Abstract
Retroposons are ubiquitous transposable elements found in the genomes of most eukaryotes, including trypanosomatids. The African and American trypanosomes (Trypanosoma brucei and Trypanosoma cruzi) contain long autonomous retroposons of the ingi clade (Tbingi and L1Tc, respectively) and short nonautonomous truncated versions (TbRIME and NARTc, respectively), as well as degenerate ingi-related retroposons devoid of coding capacity (DIREs). In contrast, Leishmania major contains only remnants of extinct retroposons (LmDIREs) and of short nonautonomous heterogeneous elements (LmSIDERs). We extend this comparative and evolutionary analysis of retroposons to the genomes of two other African trypanosomes (Trypanosoma congolense and Trypanosoma vivax) and another Leishmania sp. (Leishmania braziliensis). Three new potentially functional retroposons of the ingi clade have been identified: Tvingi in T. vivax and Tcoingi and L1Tco in T. congolense. T. congolense is the first trypanosomatid containing two classes of potentially active retroposons of the ingi clade. We analyzed sequences located upstream of these new long autonomous ingi-related elements, which code for the recognition site of the retroposon-encoded endonuclease. The closely related Tcoingi and Tvingi elements show the same conserved pattern, indicating that the Tcoingi- and Tvingi-encoded endonucleases share site specificity. Similarly, the conserved pattern previously identified upstream of L1Tc has also been detected at the same relative position upstream of L1Tco elements. A phylogenetic analysis of all ingi-related retroposons identified so far, including DIREs, clearly shows that several distinct subfamilies have emerged and coexisted, though in the course of trypanosomatid evolution, only a few have been maintained as active elements in modern trypanosomatid (sub)species.
Retroposons, also called non-long-terminal-repeat retrotransposons, are ubiquitous elements that transpose through an RNA intermediate and are found in the genomes of most eukaryotes (10, 17). The current model for transposition of retroposons predicts that an element-encoded endonuclease performs a single-strand nick of the target DNA, generating an exposed 3′ hydroxyl that serves as a primer for reverse transcription of the element's RNA. Synthesis of the complementary strand of the new DNA copy of the retroposon is performed by the element-encoded reverse transcriptase (RT). The second single-strand nick is carried out on the other strand, a few base pairs downstream of the first nick, by the same element-encoded endonuclease, generating a primer for the second-strand synthesis of the retroelement. Retroposons are therefore flanked by a direct repeat called a target site duplication (TSD), which corresponds to the sequence between the two single-strand nicks. They also have a variable-length poly(A)- or A-rich 3′ tail, due to the involvement of an RNA intermediate (25).
Trypanosomatids are protozoan parasites of major medical and veterinary significance. They not only cause serious diseases, such as sleeping sickness (Trypanosoma brucei gambiense and T. brucei rhodesiense), Chagas disease (Trypanosoma cruzi), and leishmaniasis (Leishmania spp.) in humans, but also are a serious impediment to socioeconomic development by causing disease in domestic animals (T. brucei brucei, Trypanosoma congolense, and Trypanosoma vivax). T. brucei subspecies and T. cruzi belong to the genus Trypanosoma and constitute a monophyletic group distantly related to the Leishmania spp. (18, 26). The mammalian parasites of the genus Trypanosoma are further divided into the Salivaria (African trypanosomes, including T. brucei subspecies, T. congolense, and T. vivax) and the Stercoraria (South American trypanosomes, including T. cruzi). This division is based on their modes of transmission: predominantly inoculative by tsetse flies, as is the case in the African trypanosomes, or contaminative by a variety of bloodsucking insects, as is the case in the South American trypanosomes. T. vivax is placed at the most basal position of the Salivarian group, while separation between T. brucei and T. congolense is more recent (16, 35). The year 2005 saw the publication of the nuclear genomes of T. brucei, T. cruzi, and Leishmania major (1, 14, 15, 21), which also provided a remarkable opportunity to comprehensively analyze retroposons from lower eukaryotes in their genomic context (8, 14). In this study, we extend the analysis of retroposons into a wider comparative and evolutionary context using three other trypanosomatid genomes: those of Leishmania braziliensis (31), T. congolense, and T. vivax (our unpublished data).
Retroposons of the ingi clade constitute the most abundant transposable elements described in the genomes of T. cruzi and T. brucei (∼3% of the nuclear genome) (Table 1). The T. brucei ingi (5.25 kb) (here renamed Tbingi) and T. cruzi L1Tc (4.74 kb) elements are potentially functional and autonomous retroposons that encode a single large multifunctional protein containing the N-terminal apurinic/apyrimidinic-like endonuclease, the RT, the RNase H, and the C-terminal DNA-binding domains (24, 28, 30). Tbingi and L1Tc are dispersed in the genomes, although they show relative site specificity for insertion (4, 5). The trypanosome genome also contains small nonautonomous retroposons, namely, NARTc (0.26 kb) and RIME (here renamed TbRIME; 0.5 kb), respectively, that are related to the autonomous L1Tc and Tbingi but that do not encode their own mobilization machinery. Instead, their transposition requires the enzymatic activities of L1Tc or Tbingi, with which the elements form Tbingi/TbRIME and L1Tc/NARTc pairs of retroposons, similar to pairings that have been previously described in the case of human LINE1/Alu, eel UnaL2/UnaSINE1, and plant LINE/S1 retroposon pairs (12, 22, 23, 37). The trypanosome and Leishmania genomes also contain highly degenerate elements related to retroposons of the ingi clade named DIREs (for degenerate ingi-related elements) (7). Tbingi/TbRIME, L1Tc/NARTc, and DIREs share the first 79 residues, which constitute the hallmark of trypanosomatid retroposons (“79-bp signature”). Recently, small degenerate retroposons (∼0.55 kb) containing the “79-bp signature,” named LmSIDERs (for short interspersed degenerate retroposons), have also been identified in the genomes of L. major (9), Leishmania infantum, and L. braziliensis (34). LmSIDER constitutes the largest retroposon family described so far in trypanosomatids, and members are located in the 3′ untranslated regions of genes, where they play a role in the regulation of gene expression (3, 9, 29). In this paper, we report the identification and characterization of the full complement of ingi-related elements (potentially active retroposons, DIREs, and truncated elements) in the genomes of T. congolense, T. vivax, and L. braziliensis. We also analyzed the genomic environments of these retroelements to compare their mechanisms of retrotransposition. Our analysis shows that at least six retroposon families (the ingi1 to -6 subclades) belonging to the ingi clade are represented across trypanosomatid genomes. However, most of these families have been lost in individual genomes. T. congolense is the most retroposon-rich trypanosomatid, with two potentially active families belonging to the ingi1 and ingi6 subclades. None of the Leishmania spp. analyzed contain active ingi-related retroposons.
TABLE 1.
Retroposons of the ingi clade identified in the trypanosomatid genomes
| Species | Name | Size (bp) | Gene producta | Autonomous-activeb | Copy no.c | Reference |
|---|---|---|---|---|---|---|
| T. brucei | Tbingi | 5,250 | 1,657 | Autonomous-active | 115 | 5 |
| TbDIRE | ∼50,000 | NDd | Autonomous | 73 | 7 | |
| TbRIME | 500 | NCe | Active | 86 | 5 | |
| TbSIDER | ∼570 | NC | 22 | 9 | ||
| T. congolense | Tcoingi | 5,404 | 1,751 | Autonomous-active | 56f | |
| LITco | 4,733 | 1,505 | Autonomous-active | 12f | ||
| TcoDIRE | ∼50,000 | ND | Autonomous | 173f | ||
| T. vivax | Tvingi | 5,419 | 1,752 | Autonomous-active | 756f | |
| TvDIRE | ∼50,000 | ND | Autonomous | 108f | ||
| TvRIME | 1,030 | NC | Active | 58f | ||
| T. cruzi | L1Tc | 4,736 | 1,524 | Autonomous-active | 320 | 4 |
| TcDIRE | ∼50,000 | NC | Autonomous | 257 | 7 | |
| NARTc | 260 | NC | Active | 133 | 4 | |
| L. major | LmDIRE | ∼50,000 | ND | Autonomous | 52 | 7 |
| LmSIDER | ∼550 | NC | 1,858 | 9 | ||
| L. braziliensis | LbDIRE | ∼50,000 | ND | Autonomous | 65 | |
| LbSIDER | ∼550 | NC | 1,986 | 34 |
Number of amino acids contained in the multifunctional protein encoded by the consensus sequence of autonomous and active retroposons.
Autonomous retroposons potentially code for a protein responsible for their retrotransposition. Retroposons are considered active when bioinformatics analyses suggest recent retrotransposition events for most of the elements composing the family.
Copy number per haploid genome.
ND, not determined due to high sequence heterogeneity.
NC, noncoding retroposons.
The copy number corresponds to the number of each retroposon in the analyzed data sets, which total 41.8 Mb and 47.7 Mb for T. congolense and T. vivax, respectively. The size of the haploid genome is not known but should be in the range of 25 to 35 Mb; consequently, the copy number per haploid genome is probably overestimated.
MATERIALS AND METHODS
Detection of ingi-related sequences.
A comprehensive analysis of retroposons in the genomes of T. brucei, T. cruzi, and L. major has been reported previously (1, 4-7, 9, 14, 15, 21). We used TBLASTN with Tbingi and T. cruzi L1Tc amino acid sequences as queries to detect ingi-related sequences in the T. congolense (strain IL-3000, version 1), T. vivax (strain Y486, version 2), and L. braziliensis (clone MHOM/BR/75M2904, version 2) (31) genome data sets. The T. congolense genome assembly consisted of 3,181 contigs, totaling 41.8 Mb, while the T. vivax assembly contained 10,250 contigs, totaling 47.4 Mb, including 8,279 T. vivax contigs not assigned to chromosomes. To identify further ingi-related sequences and to precisely determine the element boundaries, several rounds of BLASTN and TBLASTN searches were performed, including at each step new retroposon sequences identified in the subject data set by these reiterative BLAST searches.
Reconstitution of the DIRE coding sequences.
To determine the approximate coordinates of degenerate ingi-related sequences (DIREs), an initial TBLASTN search was performed against the T. congolense, T. vivax, and L. braziliensis contigs using the Tbingi, Tcoingi (T. congolense ingi), Tvingi (T. vivax ingi), L1Tco, and L1Tc peptide sequences as queries. The models were refined and extended by approximately 300 nucleotides by searching with the corresponding peptide sequences against a protein database composed of previously identified Tbingi, Tcoingi, Tvingi, L1Tco and/or L1Tc peptides using the BLAST-extend-repraze algorithm developed at the J. Craig Venter Institute (JCVI; formerly The Institute for Genomic Research). A subsequent Smith-Waterman alignment between the proteins, including the translation of the extensions, allowed the examination of all translation frames. To tentatively reconstitute proteins from the analyzed DIREs, frameshifts were removed manually from the DNA sequences using the BLAST-extend-repraze output. This approach generated a pseudogene for each DIRE element, encoding a single ingi-like sequence, which in most cases contained numerous stop codons.
Phylogenetic analyses.
The RT, endonuclease, and RNase H amino acid domains were aligned using the multiple-alignment software CLUSTAL X (38), followed by minor manual adjustments using MacClade version 4.06 (Sinauer Associates, Inc.). The alignments of the RT domains are shown in Fig. S1 in the supplemental material. Phylogenetic trees were generated by the neighbor-joining method as implemented in PAUP version 4.0b10 (Sinauer Associates, Inc.), using the default parameters. Bootstrapping was also carried out using PAUP.
RESULTS
Identification of potentially active ingi-related retroposons in the T. vivax and T. congolense genomes.
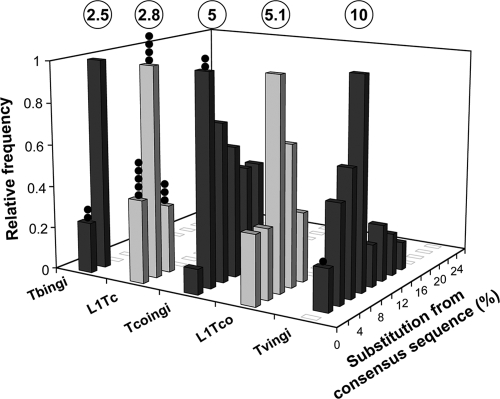
Retroposons in the T. brucei (strain TREU927/4), T. cruzi (strain CL), and L. major (strain Friedlin) genomes have previously been annotated and analyzed (4, 5, 9). Here, we identified and analyzed ingi-related retroposons in the draft genomes of T. congolense, T. vivax, and L. braziliensis (Table 1) using an iterative BLAST-based approach. The L. braziliensis genome contains 65 heterogeneous sequences (named LbrDIRE) with numerous frameshifts and/or in-frame stop codons, which inactivate their ingi-related coding sequences. In the T. vivax genome, 864 sequences were identified, including 108 TvDIREs. Based on nucleotide sequence analysis, the other 756 ingi-related sequences form a group of closely related elements, with the percentage of divergence between aligned nucleotide sequences larger than 3.5 kb (46 elements) ranging between 4.2% and 18.6% (median, 10%) (Fig. 1). The consensus sequence of this retroposon family, called Tvingi, is 5,419 bp long and encodes a 1,752-amino-acid protein sharing 31.6% identity with the Tbingi protein (Fig. 2 and Table 2). Only one Tvingi retroposon among the 11 full-length elements identified encodes a potentially functional protein.
FIG. 1.
Divergence between members of potentially active retroposons belonging to the ingi clade. Bases covered by the T. brucei (Tbingi), T. cruzi (L1Tc), T. congolense (Tcoingi and L1Tco), and T. vivax (Tvingi) retroposons were sorted by their divergence from their consensus sequences. The consensus sequences were determined from the alignment of full-length retroposons (Tbingi, 63 copies; L1Tc, 48 copies; Tcoingi, 27 copies) and all of the annotated elements larger than 300 bp (L1Tco, 8 copies) or 3.5 kb (Tvingi, 46 copies). The number of retroposons per fraction of 2% divergence is expressed as a fraction of the highest value, to which an arbitrary value of 1 was assigned. The percentage of divergence was calculated using the matching regions of the consensus sequences. The circled numbers at the top indicate the median value of each graph, and the closed circles represent the number of potentially active elements in each column.
FIG. 2.
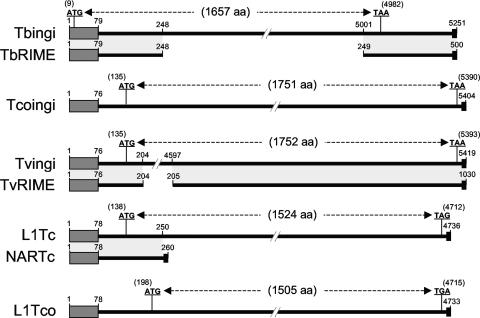
Schematic representation and comparison of potentially active autonomous and nonautonomous families of retroposons belonging to the ingi clade. The schematic maps are based on the consensus sequences generated from alignments of the Tbingi and TbRIME (T. brucei), Tcoingi and L1Tco (T. congolense), Tvingi and TvRIME (T. vivax), and L1Tc and NARTc (T. cruzi) retroposon sequences. The autonomous Tbingi, Tcoingi, Tvingi, and L1Tc consensus sequences contain a single long open reading frame coding for a multifunctional protein containing the endonuclease, RT, RNase H, and leucine zipper domains (the number of amino acids [aa] composing the encoded protein is indicated in parentheses). For L1Tco, four frameshifts were introduced into the consensus sequence to restore the putative coding sequence. The positions of the start and stop codons are indicated. Matches between autonomous and nonautonomous (TbRIME, TvRIME, and NARTc) retroposons are shown by light-gray boxes. The black boxes at the ends of both maps represent the poly(dA) terminal sequence. The N-terminal conserved motif, representing the trypanosomatid retroposon signature, is indicated by a dark-gray box.
TABLE 2.
Comparison of the proteins encoded by potentially active retroposons of the ingi clade
| Protein | % Identity (% similarity)
|
||||
|---|---|---|---|---|---|
| Tbingi | Tcoingi | Tvingi | L1Tc | L1Tco | |
| Tbingi | 100 | 32.4 (45.9) | 31.6 (45.8) | 20.9 (31.8) | 18.7 (28.9) |
| Tcoingi | 100 | 88.1 (92.8) | 18.4 (28.9) | 17.6 (26.7) | |
| Tvingi | 100 | 18.8 (28.9) | 19.1 (28.4) | ||
| L1Tc | 100 | 50.1 (66.1) | |||
| L1Tco | 100 | ||||
We also identified 241 ingi-related sequences in the T. congolense genome, which can be divided further into three groups. The first group is composed of 173 sequences containing highly degenerate coding sequences, called TcoDIREs. The other 68 sequences form two groups of closely related sequences, called Tcoingi (56 sequences) and L1Tco (12 sequences), showing a low percentage of divergence between their consensus sequences, each with a median value of ∼5% (Fig. 1). The Tcoingi consensus sequence is 5,404 bp long and encodes a 1,751-amino-acid protein sharing 32.4% and 88.1% identity with the Tbingi and Tvingi proteins, respectively (Fig. 2 and Table 2). The L1Tco consensus sequence is 4,733 bp long and, after four frame shifts were removed, coded for a 1,505-amino-acid protein sharing 50.1% identity with the L1Tc product (Fig. 2 and Table 2). Among the 11 full-length Tcoingi elements, 2 encode a potentially functional protein. The only full-length L1Tco element identified does not appear to code for a functional protein and therefore probably no longer encodes the retroposon machinery required for activation. However, one cannot exclude the possibility that the L1Tco subfamily is still active in the T. congolense genome. Indeed, the other 11 L1Tco sequences identified in the genome are truncated due to their positions at contig boundaries and could yet turn out to be potentially functional.
In contrast to the African trypanosome genomes and consistent with the genomes of L. major (7, 21) and L. infantum (data not shown), no potentially active ingi-related retroposons were detected in the L. braziliensis genome.
Phylogenetic analysis of the ingi clade retroposons.
Phylogenetic analyses of retrotransposons are commonly performed on the RT domain, which is the trademark of these mobile elements. As it is the most conserved retrotransposon domain (27), RT phylogeny is statistically more robust than phylogenetic trees generated with the endonuclease and RNase H domains. In order to perform a comprehensive phylogenetic analysis of all retroposons, we reconstituted DIRE proteins using matches to Tbingi, Tcoingi, Tvingi, L1Tc, and/or L1Tco proteins. Among the DIREs identified in the genomes of T. congolense (TcoDIRE; 173 copies), T. vivax (TvDIRE; 108 copies), and L. braziliensis (LbrDIRE; 65 copies), approximately half were successfully reconstituted, with gene products ranging between 199 and 1,702 amino acids. As observed before, all the ingi-related elements (113 DIREs from across the five species and the consensus sequences of Tbingi, Tcoingi, Tvingi, L1Tc, and L1Tco) form a monophyletic clade distinct from all the other retroposons, supported by a high bootstrap value (99%) (Fig. 3) (7). A pattern equivalent to the RT analysis was observed with the RNase H and endonuclease domains, using cellular domains as an outgroup (data not shown).
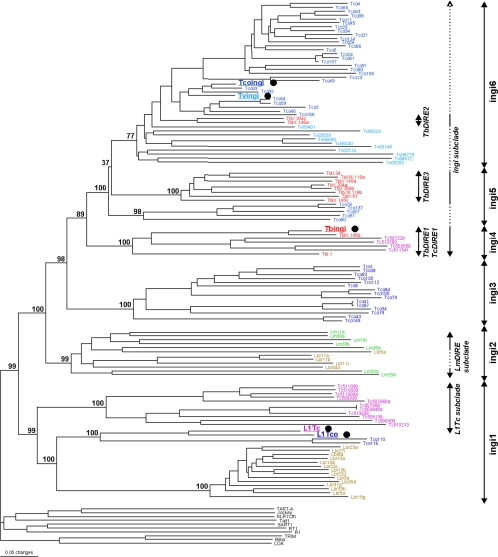
FIG. 3.
Phylogenetic analysis of the RT domain. The phylogeny is based on approximately 450 aligned amino acid residues (see Fig. S1 in the supplemental material for the alignment) corresponding to the entire RT domain of nontrypanosomatid retroposons, Tbingi, Tcoingi, Tvingi, L1Tc, L1Tco, and DIREs, which all belong to the ingi clade. The names of retroposons from T. brucei, T. congolense, T. vivax, T. cruzi, L. major, and L. braziliensis are in red, dark-blue, light-blue, magenta, green, and brown letters, respectively. The 10 nontrypanosomatid retroposons are representatives of the Jockey, Tad1, R1, CR1, and LOA retroposon clades (13). The consensus tree was generated by the neighbor-joining method and rooted on the RT sequences of nontrypanosomatid retroposons. The number next to each node indicates the bootstrap value as a percentage of 100 replicates corresponding to the tree generated by the neighbor-joining method. The black dots indicate potential functional autonomous retroposons. The names in italics on the right correspond to groups or subclades identified in a previous analysis (7), while the ingi1 to -6 subclades were defined from this phylogenetic analysis.
A previous phylogenetic analysis of the T. brucei, T. cruzi, and L. major sequences prompted us to consider three ingi subclades (Fig. 3) named L1Tc, LmDIRE, and ingi; the last was divided into three further groups, two composed of TbDIREs and the third containing TbDIREs, Tbingi, and TcDIREs (7). Although the inclusion of the T. congolense, T. vivax, and L. braziliensis ingi-related retroposons in this phylogenetic analysis did not change the overall structure of the tree, it considerably changed the complexity of each group/subclade, called here the ingi1 to ingi6 subclades (Fig. 3). The former T. cruzi L1Tc subclade (now called ingi1) is enriched with T. congolense (L1Tco and TcoDIREs) and L. braziliensis (LbrDIREs) sequences. The other LbrDIRE sequences belong to the ingi2 subclade (the former LmDIRE subclade), together with the LmDIRE elements. Among the three TbDIRE groups previously identified (7), (i) TbDIRE1 forms a monophyletic group with Tbingi and some of the TcDIRE sequences (ingi4 subclade); (ii) TbDIRE2 sequences are closely related to Tvingi, TvDIREs, Tcoingi, and the majority of the TcoDIREs (ingi6 subclade); and (iii) TbDIRE3 sequences are grouped with TcoDIRE sequences (ingi5 subclade). Finally, a subset of the TcoDIRE sequences forms a distinct group, called the ingi3 subclade. These data suggest that the evolution of the ingi-related retroposons in the trypanosomatid genomes is quite complex, with the contraction and expansion of many subfamilies during the evolution of these parasites. Among the six identified subfamilies, only three have retained potentially retrotransposition-competent retroposons, ingi1 in T. cruzi (L1Tc) and possibly T. congolense (L1Tco), ingi2 in T. brucei (Tbingi), and ingi6 in T. congolense (Tcoingi) and T. vivax (Tvingi). There is, however, no evidence of active elements in the Leishmania genomes.
Identification of conserved short nonautonomous retroposons in the African trypanosome genomes.
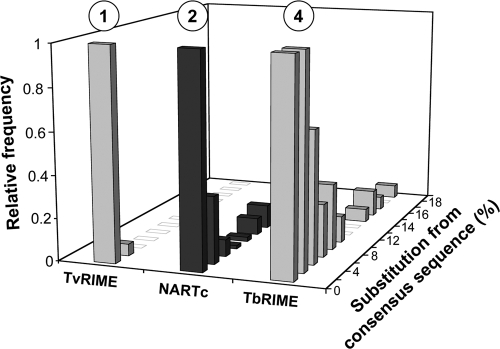
The T. brucei and T. cruzi genomes are rich in short nonautonomous retroposons, called TbRIME and NARTc, respectively, which are truncated versions of the long autonomous Tbingi and L1Tc sequences (6, 19) (Table 1 and Fig. 2). Several lines of evidence indicate that the T. vivax genome contains a short nonautonomous retroposon family (TvRIME) that corresponds to a truncated version of Tvingi: first, the TvRIME consensus sequences (1,030 bp long) share the first 204 and the last 826 residues with Tvingi (Fig. 2); second, the matching 1,030-bp residues between the TvRIME and Tvingi consensus sequences are 92% identical; third, all 58 of the identified TvRIME sequences are highly conserved, with the percentages of divergence from their deduced consensus sequence ranging between 0.1% and 2.3% (median, 1%) (Fig. 4), suggesting that they have been relatively recently mobilized and consequently are still active. It is noteworthy that all 58 of the TvRIME sequences identified in 15 contigs show a tandem arrangement, with the longest TvRIME cluster composed of 10 elements (Fig. 5). In contrast, the T. congolense genome does not contain retroposon families corresponding to shorter versions of the Tcoingi or L1Tco elements.
FIG. 4.
Divergence between members of short nonautonomous retroposons. Bases covered by the T. cruzi (NARTc), T. brucei (TbRIME), and T. vivax (TvRIME) retroposons were sorted by their divergence from their consensus sequences. The consensus sequences, determined from the alignment of full-length retroposons (NARTc, 115 copies; TbRIME, 70 copies; TvRIME, 26 copies) approximate the element's original sequence at the time of insertion. The number of retroposons per fraction of 2% divergence is expressed as a fraction of the highest value, to which an arbitrary value of 1 was assigned. The percentage of divergence was calculated using the matching regions of the consensus sequences. The circled numbers at the top indicate the median value of each graph.
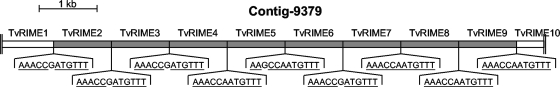
FIG. 5.
Organization of TvRIME clusters. Contig 9379 is composed of tandemly arranged TvRIME retroposons separated by the 12-bp TSD. This contig contains eight full-length TvRIME sequences (gray boxes) plus two truncated elements (white boxes) located at the extremities of the contig. The vertical double lines highlight both ends of the contig. The 12-bp TSDs located at the junctions between the TvRIME retroposons are shown, with the underlined residues corresponding to the consensus TSD sequence located between 41 TvRIMEs.
The closely related Tcoingi/Tvingi and L1Tc/L1Tco retroposons are preceded by the same conserved pattern.
Retroposons of the ingi clade show relative site specificity for insertion and are preceded by a conserved motif recognized by the element-encoded endonuclease domain, which performs two strand nicks at the target site of insertion (4, 5). In order to study the insertion sites of the Tcoingi, L1Tco, and Tvingi elements, we first considered all of the full-length elements identified in the T. congolense and T. vivax genomes. Only two of each showed a duplicated sequence (TSD) flanking the element, which constituted too small a data set to determine the target site consensus sequence (Fig. 6). As most of the T. brucei (Tbingi/TbRIME) and T. cruzi (L1Tc/NARTc) retroposons are flanked by a 12-bp TSD (4, 5), we extended this analysis to all of the Tcoingi, L1Tco, and Tvingi elements with an intact extreme 5′ end, considering that T. congolense and T. vivax TSDs may have the same conserved length (12 bp), as observed for the six full-length retroelements mentioned above (Fig. 6).
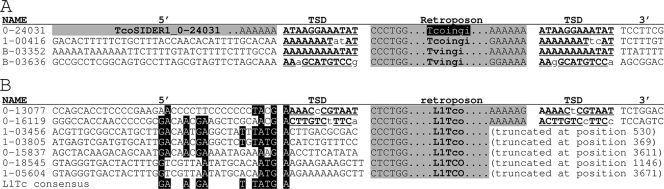
FIG. 6.
Comparison of the 5′ and 3′ adjacent sequences flanking the Tvingi and Tcoingi retroposons. (A) Only full-length Tcoingi and Tvingi flanked by a TSD presenting two mismatches at the most are considered. (B) The 5′ adjacent regions of L1Tco elements, which contain residues of the conserved motif located at the same relative position upstream of the T. cruzi L1Tc retroposon (4). Conserved residues are indicated by white letters on a black background. On the right, the names of the elements are indicated. The Tcoingi or L1Tco numbers with six characters correspond to the scaffold number (0 or 1) followed by the position (without the last three numbers) of the retroposon in the T. congolense scaffold. For the T. vivax retroposons (Tvingi), the numbers correspond to the contig numbers. The alignment of all the selected sequences was based on the retroposon sequences (gray column with “Tcoingi,” “Tvingi,” or “L1Tco”), from which only the first and last 6 bp, separated by the type of retroposon, are shown. The potentially functional Tcoingi elements that code for a full-length protein (1,751 amino acids) are indicated by white characters on a black background. The TSD flanking the retroposons is indicated by boldface letters, with underlined capital letters for the conserved residues; the lowercase letters in the TSD correspond to nonconserved residues. A gray background in the 5′ adjacent sequences (5′) means that the retroposon is preceded by another retroposon. For truncated retroposons, the position of end of the element is given.
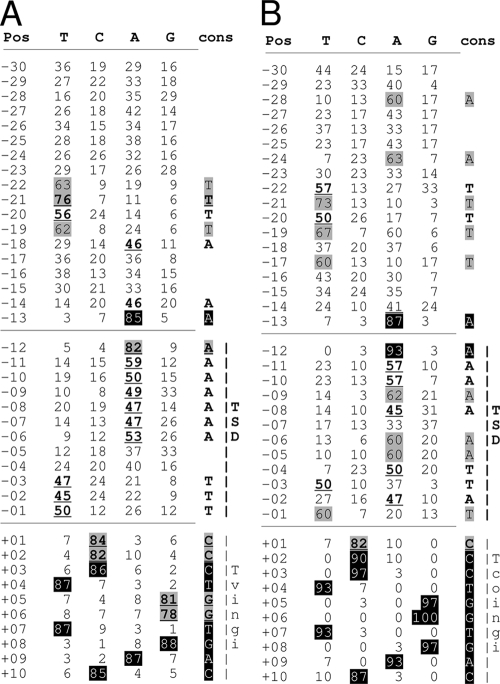
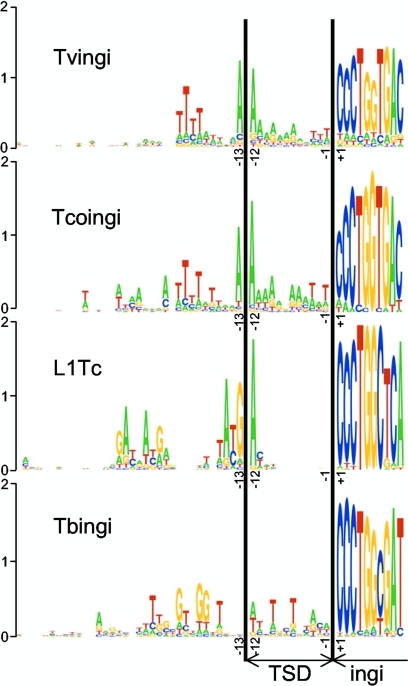
To determine the sequence conservation upstream of the Tvingi and Tcoingi elements, we considered only a single representative sequence of each group of nearly identical 5′ flanking sequences (110 and 30 sequences for Tvingi and Tcoingi out of 193 and 44 retroposons, respectively, with a 5′ extremity). Both the Tvingi and Tcoingi retroelements are preceded by well-conserved patterns (Fig. 7 and 8), which are similar (5′-TTTTAXXXAA↑AAAAAAAXXTTT-3′ and 5′-AXXXAXTTTTXTXXXA↑AAAAAXAATTAT-3′, respectively; the arrows indicate the putative first-strand cleavage sites; boldface residues show 60 to 75% conservation; boldface and underlined residues show 76 to 100% conservation). The most conserved residues are 2 adenine residues at positions −12 and −13 upstream of the Tvingi (82% and 85% conservation) and Tcoingi (93% and 87% conservation) elements. Interestingly, the most conserved residues upstream of L1Tc sequences are also located at positions −12 and −13, which flank the characterized first-strand cleavage site (Fig. 8). This strongly supports the view that the TSD size for Tvingi and Tcoingi elements is also 12 bp, as previously observed for Tbingi and L1Tc (4, 5). The conservation of the upstream pattern between the closely related Tcoingi and Tvingi retroposons suggests that the two retroelements recognize similar target sites for insertion.
FIG. 7.
Base frequencies at different positions (Pos) upstream of the Tvingi (A) and Tcoingi (B) retroposons. The frequencies were analyzed in the 30 bp upstream of the retroposon sequence (positions −30 to −01), including the duplicated 12-bp residues (TSD), of 110 Tvingi and 30 Tcoingi sequences. When upstream sequences showed more than 95% identity, only one sequence was retained for this analysis. The values in columns T, C, A, and G represent the percentages of the T, C, A, and G residues, respectively, at individual positions. Values greater than 45% are indicated as follows: 45 to 59%, underlined and boldface; 60 to 74%, gray shaded; 75 to 84%, underlined, boldface, and gray shaded; and 85 to 100%, white numbers on a black background. The last column (cons) shows the conserved residues.
FIG. 8.
Comparison of consensus sequences flanking ingi/L1Tc retroposons. The consensus sequence located upstream of the T. vivax (Tvingi), T. congolense (Tcoingi), T. brucei (Tbingi), and T. cruzi (L1Tc) retroelements is shown using the “sequence logo” graphic representation (11, 32). The analysis of the T. cruzi and T. brucei sequences was performed previously (4, 5). For T. vivax and T. congolense retroposons, the analysis was performed on the same data set used in Fig. 7. The logo representation displays the frequencies of bases at each position as the relative heights of letters, along with the degree of sequence conservation as the total height of a stack of letters, measured in bits of information. The vertical scale is in bits, with a maximum of 2 bits possible at each position. For the numbering, position +1 corresponds to the first residue of the retroelement. The first 10 residues of the retroelement sequence and the TSD are indicated.
The T. brucei and T. cruzi long autonomous and short nonautonomous retroposons of the same pair (Tbingi/TbRIME and L1Tc/NARTc, respectively) show exactly the same site specificity for insertion, as both members of the pair use the same retrotransposition machinery, which is encoded by the autonomous element (4, 5). Thus, one may expect that the same holds true for the Tvingi/TvRIME pair. The TvRIME retroposons form large clusters of tandemly repeated elements separated by the 12-bp TSD. Unfortunately, owing to the state of the draft genome assembly, all arrays of TvRIME sequences were truncated by contig boundaries. We were therefore unable to determine the conserved pattern upstream of TvRIME sequences. However, all 41 of the TSD sequences identified between TvRIMEs show at most two differences from the consensus sequences (−12 AAACCAATGTTT −1; boldface and underlined residues are shared with the consensus TSD flanking Tvingi sequences), suggesting that all of the TvRIME clusters are flanked by similar sequences and consequently are in the same genomic environment. The TSD sequences flanking TvRIMEs are similar to the consensus TSD flanking Tvingi (−12 AAAAAAAxxTTT −1; boldface and underlined residues are shared with the consensus TSD flanking TvRIME sequences), which supports the assumption that TvRIME also uses the Tvingi retrotransposition machinery.
Among the 12 L1Tco retroposons identified in the T. congolense genome data set, only 7 start with their 5′ extremities. Interestingly, two of them (1-03436 and 1-03805 in Fig. 6B) are preceded by all of the residues constituting the conserved motif located upstream of the L1Tc retroposons (5′-GAXXAXGAXXXTXTATG↑AXXXXXXXXXXX-3′) (4). At least half of these residues are also conserved upstream of the other five L1Tco sequences (Fig. 6B). These data suggest that the L1Tc and L1Tco retroposons, which are closely related in the phylogenetic tree (Fig. 3), also show the same site specificity for insertion.
DISCUSSION
A previous analysis of trypanosomatid ingi-related retroposons showed that all the retroelements identified in the T. brucei, T. cruzi, and L. major genomes (Tbingi, L1Tc, and DIREs) are grouped according to their species origin, with the exception of a few T. cruzi DIRE elements (the TcDIRE1 family) (Fig. 3), which are closely related to the T. brucei Tbingi and TbDIRE elements. We have previously interpreted these data as an indication of a lower rate of evolution for the TcDIRE1 sequences than for the other retroelements (7), as horizontal transfer of retroposons offers an unlikely explanation (27). The extension of this retroposon analysis to three other trypanosomatid genomes now provides a novel insight into the evolution of trypanosomatid retroposons and enables us to revise the previous interpretations. Indeed, retroposons of the ingi clade can be divided into six subclades, each subclade in turn containing members belonging to up to three different trypanosomatid (sub)species (ingi1 and ingi6 subclades in Fig. 3). This phylogenetic analysis clearly demonstrates that ingi subfamilies arose and disappeared in individual (sub)species during the evolution of trypanosomatid species.
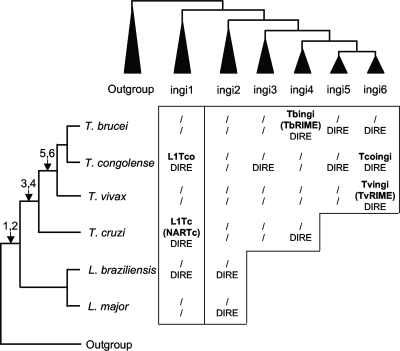
Two lines of evidence indicate that the ingi1 subclade was present in the trypanosomatid genome before Trypanosoma and Leishmania speciation. First, members of this subclade are present in the genomes of both Trypanosoma (T. congolense and T. cruzi) and Leishmania (L. braziliensis) (Fig. 9). Second, ingi1 sequences branch at the very base of the trypanosomatid RT tree and thus represent one of the most ancient ingi subclades identified (Fig. 3). The ingi2 subclade, which is composed of Leishmania DIREs (L. major and L. braziliensis), also has a basal position in the retroposon tree. The positions of the ingi1 and ingi2 sequences in the RT tree were also confirmed by the phylogenetic analyses of the endonuclease and RNase H domains (reference 7 and data not shown). Interestingly, ingi2 sequences are more closely related to ingi3 to -6 than ingi1 sequences, suggesting that the ingi1 subclade on one hand and all of the other sequences on the other hand form two different groups of retroposons. This separation of the ingi-related retroposons into two groups also clearly appears on the phylogenetic tree (Fig. 3). We therefore propose that the genome of the ancestral trypanosomatid (before Trypanosoma and Leishmania speciation) contained at least two ingi-related families, the ancestors of ingi1 and ingi2 to -6 (Fig. 9). The ingi5 and -6 subclades are present only in the genomes of African trypanosomes, while ingi4 sequences are also present in T. cruzi. This suggests that ingi4 sequences appeared in the trypanosome ancestor, followed by ingi5 and ingi6 in the African trypanosome ancestor. According to its position in the retroposon trees, the ingi3 subclade probably appeared before the Stercoraria/Salivaria speciation. We cannot exclude the possibility that ingi3 to -6 sequences were also present in the trypanosomatid genome before Trypanosoma/Leishmania speciation, although this hypothesis is unlikely, as (i) the phylogenetic analysis is in agreement with a relatively recent appearance of the ingi3 and -4 subclades, followed by ingi5 and -6 subclades, and (ii) the Leishmania and T. cruzi genomes are devoid of ingi3 to -6 and ingi5 and -6 sequences, respectively. According to the model presented in Fig. 9, only very few retroposon subfamilies are maintained in individual trypanosomatid genomes. Among the six ingi subclades identified, only one was identified in the genome of T. vivax. T. congolense is the most retroposon-rich trypanosomatid, with four identified subclades, two of which show potentially active elements (L1Tco and Tcoingi). In conclusion, Leishmania subspecies lost active retroposons of the ingi clade rapidly after their speciation, while trypanosomes maintained active retroposon families in their genomes. However, in the course of the evolution of trypanosome species, at least six subclades appeared, and elements of each clade remained in individual genomes as active families, as well as vestigial sequences. We can anticipate that the complexity of ingi evolution will increase with the number of trypanosomatid genomes analyzed.
FIG. 9.
Evolution of ingi retroposons in the trypanosomatid genomes. On the left is shown the phylogenetic relationship between trypanosomatid species and subspecies analyzed here. The tree was drawn according to data from Stevens et al. (36) and Gibson et al. (16). The arrows indicate the deduced times of appearance of the ingi1 to -6 subclades. At the top is a schematic representation of the retroposon phylogenetic tree shown in Fig. 3, and the central table shows the retroposons belonging to the ingi1 to -6 subclades identified in the genomes of these trypanosomatids. The names of potential active ingi-related retroposons are in boldface. Also indicated are vestiges of ingi-like retroposons (DIRE) and, in parentheses, the short nonautonomous retroposons (TbRIME, TvRIME, and NARTc) corresponding to active truncated versions of the long and autonomous elements mentioned above (Tbingi, Tvingi, and L1Tc, respectively). The slashes indicate loss of the active retroposon and/or the corresponding vestigial sequences.
So far, five potentially active retroposons of the ingi clade have been identified in T. brucei (Tbingi), T. congolense (Tcoingi and L1Tco), T. vivax (Tvingi), and T. cruzi (L1Tc). The closely related Tcoingi and Tvingi (86% and 88.1% identity at the nucleotide and amino acid levels, respectively) show the same conserved patterns upstream of the retroposons at positions −1 to −22 (Fig. 7 and 8). According to the current model of retrotransposition, this conserved motif represents the binding site of the retroposon-encoded endonuclease, which performs the site strand cleavage required to initiate target-primed reverse transcription of the retroelement (25). This indicates that the Tcoingi and Tvingi endonuclease domains, which are 93% identical, show the same site specificity. The L1Tc- and L1Tco-encoded endonucleases also share the same site specificity (Fig. 6B); however, they are not as closely related as the Tcoingi and Tvingi elements. Although L1Tc and L1Tco are closely related in the phylogenetic tree, they are poorly conserved at the nucleotide level and are only 50.1% identical at the amino acid level (the respective endonuclease domains are 43.5% identical). In contrast, the Tbingi endonuclease domain shows 44.7% and 43.4% identity with the Tcoingi and Tvingi endonuclease domains, respectively. However, the 5′-flanking regions do not share conservation (Fig. 8). Altogether, this comparative analysis of endonuclease domains and putative recognition sites of retroposons suggests that the trypanosomatid ingi elements may provide a good model to study the structure-function relationship of the retroposon endonuclease domains.
The rise and fall of retroelement families are well documented in eukaryotes, and it has been recently proposed that the extinction of transposable element families might be linked to molecular domestication events (2, 20, 33, 39). The expansion and domestication of two large families of short ingi-related retroposons have recently been described in the genomes of all Leishmania spp., which contain only the extinct retroposon families LmSIDER1 and LmSIDER2 (9, 34). Interestingly, members of both LmSIDER families have been domesticated by Leishmania to play a role in the regulation of gene expression at the posttranscriptional and/or posttranslational level (3, 9, 29). This massive expansion followed by domestication of transposable elements is seemingly confined to Leishmania spp. Indeed, large families of short ingi-related retroposons have not been identified in the genomes of T. brucei and T. cruzi (9) or in those of T. vivax and T. congolense.
T. brucei TbRIME (500 bp) and T. cruzi NARTc (260 bp) are short-retroposon families that have been successfully expanded in the respective genomes. Nucleotide comparisons of these retroelements and their flanking regions clearly demonstrated that TbRIME and NARTc are truncated versions of Tbingi and L1Tc, respectively, and that they use the retrotransposition machinery of the long autonomous elements for their own retrotransposition, thus forming the Tbingi/TbRIME and L1Tc/NARTc pairs (4, 5). Similarly, we identified in the T. vivax genome a truncated version of Tvingi (TvRIME), which is probably active to constitute the Tvingi/TvRIME pair. Clearly, the production of active truncated ingi-related elements occurred independently in the trypanosome genomes. First, the high level of sequence conservation between autonomous and nonautonomous members of each pair suggests that the deletion occurred quite recently in the evolution of trypanosomes, after T. brucei, T. vivax, and T. cruzi speciation. Second, the Tbingi/TbRIME, Tvingi/TvRIME, and L1Tc/NARTc pairs are distantly related and belong to different ingi subclades (Fig. 9). Third, the position of the deleted DNA fragment implies that different events led to the production of TbRIME, TvRIME, and NARTc elements (Fig. 2). In contrast, we did not detect any equivalent short versions of Tcoingi and L1Tco retroposons, suggesting that no active truncated elements appeared in the T. congolense genome. However, we cannot exclude the possibility that such a short active retroposon element evolved in the T. congolense genome and was subsequently lost in the course of the parasite's evolution.
Supplementary Material
Acknowledgments
We thank the core sequencing and informatics teams at the Wellcome Trust Sanger Institute for their assistance and the Wellcome Trust for its support of the Sanger Institute Pathogen Genomics and Pathogen Informatics groups. We are grateful to H. Valeins and P. Thebault for informatics support and bioinformatics advice.
F.B. was supported by the CNRS, the University Victor Segalen Bordeaux 2, and the Fondation de la Recherche Médicale. M.B. and C.H.-F. were funded by the Wellcome Trust (grant number WT085775/Z/08/Z).
Footnotes
Published ahead of print on 7 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, S. E. Melville, and N. M. El-Sayed. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 2.Bohne, A., F. Brunet, D. Galiana-Arnoux, C. Schultheis, and J. N. Volff. 2008. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 16:203-215. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, N., Y. Wu, C. Dumas, M. Dube, D. Sereno, M. Breton, and B. Papadopoulou. 2002. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 277:19511-19520. [DOI] [PubMed] [Google Scholar]
- 4.Bringaud, F., D. C. Bartholomeu, G. Blandin, A. Delcher, T. Baltz, N. M. El-Sayed, and E. Ghedin. 2006. The Trypanosoma cruzi L1Tc and NARTc non-LTR retrotransposons show relative site-specificity for insertion. Mol. Biol. Evol. 23:411-420. [DOI] [PubMed] [Google Scholar]
- 5.Bringaud, F., N. Biteau, E. Zuiderwijk, M. Berriman, N. M. El-Sayed, E. Ghedin, S. E. Melville, N. Hall, and T. Baltz. 2004. The ingi and RIME non-LTR retrotransposons are not randomly distributed in the genome of Trypanosoma brucei. Mol. Biol. Evol. 21:520-528. [DOI] [PubMed] [Google Scholar]
- 6.Bringaud, F., J. L. Garcia-Perez, S. R. Heras, E. Ghedin, N. M. El-Sayed, B. Andersson, T. Baltz, and M. C. Lopez. 2002. Identification of non-autonomous non-LTR retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 124:73-78. [DOI] [PubMed] [Google Scholar]
- 7.Bringaud, F., E. Ghedin, G. Blandin, D. C. Bartholomeu, E. Caler, M. J. Levin, T. Baltz, and N. M. El-Sayed. 2006. Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements. Mol. Biochem. Parasitol. 145:158-170. [DOI] [PubMed] [Google Scholar]
- 8.Bringaud, F., E. Ghedin, N. M. El-Sayed, and B. Papadopoulou. 2008. Role of transposable elements in trypanosomatids. Microbes Infect. 10:575-581. [DOI] [PubMed] [Google Scholar]
- 9.Bringaud, F., M. Muller, G. C. Cerqueira, M. Smith, A. Rochette, N. M. El-Sayed, B. Papadopoulou, and E. Ghedin. 2007. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 3:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capy, P., C. Bazin, D. Higuet, and T. Langin. 1998. Dynamics and evolution of transposable elements. Landes Bioscience, Austin, TX.
- 11.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewannieux, M., C. Esnault, and T. Heidmann. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41-48. [DOI] [PubMed] [Google Scholar]
- 13.Eickbush, T. H., and H. S. Malik. 2002. Origins and evolution of retrotransposons, p. 1111-1144. In A. G. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 14.El-Sayed, N. M., P. J. Myler, D. C. Bartholomeu, D. Nilsson, G. Aggarwal, A. N. Tran, E. Ghedin, E. A. Worthey, A. L. Delcher, G. Blandin, S. J. Westenberger, E. Caler, G. C. Cerqueira, C. Branche, B. Haas, A. Anupama, E. Arner, L. Aslund, P. Attipoe, E. Bontempi, F. Bringaud, P. Burton, E. Cadag, D. A. Campbell, M. Carrington, J. Crabtree, H. Darban, J. F. da Silveira, P. de Jong, K. Edwards, P. T. Englund, G. Fazelina, T. Feldblyum, M. Ferella, A. C. Frasch, K. Gull, D. Horn, L. Hou, Y. Huang, E. Kindlund, M. Klingbeil, S. Kluge, H. Koo, D. Lacerda, M. J. Levin, H. Lorenzi, T. Louie, C. R. Machado, R. McCulloch, A. McKenna, Y. Mizuno, J. C. Mottram, S. Nelson, S. Ochaya, K. Osoegawa, G. Pai, M. Parsons, M. Pentony, U. Pettersson, M. Pop, J. L. Ramirez, J. Rinta, L. Robertson, S. L. Salzberg, D. O. Sanchez, A. Seyler, R. Sharma, J. Shetty, A. J. Simpson, E. Sisk, M. T. Tammi, R. Tarleton, S. Teixeira, S. Van Aken, C. Vogt, P. N. Ward, B. Wickstead, J. Wortman, O. White, C. M. Fraser, K. D. Stuart, and B. Andersson. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409-415. [DOI] [PubMed] [Google Scholar]
- 15.El-Sayed, N. M., P. J. Myler, G. Blandin, M. Berriman, J. Crabtree, G. Aggarwal, E. Caler, H. Renauld, E. A. Worthey, C. Hertz-Fowler, E. Ghedin, C. Peacock, D. C. Bartholomeu, B. J. Haas, A. N. Tran, J. R. Wortman, U. C. Alsmark, S. Angiuoli, A. Anupama, J. Badger, F. Bringaud, E. Cadag, J. M. Carlton, G. C. Cerqueira, T. Creasy, A. L. Delcher, A. Djikeng, T. M. Embley, C. Hauser, A. C. Ivens, S. K. Kummerfeld, J. B. Pereira-Leal, D. Nilsson, J. Peterson, S. L. Salzberg, J. Shallom, J. C. Silva, J. Sundaram, S. Westenberger, O. White, S. E. Melville, J. E. Donelson, B. Andersson, K. D. Stuart, and N. Hall. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404-409. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, W. C., J. R. Stevens, C. M. Mwendia, J. N. Ngotho, and J. M. Ndung'u. 2001. Unravelling the phylogenetic relationships of African trypanosomes of suids. Parasitology 122:625-631. [DOI] [PubMed] [Google Scholar]
- 17.Goodier, J. L., and H. H. Kazazian, Jr. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23-35. [DOI] [PubMed] [Google Scholar]
- 18.Haag, J., C. O'Huigin, and P. Overath. 1998. The molecular phylogeny of trypanosomes: evidence for an early divergence of the Salivaria. Mol. Biochem. Parasitol. 91:37-49. [DOI] [PubMed] [Google Scholar]
- 19.Hasan, G., M. J. Turner, and J. S. Cordingley. 1984. Complete nucleotide sequence of an unusual mobile element from Trypanosoma brucei. Cell 37:333-341. [DOI] [PubMed] [Google Scholar]
- 20.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 21.Ivens, A. C., C. S. Peacock, E. A. Worthey, L. Murphy, G. Aggarwal, M. Berriman, E. Sisk, M. A. Rajandream, E. Adlem, R. Aert, A. Anupama, Z. Apostolou, P. Attipoe, N. Bason, C. Bauser, A. Beck, S. M. Beverley, G. Bianchettin, K. Borzym, G. Bothe, C. V. Bruschi, M. Collins, E. Cadag, L. Ciarloni, C. Clayton, R. M. Coulson, A. Cronin, A. K. Cruz, R. M. Davies, J. De Gaudenzi, D. E. Dobson, A. Duesterhoeft, G. Fazelina, N. Fosker, A. C. Frasch, A. Fraser, M. Fuchs, C. Gabel, A. Goble, A. Goffeau, D. Harris, C. Hertz-Fowler, H. Hilbert, D. Horn, Y. Huang, S. Klages, A. Knights, M. Kube, N. Larke, L. Litvin, A. Lord, T. Louie, M. Marra, D. Masuy, K. Matthews, S. Michaeli, J. C. Mottram, S. Muller-Auer, H. Munden, S. Nelson, H. Norbertczak, K. Oliver, S. O'Neil, M. Pentony, T. M. Pohl, C. Price, B. Purnelle, M. A. Quail, E. Rabbinowitsch, R. Reinhardt, M. Rieger, J. Rinta, J. Robben, L. Robertson, J. C. Ruiz, S. Rutter, D. Saunders, M. Schafer, J. Schein, D. C. Schwartz, K. Seeger, A. Seyler, S. Sharp, H. Shin, D. Sivam, R. Squares, S. Squares, V. Tosato, C. Vogt, G. Volckaert, R. Wambutt, T. Warren, H. Wedler, J. Woodward, S. Zhou, W. Zimmermann, D. F. Smith, J. M. Blackwell, K. D. Stuart, B. Barrell, and P. J. Myler. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 94:1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajikawa, M., and N. Okada. 2002. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111:433-444. [DOI] [PubMed] [Google Scholar]
- 24.Kimmel, B. E., O. K. Ole-MoiYoi, and J. R. Young. 1987. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol. Cell. Biol. 7:1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 26.Lukes, J., M. Jirku, D. Dolezel, I. Kral'ova, L. Hollar, and D. A. Maslov. 1997. Analysis of ribosomal RNA genes suggests that trypanosomes are monophyletic. J. Mol. Evol. 44:521-527. [DOI] [PubMed] [Google Scholar]
- 27.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 28.Martin, F., C. Maranon, M. Olivares, C. Alonso, and M. C. Lopez. 1995. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 247:49-59. [DOI] [PubMed] [Google Scholar]
- 29.McNicoll, F., M. Muller, S. Cloutier, N. Boilard, A. Rochette, M. Dube, and B. Papadopoulou. 2005. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 280:35238-35246. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, N. B., A. Pays, P. Tebabi, H. Coquelet, M. Guyaux, M. Steinert, and E. Pays. 1987. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol. 195:855-871. [DOI] [PubMed] [Google Scholar]
- 31.Peacock, C. S., K. Seeger, D. Harris, L. Murphy, J. C. Ruiz, M. A. Quail, N. Peters, E. Adlem, A. Tivey, M. Aslett, A. Kerhornou, A. Ivens, A. Fraser, M. A. Rajandream, T. Carver, H. Norbertczak, T. Chillingworth, Z. Hance, K. Jagels, S. Moule, D. Ormond, S. Rutter, R. Squares, S. Whitehead, E. Rabbinowitsch, C. Arrowsmith, B. White, S. Thurston, F. Bringaud, S. L. Baldauf, A. Faulconbridge, D. Jeffares, D. P. Depledge, S. O. Oyola, J. D. Hilley, L. O. Brito, L. R. Tosi, B. Barrell, A. K. Cruz, J. C. Mottram, D. F. Smith, and M. Berriman. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 39:839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit, A. F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 34.Smith, M., F. Bringaud, and B. Papadopoulou. 2009. Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genomics 10:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens, J., and A. Rambaut. 2001. Evolutionary rate differences in trypanosomes. Infect. Genet. Evol. 1:143-150. [DOI] [PubMed] [Google Scholar]
- 36.Stevens, J. R., H. A. Noyes, C. J. Schofield, and W. Gibson. 2001. The molecular evolution of Trypanosomatidae. Adv. Parasitol. 48:1-56. [DOI] [PubMed] [Google Scholar]
- 37.Tatout, C., L. Lavie, and J. M. Deragon. 1998. Similar target site selection occurs in integration of plant and mammalian retroposons. J. Mol. Evol. 47:463-470. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volff, J. N. 2006. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28:913-922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.