Abstract
The function of human TFIIH-associated Cdk7 in RNA polymerase II (Pol II) transcription and C-terminal domain (CTD) phosphorylation was investigated in analogue-sensitive Cdk7as/as mutant cells where the kinase can be inhibited without disrupting TFIIH. We show that both Cdk7 and Cdk9/PTEFb contribute to phosphorylation of Pol II CTD Ser5 residues on transcribed genes. Cdk7 is also a major kinase of CTD Ser7 on Pol II at the c-fos and U snRNA genes. Furthermore, TFIIH and recombinant Cdk7-CycH-Mat1 as well as recombinant Cdk9-CycT1 phosphorylated CTD Ser7 and Ser5 residues in vitro. Inhibition of Cdk7 in vivo suppressed the amount of Pol II accumulated at 5′ ends on several genes including c-myc, p21, and glyceraldehyde-3-phosphate dehydrogenase genes, indicating reduced promoter-proximal pausing or polymerase “leaking” into the gene. Consistent with a 5′ pausing defect, Cdk7 inhibition reduced recruitment of the negative elongation factor NELF at start sites. A role of Cdk7 in regulating elongation is further suggested by enhanced histone H4 acetylation and diminished histone H4 trimethylation on lysine 36—two marks of elongation—within genes when the kinase was inhibited. Consistent with a new role for TFIIH at 3′ ends, it was detected within genes and 3′-flanking regions, and Cdk7 inhibition delayed pausing and transcription termination.
Dynamic modification of the RNA polymerase II (Pol II) C-terminal domain (CTD) by phosphorylation and dephosphorylation plays important roles in controlling both transcription and cotranscriptional RNA processing (9, 31). There are 52 heptad repeats with the consensus sequence Y1S2P3T4S5P6S7 in the human CTD that can be phosphorylated cotranscriptionally on serines 2, 5, and 7 (S2, S5, and S7, respectively) (6, 31). S5 phosphorylation normally occurs early in the transcription cycle coincident with initiation, whereas S2 phosphorylation predominates later, during elongation and termination (13, 20). The complex pattern of heptad repeat phosphorylation serves in part to control binding of partner proteins, including elongation factors, RNA processing factors (31), and chromatin modifiers (25, 39). Ser5 phosphorylation enhances cotranscriptional mRNA capping (17), and Ser2 facilitates 3′-end formation by cleavage/polyadenylation (1, 4, 29). Ser7 phosphorylation has been specifically implicated in formation of U snRNA 3′ ends but is also found on mRNA coding genes (10) (6). Exactly how S2, S5, and S7 phosphorylation affect initiation, elongation, and termination remains poorly understood.
It is generally thought that the positive transcription elongation factor PTEFb (Cdk9/cyclin T [CycT]) is the principal S2 kinase and that Cdk7 associated with TFIIH is the principal S5 kinase (31, 32, 38). How Cdk7 affects CTD phosphorylation on metazoan genes in vivo is still an unresolved question, and the S7 kinase is yet to be identified. In the only previous investigation of Cdk7 effects on CTD phosphorylation at the site of transcription in a multicellular organism, phospho-S5 and total Pol II levels were reduced equivalently at the Drosophila melanogaster Hsp70 gene in a Cdk7 temperature-sensitive (TS) mutant, and it was suggested that multiple S5 kinases may operate in vivo (36). Indeed, there is evidence that Cdk9 can phosphorylate CTD S5 from RNA interference knockdown of this kinase in Drosophila (11) and from the fact that the Cdk9 inhibitor 5,6-5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) reduced S5 phosphorylation on the human p21 gene (14).
Promoter-proximal pausing is an obligate step in the RNA Pol II transcription cycle for thousands of human genes and constitutes a rate-limiting step in mRNA synthesis with particular importance for inducible genes and in stem cells (8, 16, 21). Paused Pol II complexes have recently been shown to regulate enhancer function by acting as insulators (7). The mechanisms responsible for establishment of the pause are not fully understood; however, two key regulators have been identified: negative elongation factor NELF and DRB sensitivity-inducing factor DSIF (Spt4/5) (30, 33, 41). Knockdown of NELF inhibits transcription of some genes, suggesting that the promoter-proximal pause can facilitate gene activation (12). Pausing is antagonized by the positive transcription elongation factor PTEFb (Cdk9/CycT), which can phosphorylate DSIF, NELF, and the Pol II CTD and is sensitive to the inhibitor, DRB (27, 30, 33, 41-43). Exactly how the TFIIH-associated Cdk7 activity affects Pol II transcription remains unclear. The kinase is dispensable for initiation in vitro in many cases (26, 37, 40) but has been implicated in early elongation of dihydrofolate reductase transcripts (3). Interestingly, in Drosophila, inactivation of TS Cdk7 reduced the amount of Pol II accumulated at the 5′ end prior to gene activation, suggesting that TFIIH helps establish promoter-proximal pausing (36). However, it is difficult to distinguish in this case whether reduced amounts of paused Pol II are specifically caused by loss of Cdk7 activity or by a secondary effect on initiation due to destabilization of TFIIH, which requires Cdk7 for its integrity (40).
Pol II encounters a second prominent pause site on many human genes in the 3′-flanking region several hundred base pairs past the poly(A) site (13, 15), and following this pause, termination occurs. S2 phosphorylation is maximal at the 3′ pause, and DRB inhibition of the S2 kinase Cdk9 inhibits 3′ pausing (13). Whether TFIIH-associated Cdk7 might also function in 3′ pausing and termination is unknown.
In this report we investigated the roles of Cdk7 in CTD phosphorylation and polymerase pausing on human genes using mutant HCT116 cells in which both copies of the Cdk7 gene driven by its endogenous promoter were replaced by homologous recombination with an analogue-sensitive (as) mutant of the ATP-binding site (23). This strategy permits specific inhibition of kinases with cell-permeable ATP analogues and has been used to target budding yeast Cdk7 (18), human Cdk7 (23), and many other kinases in vivo (19). This strategy is preferable to TS or deletion mutations or RNA interference-mediated knockdown because it permits one to distinguish between the catalytic and structural functions of kinases (19). We combined targeted inhibition of Cdk7(as) with high-resolution chromatin immunoprecipitation (ChIP) analysis of Pol II and associated elongation factors to show that this kinase can phosphorylate S7 and S5 in vivo and affects both the 5′ and 3′ pausing events that are characteristic features of transcription on many human genes. While this work was in progress, it was independently reported that budding yeast TFIIH phosphorylates CTD Ser7 residues (2).
MATERIALS AND METHODS
ChIP.
Wild-type HCT116 or Cdk7as/as cells were treated with 3-methylbenzyl-pyrazolopyrimidine (3-MB-PP1) (1 μM), doxorubicin (0.4 μM), and DRB (50 μM, Sigma) for 8 h (except for Fig. S1B in the supplemental material), and ChIP was performed as described previously (13, 14, 23) except that 1 ml of extract at 1.5 mg/ml protein was used per IP. Control samples were all treated for the same period with dimethyl sulfoxide (DMSO) solvent. The average maximum ChIP signals obtained on c-myc in arbitrary fluorescence units are 101 for anti-total Pol II (pan-CTD), 35 for anti-Spt5, 6.5 for p62, 2.2 for Cdk7, 1.2 for Cdk9, 45 for phosphorylated Ser2 (Ser2-PO4), and 40.5 for Ser7-PO4 compared to less than 0.1 for the no-antibody control and less than 0.05 for the mitochondrial CoxIII gene.
Antibodies.
Antibodies against the following antigens have been previously described: pan-CTD and CTD Ser5-PO4 (35), histone 3 (H3) C terminus, acetylated histone 4 (H4), histone H3 trimethylated at lysine 4 (H3K4me3), and CTD Ser2-PO4 (46), rabbit TFIIH p62 and Cdk7 (44), TFIIB, Spt5 (13), and ERCC2 (34). Monoclonal anti-Cdk7 (Zymed) (see Fig. 1B) and anti-CTD Ser7-PO4 (4E12) (6) were used. Anti-NELF-A was from Santa Cruz (sc-23599), anti-Cdk9 was from Santa Cruz (sc-8338), and H3K36me3 was from Abcam (antibody 9050). Anti-CTD and Ser5-PO4 antibodies used in Fig. 6A were from Bethyl Labs.
FIG. 1.
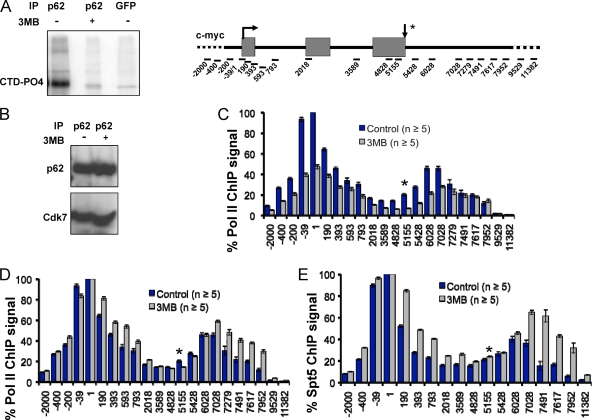
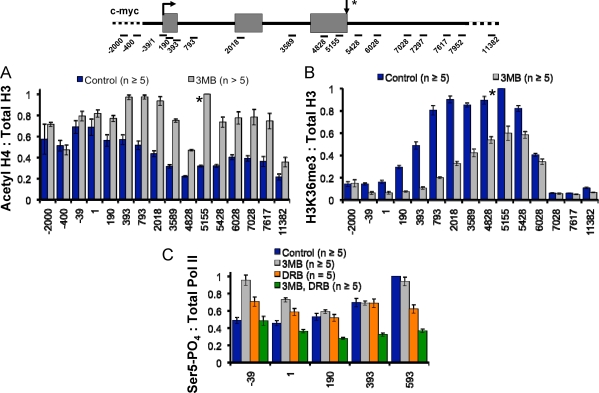
(A) Inhibition of analogue-sensitive TFIIH-associated kinase by 3-MB-PP1. IP kinase assays from nuclear extract of HCT116 Cdk7as/as cells immunoprecipitated with anti-TFIIH p62 or anti-green fluorescent protein (anti-GFP) as a control were performed with GST-CTD and [γ-32P]ATP. An autoradiograph of a sodium dodecyl sulfate-polyacrylamide gel is shown, and the position of phosphorylated GST-CTD (CTD-PO4) is indicated. For the two p62 lanes, a single IP reaction mixture was divided in two for the kinase reactions with 3-MB-PP1 (3MB) (2 μM) (+) or an equal volume of DMSO (−). (B) Cdk7(as) remains associated with TFIIH in the presence of kinase inhibitor. TFIIH was precipitated with anti-p62 from nuclear extracts prepared from control HCT116 Cdk7as/as cells (−) and cells treated with 3-MB-PP1 (1 μM) for 8 h (+). Immunoprecipitates were analyzed by Western blotting with anti-p62 (top) and anti-Cdk7 (bottom). (C and D) Promoter-proximal pausing is depressed by inhibition of Cdk7 kinase. ChIP signals for total Pol II along c-myc in control (DMSO) and 3-MB-PP1 (3MB)-treated Cdk7as/as cells. Values are normalized to the maximum signal for the control data set. Note the broadening of the 5′ peaks of Pol II and the shift of the 3′ peaks into the downstream flanking sequences when Cdk7 was inhibited. In panel D, Pol II ChIP signals from panel C are normalized to the maximum signal for each data set. (E) Cdk7 inhibition alters the 5′-3′ profile of Spt5 in parallel with Pol II. ChIP signals are normalized to the maximal signal for each data set. In this and all subsequent figures, ChIP signals are represented by the means ± SEMs (error bars). The arrow pointing down and asterisk indicate RNA 3′ ends in all subsequent figures.
FIG. 6.
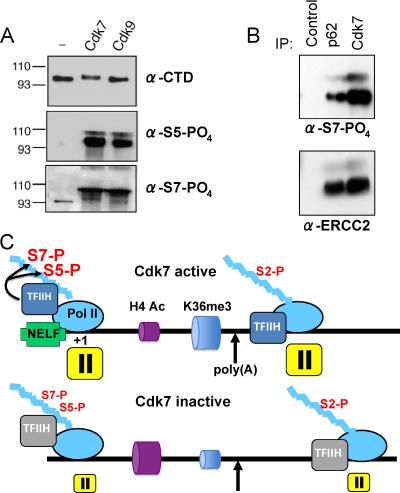
(A) Recombinant Cdk7 and Cdk9 both phosphorylate CTD S5 and S7. Kinase assays were performed with GST-CTD substrate, and the products were immunoblotted with antibodies specific to total CTD, phospho-S2, phospho-S5 (Bethyl), or phospho-S7 (4E12). α-CTD, antibody to total CTD; α-S5-PO4, antibody to phosphorylated serine 5. The positions of molecular size markers (in kilodaltons) are shown to the left of the gels. (B) Immunoprecipitated TFIIH phosphorylates CTD S7. Immunoprecipitates from HeLa nuclear extract with control antibody, anti-p62 or anti-Cdk7 were incubated with GST-CTD substrate and ATP and immunoblotted for phospho-S7 (4E12) and the ERCC2 subunit of TFIIH as a loading control. S7 phosphorylation was dependent on the addition of ATP (not shown). α-S7-PO4, antibody to phosphorylated serine 7. (C) Summary of Cdk7 effects on CTD phosphorylation, pausing, and chromatin modification. When Cdk7 is inhibited, CTD S5 and S7 phosphorylation (S5-P and S7-P, respectively) is reduced to different extents on different genes. In addition, promoter-proximal pausing is suppressed and the 3′-flanking region pause, where CTD S2 phosphorylation is maximal, shifts further downstream. During elongation within the gene, H4 acetylation (H4 Ac) is enhanced and H3K36 trimethylation (K36me3) is reduced when Cdk7 is inhibited.
Real-time PCR.
PCRs (10 μl) were performed with Sybr green using the Roche LC-480 (Roche Applied Science) in 384-well plates as described previously (13). Primer pairs with amplification efficiencies of >2.2 or <1.8 were discarded. Primer sequences were described previously (13) and are shown in Table 1. ChIP DNA samples were quantified by extrapolation from standard curves of input chromatin for each amplicon using the absolute quantification second derivative maximum method with Lightcycler 480 1.2 software (Roche). To generate 5′-3′ profiles of occupancy, ChIP values were normalized relative to the amplicon with the highest fluorescence value for each gene. The values were then averaged, and the standard errors of means (SEMs) were calculated for each primer set. Each PCR determination was made on an independent plate relative to a standard curve on the same plate. n values refer to the number of PCR determinations from at least three independent IPs.
TABLE 1.
Previously unpublished PCR primer pairs for human c-myc, GAPDH, U1, and p21 genesa
| Gene | Positionb | Primer sequence
|
|
|---|---|---|---|
| Forward | Reverse | ||
| GAPDH | 946 | GCC CGA TTT CTC CTC CG | GGA CCT CCA TAA ACC CAC TT |
| 4157 | GCC AAA CAT GGA AGA AGC TAT T | GTC AGA GCC CAG TGC GA | |
| c-myc | 7279 | TGG CCC TTA GAG CCT ATA CAG | CCC TTC ACG TCC GAT TCT T |
| 7952 | GGA GAA GAG AGG CCG GTC | TCA TGA TCC GCC CAC TTC G | |
| p21 | 804 | GGT ACA GCG GGA CTC CA | GCC AAA TAG GTC ACT GTG C |
| GAPDH | 34 | CTT CGC TCT CTG CTC CTC | TCT CTC CGC CCG TCT TCA |
| U1 | 1 | GCC CAA GAT CTC ATA CTT ACC TGG CA | GAT AAG CCT CGC CCT GGG AAA |
| 48 | GGG GAG ATA CCA TGA TCA CGA AGG | AGC ACA TCC GGA GTG CAA T | |
| 100 | CAT TGC ACT CCG GAT GTG CTG A | CAC TAC CAC AAA TTA TGC AGT CGA GTT | |
| 175 | CTG CGT TCG CGC TTT CCC | CCT TGG CGT ACG GTC TGT T | |
| 232 | ACG CCA AGG GTC ATG TCT TT | ATC AGG CCT CCA CTG TAG GAT | |
| 399 | GTC CGC TCA GCT CTT CCA TT | GAC TGG AGA CCA CGG ACC AA | |
All other primers used for real-time quantitative PCR have been described previously (13).
The positions correspond to the middle of the amplicon relative to the transcription start site.
Transient transfections and RNA analysis.
HCT116 Cdk7as/as cells were transfected with 7 μg of CMV-β-globin reporter (CMV stands for cytomegalovirus) (5) (see Fig. 3D) using polyethyleneimine (molecular weight, 25,000). 3-MB-PP1 (1 μM) or DMSO solvent control was added at the time of transfection. Total RNA was harvested after 24 h, digested with DNase I, and analyzed by RNase protection (5). Results were quantified by using a Molecular Dynamics PhosphorImager. Ratios of processed/unprocessed RNA were calculated after compensating for [32P]U content.
FIG. 3.
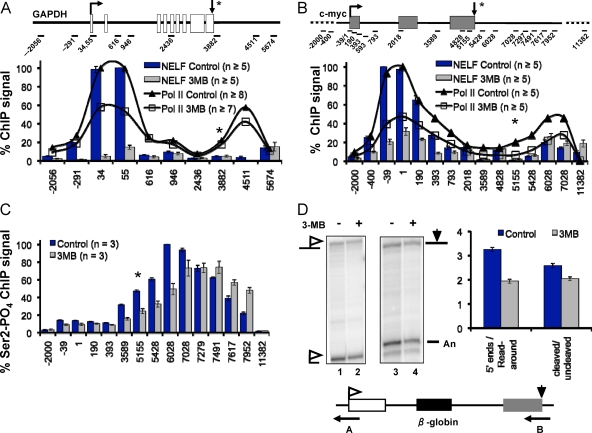
Cdk7 is required for recruitment of the negative elongation factor NELF. (A and B) ChIP signals of NELF-A along GAPDH and c-myc, respectively, in control Cdk7as/as cells and Cdk7as/as cells treated with 3-MB-PP13 (3MB). For reference, total Pol II profiles are shown as lines. (C) Cdk7 inhibition causes a downstream shift in the peak of CTD S2-phosphorylated Pol II. Phospho-S2 (Ser2-PO4) ChIP signals along the c-myc gene in control and 3-MB-PP1-treated Cdk7as/as cells normalized to the highest value in the control data set are shown. (D) Cdk7 inhibition inhibits poly(A) site cleavage and termination of β-globin transcripts. RNase protection analysis of total RNA from Cdk7as/as cells transfected with CMV-β-globin plasmid in the presence (+) and absence (−) of 3-MB-PP1. Antisense RNA probe A (lanes 1 and 2) distinguishes correctly initiated 5′ ends and transcripts that read around the plasmid. Probe B (lanes 3 and 4) distinguishes cleaved and uncleaved transcripts at the poly(A) site. The ratios of correct 5′ ends/read-around 5′ ends and cleaved/uncleaved transcripts from three RNase protection assay experiments are summarized in the graph.
Reverse transcription (RT) reactions were performed with SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions with random hexamer primers.
IP kinase assays and Western blotting.
TFIIH was immunoprecipitated from precleared HeLa or HCT116 Cdk7as/as nuclear extract in buffer D (20 mM HEPES [pH 7.9], 0.1 M KCl, 0.2 mM EDTA, 10% glycerol, 0.2 mM dithiothreitol) with affinity-purified rabbit anti-p62 and washed four times with buffer D containing 0.5% NP-40 and 0.3 M KCl. Kinase reactions (100 μl) contained 10 μl washed beads, 0.4 mM ATP, 0.5 mM microcystin, 55 μg/ml glutathione S-transferase (GST)-CTD (52 heptads) in kinase buffer (20 mM Tris-HCl [pH 8.0], 50 mM KCl, 7 mM MgCl2, 2 mM dithiothreitol, 5 mM β-glycerophosphate, 100 μg/ml bovine serum albumin) for 45 min at 30°C. Cdk7-CycH-Mat1 and Cdk9-CycT1 complexes expressed by baculovirus infection of insect Sf9 cells were purified as described previously (22). Kinase reactions (see Fig. 6A) contained ∼50 ng Cdk9-CycT1 or Cdk7-CycH-Mat1 and ∼2 μg GST-CTD in 25 mM HEPES (pH 7.4), 150 mM NaCl, 10 mM MgCl2, and 1 mM ATP and were incubated at 25°C for 15 min.
Western blots (see Fig. 1B) of immunoprecipitated TFIIH were reacted with rabbit anti-p62 and monoclonal anti-Cdk7 and developed with horseradish peroxidase-coupled protein A and anti-mouse immunoglobulin G, respectively.
RESULTS
Promoter-proximal pausing is suppressed by inhibiting Cdk7.
To address the function of TFIIH-associated Cdk7 activity, we used HCT116 Cdk7as/as cells in which Cdk7 is susceptible to the inhibitor 3-MB-PP1, which binds the expanded ATP-binding pocket of the mutant protein (23, 28). As expected, the TFIIH-associated kinase immunoprecipitated from Cdk7as/as cells was strongly inhibited by 3-MB-PP1 when assayed by 32P incorporation into a GST-CTD substrate (Fig. 1A). Because Cdk7 has a structural role within TFIIH (40), it was important to determine whether binding of 3-MB-PP1 to Cdk7(as) affected the integrity of TFIIH in addition to inhibiting its kinase activity. We therefore immunoprecipitated TFIIH from Cdk7as/as cells with antibody against the core p62 subunit and showed that Cdk7(as) coprecipitated equally well before and after treatment with 3-MB-PP1 (Fig. 1B). We conclude that holo-TFIIH remains intact after inhibition of the Cdk7 kinase.
Having established that 3-MB-PP1 inhibits Cdk7(as) without disrupting TFIIH, we asked how the kinase affects Pol II transcription in vivo. We mapped the distribution of Pol II on c-myc by ChIP with a pan-specific anti-CTD antibody before and after an 8-h 3-MB-PP1 treatment of HCT116 Cdk7as/as cells. Polymerase occupancy was assayed at 15 amplicons spanning the c-myc gene and its flanking sequences (13). For negative controls, we included amplicons from the 5′- and 3′-flanking regions in all ChIP experiments reported here. Remarkably, when Cdk7 was inhibited, Pol II occupancy at the promoter-proximal pause site was specifically suppressed relative to regions downstream (Fig. 1C, compare amplicons −39 and +1 with +190 to +593). This broadening of the polymerase distribution at the 5′ end is made obvious by normalizing Pol II ChIP signals to the highest value in each profile, revealing a pronounced shoulder of increased Pol II signals extending downstream in the 3-MB-PP1-treated sample (Fig. 1D). This change in the distribution of polymerase after Cdk7 inhibition is mimicked by a similar broadening of the peak of the Pol II-associated elongation factor, Spt5, at the c-myc 5′ end (Fig. 1E) and is most consistent with a defect in promoter-proximal pausing. We note that Cdk7 inhibition also altered the position of the 3′ pause downstream of the poly(A) site (Fig. 1C, D, and E), and the significance of this observation is discussed below. No effect on Pol II occupancy at the 5′ or 3′ end was observed when wild-type HCT116 cells were treated with 3-MB-PP1 (see Fig. S1A in the supplemental material), showing that the effect on transcription is caused by Cdk7 inhibition rather than an off-target effect of the inhibitor.
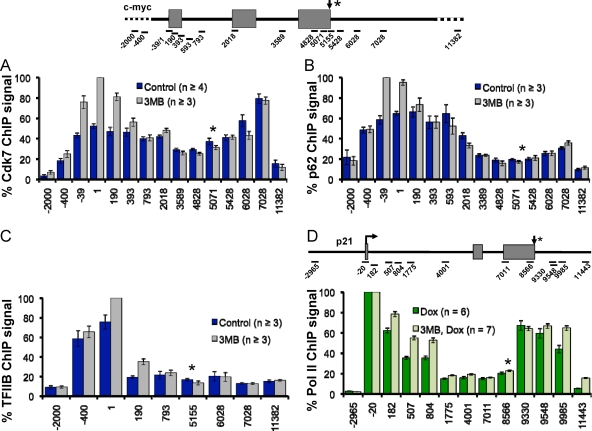
We asked whether Cdk7 inhibition affected recruitment of the general transcription factors TFIIH and TFIIB to the c-myc gene. Surprisingly, ChIP analysis of the Cdk7(as) and p62 subunits of TFIIH showed that this factor is distributed throughout the c-myc coding region and the 3′-flanking region (Fig. 2A and B), suggesting that this factor is a relatively ubiquitous component of the elongation complex. 3-MB-PP1 did not reduce occupancy of either Cdk7(as) or p62 on c-myc. In fact, the inhibitor appeared to stabilize TFIIH at the 5′ end of the gene (Fig. 2A and B). These experiments therefore show that the effects of 3-MB-PP1 on transcription in Cdk7as/as cells are due to reduced kinase activity, rather than reduced TFIIH recruitment to transcribed genes.
FIG. 2.
(A and B) TFIIH is distributed throughout the c-myc gene before and after inhibiting Cdk7. Relative ChIP signals along the c-myc gene are plotted for TFIIH subunits Cdk7 and p62 in HCT116 Cdk7as/as cells in the presence of 3-MB-PP1 (3MB) and in the absence of 3-MB-PP1 (Control). Values are normalized to the maximum signal for the 3MB data sets. (C) Cdk7 inhibition does not reduce TFIIB recruitment to the c-myc promoter. Relative TFIIB ChIP signals on c-myc in the presence and absence of 3-MB-PP1 in HCT116 Cdk7as/as cells are normalized to the maximal signal for the 3MB data set. Means ± SEMs (error bars) are shown. (D) Cdk7 inhibition reduces polymerase pausing on activated p21. Relative Pol II ChIP signals in the presence and absence of 3-MB-PP1 in Cdk7as/as cells treated with doxorubicin (Dox) normalized to the maximal signals in each data set are shown.
TFIIB localized exclusively to the c-myc promoter region, and like TFIIH, recruitment of this initiation factor was slightly enhanced following Cdk7 inhibition (Fig. 2C). We conclude that Cdk7 inhibition does not impair recruitment of either initiation factor TFIIH or TFIIB. Although it is difficult to absolutely exclude an effect on transcription initiation, these results suggest that the most important effect of Cdk7 kinase inhibition on transcription at the c-myc 5′ end is to prevent normal promoter-proximal pausing.
It is unlikely that the altered pattern of promoter-proximal Pol II is a result of cell cycle arrest at either G1/S or G2/M because 3-MB-PP1 treatment for 8 h does not alter the cell cycle distribution of unsynchronized cells (23) (S. Larochelle and R. P. Fisher, unpublished observations) and because a similar pausing defect on c-myc was observed whether the cells were treated for 8 h or 2 h with 3-MB-PP1 (Fig. 1D) (see Fig. S1B in the supplemental material). Furthermore, neither hydroxyurea, which blocks at G1/S, nor doxorubicin, which blocks at G2/M (14), caused a detectable change in Pol II distribution on c-myc in wild-type HCT116 cells (see Fig. S1C and S2A in the supplemental material). In summary, these results show that the effect of Cdk7 inhibition on the Pol II profile at the 5′ end of c-myc is not an indirect effect of cell cycle arrest.
We investigated whether the suppression of Pol II accumulation at the c-myc 5′ end applies to other genes with a promoter-proximal pause. On the p21 gene both before and after activation with doxorubicin, inhibition of Cdk7 specifically broadened the 5′ peak of Pol II consistent with diminished promoter-proximal pausing (Fig. 2D) (see Fig. S2B in the supplemental material). In contrast, treatment of wild-type HCT116 cells with 3-MB-PP1 did not affect Pol II distribution on p21 (see Fig. S2A in the supplemental material). A similar broadening in the peaks of paused Pol II at the transcription start site was also observed at c-fos and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) following inhibition of Cdk7 (see Fig. S3A and S3B in the supplemental material). Interestingly, Cdk7 inhibition over an 8-hour period reduced c-myc mRNA abundance by 50% relative to a mitochondrial control transcript, COXIII, as determined by real-time RT-PCR (see Fig. S3C in the supplemental material). Unlike c-myc, Cdk7 inhibition had little or no effect on activated or nonactivated p21 or GAPDH mRNA levels as determined by quantitative RT-PCR (see Fig. S3C in the supplemental material). This observation is consistent with microarray analysis of mRNA abundance in HCT116 Cdk7as/as cells showing that only a fraction of genes are downregulated by Cdk7 inhibition (L. Wohlbold and R. P. Fisher, unpublished data). We conclude from the results in Fig. 1 and 2, and in Fig. S2B, S3A, and S3B in the supplemental material, that normal promoter-proximal pausing on multiple human genes depends on Cdk7 kinase activity.
Cdk7 requirement for NELF recruitment.
Pol II stalling at 5′ ends is normally promoted by NELF. Because Cdk7 inhibition appears to reduce 5′ pausing, we monitored recruitment of this elongation factor before and after Cdk7 inactivation in Cdk7as/as cells by ChIP of the NELF-A subunit. Inhibition of Cdk7 almost completely eliminated NELF-A occupancy at the 5′ end of the GAPDH gene and reduced it by about 75% at the 5′ end of the c-myc gene (Fig. 3A and B). For both genes, the reduction in NELF exceeded the reduction in Pol II at the start site (Fig. 3A and B). Together, these experiments show that Cdk7 kinase activity promotes the recruitment or stabilization of NELF at these pause sites. Furthermore, Cdk7 inhibition caused a modest increase in recruitment of the positive transcription elongation factor Cdk9 to the c-myc gene (see Fig. S3D in the supplemental material). Together, the effects of Cdk7 inhibition on NELF and Cdk9 localization support the interpretation that reduced accumulation of Pol II at start sites under these conditions is caused at least partly by reduced promoter-proximal pausing.
Cdk7 inhibition delays transcription pausing and termination at the 3′ ends of genes.
The effects of Cdk7 inhibition on Pol II transcription are not limited to the promoter-proximal pause but are also felt at 3′ ends where there is a second pause that precedes termination. Pol II ChIP revealed that the 3′ pause site on c-myc shifted by about 1 kb in the downstream direction when Cdk7 was inhibited (Fig. 1C and D, compare amplicons 6028 to 7952). In parallel with Pol II, another component of the transcriptional elongation complex, Spt5, also shifted in the downstream direction (Fig. 1E). A subtle broadening of the peak of Pol II density in the 3′-flanking region was also observed at the activated p21 gene following Cdk7 inhibition (Fig. 2D). This effect of the TFIIH-associated kinase on transcription at the 3′ end of a gene is unexpected but consistent with our finding that Cdk7 and p62 are present in the c-myc 3′-flanking region (Fig. 2A and B). To confirm the effect of Cdk7 at 3′ ends, we assayed the localization of Ser2-phosphorylated Pol II, which is characteristic of the 3′ pause. Similar to the pattern of total Pol II, the peak of phospho-S2 signal shifted downstream about 1,000 bp when Cdk7 was inhibited relative to the control (Fig. 3C).
The shift in localization of total Pol II and S2-phosphorylated Pol II in the 3′-flanking region as a result of Cdk7 inhibition suggests that 3′-end processing and coupled termination could be impaired. To test this possibility, we transfected a β-globin reporter plasmid into HCT116 Cdk7as/as cells with and without 3-MB-PP1 treatment and quantified transcripts by RNase protection assay with probes spanning the poly(A) site and the initiation site (Fig. 3D). We detected a small but consistent decrease in the efficiency of poly(A) site cleavage and an increase in the level of transcripts that read around the plasmid as a result of poor termination. These results are therefore consistent with the ChIP experiments that show a delay in CTD S2 phosphorylation and termination in 3′-flanking regions. In summary, the changes in localization of paused, S2-phosphorylated Pol II and Spt5 to positions further downstream in the 3′-flanking sequences and impaired 3′-end processing and termination when Cdk7 was inhibited reveal a new function for TFIIH at the 3′ ends.
Cdk7 inhibition alters chromatin modifications within genes.
We sought independent confirmation that Cdk7 affects transcription in vivo by asking whether inhibition of kinase activity affected covalent chromatin marks associated with transcription elongation. Inhibition of the kinase in Cdk7 as/as cells enhanced H4 acetylation by approximately twofold across most of the c-myc gene and in the 3′-flanking region (amplicons +190 to +7617, Fig. 4A, gray bars) without affecting total histone levels (data not shown). A similar enhancement of H4 acetylation occurred throughout the GAPDH gene and at the 5′ end of the uninduced p21 gene (see Fig. S4A and S4C in the supplemental material) following Cdk7 inactivation. Conversely, Cdk7 inhibition decreased H3K36 trimethylation (me3) at the c-myc and GAPDH genes and shifted the profile of this chromatin mark toward the 3′ ends of these genes (Fig. 4B) (see Fig. S4B in the supplemental material). These results are consistent with the previously reported link between histone deacetylation and H3K36 methylation (25). In control experiments, 3-MB-PP1 did not affect acetyl-H4 or H3K36me3 in wild-type cells (data not shown). A similar enhancement of H4 acetylation and loss of H3K36me3 was reported on c-myc in response to knockdown of Iws1 which is implicated in transcription elongation and mRNA export (45). We note that although CTD S2 phosphorylation has been linked to K36 trimethylation by Set2 methyltransferases (45), the profile of K36me3 on c-myc does not closely coincide with S2-phosphorylated Pol II (compare Fig. 3C and 4B, blue bars). Together, the altered 5′-3′ profiles of Pol II and covalent chromatin marks provide independent lines of evidence that Cdk7 kinase affects transcription elongation at both 5′ and 3′ ends.
FIG. 4.
(A and B) Cdk7 inhibition increases H4 acetylation and decreases H3 K36 trimethylation. Ratios of acetylated H4 (Acetyl H4) and trimethylated H3 K36 to total H3 ChIP signals on c-myc in Cdk7as/as cells in the presence and absence of 3-MB-PP1 (3MB) are shown. (C) Cdk7 and Cdk9 are both CTD Ser5 kinases on c-myc. Ratios of phospho-S5 (Ser5-PO4) to total Pol II ChIP signals along c-myc for control Cdk7as/as cells or Cdk7as/as cells treated with 3-MB-PP1, DRB, or 3-MB-PP1 plus DRB are depicted. Mean values normalized to the maximal signal of the control at amplicon +593 are shown. Note the partial reduction in phospho-S5 when both Cdk7 and Cdk9 are inhibited.
Cdk7 and Cdk9 are both CTD S5 kinases on transcriptionally active Pol II.
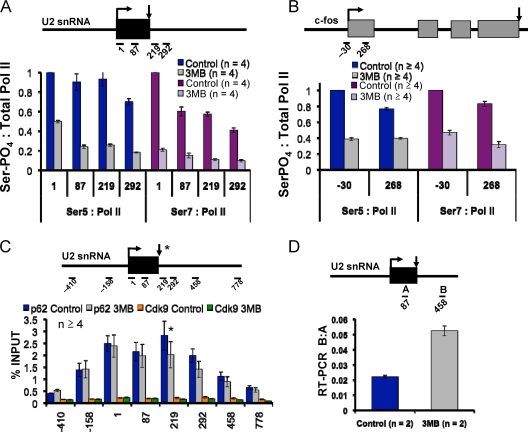
We tested whether Cdk7 inhibition altered CTD S5 phosphorylation of transcriptionally engaged Pol II. ChIP signals with polyclonal phospho-S5-specific antibody were normalized to total Pol II monitored with a pan-specific anti-CTD antibody to obtain a measure of the relative extent of modification. Cdk7 inhibition did not decrease the extent of S5 phosphorylation at the 5′ ends of the c-myc or GAPDH genes (Fig. 4C and data not shown) consistent with the fact that total cellular CTD S5 phosphorylation is unaffected (23). The lack of an effect of Cdk7 inhibition on S5 phosphorylation at c-myc was confirmed by ChIP with the H14 monoclonal antibody (data not shown). Although the extent of S5 phosphorylation on c-myc and GAPDH was not detectably reduced, it remains possible that the pattern of S5 phosphates on the CTD heptad repeats could be altered by Cdk7 inhibition. We conclude that either Cdk7 is not a major S5 kinase on these genes or, more likely, that another kinase such as Cdk9 has an overlapping function. In support of this idea, when Cdk7 and Cdk9 were simultaneously inhibited in Cdk7as/as cells with 3-MB-PP1 and DRB, S5 phosphorylation was partially reduced in the regions downstream of the c-myc start site (amplicons +190 to +593) (Fig. 4C, green bars), whereas inhibition of either kinase alone had little effect (gray bars and orange bars). A direct role of Cdk9 in Ser5 phosphorylation is further suggested by the fact that recombinant baculoviral Cdk9-CycT1 readily phosphorylated GST-CTD on S5 residues in vitro (see Fig. 6A). Even when Cdk7 and Cdk9 were both inhibited by a combination of 3-MB-PP1 and DRB, S5 phosphorylation was not completely abolished on c-myc and GAPDH (Fig. 4C and data not shown), suggesting a role for additional kinases such as Cdk8. In contrast to c-myc and GAPDH, at the U2 snRNA and c-fos genes, Cdk7 inhibition reduced S5 phosphorylation by over 50% (Fig. 5A and B). The large relative effect of Cdk7 on S5 phosphorylation at these genes is probably linked to low Cdk9 activity. Cdk9 occupancy relative to Pol II was about 30-fold lower on the U2 snRNA gene than on c-myc. In contrast, high levels of TFIIH are present at U2 (Fig. 5C) (see Fig. S5A in the supplemental material). We conclude that the relative contribution of TFIIH-associated Cdk7 to CTD S5 phosphorylation varies between genes and that Cdk9 also makes important contributions to S5 phosphorylation.
FIG. 5.
(A and B) Cdk7 inhibition reduces CTD S5 and S7 phosphorylation on U2 snRNA and c-fos genes. Ratios of ChIP signals for phospho-S5 and -phospho-S7 relative to total Pol II are shown in Cdk7as/as cells in the presence and absence of 3-MB-PP1 (3MB). Values are normalized to the maximum in the control data set. Ser-PO4, phosphorylated serine. (C) Detection of TFIIH, but not Cdk9, on the U2 snRNA gene. ChIP signals for TFIIH subunits p62 and Cdk9 are shown. (D) Cdk7 inhibition causes readthrough transcription of the U2 snRNA gene. Ratios of real-time RT-PCR products for amplicon B at +458 downstream of the gene relative to amplicon A at +87 within the gene are shown for random-primed cDNA from control and 3MB-PP1-treated HCT116 Cdk7as/as cells.
TFIIH phosphorylates Pol II CTD S7 residues.
S7 phosphorylation of the CTD heptad repeats occurs on mRNA and U snRNA genes where it facilitates cotranscriptional 3′-end formation (6, 10), but the S7 kinase is unknown. We tested whether Cdk7 inhibition affected CTD S7 phosphorylation at U snRNA genes in Cdk7as/as cells by ChIP with the 4E12 monoclonal antibody (6, 10). The relative level of S7 phosphorylation normalized to total Pol II was reduced by 80 to 90% on the U1 and U2 snRNA genes following Cdk7 inhibition (Fig. 5A) (see Fig. S5B in the supplemental material). S7 phosphorylation was also reduced by 60% on c-fos when Cdk7 was inhibited (Fig. 5B). S7 phosphorylation is important for snRNA 3′-end formation (10); therefore, we predicted that Cdk7 inhibition may induce formation of U2 transcripts with abnormal 3′ ends. RT-PCR analysis of U2 snRNAs confirmed that Cdk7 inhibition did indeed modestly increase the fraction of transcripts that extend beyond the normal 3′ end (Fig. 5D). On the other hand, like S5, S7 phosphorylation on c-myc was not significantly reduced by inhibition of Cdk7 (see Fig. S5C in the supplemental material). Other kinases are therefore likely to contribute to S7 phosphorylation on c-myc. One possibility is that Cdk9 also phosphorylates S7 on c-myc as this kinase can phosphorylate S7 in vitro (Fig. 6A). Our results therefore do not implicate S7 phosphorylation in the altered promoter-proximal pausing observed on c-myc when Cdk7 is inhibited (Fig. 1C to E).
To determine whether TFIIH can phosphorylate CTD S7 residues directly, it was immunoprecipitated from HeLa nuclear extract with anti-p62 or anti-Cdk7, and kinase assays were performed with GST-CTD as a substrate. The experiment in Fig. 6B revealed robust S7 phosphorylation of GST-CTD by Cdk7-associated TFIIH. As expected, these IP kinase assays were also positive for phospho-S5 and negative for phospho-S2 by Western blotting (data not shown). S7 phosphorylation of full-length GST-CTD by Cdk7 was confirmed using recombinant Cdk7-CycH-Mat1 (Fig. 6A) in agreement with the recent report of phosphorylation of a CTD peptide by recombinant Cdk7-CycH (2). Recombinant Cdk9-CycT1 was also capable of phosphorylating CTD S7 residues, suggesting that this kinase may also contribute to S7 phosphorylation on some genes (Fig. 6A). Together, the results in Fig. 4C, 5A to D, and 6B show that Cdk7-associated TFIIH can phosphorylate both S5 and S7 residues of the CTD and that it is a major kinase of S7 residues on the c-fos and U snRNA genes.
DISCUSSION
We report the first study of how Cdk7 activity affects the mammalian RNA Pol II transcription cycle in vivo using an analogue-sensitive HCT116 Cdk7as/as cell line. The kinase can be dispensable for transcription in vitro (26, 37, 40), and in some cases, Cdk7 inactivation has only minor effects on gene expression in vivo (18, 24). Consistent with these previous results, Cdk7 inhibition in vivo did not strongly inhibit general transcription, but it did markedly reduce Pol II accumulation near start sites relative to positions downstream. This effect indicates a loss of definition of the promoter-proximal pause or leaking of polymerase from the pause into downstream sequences that is consistent with a previous report on Drosophila Hsp70 (36). We observed a similar flattening of the Pol II density profile at the 5′ ends of all mRNA coding genes we examined, including c-myc, p21, GAPDH, c-fos, and Hsp70 (Fig. 1C, D, and 2D) (see Fig. S3A and S3B in the supplemental material) (data not shown). Together, the effects of Cdk7 inhibition on 5′-3′ profiles of Pol II, Spt5, H4 acetylation, and H3K36 methylation all point to a function of this kinase in regulating transcriptional elongation/pausing. Cdk7 control of pausing is probably influenced by both the number and distribution of phosphorylation sites on the CTD heptad repeats as well as other substrates such as Spt5 (22). The marked inhibition of NELF recruitment when Cdk7 was inhibited (Fig. 3A and B) argues that defective promoter-proximal pausing under these conditions results from disruption of normal negative regulation of elongation. Cdk9 normally functions to antagonize pausing mediated by DSIF/NELF, and the role of Cdk7 in NELF recruitment therefore suggests a new link between Cdk7 and Cdk9 functions in transcription elongation.
Unexpectedly, Cdk7 affected not only the promoter-proximal pause but also the second major pause downstream of the poly(A) site. Upon inactivation of Cdk7, the peaks of polymerase density 3′ of the c-myc and p21 poly(A) sites were relocated further downstream, suggesting that the 3′ pause site is shifted (Fig. 1D and 2D). Relocation of the c-myc 3′ pause was confirmed by localization of S2-phosphorylated Pol II and the elongation factor, Spt5 (Fig. 3C and 1E). In addition, histone H4 acetylation was elevated in the 3′-flanking region of c-myc when Cdk7 was inactivated (Fig. 4A). While an effect of a general transcription factor that is a component of the preinitiation complex on transcription at the 3′ end of a gene seems surprising, it is consistent with our finding that Cdk7 and TFIIH p62 colocalize with Pol II throughout the c-myc gene and into the 3′-flanking region (Fig. 2A and B). In agreement with the latter result, Drosophila Cdk7 was previously localized along most of the length of the Hsp70 gene (36). It remains possible that Cdk7 could function at the 3′ end as part of the CAK subcomplex rather than intact TFIIH however. The delay in 3′ pausing and termination indicates that the signal to begin terminating is received late, either because the polymerase is moving abnormally fast and/or the signal is delivered late, perhaps due to slow recognition of the poly(A) site. This idea is supported by our finding that poly(A) site cleavage and coupled termination are inhibited on a β-globin reporter gene (Fig. 3D).
Phosphorylation of S7 residues on the CTD occurs on mRNA coding genes and U snRNA genes where it has been implicated in recruitment of the integrator complex that processes U snRNA 3′ ends (6, 10). Unlike the S2 and S5 residues of the heptad repeat (YSPTSPS), S7 does not conform to the SP recognition site for Cdk enzymes. Cdk7 is known to phosphorylate sites lacking Pro residues in the +1 position in the activation loops of the Cdk enzymes through recognition of substrate determinants away from the actual phosphorylation site (22). Remarkably, most S7 phosphorylation of Pol II at the uninduced c-fos promoter and the U1 and U2 snRNA genes, but not c-myc, was eliminated by Cdk7 inhibition Cdk7as/as cells (Fig. 5A and B) (see Fig. S5B and S5C in the supplemental material). Furthermore, immunoprecipitated TFIIH and recombinant Cdk7 and Cdk9 phosphorylated S7 residues of the CTD in vitro (Fig. 6A and B). Together with the recent report that TFIIH phosphorylates S7 in budding yeast (2), these results reveal a new function of TFIIH-associated Cdk7 in phosphorylation of the CTD. In the future, it will be of interest to investigate how Cdk7 recognizes S7 and how the S5 and S7 kinase activities of TFIIH on the same substrate might influence one another.
Taken together, the effects of Cdk7 inhibition at multiple stages of the transcription cycle reveal a new picture of how this kinase functions in transcription of human genes that differs from the conventional view in three principal ways summarized in Fig. 6C. (i) Cdk7 phosphorylates CTD S7 as well as S5 residues in vivo, and there can be extensive overlap between Cdk7 and Cdk9 for phosphorylation of S5 and potentially also S7. (ii) TFIIH-associated Cdk7 is an important component of the mechanism that establishes promoter-proximal pausing, along with DSIF and NELF. (iii) Rather than acting exclusively at the promoter, TFIIH-associated Cdk7 also affects pausing and termination downstream of the gene.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM063873 to D.L.B., GM056985 to R.P.F., and EB001987 to K.S. and by NIH fellowship 5F31 GM072099 to K.G.-C.
We thank J. Espinosa (University of Colorado), S. Kim, S. Johnson, and R. Perales for valuable suggestions, D. Eick (Helmholtz Center, Munich, Germany) for 4E12 antibody, and N. Fong for GST-CTD.
Footnotes
Published ahead of print on 10 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar, M., M. Heidemann, J. Tietjen, D. Zhang, R. D. Chapman, D. Eick, and A. Ansari. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34:387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akoulitchev, S., T. P. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17:251-256. [DOI] [PubMed] [Google Scholar]
- 5.Bird, G., D. A. Zorio, and D. L. Bentley. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 24:8963-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, R. D., M. Heidemann, T. K. Albert, R. Mailhammer, A. Flatley, M. Meisterernst, E. Kremmer, and D. Eick. 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318:1780-1782. [DOI] [PubMed] [Google Scholar]
- 7.Chopra, V. S., J. Cande, J. W. Hong, and M. Levine. 2009. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 23:1505-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core, L. J., and J. T. Lis. 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319:1791-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egloff, S., and S. Murphy. 2008. Cracking the RNA polymerase II CTD code. Trends Genet. 24:280-288. [DOI] [PubMed] [Google Scholar]
- 10.Egloff, S., D. O'Reilly, R. D. Chapman, A. Taylor, K. Tanzhaus, L. Pitts, D. Eick, and S. Murphy. 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318:1777-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eissenberg, J. C., A. Shilatifard, N. Dorokhov, and D. E. Michener. 2007. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol. Genet. Genomics 277:101-114. [DOI] [PubMed] [Google Scholar]
- 12.Gilchrist, D. A., S. Nechaev, C. Lee, S. K. Ghosh, J. B. Collins, L. Li, D. S. Gilmour, and K. Adelman. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22:1921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glover-Cutter, K., S. Kim, J. Espinosa, and D. L. Bentley. 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 15:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromak, N., S. West, and N. J. Proudfoot. 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 26:3986-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, C., and S. Shuman. 1999. Distinct effector roles for Ser2 and Ser5 phosphorylation of the RNA polymerase II CTD in the recruitment and allosteric activation of mammalian capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 18.Kanin, E. I., R. T. Kipp, C. Kung, M. Slattery, A. Viale, S. Hahn, K. M. Shokat, and A. Z. Ansari. 2007. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. USA 104:5812-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight, Z. A., and K. M. Shokat. 2007. Chemical genetics: where genetics and pharmacology meet. Cell 128:425-430. [DOI] [PubMed] [Google Scholar]
- 20.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krumm, A., L. Hickey, and M. Groudine. 1995. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 9:559-572. [DOI] [PubMed] [Google Scholar]
- 22.Larochelle, S., J. Batliner, M. J. Gamble, N. M. Barboza, B. C. Kraybill, J. D. Blethrow, K. M. Shokat, and R. P. Fisher. 2006. Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat. Struct. Mol. Biol. 13:55-62. [DOI] [PubMed] [Google Scholar]
- 23.Larochelle, S., K. A. Merrick, M. E. Terret, L. Wohlbold, N. M. Barboza, C. Zhang, K. M. Shokat, P. V. Jallepalli, and R. P. Fisher. 2007. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol. Cell 25:839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, K. M., I. Miklos, H. Du, S. Watt, Z. Szilagyi, J. E. Saiz, R. Madabhushi, C. J. Penkett, M. Sipiczki, J. Bahler, and R. P. Fisher. 2005. Impairment of the TFIIH-associated CDK-activating kinase selectively affects cell cycle-regulated gene expression in fission yeast. Mol. Biol. Cell 16:2734-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 26.Makela, T. P., J. D. Parvin, J. Kim, L. J. Huber, P. A. Sharp, and R. A. Weinberg. 1995. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc. Natl. Acad. Sci. USA 92:5174-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, N. F., J. M. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 28.Merrick, K. A., S. Larochelle, C. Zhang, J. J. Allen, K. M. Shokat, and R. P. Fisher. 2008. Distinct activation pathways confer cyclin-binding specificity on Cdk1 and Cdk2 in human cells. Mol. Cell 32:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13:55-65. [DOI] [PubMed] [Google Scholar]
- 30.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297-305. [DOI] [PubMed] [Google Scholar]
- 31.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922-2936. [DOI] [PubMed] [Google Scholar]
- 32.Roy, R., J. P. Adamczewski, T. Seroz, W. Vermeulen, J. P. Tassan, L. Schaeffer, E. A. Nigg, J. Hoeijmakers, and J. M. Egly. 1994. The MO15 cell-cycle kinase is associated with the TFIIH transcription DNA-repair factor. Cell 79:1093-1101. [DOI] [PubMed] [Google Scholar]
- 33.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557-567. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer, L., V. Moncollin, R. Roy, A. Staub, M. Mezzina, A. Sarasin, G. Weeda, J. H. J. Hoeijmakers, and J. M. Egly. 1994. The ERCC2/DNA repair protein is associated with the class-II BTF2/TFIIH transcription factor. EMBO J. 13:2388-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder, S., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz, B. E., S. Larochelle, B. Suter, and J. T. Lis. 2003. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol. Cell. Biol. 23:6876-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serizawa, H., J. W. Conaway, and R. C. Conaway. 1993. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature 363:371-374. [DOI] [PubMed] [Google Scholar]
- 38.Shiekhattar, R., F. Mermelstein, R. P. Fisher, R. Drapkin, B. Dynlacht, H. C. Wessling, D. O. Morgan, and D. Reinberg. 1995. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 374:283-287. [DOI] [PubMed] [Google Scholar]
- 39.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 40.Tirode, F., D. Busso, F. Coin, and J. M. Egly. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3:87-95. [DOI] [PubMed] [Google Scholar]
- 41.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 44.Yankulov, K. Y., and D. L. Bentley. 1997. Regulation of Cdk7 substrate-specificity by Mat1 and TFIIH. EMBO J. 16:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoh, S. M., J. S. Lucas, and K. A. Jones. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22:3422-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.