Abstract
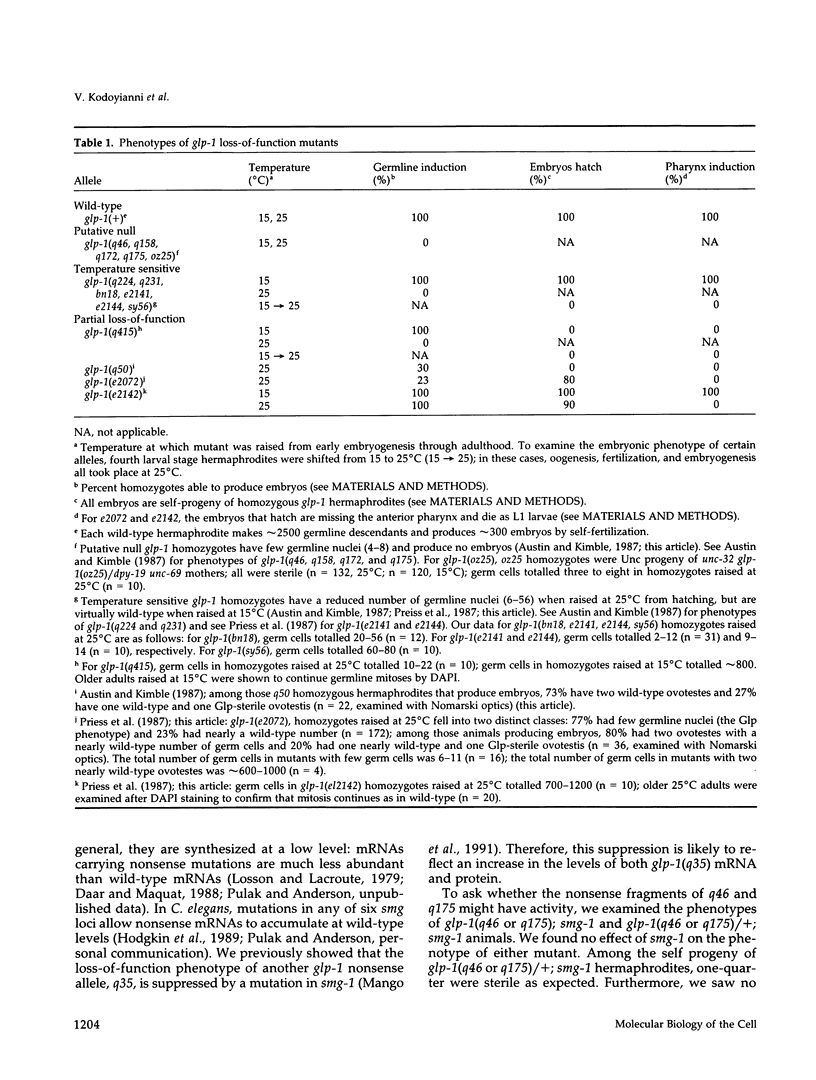
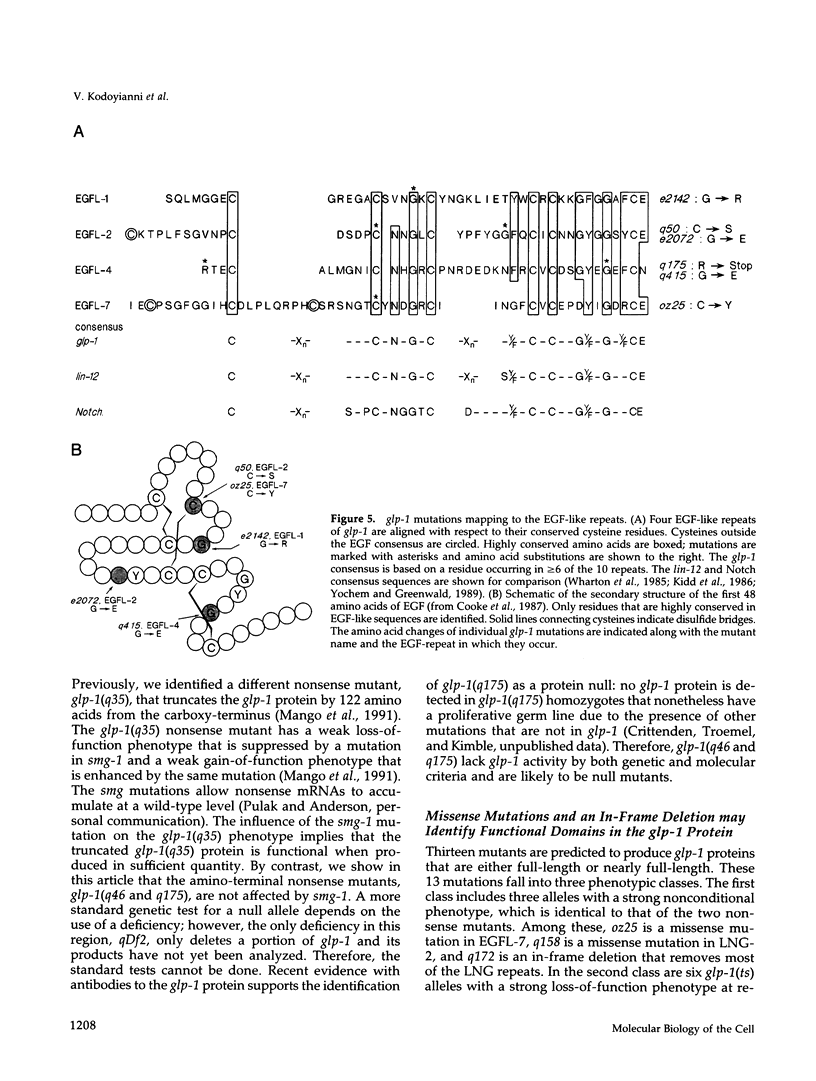
The glp-1 gene encodes a membrane protein required for inductive cell interactions during development of the nematode Caenorhabditis elegans. Here we report the molecular characterization of 15 loss-of-function (lf) mutations of glp-1. Two nonsense mutations appear to eliminate glp-1 activity; both truncate the glp-1 protein in its extracellular domain and have a strong loss-of-function phenotype. Twelve missense mutations and one in-frame deletion map to sites within the repeated motifs of the glp-1 protein (10 epidermal growth factor [EGF]-like and 3 LNG repeats extracellularly and 6 cdc10/SWI6, or ankyrin, repeats intracellularly). We find that all three types of repeated motifs are critical to glp-1 function, and two individual EGF-like repeats may have distinct functions. Intriguingly, all four missense mutations in one phenotypic class map to the N-terminal EGF-like repeats and all six missense mutations in a second phenotypic class reside in the intracellular cdc10/SWI6 repeats. These two clusters of mutations may identify functional domains within the glp-1 protein.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamson E. D. EGF receptor activities in mammalian development. Mol Reprod Dev. 1990 Sep;27(1):16–22. doi: 10.1002/mrd.1080270106. [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976 Aug 10;275(938):299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989 Apr 7;57(1):21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989 Dec 14;342(6251):830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- Appella E., Robinson E. A., Ullrich S. J., Stoppelli M. P., Corti A., Cassani G., Blasi F. The receptor-binding sequence of urokinase. A biological function for the growth-factor module of proteases. J Biol Chem. 1987 Apr 5;262(10):4437–4440. [PubMed] [Google Scholar]
- Aroian R. V., Koga M., Mendel J. E., Ohshima Y., Sternberg P. W. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature. 1990 Dec 20;348(6303):693–699. doi: 10.1038/348693a0. [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell. 1989 Aug 11;58(3):565–571. doi: 10.1016/0092-8674(89)90437-6. [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987 Nov 20;51(4):589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Aves S. J., Durkacz B. W., Carr A., Nurse P. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 'start' gene. EMBO J. 1985 Feb;4(2):457–463. doi: 10.1002/j.1460-2075.1985.tb03651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987 Feb 13;48(3):389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature. 1987 Oct 15;329(6140):651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman C., Harris W., Kintner C. Xotch, the Xenopus homolog of Drosophila notch. Science. 1990 Sep 21;249(4975):1438–1441. doi: 10.1126/science.2402639. [DOI] [PubMed] [Google Scholar]
- Cohen S., Carpenter G., King L., Jr Epidermal growth factor-receptor-protein kinase interactions. Co-purification of receptor and epidermal growth factor-enhanced phosphorylation activity. J Biol Chem. 1980 May 25;255(10):4834–4842. [PubMed] [Google Scholar]
- Cooke R. M., Wilkinson A. J., Baron M., Pastore A., Tappin M. J., Campbell I. D., Gregory H., Sheard B. The solution structure of human epidermal growth factor. 1987 May 28-Jun 3Nature. 327(6120):339–341. doi: 10.1038/327339a0. [DOI] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. G., Goldstein J. L., Südhof T. C., Anderson R. G., Russell D. W., Brown M. S. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987 Apr 23;326(6115):760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- Denton P. H., Fowlkes D. M., Lord S. T., Reisner H. M. Hemophilia B Durham: a mutation in the first EGF-like domain of factor IX that is characterized by polymerase chain reaction. Blood. 1988 Oct;72(4):1407–1411. [PubMed] [Google Scholar]
- Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., Smith S. D., Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991 Aug 23;66(4):649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L., Ruan K. S., Katzenberg D. Evidence for a transposon in Caenorhabditis elegans. Cell. 1983 Jan;32(1):55–65. doi: 10.1016/0092-8674(83)90496-8. [DOI] [PubMed] [Google Scholar]
- Graf J., Iwamoto Y., Sasaki M., Martin G. R., Kleinman H. K., Robey F. A., Yamada Y. Identification of an amino acid sequence in laminin mediating cell attachment, chemotaxis, and receptor binding. Cell. 1987 Mar 27;48(6):989–996. doi: 10.1016/0092-8674(87)90707-0. [DOI] [PubMed] [Google Scholar]
- Gray A., Dull T. J., Ullrich A. Nucleotide sequence of epidermal growth factor cDNA predicts a 128,000-molecular weight protein precursor. Nature. 1983 Jun 23;303(5919):722–725. doi: 10.1038/303722a0. [DOI] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983 Sep;34(2):435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Greenwald I., Rubin G. M. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell. 1992 Jan 24;68(2):271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- Greenwald I., Seydoux G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature. 1990 Jul 12;346(6280):197–199. doi: 10.1038/346197a0. [DOI] [PubMed] [Google Scholar]
- Hartley D. A., Xu T. A., Artavanis-Tsakonas S. The embryonic expression of the Notch locus of Drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein. EMBO J. 1987 Nov;6(11):3407–3417. doi: 10.1002/j.1460-2075.1987.tb02664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskill S., Beg A. A., Tompkins S. M., Morris J. S., Yurochko A. D., Sampson-Johannes A., Mondal K., Ralph P., Baldwin A. S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991 Jun 28;65(7):1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991 Mar 22;64(6):1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989 Oct;123(2):301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe P. E., Greenspan R. J. Local function of the Notch gene for embryonic ectodermal pathway choice in Drosophila. Cell. 1986 Aug 29;46(5):773–783. doi: 10.1016/0092-8674(86)90353-3. [DOI] [PubMed] [Google Scholar]
- Hoppe P. E., Greenspan R. J. The Notch locus of Drosophila is required in epidermal cells for epidermal development. Development. 1990 Aug;109(4):875–885. doi: 10.1242/dev.109.4.875. [DOI] [PubMed] [Google Scholar]
- Johansen K. M., Fehon R. G., Artavanis-Tsakonas S. The notch gene product is a glycoprotein expressed on the cell surface of both epidermal and neuronal precursor cells during Drosophila development. J Cell Biol. 1989 Nov;109(5):2427–2440. doi: 10.1083/jcb.109.5.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley M. R., Kidd S., Deutsch W. A., Young M. W. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell. 1987 Nov 20;51(4):539–548. doi: 10.1016/0092-8674(87)90123-1. [DOI] [PubMed] [Google Scholar]
- Kidd S., Baylies M. K., Gasic G. P., Young M. W. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev. 1989 Aug;3(8):1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- Kidd S., Kelley M. R., Young M. W. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986 Sep;6(9):3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. E., White J. G. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981 Jan 30;81(2):208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Kiyatkin N. I., Dulubova I. E., Chekhovskaya I. A., Grishin E. V. Cloning and structure of cDNA encoding alpha-latrotoxin from black widow spider venom. FEBS Lett. 1990 Sep 17;270(1-2):127–131. doi: 10.1016/0014-5793(90)81250-r. [DOI] [PubMed] [Google Scholar]
- Kurosawa S., Stearns D. J., Jackson K. W., Esmon C. T. A 10-kDa cyanogen bromide fragment from the epidermal growth factor homology domain of rabbit thrombomodulin contains the primary thrombin binding site. J Biol Chem. 1988 May 5;263(13):5993–5996. [PubMed] [Google Scholar]
- LaMarco K., Thompson C. C., Byers B. P., Walton E. M., McKnight S. L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991 Aug 16;253(5021):789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J. Genetic control of cell interactions in nematode development. Annu Rev Genet. 1991;25:411–436. doi: 10.1146/annurev.ge.25.120191.002211. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. 1991 May;112(1):231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux S. E., John K. M., Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990 Mar 1;344(6261):36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- Maine E. M., Kimble J. Identification of genes that interact with glp-1, a gene required for inductive cell interactions in Caenorhabditis elegans. Development. 1989 May;106(1):133–143. doi: 10.1242/dev.106.1.133. [DOI] [PubMed] [Google Scholar]
- Mango S. E., Maine E. M., Kimble J. Carboxy-terminal truncation activates glp-1 protein to specify vulval fates in Caenorhabditis elegans. Nature. 1991 Aug 29;352(6338):811–815. doi: 10.1038/352811a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione G. T., Wüthrich K., Nice E. C., Burgess A. W., Scheraga H. A. Solution structure of murine epidermal growth factor: determination of the polypeptide backbone chain-fold by nuclear magnetic resonance and distance geometry. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5226–5230. doi: 10.1073/pnas.84.15.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell D. J., Goodman C. S. Drosophila substrate adhesion molecule: sequence of laminin B1 chain reveals domains of homology with mouse. Cell. 1988 May 6;53(3):463–473. doi: 10.1016/0092-8674(88)90166-3. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Nolan G. P., Ghosh S., Liou H. C., Tempst P., Baltimore D. DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell. 1991 Mar 8;64(5):961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- Pikkarainen T., Eddy R., Fukushima Y., Byers M., Shows T., Pihlajaniemi T., Saraste M., Tryggvason K. Human laminin B1 chain. A multidomain protein with gene (LAMB1) locus in the q22 region of chromosome 7. J Biol Chem. 1987 Aug 5;262(22):10454–10462. [PubMed] [Google Scholar]
- Price J. V., Clifford R. J., Schüpbach T. The maternal ventralizing locus torpedo is allelic to faint little ball, an embryonic lethal, and encodes the Drosophila EGF receptor homolog. Cell. 1989 Mar 24;56(6):1085–1092. doi: 10.1016/0092-8674(89)90641-7. [DOI] [PubMed] [Google Scholar]
- Priess J. R., Schnabel H., Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987 Nov 20;51(4):601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Priess J. R., Thomson J. N. Cellular interactions in early C. elegans embryos. Cell. 1987 Jan 30;48(2):241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Rebay I., Fleming R. J., Fehon R. G., Cherbas L., Cherbas P., Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991 Nov 15;67(4):687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Rosenbluth R. E., Baillie D. L. The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics. 1981 Nov-Dec;99(3-4):415–428. doi: 10.1093/genetics/99.3-4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Kato S., Kohno K., Martin G. R., Yamada Y. Sequence of the cDNA encoding the laminin B1 chain reveals a multidomain protein containing cysteine-rich repeats. Proc Natl Acad Sci U S A. 1987 Feb;84(4):935–939. doi: 10.1073/pnas.84.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J., Urdea M., Quiroga M., Sanchez-Pescador R., Fong N., Selby M., Rutter W. J., Bell G. I. Structure of a mouse submaxillary messenger RNA encoding epidermal growth factor and seven related proteins. Science. 1983 Jul 15;221(4607):236–240. doi: 10.1126/science.6602382. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989 Jun 30;57(7):1237–1245. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Schedl T., Greenwald I. Cell-cell interactions prevent a potential inductive interaction between soma and germline in C. elegans. Cell. 1990 Jun 15;61(6):939–951. doi: 10.1016/0092-8674(90)90060-r. [DOI] [PubMed] [Google Scholar]
- Spence A. M., Coulson A., Hodgkin J. The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell. 1990 Mar 23;60(6):981–990. doi: 10.1016/0092-8674(90)90346-g. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts V. J., Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991 Sep;113(1):199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Yochem J., Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989 Aug 11;58(3):553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988 Oct 6;335(6190):547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]


