Abstract
Plant recalcitrance is the major barrier in developing Agrobacterium-mediated transformation protocols for several important plant species. Despite the substantial knowledge of T-DNA transfer process, very little is known about the factors leading to the plant recalcitrance. Here, we analyzed the basis of Hypericum perforatum L. (HP) recalcitrance to Agrobacterium-mediated transformation using cell suspension culture. When challenged with Agrobacterium, HP cells swiftly produced an intense oxidative burst, a typical reaction of plant defense. Agrobacterium viability started to decline and reached 99% mortality within 12 h, while the plant cells did not suffer apoptotic process. This is the first evidence showing that the reduction of Agrobacterium viability during co-cultivation with recalcitrant plant cells can affect transformation.
Keywords: Agrobacterium, Hypericum, Plant defense response, Plant recalcitrance
Introduction
Agrobacterium-mediated plant transformation is an indispensable tool in modern plant biology. Regardless of the availability of alternative plant transformation tools, Agrobacterium is the most preferred vehicle of gene delivery for its simplicity, cost-effectiveness and frequent single copy gene integration into the host plant genome. Having access to a wide array of useful genes in the post-genomic era, the interest of applying this system to all crops has been intensified. Amenability of different plant species, varieties, tissues and cells to Agrobacterium infection greatly varies, which makes transformation either inefficient or impossible in recalcitrant plants. Hence, there is a need to study the basis of plant recalcitrance towards this bacterium in the view of plant–pathogen interaction.
Agrobacterium has been generally recognized as a unique pathogen, which does not induce plant defense response (Robinette and Matthysse 1990; Felix et al. 1999). However, there are evidences for the induction of necrosis (Perl et al. 1996) and programmed cell death (Hansen 2000; Parrot et al. 2002) in plant cells after co-cultivation with Agrobacterium. Ditt et al. (2001, 2005, 2006) have shown that plants can modulate their gene expression pattern and trigger defense machinery in response to Agrobacterium. They also proposed that the plant defense system has an important role in controlling infection and transformation by Agrobacterium (Ditt et al. 2005). Most recently, Yuan et al. (2007) demonstrated that the plant signaling compound salicylic acid (SA) can shut down the expression of Agrobacterium vir regulon and directly affect the infection process. Taken these reports into account, the existence of plant resistance response against Agrobacterium seems evident.
Exploring the basis of plant recalcitrance to Agrobacterium-mediated transformation would be useful to improve the transformation efficiency of recalcitrant plant species. Such attempt has so for not been made in any of the recalcitrant plant species to the best of our knowledge. Hence, we analyzed the basis of Hypericum perforatum L. (HP) plant recalcitrance to Agrobacterium-mediated transformation in the current investigation. Our results show that HP plant defense response quickly reduces Agrobacterium viability and affects the transformation process during co-cultivation.
Materials and methods
Bacteria, culture conditions and inoculum preparation
Agrobacterium tumefaciens EHA105, A. rhizogenes A4 and Escherichia coli DH5α (Invitrogen, Carlsbad, CA, USA) were used in the present study. All the strains were transformed with the plasmid pCAMBIA1301 to obtain kanamycin resistance. A. tumefaciens and E. coli were cultured in Luria–Bertani (LB) medium and A. rhizogenes was cultured in YMA medium (0.4 gl−1 yeast extract, 10 gl−1 mannitol, 0.5 gl−1 K2HPO4, 0.2 gl−1 MgSO4 and 0.1 gl−1 NaCl, pH 7.0). Both LB and YMA media were augmented with 50 mgl−1 kanamycin. Broth cultures were initiated by inoculating single colonies of these bacteria into flasks (25 ml) containing 5 ml of liquid media. Cultures were kept at 200 rpm on a rotary shaker at specific temperatures (28°C for A. tumefaciens and A. rhizogenes; 37°C for E. coli) for 1 day. Subsequently, 2 ml of grown bacterial cultures were transferred to flasks (500 ml) containing 200 ml of the respective media and incubated as mentioned above. When the optical density (OD) of A. tumefaciens and A. rhizogenes cultures reached around 0.6 at 660 nm, 200 μM acetosyringone (AS) was added. Bacteria were spun down using a tabletop centrifuge (Sigma, St Louis, MO, USA) at 2,415 g, when the cultures reached approximately 1.0 OD at 660 nm. The pellets were gently suspended in 2 ml MS medium (Murashige and Skoog 1962) and used as bacterial stock.
Co-cultivation of Agrobacterium with HP cells
HP cell suspension culture was established from the variety “Helos” (Richters seeds, ON, Canada) as described earlier (Dias et al. 2001). After 5 days of subculture, HP suspended cells were aseptically collected in a sterile beaker. The cell density was estimated in an improved Neubauer haemocytometer (Neubauer, Wertheim, Germany) and adjusted to a final concentration of about 5,000 HP cells ml−1 using MS medium. A final concentration of 100 μM AS was added to the suspended cells. Flasks containing 80 ml of this HP cell suspension were inoculated with the stock of Agrobacteria (A. tumefaciens or A. rhizogenes) in a way to reach 250 × 107, 125 × 107 and 25 × 107 bacterial cells ml−1, concentrations generally used in plant transformation protocols. Flasks containing just HP cells served as HP control. Flasks containing 80 ml MS medium inoculated just with bacteria to a final concentration of 250 × 107 cells ml−1 served as the bacterial control. All the experiments were done in triplicate.
To examine the effect of HP cells on a non-pathogenic strain, E. coli DH5α was co-cultivated with HP cells. The effect of a non-recalcitrant plant on A. tumefaciens viability was scored by co-cultivating this bacterium with tobacco BY2 cells (PC-1181, DSMZ, Braunschweig, Germany).
All the treatment and control cultures were incubated under photoperiod at 25°C in a growth chamber during co-cultivation.
Intracellular reactive oxygen species (ROS) production
Intracellular reactive oxygen species (ROS) production in the control and Agrobacterium-inoculated HP cells was measured using 2′, 7′-dichlorodihydrofluorescein diacetate (H2DCFDA) probe (Molecular Probes, Invitrogen). Briefly, 1 ml of HP cell suspension taken at different post inoculation timings (0, 15, 30, 60, 120, 180, 240, 300 and 360 min) were transferred to Eppendorf tubes containing 10 μl of 200 μM H2DCFDA and mixed in a vortex for 20 s. After 15 min incubation in dark, the cells were spun down and 0.5 ml of the supernatant was transferred to a cuvette containing 2.5 ml sterile distilled water. Fluorescence of the samples was read in a Perkin Elmer LS50 spectrofluorimeter (Perkin Elmer, Buckinghamshire, UK) at 488 nm excitation and 525 nm emission with 1 s integration.
Measurement of cell viability
Control and treated plant cultures were checked for their cell viability after bacterial inoculation, for five consecutive days, by fluorescein diacetate (FDA) and propidium iodide (PI) double staining as described by Conde et al. (2007). Briefly, after thorough mixing using cut pipette tips, 1.0 ml cell suspension from each cultures were taken in Eppendorf tubes. To these samples, 10 μl FDA (500 μg μl−1, Sigma) and 1 μl PI, (500 μg μl−1 Sigma) were added, thoroughly mixed and incubated in dark at room temperature (±25°C). After 10 min of incubation, 100 μl of cell suspension was spread on a glass slide and observed under a Leica DM 5000B microscope (Leica Microsystems, Wetzlar, Germany) equipped with AF6000 fluorescent lamp (Leica Microsystems) programmed for excitation at 490 nm and emission at 510 nm, for FDA, and excitation at 543 nm and emission at 570 nm, for PI. Light microscopic and fluorescent images were acquired using a DFC350 camera (Leica Microsystems) attached to the microscope.
Additionally, 1 ml samples of HP cell suspension co-cultivated with Agrobacterium as described above were transferred to solid MS medium containing 0.5 mgl−1 NAA, and cultured to check if they grow normally.
The bacteria viability (A. tumefaciens, A. rhizogenes and E. coli) was also monitored during co-cultivation with HP cells. Briefly, 100 μl samples were taken from plant cell cultures at different time intervals (0, 6, 12, 24 and 48 h) after bacterial inoculation and spread on plates containing semi-solid LB (for A. tumefaciens and E. coli) or YMA (for A. rhizogenes) medium augmented with 50 mgl−1 kanamycin. In order to count the colony forming units (cfu), 10−6 dilution of these samples were also plated in the same manner. Plates were incubated in the dark at specific temperature mentioned before for each bacterial strain. A. tumefaciens and A. rhizogenes colonies were counted, respectively, after 2 and 4 days of incubation.
Moreover, media from control and HP cultures co-cultivated with Agrobacterium for 24 h were filtered through 0.45 μm sterile membrane filter (Schleicher and Schuell, Dassel, Germany). Each filtrate was inoculated with fresh Agrobacterium to a final concentration of 250 × 107 cells ml−1 and 100 μl aliquots were plated after 24 h incubation, and bacterial growth was checked after 2 days.
Effect of Agrobacterium co-cultivation on the stability of HP DNA
To observe whether Agrobacterium can induce DNA fragmentation of HP cells, DNA was isolated using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), from control and treatments after 1, 2 and 3 days of co-cultivation and 0.5 μg of DNA/lane were resolved in 0.8% agarose gel with ethidium bromide.
Northern-blot analysis of PAL gene expression in HP cells
Total RNA was isolated from control and A. tumefaciens treated HP cells after 0, 4, 12 and 24 h of inoculation following the procedure of Çakir et al. (2003). From each sample, 20 μg RNA was taken and separated in formaldehyde–agarose gel before transferring them to Hybond-N® membrane (Amersham Biosciences, Buckingham, UK) via capillary transfer. A PAL gene specific fragment amplified from HP cDNA library was labeled with α-[32P] dCTP (Amersham) using Prime-a-Gene® labeling kit (Promega, Madison, WI, USA) and used as probe for hybridization. Hybridized blots were washed twice with 2× SSC + 0.1% SDS (2 × 15 min) and with 0.1× SSC + 0.1% SDS (5 min) at 65°C. The blots were exposed to the imaging screen for 12 h and scanned in a Personal Molecular Imager (Bio-Rad, Hercules, CA, USA).
Assay for T-DNA transfer
The presence of T-DNA in the HP and tobacco BY2 cells after co-cultivation with A. tumefaciens was analyzed by GUS assay (Jefferson et al. 1987). Samples (1.0 ml) taken from plant cultures in successive days (1, 2, 3, 4 and 5 days) were centrifuged at 600 g. The supernatant was discarded and the pellet was thoroughly washed in sterile distilled water to get rid of the bacteria. Then the plant cells were re-suspended in 1.0 ml GUS solution and incubated at 37°C for 24 h. Cells were viewed under a light microscope for the presence of blue staining.
Statistical analysis
For all the treatments, three independent experiments (each with three replica) were done. Statistical analysis was performed using GraphPad Prism, version 4.00 for Windows (GraphPad Software, San Diego, CA, USA).
Results
Agrobacterium inoculation rapidly evokes HP defense response
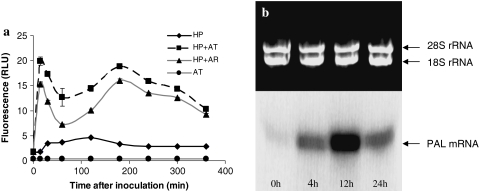
HP cells co-cultivated with A. tumefaciens or A. rhizogenes produced an intense biphasic ROS burst when compared to control cells (Fig. 1a). A first ROS peak was observed within 15 min and a second one between 2 and 5 h of co-cultivation (Fig. 1a). Moreover, PAL gene expression was up regulated as early as in 4 h after challenging with Agrobacterium and reached the peak in 12 h, declining thereafter (Fig. 1b).
Fig. 1.
Intracellular ROS production (a) in HP cells during co-cultivation with Agrobacterium. Control HP cells without bacteria (HP), HP cells co-cultivated with A. tumefaciens (HP + AT), HP cells co-cultivated with A. rhizogenes (HP + AR) and control A. tumefaciens without HP cells (AT). Northern-blot analysis (b) of PAL gene expression in HP cells after A. tumefaciens inoculation
Agrobacterium co-cultivation does not reduce HP cell viability
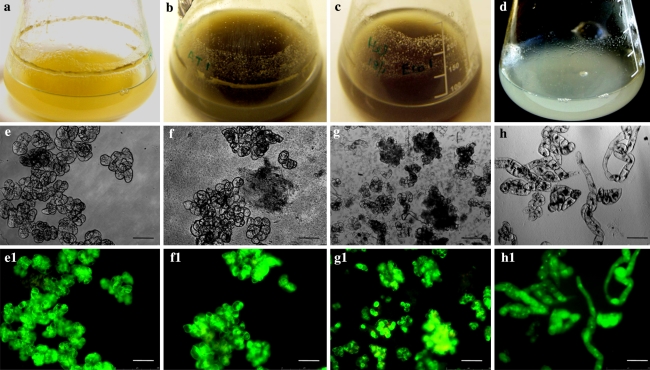
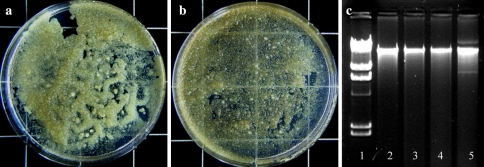
Control HP cultures remained characteristic yellowish throughout the experiment (Fig. 2a). Meanwhile, all the HP cultures inoculated with Agrobacterium and E. coli darkened within a day (Fig. 2b, c) irrespective of the bacterial concentration. Nevertheless, the intensity of darkness was directly proportional to bacterial concentration (data not shown). For example, the darkening was more intense and developed faster in the cultures inoculated with 250 × 107 bacterial cells ml−1 than in the lower bacterial densities. In the tobacco BY2 cultures co-cultivated with A. tumefaciens, no darkening was noticed (Fig. 2d). HP cells have remained viable after bacterial co-cultivation (Fig. 2f–f1, g–g1) similar to the control HP (Fig. 2e–e1) and tobacco BY2 cells (Fig. 2h–h1). These HP cells grew normally on solid MS medium as the control cells (Fig. 3a, b). Genomic DNA isolated from the HP cells co-cultivated with these bacteria did not show DNA laddering (Fig. 3c, lanes 3–5) similar to the control cells (Fig. 3c, lane 2).
Fig. 2.
Changes in HP cells in response to Agrobacterium co-cultivation. Control HP cell culture remain characteristic yellowish (a), HP cultures inoculated with A. tumefaciens (b) and with E. coli (c) became dark, while tobacco cells inoculated with A. tumefaciens (d) remain white. Light microscopic view of control HP cells (e), cells inoculated with A. tumefaciens (f) cells inoculated with E. coli (g), and tobacco cells inoculated with A. tumefaciens (h) and the corresponding epifluorescent view of the frames (e1–h1). HP cells remains viable after 3 days of co-cultivation with Agrobacterium, as shown by FDA-PI double staining (green fluorescence indicating viable cells and red fluorescence indicating dead cells). Bar = 125 μm
Fig. 3.
Growth of HP cells on solid MS medium established from control (a) and from cells co-cultivated for 5 days with A. tumefaciens (b) (pictures taken after 10 days of culture). c Agarose gel electrophorogram of HP genomic DNA isolated from control and cultures inoculated with bacteria after 3 days, lane 1HindIII digest of λ phage DNA, lane 2 control cells, lane 3 culture inoculated with A. tumefaciens, lane 4 culture inoculated with A. rhizogenes, lane 5 culture inoculated with E. coli
HP cells reduces Agrobacterium viability during co-cultivation
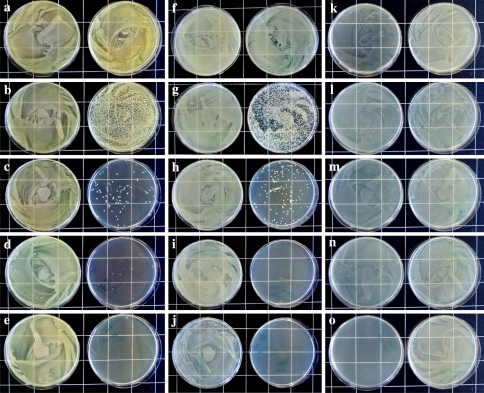
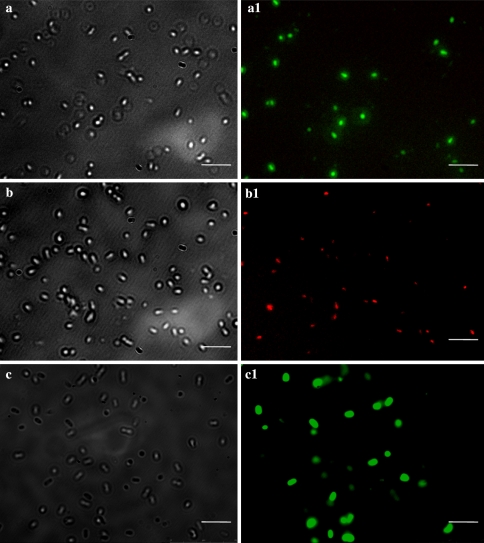
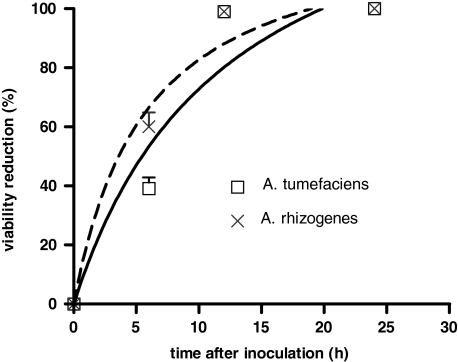
Agrobacterium viability declined quickly and reached complete mortality, when co-cultivated with HP cells (Fig. 4a–j). However, E. coli viability was not affected (Fig. 4k–o). Fluorescent staining showed that Agrobacterium retained 100% viability, when co-cultivated with tobacco BY2 cells (Fig. 5c–c1), similar to the control bacteria (Fig. 5a–a1). However, when co-cultivated with HP cells, they were completely killed in 24 h (Fig. 5b–b1). The number of colony forming units of A. tumefaciens and A. rhizogenes started to decline within 3 h and suffered 99% of viability reduction within 12 h of co-cultivation (Fig. 6).
Fig. 4.
Growth of bacteria after co-cultivation with HP cells. Plates spread with 100 μl suspension containing bacterial cells plus plant cells taken from HP cultures after 0, 6, 12, 24 and 48 h of contact with A. tumefaciens (a–e), A. Rhizogenes (f–j) and E. coli (k–o). Corresponding controls on the left side
Fig. 5.
A. tumefaciens viability of after co-cultivation with HP and tobacco cells. Bar = 25 μm. Light microscopic view of A. tumefaciens control (a), after 24 h co-cultivation with HP cells (b), and after 24 h co-cultivation with tobacco BY2 cells (c); epifluoresent view of the corresponding frames on the right (a1, b1 and c1)
Fig. 6.
Reduction of A. tumefaciens and A. rhizogenes colony forming units after 0, 6, 12 and 24 h of their inoculation into HP cells. To estimate the number of viable bacterial cells, HP culture was diluted until 10−6 and 100 μl (which contains approximately 250 bacterial cells) was spread on culture plates and incubated in dark at 28°C
The cell-free liquid media filtered from Agrobacterium-treated HP cultures inhibited the growth of Agrobacterium, whereas, the corresponding filtrate from control HP cultures did not show any negative effect.
Agrobacterium viability reduction affects HP transformation
Irrespective of the bacterial concentration and period of co-cultivation, HP cultures co-cultivated with A. tumefaciens did not show any transformed cells. Whereas, tobacco BY2 cell culture co-cultivated with A. tumefaciens had many transformed cells.
Discussion
Agrobacterium possess the unique ability of inter-kingdom gene transfer by introducing a defined piece of DNA, known as T-DNA (transferred DNA), from its tumour inducing (Ti) or root inducing (Ri) plasmid to the host plant cells by conjugal transfer. Integration of T-DNA into the plant genome and the expression of encoded genes lead to the development of crown gall or hairy roots in plants. This natural gene transfer phenomena has been exploited for the modern plant genetic engineering. Even though this transformation tool has been extended to several crops, many plant species remain recalcitrant. This might be due to their defence responses against these bacteria.
In any plant–pathogen interaction, one of the earliest events of plant stress response is an intense oxidative burst due to the excess production of ROS in the plant cells. Similar ROS burst was observed in HP cells clearly demonstrating that Agrobacterium impose an intense stress to the recalcitrant HP plant cells. Even though there is no report, the existence of a ROS burst in Agrobacterium–plant interactions could be the reason for the common utilization of antioxidants (ROS scavengers) for successful transformation (Perl et al. 1996; Frame et al. 2002). Hence, and based on our results, we assume that intense ROS production could be an event of Agrobacterium–plant interactions, which may affect transformation process.
The several fold up-regulation of PAL gene expression quickly after Agrobacterium inoculation indicates that the HP phenolic metabolism was activated immediately after sensing the bacterium. Increase in PAL synthesis and corresponding mRNA accumulation might be connected to the up-regulation of downstream products, which would either protect the plant cells or harm the bacteria. Transcriptome analysis of Arabidopsis thaliana challenged with Agrobacterium also revealed several fold up-regulation of phenolic pathway genes downstream of PAL (Ditt et al. 2006). Increased expression of PAL gene in response to microbial or endogenous elicitors in many other plant–pathogen systems (Gomez-Vasqez et al. 2004) is a known plant defense mechanism.
The time-course darkening of HP cells and corresponding PAL gene up-regulation clearly demonstrates that these two events are interrelated. Previously, it was shown that HP cells challenged with a fungal pathogen (Colletotrichum gloeosporioides), exhibited similar darkening together with change in the phenolic profile (Conceição et al. 2006). Darkening of explants frequently observed during Agrobacterium-mediated plant transformation also has generally been attributed to phenolic production, which may eventually lead to the death of plant cells (Perl et al. 1996; Hansen 2000; Parrot et al. 2002). In our case, HP cells did not lose their viability in spite of their darkening. Furthermore, the absence of DNA fragmentation in the HP cells treated with Agrobacterium confirm that the incompatibility of Agrobacterium-mediated transformation in HP is not due to apoptosis or programmed cell death, as reported in other species (Hansen 2000; Parrot et al. 2002).
The drastic reduction of Agrobacterium viability during co-cultivation with HP cells was not expected, since this bacterium is postulated not to induce plant defense response (Robinette and Matthysse 1990; Felix et al. 1999). There has been no previous evidence that these bacteria could be killed by plant cells during co-cultivation, as we have shown here. Antimicrobial potential of the cell-free liquid medium of Agrobacterium- treated HP cultures suggests that the HP cells have released some antimicrobial substance(s) to the media.
Since the A. rhizogenes strain A4 has reached 99% mortality within 12 h of co-cultivation with HP cells in the present study, the hairy root induction of HP reported previously (Vinterhalter et al. 2006) is paradoxical.
The high correlation between the time-course mortality rate of Agrobacterium and the PAL mRNA accumulation in the HP cells indicates that the activation of phenolic metabolism might play an important role in the killing. It is well known that HP plants produce certain antimicrobial secondary metabolites like hypericin and hyperforin, which could be induced in response to biotic elicitation (Sirvent and Gibson 2002). However, these compounds can accumulate only in specialized tissue glands (Pasqua et al. 2003) but not in suspended cells (Conceição et al. 2006). Moreover, the antimicrobial activity of these compounds is confined to Gram-positive bacteria (Avato et al. 2004). Altogether, these observations imply that the killing of Agrobacterium is not due to the antimicrobial activity of hypericin and/or hyperforin.
Histochemical GUS assay of HP cultures co-cultivated with A. tumefaciens revealed the absence of transformed cells, while several transformed cells could be seen in tobacco BY2 cultures, which were co-cultivated under similar conditions. T-DNA transfer failure of HP has shown to be independent of the explants types (leaf blade, petiole, stem, root etc.), Agrobacterium species (tumefaciens, rhizogenes), virulence of the bacterial strains (EHA 105, LBA4402, A4, LBA9402) and co-cultivation conditions (Franklin et al. 2007). However, when particle-bombardment was used as the method of gene delivery, HP cell suspension cultures were successfully transformed with the same plasmid (pCAMBIA1301) and transgenic plants could be obtained (Franklin et al. 2007). Taken together, these results strongly suggest that the recalcitrance of HP towards Agrobacterium-mediated transformation is not due to a block point at later steps in the transformation process (T-DNA integration or efficient transgene expression), but due to the direct effect of plant defense on Agrobacterium viability.
Conclusion
From our study, it is clear that HP recognizes Agrobacterium as a potential pathogen and rapidly evokes its defense responses, leading to the drastic reduction of Agrobacterium viability. This could be one of the main reasons for the recalcitrance of HP. Although Agrobacterium is recognized as a tool for plant transformation, naturally it is a potent pathogen causing neoplastic diseases. As the disease progresses independent of the causative agent following the initial transformation event, strategies to eliminate this pathogen must necessarily be exercised before infection to avoid T-DNA entry. In this context, further exploration of anti-Agrobacterium defense mechanism of HP and characterization of related genes would provide effective ways to control the neoplastic diseases.
Acknowledgments
We are grateful to Dr. David Tepfer (INRA, Versailles, France) for the gift of A. rhizogenes A4, Prof. Margarida Oliveira (IBET, Lisbon, Portugal) for providing A. tumefaciens EHA105, Prof. Richard Jefferson (CAMBIA, Australia) for the plasmid pCAMBIA1301 and Dr Paulo Silva (UM, Braga, Portugal) for helping in fluorescence microscopy. This work was supported by Fundação de Ciência e Tecnologia (POCTI/ AGR/ 40283/ 2001). Postdoctoral fellowship (SFRH/ BPD/ 17102/ 2004) awarded to G. Franklin by Fundação de Ciência e Tecnologia is gratefully acknowledged.
Abbreviations
- CFU
Colony forming unit
- FDA
Fluorescein diacetate
- H2DCFDA
2′,7′-Dichlorodihydrofluorescein diacetate
- HP
Hypericum perforatum
- PI
Propidium iodide
- ROS
Reactive oxygen species
References
- Avato P, Raffo F, Guglielmi G, Vitali C, Rosato A (2004) Extracts from St John’s wort and their antimicrobial activity. Phytother Res 18:230–232 [DOI] [PubMed]
- Çakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R (2003) A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 15:2165–2180 [DOI] [PMC free article] [PubMed]
- Conceição LFR, Ferreres F, Tavares RM, Dias ACP (2006) Induction of phenolic compounds in Hypericum perforatum L. cells by Colletotrichum gloeosporioides elicitation. Phytochemistry 67:149–155 [DOI] [PubMed]
- Conde C, Silva P, Agasse A, Lemoine R, Delrot S, Tavares R, Gerós H (2007) Utilization and transport of mannitol in Olea europaea and implications for salt stress tolerance. Plant Cell Physiol 48:42–53 [DOI] [PubMed]
- Dias ACP, Seabra RM, Andrade B, Ferreres F, Ferreira MF (2001) Xanthone production in calli and suspended cells of Hypericum perforatum. J Plant Physiol 158:821–827 [DOI]
- Ditt RF, Nester EW, Comai L (2001) Plant gene expression response to Agrobacterium tumefaciens. Proc Natl Acad Sci USA 11:10954–10959 [DOI] [PMC free article] [PubMed]
- Ditt RF, Nester EW, Comai L (2005) The plant cell defense and Agrobacterium tumefaciens. FEMS Microbiol Lett 247:207–213 [DOI] [PubMed]
- Ditt RF, Kerr KF, Figueiredo PD, Delrow J, Comai L, Nester EW (2006) The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant Microbe Interact 19:665–681 [DOI] [PubMed]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276 [DOI] [PubMed]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SEK, Li B, Nettleton DS, Pei D, Wang K (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129:13–22 [DOI] [PMC free article] [PubMed]
- Franklin G, Oliveira M, Dias ACP (2007) Production of transgenic Hypericum perforatum plants via particle bombardment-mediated transformation of novel organogenic cell suspension cultures. Plant Sci 172:1193–1203 [DOI]
- Gomez-Vasqez R, Day R, Buschmann H, Randles S, Beeching JR, Cooper RM (2004) Phenylpropanoids, phenylalanine ammonia lyase and peroxidases in elicitor-challenged cassava (Manihot esculenta) suspension cells and leaves. Ann Bot 94:87–97 [DOI] [PMC free article] [PubMed]
- Hansen G (2000) Evidence of Agrobacterium-induced apoptosis in maize cells. Mol Plant Microbe Interact 13:649–657 [DOI] [PubMed]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907 [DOI] [PMC free article] [PubMed]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497 [DOI]
- Parrot DL, Anderson AJ, Carman JG (2002) Agrobacterium induces plant cell death in wheat (Triticum aestivum L.). Physiol Mol Plant Pathol 60:59–69 [DOI]
- Pasqua G, Avato P, Monacelli B, Santamaria AR, Argentieri MP (2003) Metabolites in cell suspension cultures, calli, and in vitro regenerated organs of Hypericum perforatum cv. Topas. Plant Sci 165:977–982 [DOI]
- Perl A, Lotan O, Abu-Abied M, Holland D (1996) Establishment of an Agrobacterium-mediated transformation system for grape (Vitis vinifera L.): The role of antioxidants during grape-Agrobacterium interactions. Nat Biotechnol 14:624–628 [DOI] [PubMed]
- Robinette D, Matthysse AG (1990) Inhibition by Agrobacterium tumefaciens and Pseudomonas savastanoi of development of the hypersensitive response elicited by Pseudomonas syringae pv. Phaseolicola. J Bacteriol 172:5742–5749 [DOI] [PMC free article] [PubMed]
- Sirvent T, Gibson DM (2002) Induction of hypericins and hyperforin in Hypericum perforatum L. in response to biotic and chemical elicitors. Physiol Mol Plant Pathol 60:311–320
- Vinterhalter B, Ninkovi S, Cingel A, Vinterhalter D (2006) Shoot and root culture of Hypericum perforatum L. transformed with Agrobacterium rhizogenes A4M70GUS. Biol Plant 50:767–770 [DOI]
- Yuan Z-C, Edlind MP, Liu P, Saenkham P, Banta LM, Wise AA, Ronzone E, Binns AN, Kerr K, Nester EW (2007) The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci USA 104:11790–11795 [DOI] [PMC free article] [PubMed]