosophila genome sequences. We identified extensive variation within and among closely related Drosophila species in BRC repeat number, to the extent that variation within this genus recapitulates the extent of variation found across the entire animal kingdom. We describe patterns of evolution across species by documenting recent repeat expansions (sometimes in tandem arrays) and homogenizations within available genome sequences. Overall, we have documented patterns and modes of evolution in a new model system of a gene which is important to human health.
Keywords: Recombination, Drosophila, Repeat Evolution, BRCA2
Introduction
Recombination has manifold effects on biological diversity through its influences on patterns of genetic variation within a species and the formation and persistence of new species. Our understanding of the mechanistic basis of homologous recombination has increased dramatically over the past twenty years, and many genes that affect either the rate or the process of recombination have been identified (Gerton and Hawley 2005; Li and Ma 2006; Kong et al. 2008). In addition to generating biological diversity upon which evolution can act, homologous recombination also functions in the repair of double-stranded DNA breaks. This process is of fundamental importance to an organism, as mutations in genes affecting homologous recombination can cause infertility and inviability, including some forms of cancer.
Two loci that function in the repair of double-stranded breaks in DNA via homologous recombination are the tumor suppressor loci, BRCA1 and BRCA2, whose inactivation is associated with susceptibility to breast cancer (Venkitaraman 2002; Gudmundsdottir and Ashworth 2006; Nagaraju and Scully 2007). In humans, the locus BRCA2 functions in the regulation of RAD-51, the eukaryotic ortholog of RecA. RAD-51 forms nucleoprotein filaments on damaged DNA that are crucial to repair by recombination (Pellegrini and Venkitaraman 2004). BRCA2 binds to RAD-51 recombinase by association with sequence motifs, dubbed “BRC repeats” (Bork et al. 1996; Pellegrini et al. 2002), to promote DNA repair and homologous recombination. Because of this interaction with RAD-51, mutants for BRCA2 display lower rates of recombination in human cell lines, and the specific interaction of BRCA2 with RAD-51 appears essential for regulating homologous recombination (Xia et al. 2001). In the study species Trypanosoma brucei, the number of BRC repeats in BRCA2 is crucial to determining the efficiency of homologous recombination and RAD-51 localization (Hartley and Mcculloch 2008). Despite these known facts, researchers still struggle to determine how BRCA2 coordinates its Rad51- and ssDNA-binding activities to facilitate the transfer of RAD-51 onto DNA (but see Shivji et al. 2006). As a result, Pellegrini and Venkitaraman (2004) suggested that primitive organisms harboring simple versions of the BRCA2 protein will provide useful model systems.
Recently a “simpler” putative BRCA2 homolog was identified in Drosophila melanogaster using sequence fingerprints representing key residues for BRCA2-RAD51 interactions. This homolog was associated with the predicted locus name CG30169, later dubbed “dmbrca2” (Brough et al. 2008). Klovstad et. al. (2008) concluded that the Drosophila BRCA2 represents a functional homolog of the gene that can be used to characterize its human counterpart. Unlike the mammalian BRCA2, which has eight “BRC repeats,” the D. melanogaster homolog was found to contain only three repeats, fewer than any animal studied besides C. elegans (Lo et al. 2003). Functional studies of this Drosophila gene have shown that it interacts with D. melanogaster Rad-51 (spnA) and that disruption of this gene affects rates of mitotic and meiotic DNA repair and homologous recombination (Brough et al. 2008; Klovstad et al. 2008; Barnwell et al. 2008). In this study, we examine patterns of variation in this putatively “primitive” BRCA2 homolog both within and among Drosophila species to understand the evolutionary processes that have acted upon it and to evaluate the utility of this model system for examining the effects of cancer-related gene BRCA2 in humans.
Methods and Materials
Drosophila Stocks
Drosophila pseudoobscura stocks used in the present study were collected from Mather, California (three strains), Flagstaff, Arizona (two strains), Zapotitlan, Mexico (one strain), Baja, California (one strain), Mesa Verde, Colorado (one strain), Sonora, Mexico (one strain), and American Fork Canyon (near Provo), Utah (one strain). They were maintained under laboratory conditions for 5–15 years. One strain of D. miranda (number 28) derived from a single female collected in Mather, California, was used as an outgroup for the sequencing portions of this study, though the same repeat number was inferred in three additional strains via PCR and gel visualization (another from Mather, California, and two from Mount St. Helena, California).
DNA Isolation, PCR Amplification and Sequencing
Genomic DNA was isolated from adult males of D. pseudoobscura and D. miranda with a single fly squish protocol (Gloor and Engels 1992). Multiple primers throughout the dmbrca2 region were used to PCR amplify overlapping segments of the gene in 25 μL reaction volumes. Primers for PCR amplification were designed from the published genome sequence assembly (Richards et al. 2005). Sizes of PCR products were confirmed by electrophoresis on a 1% agarose gel. PCR products were purified using ExoSAP-It (USB Corp) and sequenced using ABI BigDye at the Duke University IGSP sequencing facility. Sequences were deposited in the GenBank/EMBL databases under accession numbers FJ823264–FJ823275.
Interspecific Variation
To examine variation in number and amino acid composition of the dmbrca2 BRC repeat regions of various Drosophila species, we obtained the full assembled sequences of this region from 12 twelve Drosophila species (Clark et al. 2007). We translated the DNA sequence 3′ of the introns ourselves to identify regions bearing the proposed eukaryotic sequence fingerprints of the BRC repeats (Lo et al. 2003). We categorized the repeats as starting with the highly conserved F-TAS amino acid fingerprint identified in that study. The distinct repeat units within each published sequence were then aligned manually.
Data Analyses
DNA sequences were aligned computationally using BioEdit 7.0.9 (Hall 1994), and then modified by manual alignment. Once aligned, phylogenetic estimates were generated with ProML protein maximum likelihood software within BioEdit, but the topology was generally consistent with a tree generated using the Fitch-Margoliash (1967) method. DNAsp (Rozas et al. 2003) was used for various tests of natural selection including McDonald-Kreitman (Mcdonald and Kreitman 1991), Tajima’s D (Tajima 1989), Fu and Li’s D (Fu and Li 1993), and Fay and Wu’s H (Fay and Wu 2000) for the dmbrca2 region.
Results
The Drosophila melanogaster dmbrca2 (CG30169) gene is annotated in GenBank to encode a 971 amino acid protein with two small introns (60 and 59bp, respectively). The final exon of this gene bears three BRC repeat units (Lo et al. 2003). In contrast, we found that the D. pseudoobscura homolog of this gene is predicted to encode a 1269 amino acid protein with eleven BRC repeat units, based on the sequence signature described by Lo et al (2003). As a result of its larger size, we examined intraspecific variation in repeat numbers in D. pseudoobscura and interspecific variation in repeat numbers across all the sequenced species of this genus.
Intraspecific variation in BRC repeats
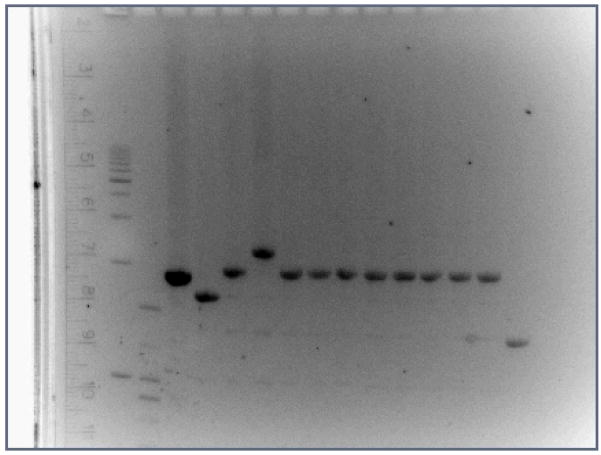
Using gel visualizations of amplified PCR products (see Fig. 1), we found that the lengths of the dmbrca2 gene region varied among our 10 inbred D. pseudoobscura strains. This intraspecific variation in numbers of BRC repeats was confirmed by examining the DNA sequences for the 10 lines, and we showed that each strain had 7, 9, or 11 BRC repeats. Further, we observed a repetitive pattern in the translated DNA sequences where the first BRC repeat resembled the third and fifth in amino acid sequence, and the second resembled the fourth and sixth rather than the first. This observation suggests that the expansion of the motif occurred by an even number of units.
Fig. 1.
PCR gel picture showing the lengths of the amplifications from the dmbrca2 gene in each of 10 lines of D. pseudoobscura and 1 of D. miranda. Lanes 1 and 2 are ladders, lanes 3–14 are amplicons from strains of D. pseudoobscura (most have 9 “BRC” repeats), and lane 15 is D. miranda (5 BRC repeats). Lane 4 represents strain MV-17 which bears the smallest BRC repeat region of any D. pseudoobscura strain sampled in this experiment (7 repeats), and lane 6 represents strain AFC12, which bears the longest (11 repeats)
Interspecific variation in BRC repeats
We aligned the published genome sequences for species across the genus Drosophila and evaluated the number of BRC repeats present in the 3′ region of dmbrca2 (see Supplementary Tables 1–2). Based on these assemblies, we found that Drosophila species have between three and eleven copies of the 55 amino acid BRC repeat (see Fig. 2). Evolution appeared sometimes rapid, as illustrated by the difference in number of repeats between the multiple lines of D. pseudoobscura (7–11) and closely related species D. miranda, which bore only 5 repeats in all four isolates surveyed (see Fig. 1 for amplicon image from one isolate). However, we cannot exclude the possibility that the D. miranda allele is present at low frequency in D. pseudoobscura. Nonetheless, by placing the numbers of repeats onto the established phylogeny of the sequenced species (see Fig. 2), we observe that closely related species tend to have similar numbers of BRC repeats overall.
Fig. 2.
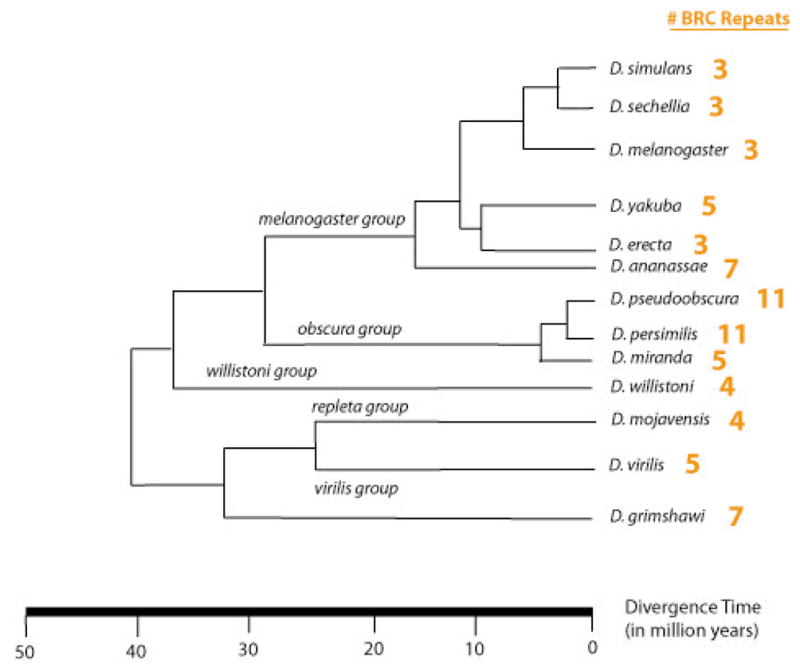
Variability in number of “BRC” repeats across Drosophila species
We confirmed the phylogenetic trend in repeat number using two tests. First, we observed a significant positive correlation between difference in number of BRC repeats and difference in amino acid sequence of the nonrepetitive portion of dmbrca2 (r=0.55, resampling p=0.0007). Second, if the repetitive portion of this gene evolves via a stepwise mutation model similar to other repetitive sequences (see Discussion), we predicted there should be greater variance in interspecies difference in repeat number (IDRN) among more distantly related species than among closely related species. We tested this by dividing all 66 possible between-species pairwise comparisons into either a “high” or “low” subset based on DNA sequence similarity at another locus (Adh) and comparing the high and low subsets for variance in IDRN. As predicted, the subset with higher divergence at Adh bore a much greater variance in IDRN for dmbrca2 (high subset, variance= 6.27; low subset, variance= 3.25). Moreover, the observed difference between the high and low subsets for IDRN was greater than in 2328 of 100,000 resamplings of the dataset (hence, p=0.023).
Among the species groups, we observed much variability in the number of amino acids between the BRC repeat conserved F-TAS amino acid fingerprints. While virtually all of the species displayed sequences resembling the primary sequence fingerprint over 25 amino acids, some species had repeat units with extensive additional intervening sequence. For instance, D. ananassae had five F-TAS start-points separated by 44–50 amino acids, but one pair was separated by 149 amino acids, perhaps indicating two degenerated ancestral repeat units. In contrast, D. pseudoobscura’s eleven repeat start-points are all separated by 55–57 amino acids. D. melanogaster’s arrays are less consistent in size than either species’, having three start-points separated by 101 and 75 amino acids.
Because of the extensive variation in repeat unit size among species, we further examined the mechanisms of BRC repeat evolution by generating a phylogeny using the first 25 amino acids of each BRC repeat within each species (see Supplementary Fig. 1). The D. melanogaster subgroup’s three repeats reflect high conservation among species: for example, D. melanogaster’s repeat 1 is more similar in sequence to repeat 1 of D. simulans, D. sechellia, D. yakuba, and D. erecta than it is to repeat 2 or 3 (Supplementary Table 2). D. yakuba’s two additional repeat units resemble (and perhaps originated from) the ancestral repeats 2 and 3 found in the other D. melanogaster subgroup species. However, evolutionary patterns were somewhat different within and among the other Drosophila species. As mentioned previously, D. pseudoobscura’s even and odd repeats resemble each other, but the resemblance for the odd-numbered repeats is stronger than their resemblance to other species’ repeats, suggesting they share closer relationships to each other than to other species. Similarly, D. willistoni’s repeats all closely resemble each other more than they resemble those from other species, as illustrated by their repeats all sharing a tryptophan in the second position rather than arginine, as in all the other species (Supplementary Table 2). The D. mojavensis repeats also all closely resembled each other, but also resemble one pair of repeats in D. virilis. The repeats of the other species appeared widespread across the phylogeny, reflecting a potentially complex history of gain and loss.
Tests for Selection Within the D. pseudoobscura Species Group
Because we observed variation within D. pseudoobscura in BRC repeat number and because of the unusually long size of the repeat region of this BRCA2 homolog, we tested for natural selection acting on DNA sequence variation within this species group. We focus on the non-repetitive 5′ coding region of the gene (see Table 1 for basic statistics).
Table 1.
Description of Drosophila pseudoobscura dmbrca2 sequences studied
| Total length of DNA sequence obtained | 1744 bp |
| Length of DNA sequence used for statistics below* | 1236 bp (of which 109 bp are within 2 introns) |
| D. pseudoobscura alignment start position | Chromosome 3, position 18254450 |
| Number of synonymous variable sites | 2 |
| Number of non-synonymous variable sites | 5 |
| Number of intronic variable sites | 0 |
| Number of singleton sites | 5 |
| Minimum number of recombination events(Hudson & Kaplan 4-gamete test) | 0 |
| Pi overall | 0.00162 |
| Pi synonymous | 0.00296 |
| Pi non-synonymous | 0.00144 |
| Theta overall | 0.00200 |
| Theta synonymous | 0.00285 |
| Theta non-synonymous | 0.00201 |
This length sequence was used for analysis to remain outside the repetitive region. It ends immediately before the first “LRTAS” amino acid motif.
Within this region, we observed two synonymous and five non-synonymous variable sites within D. pseudoobscura and sixteen synonymous and twenty-two non-synonymous changes between it and D. miranda, suggesting a non-significant excess of non-synonymous divergence via the McDonald-Kreitman test (P= 0.68). Tajima’s D test for natural selection was also non-significant (D= −0.812) (P > 0.10), and indeed, matched the average value observed among 18 random autosomal loci studied by Machado et al (2007) perfectly (average D= −0.820). Fu and Li’s D test (D= −1.274) (P > 0.10) and Fay and Wu’s H test (H= 0.711) were also non-significant. However, this nonsignificance may reflect the low statistical power of these tests on these sequences, given the number of segregating sites available.
Discussion
In this report, we examine patterns of evolution of a homologue of BRCA2 within and among Drosophila species, focusing especially on a region of this protein that operates in homologous recombination. BRCA2 functions in the pathway for double-strand break repair, and this process is mediated by interaction between a repetitive region of the BRCA2 gene, the “BRC repeats”, and RAD-51 complexes (Pellegrini and Venkitaraman 2004). The number of repeats determines the efficiency of homologous recombination in Trypanosoma brucei (Hartley and Mcculloch 2008), yet repeat number is highly variable across diverse taxa (Lo et al. 2003). Understanding the patterns of evolution of these repeats within and across species necessitates the use of a suitable model system. Recently, the Drosophila BRCA2 homolog (dmbrca2) has been utilized in the study of mitotic and meiotic DNA repair and homologous recombination (Brough et al. 2008; Klovstad et al. 2008; Barnwell et al. 2008). This Drosophila homologue bears a simpler structure than vertebrates, harboring only three repeats. We leveraged existing genome sequences (Clark et al 2007) and sequenced this gene from multiple strains within one Drosophila species to examine the evolutionary history of BRCA2 in Drosophila, focusing on the repetitive region.
Based on the available published genome sequences (Richards et al. 2005), the species D. pseudoobscura bears the largest BRC repeat region among Drosophila species. We further documented variation in the number of BRC repeats (7 to 11) between interfertile strains of this species. Among Drosophila species, we also observed a range of BRC repeat numbers, from the D. melanogaster subgroup species having only 3 repeats up to D. persimilis and D. pseudoobscura having 11 repeats. The variation we observed within the genus Drosophila recapitulates the variation documented previously across the entire animal kingdom (Lo et al. 2003). Further, the differences among strains within species and between some very closely related species indicates a sometimes rapid rate of change over a short evolutionary time. These findings contradict the generalization of Hartley and McCulloch (2008) that simple organisms harbor smaller numbers of BRC repeats and emphasizes the importance of examining variation within genera and species.
Although there is not yet a definitive explanation for the variation in number of repeats, it might be beneficial for an organism to have more control over homologous recombination. For example, having a larger number of BRC repeats could allow the cell’s RAD-51 protein complexes to be held until damage to the genome is detected, thereby preventing excessive recombination that could be detrimental to an organism’s genome (Hartley and McCulloch 2008). Despite the essential function of this gene and differences among species in the repetitive portion, we did not detect evidence for directional selection operating on the non-repetitive portion of the gene. However, we cannot be certain that this lack of evidence for selection does not reflect low power in our tests. Because we could not guarantee homology of particular repeats, we did not specifically test for selection on BRC repeat number or within the BRC repeats themselves.
Examining the sequence and number of repeats within the different Drosophila species allowed us to interpret evolutionary patterns of repeat number. Within the D. melanogaster subgroup, individual repeats exhibited greater similarity to the same repeat in other species than to other repeats within the same genome sequence. This observation suggests that these repeats are relatively stable and not being homogenized within a species. In contrast, the individual repeats for D. willistoni and D. mojavensis were more similar to each other than to the repeats of other species. This similarity within a genome sequence could result from gene conversion or non-homologous recombination creating a pattern of concerted evolution. Alternatively, there could have been repeat “births” and “deaths” to the point that the present panel of repeats all coalesce to a single common ancestor repeat. We identified some likely cases of tandem repeat births, indicated by the high similarity between the D. yakuba fourth and fifth repeat to the D. erecta and other D. melanogaster subgroup species’ second and third repeats. Similarly, the even-numbered D. pseudoobscura repeats all resemble each other, while the odd repeats are more similar. In some of the species, such as D. ananassae and D. grimshawi, it seems that complex processes, or multiple phases of the processes described above have occurred because their patterns of repeat evolution defy simple explanations.
Evolution of the dmbrca2 repetitive sequence across the genus Drosophila may reflect patterns inferred from microsatellites and other repetitive sequences, thought to evolve roughly in accordance with a modified “stepwise mutation model” (e.g., Valdes et al. 1993). According to this model, gains or losses of a single repeat unit are more common than larger changes, and the variance in number of repeats increases with phylogenetic divergence (Goldstein et al. 1995). We leveraged this expectation to provide statistical support that the published sequences of closely related species tend to have more similar numbers of BRC repeats than distantly related species. Variability observed among species was not as great as for some other repetitive motifs in Drosophila genes, such as the smaller threonine-glycine repeat motif of period (Peixoto et al. 1992). However, we observed that the species with the largest BRC repeat array bore multiple alleles within species, consistent with the documented correlation between repeat variability and number of units in the repeat array in satellite sequences (e.g., Wierdl et al. 1997; Legendre et al. 2007).
This study’s examination of the sequences of the breast cancer susceptibility gene, BRCA2, in the Drosophila model system sheds light on the molecular evolution of an important gene related to the development of breast and ovarian cancers (Pellegrini and Venkitaraman 2004). Disruption in BRCA2’s recombinational function may be associated with its oncogenic effects in humans (Xia et al. 2001), so understanding the evolutionary processes acting on its BRC repeats that mediate this function can assist with evaluating their effects on cancer. Future work in this area will determine whether repeat number modifies the interaction with RAD-51. By inserting different dmbrca2 alleles into a common background, we are testing for an association between numbers of BRC repeats and recombination rate in this model system. This information could impact our understanding of the rapid evolution of BRCA2 in terms of the functional significance of the initial expansion of the BRC repeats and its continuing evolution of repeat number.
Supplementary Material
Supplementary Fig. 1 Unrooted tree illustrating sequence similarity of 25 amino acids from different BRC repeats of various sequenced Drosophila species. Sibling species (D. simulans, D. sechellia, and D. persimilis) were excluded from the figure.
Supplementary Table 1 Drosophila species dmbrca2 BRC repeat amino acid sequences, FASTA-formatted.
Supplementary Table 2 Pairwise amino acid sequence differences among dmbrca2 BRC repeats within/between Drosophila species (Microsoft Excel formatted)
Acknowledgments
The authors thank Callie Barnwell and Lisa Bukovnik (IGSP sequencing center) for technical assistance. Funding was provided by a Research Experience for Undergraduates (REU) supplement from the National Science Foundation (to award 0509780) as well as NSF grant 0715484 and NIH grant GM076051.
Literature Cited
- Barnwell CV, Kommaraju KR, Noor MAF. Disruption of Drosophila melanogaster dmbrca2 (CG30169) affects rates of female meiotic crossing over. Drosoph Inf Serv. 2008;91:1–4. [Google Scholar]
- Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- Brough R, Wei D, Leulier S, et al. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair. 2008;7:10–19. doi: 10.1016/j.dnarep.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, Hawley RS. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nature Reviews Genetics. 2005;6:477–488. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Engels WR. Single-fly DNA preps for PCR. Drosoph Inf Serv. 1992;71:148–149. [Google Scholar]
- Goldstein DB, Linares AR, Cavalli-Sforza LL, Feldman MW. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1994;41:95–98. [Google Scholar]
- Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovstad M, Abdu U, Schupbach T. Drosophila brca2 Is Required for Mitotic and Meiotic DNA Repair and Efficient Activation of the Meiotic Recombination Checkpoint. PLoS Genet. 2008;4:e31. doi: 10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma H. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Research. 2006;16:401–412. doi: 10.1038/sj.cr.7310052. [DOI] [PubMed] [Google Scholar]
- Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair. 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MAF. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nagaraju G, Scully R. Minding the gap: The underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst) 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto AA, Costa R, Wheeler DA, Hall JC, Kyriacou CP. Evolution of the threonine-glycine repeat region of the period gene in the melanogaster species subgroup of Drosophila. J Mol Evol. 1992;35:411–419. doi: 10.1007/BF00171819. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Davies OR, Savill JM, et al. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Slatkin M, Freimer NB. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Wierdl M, Dominska M, Petes TD. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F, Taghian DG, DeFrank JS, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci U S A. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Unrooted tree illustrating sequence similarity of 25 amino acids from different BRC repeats of various sequenced Drosophila species. Sibling species (D. simulans, D. sechellia, and D. persimilis) were excluded from the figure.
Supplementary Table 1 Drosophila species dmbrca2 BRC repeat amino acid sequences, FASTA-formatted.
Supplementary Table 2 Pairwise amino acid sequence differences among dmbrca2 BRC repeats within/between Drosophila species (Microsoft Excel formatted)