Abstract
The importance of DNA supercoiling in transcriptional regulation has been known for many years, and more recently, transcription itself has been shown to be a source of this superhelicity. To mimic the effect of transcriptionally induced negative superhelicity, the G-quadruplex/i-motif-forming region in the c-Myc promoter was incorporated into a supercoiled plasmid. We show, using enzymatic and chemical footprinting, that negative superhelicity facilitates the formation of secondary DNA structures under physiological conditions. Significantly, these structures are not the same as those formed in single-stranded DNA templates. Together with the recently demonstrated role of transcriptionally induced superhelicity in maintaining a mechanosensor mechanism for controlling the firing rate of the c-Myc promoter, we provide a more complete picture of how c-Myc transcription is likely controlled. Last, these physiologically relevant G-quadruplex and i-motif structures, along with the mechanosensor mechanism for control of gene expression, are proposed as novel mechanisms for small molecule targeting of transcriptional control of c-Myc.
INTRODUCTION
The c-Myc gene product is a potent oncoprotein and a transcription factor that plays an essential role in the control of cell growth, as well as in cell fate determinations related to the induction of apoptosis.1-3 The activation of c-Myc is known to induce transcription of growth-stimulating genes in many types of human cancer, including breast, stomach, lung, prostate, colorectal, and pancreatic cancers and leukemia.4,5 c-Myc overexpression can be caused by different mechanisms, including gene amplification,6-8 translocation,9,10 and simple upregulation of transcription.4,5 In a very recent paper, the validity of c-Myc as a molecular target in oncology has been demonstrated.11
Polypurine/polypyrimidine tracts are overly represented sequences in the mammalian genome and are often found in the proximal promoter region of many growth-related genes, including many oncogenes.12-19 Previous studies have shown that DNase I or S1 nuclease hypersensitive sites are often found within the regions of DNA that contain these tracts in both chromatin and negatively supercoiled plasmid DNA.13-16 These studies also suggest that these tracts are structurally dynamic and can be converted easily into secondary structures. The structural transition of B-DNA to non-B-DNA secondary structures must be preceded or accompanied by localized unwinding or melting of the double helix.20-23 Local unwinding or melting of duplex DNA is known to be facilitated by negative supercoiling stress, which is naturally generated behind the translocating RNA polymerase complex during gene transcription.24,25
The promoter of the c-Myc oncogene contains a number of elements that have either been shown to assume,26 or are suspected to assume, non-duplex regions under negative superhelical stress. We have been particularly interested in a polypurine/polypyrimidine tract known as the nuclease hypersensitive element III1 (NHE III1) or CT element (Figure 1A),27 because under conditions when both strands are single-stranded, they assume secondary DNA structures, namely, a G-quadruplex on the purine-rich strand28-30 and an i-motif on the pyrimidine-rich strand.31 In vivo, the single-stranded DNA-binding proteins CNBP32 and hnRNP K33 bind to these regions to affect transcription, suggesting that the NHE III1 can assume a single-stranded form. However, several important questions remain unanswered. Are these elements capable of forming specific secondary DNA structures, and if so, are they the same as those found in the single-stranded DNA templates? Can negative superhelicity provide the torsional stress necessary to induce these structures? These questions have been addressed in this contribution.
Figure 1.
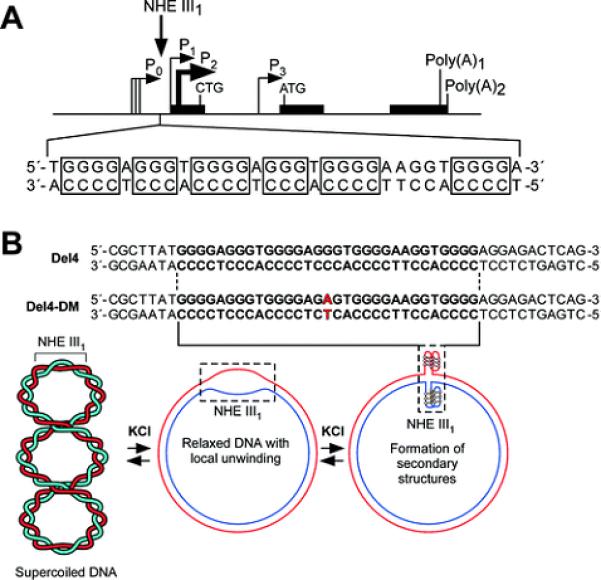
(A) Promoter structure of the c-Myc gene and location of the NHE III1 region. (B) Sequences of the wild-type (Del4) and mutant (Del4-DM) plasmids and proposed equilibrium between supercoiled and partially relaxed forms of the plasmid.
G-quadruplexes have become of considerable interest in medicinal chemistry because they represent alternative DNA structures that are folded and as such assume globular structures and are thus potentially targeted by small molecules. In this respect they mimic protein structures that are specifically assembled into their precise folding patterns determined by the primary amino acid sequence. Consequently a 30-base sequence can assume a specific folding pattern and globular structure determined by the precise DNA sequence in the promoter element. In eukaryotic promoter sequences of such genes as c-Myc, Bcl-2, c-Kit, RET, VEGF, Hif-1α, PDGFA, c-Myb, and KRAS,34 a variety of G-quadruplex folding patterns and structures have been shown to form from single-stranded DNA, and there is accumulating evidence that these structures can be targeted by small molecules to modulate gene transcription.34 The C-rich strand has received much less attention because it would not be expected to form i-motif structures under physiological conditions. Thus the questions posed at the end of the previous paragraph become critical for future drug discovery efforts.
In a recent paper by Levens et al., the effect of transcriptionally generated negative supercoiling in vivo has been demonstrated in the c-Myc promoter.26 This supercoiling is not relieved by topoisomerase I or II and directs the melting of the Far Upstream Element (FUSE) 1.4 Kb upstream of the P1 promoter, which in turn binds to the transcriptional activating and extinguishing factors FUSE Binding Protein (FBP) and FBP Interacting Repressor (FIR), a process that is likened to a “cruise control” mechanism, in which undulating superhelical stress controls the rate of promoter firing through a feedback loop. The NHE III1 is much nearer the P1 promoter and thus is likely to be subjected to greater torsional stress than the FUSE element. To mimic this transcriptionally induced negative superhelicity, we have used a supercoiled plasmid system in which either the wild-type (Del4) or mutant (Del4-DM) NHE III1 element has been included (Figure 1B). The supercoiled plasmid used in this study retained the native level of DNA supercoiling. The footprinting agents used in this study include S1 nuclease, DNase I, dimethyl sulfate (DMS), KMnO4, and bromine (Br2), and the reactivity of these probes is very sensitive to the conformation of DNA molecules.35-39 Thus, these reagents have often been used to probe the structural transition from BDNA to non-B-DNA secondary structures, such as melted DNA, hairpin, G-quadruplexes, or i-motifs.36-39 The results of this study, alongside the data from the Levens laboratory, provide compelling data for a dynamic equilibrium between duplex, unwound duplex, single-stranded DNA, and secondary DNA structures within and around the NHE III1 that determines the transcriptional state of the c-Myc promoter in cells.
RESULTS AND DISCUSSION
Under Conditions of Negative Superhelicity, Enzymatic Footprinting with S1 Nuclease and DNase I Reveals That the NHE III1 and Its Flanking Regions Are Highly Dynamic and Exist in an Equilibrium Mixture of Duplex, Partially Melted, and Secondary DNA Structures
(a) Comparison of S1 nuclease footprinting of the wild-type and mutant NHE III1 and flanking regions of the c-Myc promoter in supercoiled form implies that while the G- and C-rich strands in the NHE III1 exist in secondary structures, the flanking regions are in a dynamic equilibrium, which is dampened by the presence of KCl, concomitantly stabilizing the G-quadruplex structures
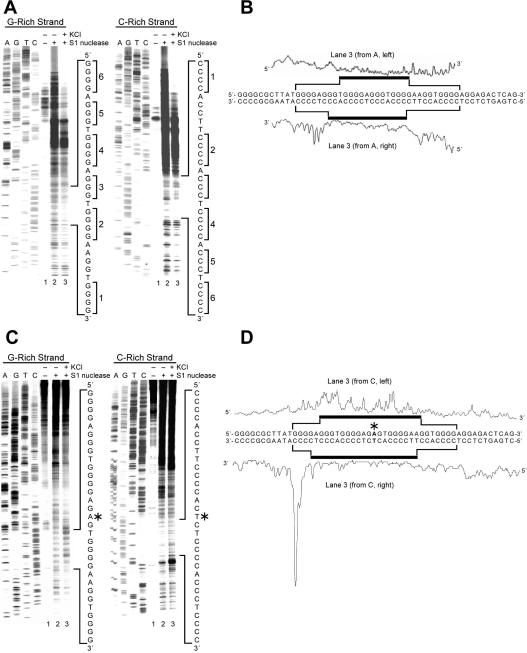
S1 nuclease preferentially cleaves single-stranded regions over locally unwound regions, normal duplex regions, or secondary structures, such as cruciform DNA.37,40 Thus, S1 nuclease footprinting experiments were carried out to examine the possibility of secondary structures forming in the NHE III1 within the c-Myc promoter under the influence of negative supercoiling. Our conditions were optimized so that there was one hit or cleavage event per DNA molecule. Once cleavage occurred, the experiment was over, but this would not invalidate the experiment under the conditions used. As shown in Figure 2, A and B, the G-rich and C-rich strands of the wild-type NHE III1 both show overall increased reactivity to S1 nuclease on the 5′-side of the NHE III1, particularly on the C-rich strand, but reduced reactivity within the NHE III1. The increased reactivity to S1 nuclease on the 5′-side could be explained either by a highly dynamic region exhibiting single-stranded characteristics or by the formation of hairpin structures, which are commonly formed under negative supercoiling conditions.20,21 Examination of the sequence of the flanking region did not reveal any palindromic sequences, ruling out hairpins as the basis for this enhanced reactivity to S1 nuclease. The effect of 100 mM KCl on both strands of the NHE III1 and the flanking regions is to reduce the reactivity to S1 nuclease (compare lanes 2 and 3 in Figure 2A). Since KCl is known to stabilize G-quadruplex structures, this suggests that the reduced reactivity to S1 nuclease within the NHE III1 region is due to the enhanced stability of these structures, which also affects both the C-rich strand in the NHE III1 region and the flanking regions. We propose that the G-quadruplex and an accompanying structure on the C-rich strand (the i-motif) act as a buffer for the dynamic stress induced by negative supercoiling and that this then dampens the dynamic nature of the flanking regions on both strands, but particularly on the 5′-sides of both strands.
Figure 2.
In vitro footprinting of the wild-type and mutant c-Myc promoter regions with S1 nuclease. (A) Autoradiograms showing S1 nuclease cleavage sites on the G- and C-rich strands of the supercoiled Del4 plasmid. Plasmid DNA was incubated in the absence of salt (lanes 2) and in the presence of 100 mM KCl (lanes 3) at 37 °C overnight before digesting with S1 nuclease. (B) Densitometric scans of lanes 3 in (A), left and right panels. (C) Autoradiograms showing S1 nuclease cleavage sites on the G- and C-rich strands of the supercoiled Del4-DM plasmid. Plasmid DNA was incubated in the absence of salt (lanes 2) and in the presence of 100 mM KCl (lanes 3) at 37 °C overnight before digesting with S1 nuclease. (D) Densitometric scans of lanes 3 in (C), left and right panels.
To further investigate the potential significance of the formation of G-quadruplex structures in the structural transition of the NHE III1 region, particularly in the presence of KCl, S1 nuclease footprinting experiments were carried out using both linear DNA and a mutant plasmid, where a single GC-to-AT mutation (Figure 2, C and D) was introduced into the NHE III1 of the wild-type plasmid by site-directed mutagenesis to abolish the capability of the G-rich strand to form G-quadruplex structures.28 For linear DNA, S1 nuclease activity in the Del4 DNA was extremely low compared to supercoiled DNA, suggesting the importance of negative supercoiling in promoting local unwinding of DNA. To more accurately define the effect of negative supercoiling on the NHE III1 and surrounding region, the mutant plasmid was used. As shown in Figure 2C, the S1 nuclease cleavage pattern on the NHE III1 exhibits selective enhanced reactivity of S1 nuclease toward the G-rich strand of the mutant NHE III1 occurred in the presence of KCl (Figure 2C, left panel, and Figure 2D, upper trace). This result suggests that this sequence exists partly in a single-stranded form rather than the normal double-stranded form or as a secondary structure. The 5′ flanking region shows reduced reactivity to S1 nuclease in comparison to the wild-type (Figure 2A), suggesting that this region is far less dynamic. In contrast, the complementary C-rich strand showed reduced reactivity to S1 nuclease, which was maintained in the presence of KCl (Figure 2C, right panel, and Figure 2D, lower trace), suggesting that secondary structures may still exist in this region and are less sensitive to the C-to-T mutation. However, the identify of these structures cannot be ascertained from these experiments but are addressed later in sections related to chemical footprinting.
(b) DNase footprinting of the C-rich strand of the wild-type plasmid reveals that the flanking regions of the NHE III1 are duplex structures, while the NHE III1 region is non-duplex in character, and this effect is more pronounced upon addition of KCl
DNase I is known to preferentially cleave locally unwound or normal duplex regions over single-stranded regions or secondary structures, such as the hairpin.40 In this study, we performed in vitro footprinting experiments with DNase I to probe the possible structural transition of the NHE III1 of the c-Myc promoter in supercoiled plasmid DNA and the nature of the flanking regions (Figure 3). Since the S1 nuclease experiments (Figure 2) had shown a partially single-stranded character of the 5′ flanking sequence, which was dampened in the presence of KCl, we used DNase I to determine whether this single-stranded region was in equilibrium with partially unwound or duplex DNA. The results (Figure 3) show that strong cleavage of the 5′ flanking region occurs, which is significantly dampened in the presence of 100 mM KCl. Taken together with the S1 nuclease results, this strongly suggests that the 5′ flanking region on the C-rich strand is highly dynamic in nature and consists of an equilibrium between duplex DNA, partially unwound duplex, and single-stranded DNA. Furthermore, the addition of 100 mM KCl, which further stabilizes the G-quadruplex, severely dampens the enzymatic cleavage in the flanking regions by both DNase I and S1 nuclease. This suggests that the stabilization of the secondary DNA structures in the NHE III1 acts as a dynamic buffer to absorb the torsional stress associated with the negative superhelicity.
Figure 3.
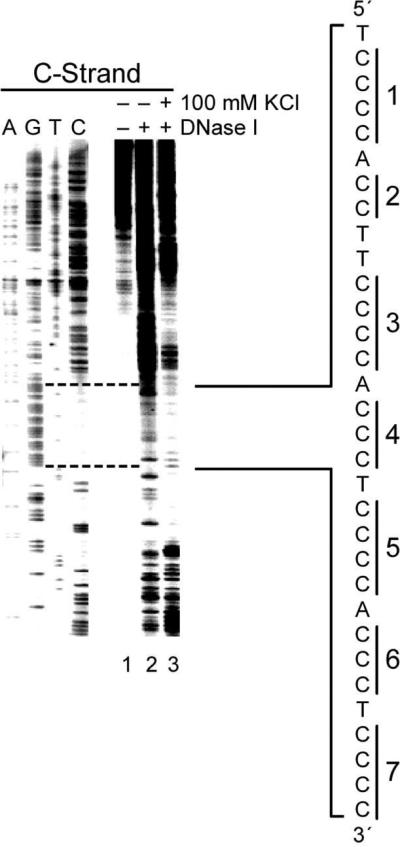
DNase I footprinting of the wild-type c-Myc promoter region using the supercoiled Del4. The autoradiogram represents DNase I cleavage sites on the C-rich strand of the supercoiled Del4 plasmid in the absence of salt (lane 2) and in the presence of 100 mM KCl (lane 3). The vertical bar next to the gel indicates the polypyrimidine tracts. To the right of the gel is the sequence of the NHE III1 with cytosine tracts 1−6 indicated by brackets.
Chemical Footprinting with DMS, KMnO4, and Br2 Provides Insight into the Secondary DNA Structures Formed in the NHE III1
(a) DMS footprinting of the G-rich strand shows that a G-quadruplex structure is formed from the four 5′-end guanine tracts, and that this is dependent upon superhelicity and requires the wild-type sequence
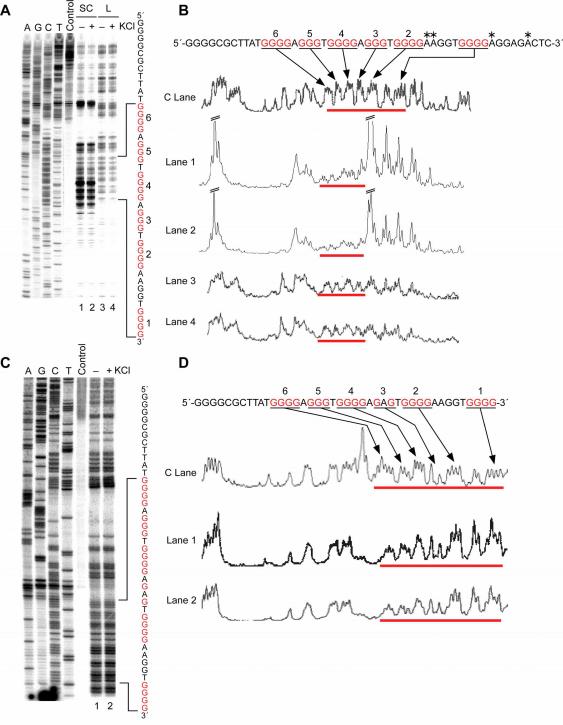
DMS footprinting is particularly useful for probing G-quadruplexes in vitro, because these structures require N7 of guanine and there is protection of the guanine residues involved in G-quadruplex formation from N7 methylation by DMS, as we have previously demonstrated.28 Thus, DMS footprinting is useful for fine-mapping the presence of G-quadruplex structures or single-stranded regions within the NHE III1 of the c-Myc promoter. As shown in Figure 4, A and B (lanes 1 and 2), under supercoiled conditions DMS shows significantly reduced reactivity within the four 5′-end guanine tracts on the G-rich strand of the NHE III1, where G-quadruplex structures are proposed to be present. Significantly, the immediate flanking regions of the proposed G-quadruplex-forming sequence show enhanced reactivity to DMS (Figure 4, A and B), presumably because of the presence of locally unwound structures at the junctions between normal duplex regions and stable secondary structures. It is the relative reactivity of the protected guanine tracts (3−6) to those in the adjacent region (tracts 1−2) that is critical in making the conclusion as to which guanines are involved in G-quadruplex formation. It is important to note that some adenine residues at these flanking regions also show unusual reactivity toward DMS, suggesting that the N3 position of adenine is sterically accessible to electrophilic attack by DMS, as has been previously observed at or near B-DNA–Z-DNA junctions.41
Figure 4.
In vitro footprinting of the wild-type and mutant c-Myc promoter regions with DMS. (A) Autoradiogram showing DMS modification sites on the G-rich strand of a supercoiled (SC) or linearized (L) form of the wild-type plasmid. Plasmid DNA was incubated in the absence of salt (lanes 1 and 3) and in the presence of 100 mM KCl (lanes 2 and 4) at 37 °C overnight before treatment with DMS. DMS modification sites were mapped using linear amplification by PCR with 32P-labeled gene-specific primers. (B) Densitometric scans of the autoradiogram in (A). Asterisks indicate the adenine residues that are hypermethylated with DMS. The bars indicate the guanine repeats, which are numbered 1−6 from the 3′-end. (C) Autoradiogram showing DMS modification sites on the G-rich strand of a supercoiled mutant plasmid in the absence of salt (lane 1) and in the presence of 100 mM KCl (lane 2) at 37 °C overnight before treatment with DMS. (D) Densitometric scans of the autoradiogram in (C). The bars indicate the guanine repeats, which are numbered 1−6 from the 3′-end.
We were initially surprised to observe that DMS reactivity toward guanine or adenine residues within the G-rich region was not affected by the presence of KCl (compare lanes 1 and 2 in Figure 4, A and B), suggesting the ability of the c-Myc promoter region in supercoiled form to form G-quadruplexes in vitro irrespective of the presence of KCl. This contrasts with the differential reactivity of the same region to enzymatic probes in the absence or presence of KCl (see Figures 2 and 3). The reactivity of DMS toward the guanine residues within the complete G-rich region is normal in the linearized form of the wild-type plasmid (Figure 4A and B, lanes 3 and 4), showing that negative supercoiling is required to drive the local unwinding of the NHE III1 region, allowing a specific G-rich region (tracts 3−6) to form a G-quadruplex.
The mutant plasmid was used to further investigate the potential significance of the formation of G-quadruplex structures by the G-rich strand and the unusual reactivity of the NHE III1 region toward DMS under negative supercoiling stress by carrying out a footprinting experiment. As noted previously, we had demonstrated that the single-base mutation in this plasmid is able to dramatically reduce the propensity of the sequence to form a G-quadruplex structure.28 As shown in Figure 4, C and D, the DMS reactivity pattern on the G-rich sequence of the mutant plasmid is very different from that of the wild-type plasmid and more closely resembles that associated with duplex or single-stranded DNA. This observation strongly suggests that the secondary structure formed under negative superhelicity is a G-quadruplex, because it involves just four runs of guanines, and a single-base mutation known to destabilize the G-quadruplex results in loss of this DMS protection.
(b) Chemical footprinting of the C-rich strand of the c-Myc promoter with KMnO4 and Br2 shows that this region forms a secondary DNA structure consistent with an i-motif
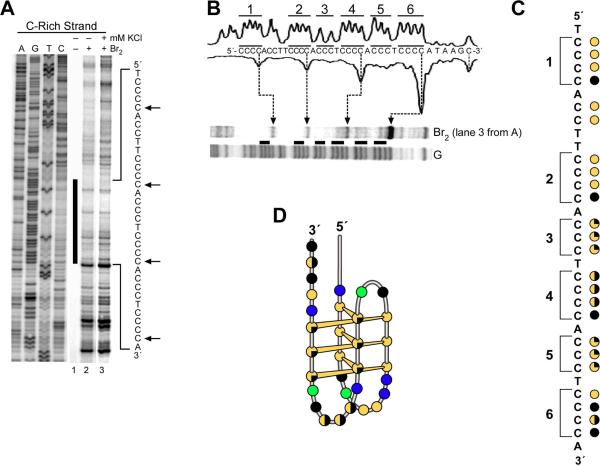
KMnO4 and Br2 were used in further footprinting experiments to probe for secondary structures that can be formed specifically in the C-rich strand in the NHE III1 region of the c-Myc promoter. KMnO4 is a chemical probe that reacts with the C5–C6 double bond of thymidine and is specific for thymine residues in single-stranded regions in which the C5–C6 bond is more exposed than usual.38-40 Thus we would expect thymine residues in the 5′ flanking region to be more reactive to KMnO4 than thymines in the NHE III1 if they are contained in a secondary structure such as an i-motif. Furthermore, their reactivity to KMnO4 in both the flanking region and NHE III1 element should be reduced by stabilization of the G-quadruplex upon addition of KCl. The results shown in Figure 5 are precisely in accord with these expectations. While there is more pronounced KMnO4 cleavage of the thymine residues in the flanking region relative to the potential i-motif region, both regions of cleavage are suppressed upon addition of KCl. The lower reactivity of thymines in the polypyrimidine tract to KMnO4 in both the absence and presence of KCl suggests that this region adopts a secondary structure that may be in part stabilized by stacking of the thymidine residues into the secondary structure. Significantly, the 5′ thymine between cytosine tracts 1 and 2 (asterisk in Figure 5) showed no reactivity to KMnO4, suggesting that it is sterically inaccessible to KMnO4, in contrast to its 3′ neighbor.
Figure 5.
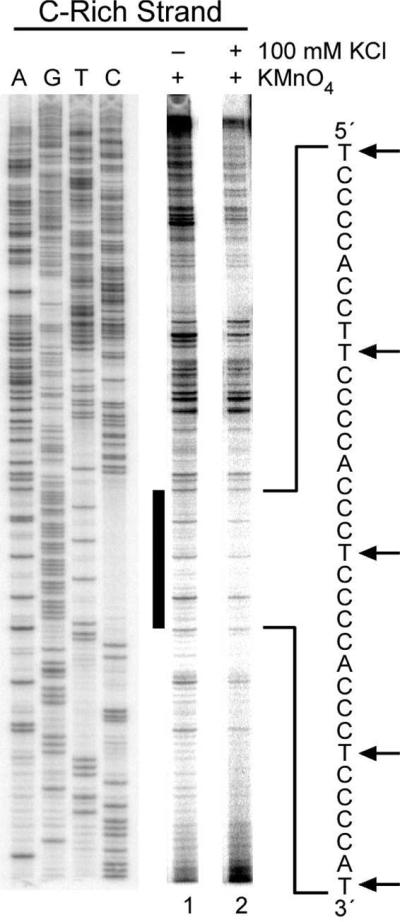
In vitro footprinting of the c-Myc promoter region with KMnO4. Autoradiogram showing KMnO4-reactive sites on the C-rich strand of the supercoiled Del4 plasmid. Plasmid DNA was incubated in the absence of salt (lane 1) and in the presence of 100 mM KCl (lane 2) at 37 °C overnight before incubation with KMnO4. To the right of the gel is the sequence of the NHE III1. Dotted lines indicate the KMnO4 cleaved thymines, an asterisk marks the protected thymine site, and the numbered brackets denote the cytosine tracts.
Next Br2 protection experiments were used to probe for secondary structures that can be formed specifically by the C-rich strand in the NHE III1 region of the c-Myc promoter (Figure 6A). Br2 is known to react selectively with the C5–C6 double bond of the pyrimidine base within DNA, resulting in 5-bromocytosine and 6-bromothymidine.39 In particular, the Br2 reaction with the cytosine residues in a single-stranded region is at least 10-fold higher than that in duplex DNA.39 Therefore, we presumed that the cytosine residues in the loop regions of i-motif structures would be more reactive to Br2 than the cytosine residues involved in base pairing and intercalation, allowing us to deduce specific cytosine residues required for base pairing and intercalation in these structures.19,42 Indeed, we have previously shown that the Br2 reactivity of the i-motifs in VEGF42 and RET19 shows a clear pattern of protection for cytosines involved in these structures. Several cytosines that are more reactive to Br2 occur within four of the six cytosine tracts in the NHE III1 (arrows in Figure 6A).
Figure 6.
(A) In vitro footprinting of the c-Myc promoter region with Br2. The plasmid DNA was incubated in the absence of salt (lanes 1 and 2) and in the presence of 100 mM KCl (lane 3) at 37 °C overnight before incubation with Br2. Arrows indicate enhanced reactivity to Br2. (B) Expansion of the NHE III1 region from (A) together with the densitometer scans of lane 3 and the G-sequencing lane. The horizontal bars indicate the six cytosine tracts numbered from the 5′ end. (C) Cartoon representation of the relative intensity of Br2 cleavage of lane 3 from (A). Filled black circles = fully cleaved; half-filled black circles = moderately cleaved; quarter-filled black circles = weakly cleaved; filled yellow circles = fully protected. The brackets indicate the six cytosine tracts. (D) The major i-motif folding pattern deduced from the data in (B) and (C).
This same region and its reactivity to Br2 are examined in more detail in Figure 6, B and C. First, only in the tracts of four cytosines is there enhanced cleavage of the 3′-cytosine by Br2, as was previously shown for similar cytosine tracts in the RET and VEGF promoters that form i-motifs.19,42 The two tracts of three cytosines (3 and 5 in Figure 6, B and C) do not show 3′-enhanced cleavage. Second, the extent of cleavage of the cytosine tracts is biased toward the 3′-end of the NHE III1, with cytosine tracts 1, 2, and 3 showing noticeably less cleavage than the other three tracts. The pattern of cleavage of cytosine tracts 4 and 6 is more complex than the other cytosine tracts. Tracts 4 and 6, which have four bases each, show pronounced cleavage not only at the 3′-ends but also at intermediate bases, suggesting that they may reside in loop regions. Tract 5, which has three bases, is also moderately cleaved but without a bias toward the 3′-end. Overall, this analysis implies that the three 5′ tracts of cytosines (1, 2, and 3) are clearly preferred for providing three of the four tracts of cytosines necessary for i-motif formation. A fourth tract of three cytosines is needed to complete an intramolecular i-motif formed from six cytosine–cytosine base pairs. The relatively uniform and moderate cleavage of cytosine tract 5 and the uneven cleavage patterns of tracts 4 and 6 both suggest that this fourth tract is most likely provided by tract 5, leaving tract 4 in a loop region and tract 6 at a junction site. We speculate that the 3′ cytosine in run 6 is much more accessible than any of the other cytosines, because that residue might be located at the junction between normal duplex and stable secondary structures formed by the C-rich sequence of the NHE III1 region. We believe that the loop regions of the i-motif structure proposed in Figure 6D could form hydrogen-bonded capping structures to further stabilize i-motif structures, thus inhibiting cleavage by single-strand-specific probes such Br2.
This overall pattern of cytosine protection and cleavage is best described by the i-motif 6:2:6 loop isomer shown in Figure 6D. This model predicts the marked Br2 cleavage of the 3′ cytosines in tracts 1 and 2 and the relatively even but modest cleavage of tracts 3 and 5. Tracts 4 and 6 and the five-base sequence between tracts 1 and 2 then provide the loop regions. The less predictable Br2 cleavage patterns of tracts 4 and 6 and the two cytosines between tracts 1 and 2 (protected) are most likely explained by either stable capping structures or loop slippage, the latter of which would give rise to alternative loop isomers. Last, the relatively protected cleavage of thymines to KMnO4 in the loop regions also suggests that these bases are involved in stable capping structures. Significantly, we observed that 5 mM bromine did not produce any cleavage above background levels of the NHE III1 in a linearized plasmid DNA, even after 1 h incubation (see Supporting Information). This confirms the critical importance of negative supercoiling in inducing transient single-stranded regions and the secondary DNA structures in duplex DNA, as shown in Figure 7A.
Figure 7.
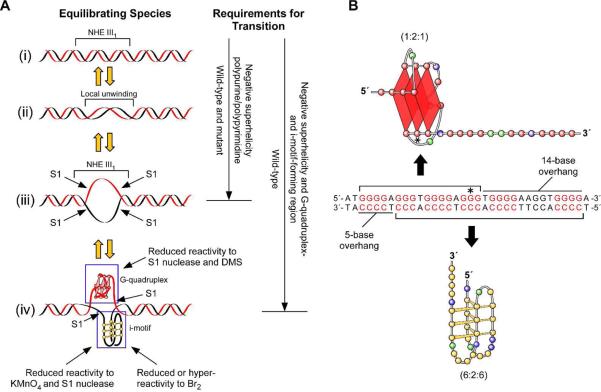
(A) Proposed equilibrating forms of the NHE III1 produced under negative superhelicity. The resistance/sensitivity to S1 nuclease, DMS, KMnO4, and Br2 of the various forms is shown in the left panel. Requirements for transition to the single-stranded form or G-quadruplex/i-motif species are shown in the right panel. (B) Asymmetric positioning of the DMS-protected G-quadruplex (top bracket) and i-motif (bottom bracket) together with 14- and 5base overhangs. An asterisk marks the position of the G-to-A mutant base-pairs in the G-quadruplex loop isomer.
Dynamic Equilibrium in the NHE III1 and Flanking Region of the c-Myc Promoter
The enzymatic (S1 nuclease and DNase I) and chemical (DMS, KMnO4) footprinting experiments provide a picture of dynamic regions that extend well beyond the polypurine/polypyrimidine region of the NHE III1. In both flanking regions, S1 nuclease and DNase I show evidence for a dynamic equilibrium between duplex, partially unwound duplex, and single-stranded regions. As anticipated, addition of 100 mM KCl, which is expected to stabilize the G-quadruplex, dampens the dynamics of these flanking regions, presumably because the more stable secondary structures act as a buffer for the absorption of the negative superhelicity. However, these enzymatic probes do not provide direct insight into the identity of the secondary DNA structures.
Significantly, G-quadruplex formation was shown to be dependent on both negative superhelicity and the wild-type sequence; however, the polypurine/polypyrimidine tract still present in the mutated NHE III1 shows evidence of a partially single-stranded region under conditions of negative superhelicity. In this case, the addition of KCl does not have as nearly a significant effect on the S1 nuclease cleavage of the flanking regions as with the wild-type sequence, again supporting the role of the secondary DNA structures in absorption of the negative superhelicity (see Figure 2, C and D). Figure 7A summarizes the equilibrating species in the NHE III1 with the requirements for transition to a partially single-stranded form (iii) and the secondary DNA structures (iv).
In sharp contrast to the enzymatic probes S1 nuclease and DNase I, the reactivity of the chemical probes DMS, KMnO4, and Br2 toward the NHE III1 region of the c-Myc promoter was little affected by the presence of potassium ions, suggesting that secondary structures already exist within this region in supercoiled form in vitro, even without additional potassium ions (compare Figures 2A and 3 with Figures 4A, 5, and 6A). Enzymatic nucleases are known to bind to and interact with DNA to induce large conformational changes in both the enzyme and the DNA, bringing the scissile phosphate backbone in closer proximity to the enzyme-active site.40,43 Therefore, the enhanced stability of the secondary structures in the presence of KCl is anticipated to have more influence on the extent and sites of DNA cleavage by enzymatic nucleases than by chemical DNA-cleaving or DNA-modifying agents such as DMS, KMnO4, and Br2, which do not require conformational changes of the DNA for cleavage and, correspondingly, are predicted to be less sensitive to KCl-dependent stabilization.
G-Quadruplex and i-Motif Formation
DMS and Br2 footprinting experiments provide direct insight into the G-quadruplex and i-motif folding patterns in the NHE III1. These footprinting experiments suggest that there is a single G-quadruplex isomer (1:2:1 loop isomer) and also one major form of the i-motif (6:2:6 loop isomer) (Figure 7B). These two structures are offset on both strands, showing a common use of only three purine/pyrimidine tracts (2−4) and leaving 14- and 5-base overhangs on the purine and pyrimidine strands, respectively (Figure 7B). Significantly, these single-stranded overhang regions are extensively cleaved by DMS (purine strand; see Figure 4, A and B) and Br2 (pyrimidine strand; see Figure 6, A and B). As anticipated, a specific G-to-A mutation of the central tetrad of the G-quadruplex (asterisk in Figure 7B) results in destabilization of this structure, as shown in Figure 4, C and D. While we had previously correctly identified the five guanine tracts at the 5′-end of the NHE III1 as being involved in the biologically relevant G-quadruplex structure,28 the DMS footprinting analysis of the single-stranded G-rich strand had identified tracts 2−5 rather than tracts 3−6 as forming the more stable G-quadruplex. In the context of duplex DNA under negative superhelicity, it is perhaps not too surprising that the result might be different from that in a single-stranded substrate.
One of the most significant conclusions of our study is that the i-motif is able to form under neutral pH conditions, provided negative superhelicity is maintained. This is in direct contrast to single-stranded DNA, where acidic pHs are required to form the hemiprotonated cytosine–cytosine base pair. In a previous study using a single-stranded G-rich 33-mer corresponding to the full NHE III1 sequence at a pH of 5.0−6.0, CD titration analysis showed eight cytosine–cytosine base pairs, which must arise from cytosine tracts 1, 2, 4, and 6.31 Presumably the acidic pH–determined stability is maximized in this form where all four tracts of four cytosines are utilized. In contrast, under negative superhelicity, the 6:2:6 isomer utilizes four tracts of three cytosines, thereby increasing the loop sizes relative to those found under acidic conditions. This is perhaps why a 14-base overhang on the G-rich strand is required to permit simultaneous formation of a specific 6-base capping loop in the i-motif. The partial or complete protection of many of the cytosine and thymine bases in the loops to chemical reagents is also supportive of the idea that the specific capping structures are an important component of the conformational stability of the i-motif induced under negative superhelicity. This important result implies that i-motifs as well as G-quadruplexes can form in promoter regions under conditions of transcriptionally induced negative superhelicity and may therefore displace transcriptional factors, such as CNBP or hnRNP K, that bind to single-stranded CT elements to activate transcription of c-Myc.32,33
c-Myc Transcriptional Regulation
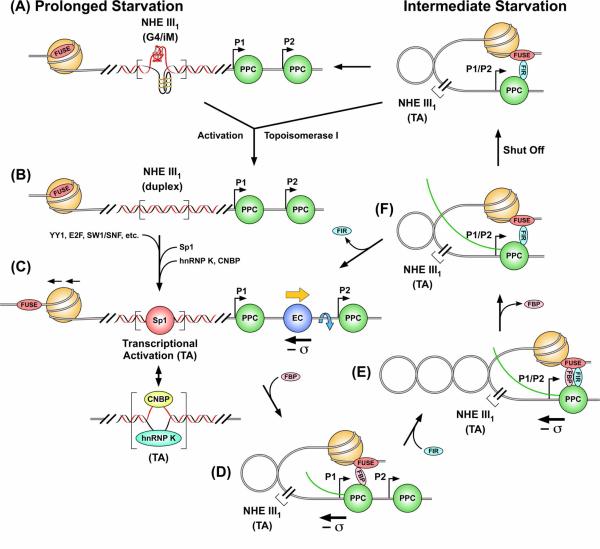
A long-neglected aspect of transcriptional regulation is the potential importance of torsional stress, which is propagated through DNA as a consequence of the forward movement of polymerase relative to the active promoter.44 The previously described mechanism for control of the rate of transcriptional firing from the c-Myc promoter involves transcription-generated supercoiling together with melting of the FUSE 1.7 Kb upstream of the P2 promoter, which has been termed a “molecular servomechanism.”45 This servomechanism, which is controlled by the magnitude of ongoing transcription, has been aptly called a “cruise control,” but this mechanism does not constitute a “starter” mechanism, since the FUSE is unable to modulate transcriptional output until transcription is initiated.45 Two questions arise from this observation: what are the elements that constitute the starter and how can these control mechanisms coordinate their activities? The local topology of the c-Myc promoter with P1 and P2 promoters, together with the NHE III1 and the FUSE, is shown in Figure 8A. The P1 promoter is 117 base pairs downstream of the NHE III1 and 1.5 Kb downstream of the FUSE. As demonstrated here, supercoiling has the ability to generate the formation of G-quadruplex and i-motif structures within the NHE III1. These structures have been shown to be silencer elements in a luciferase reporter system, because when destabilized in a mutant plasmid, they enhance transcriptional activity.28 Conversely, stabilization by G-quadruplex-interactive compounds reduces transcriptional activity.28 In the serum-starved condition, we propose that these structures exist in NHE III1, which would prevent transcriptional factors from binding to this region. These structures would also provide a dynamic buffer able to absorb transcriptionally induced negative superhelicity, thus maintaining the FUSE in a nucleosomal state (Figure 8A). Additionally, the superhelicity stored in the G-quadruplex and i-motif structures might be used for DNA melting at the start point and then to assist in polymerase escape. Removal of the negative superhelicity by topoisomerase I provides conditions more amenable to conversion back to duplex and, subsequently, single-stranded DNA (Figure 8B). Both forms are binding sites for transcriptional factors (Sp1 for duplex and hnRNP K and CNBP for single-stranded DNA), which are known to bind to this element to activate transcription (Figure 8C).32,33 Protein-facilitated removal of the secondary DNA structures, which are buffers for absorption of negative superhelicity, also provides a permissive topology for this transcriptionally induced supercoiling to propagate and move the FUSE into a nonnucleosomal configuration (Figure 8C).26 The FUSE “mechanosensor” function then takes over to provide a real-time feedback from the P2 promoter involving the competitive binding of FBP (promoter firing; Figure 8D) and FIR (promoter extinction; Figure 8, E and F) until a time when starvation triggers the FUSE once more to become nucleosomal, P1 transcriptionally induced torque rejects the transcriptional factor, and the G-quadruplex and i-motif are reinstated to silence transcription (Figure 8A).
Figure 8.
Cartoon showing a model for the mechanosensor mechanism and activation/silencing of the c-Myc promoter involving the FUSE and NHE III1 in association with transcriptionally generated negative superhelicity (adapted from Figure 7 in ref. 44). (A) In the absence of serum, c-Myc transcription is turned off and the FUSE is masked by the nucleosome, and the binding sites for Sp1, CNBP, and hnRNP K are sequestered in the G-quadruplex/i-motif forms (G4/iM). The Pre-Promoter Escape Complex (PPC) remains paused within the promoter proximal regions (P1 and P2). (B) Upon addition of serum, topoisomerase I removes negative superhelicity to provide conditions amenable to conversion of secondary DNA structures in the NHE III1 back to duplex or single-stranded forms. (C) Sp1 binds to the duplex NHE III1, the FUSE-masking nucleosome is remodeled by SWI/SNF to expose FUSE, and the earliest factors bind to the promoter (YY1, E2F) to advance the PPC to a full Elongation Complex (EC). Negative superhelicity (– σ) generated by EC then melts the FUSE after nucleosomal remodeling. As negative superhelicity increases, duplex DNA is replaced by single-stranded DNA, and CNBP and hnRNP K bind to the NHE III1. (D) Melted FUSE recruits FBP, which drives transcription up to peak levels by looping with TFIIH. (E) FBP activity leads to negative supercoil accumulation within the topologically closed FBP–TFIIH loop. High negative superhelicity fully melts FUSE and conscripts FIR through protein–protein and protein–DNA contacts. (F) As FIR represses transcription, the dynamic stress at FUSE falls, ejecting FBP. The FIR–TFIIH connection allows only basal transcription, so the polymerase at a low rate dissociates from TFIIH to become an EC. Without sufficient activation, the superhelical stress eventually dissipates, and the c-Myc promoter reverts back to the silent state in (A). For illustrative purposes, only the FUSE-masking nucleosome is shown.
In a more general sense, putative G-quadruplex-forming elements have been found to be concentrated in regions immediately upstream of the transcription initiation site,12 and conservation of the sequences that form stable intramolecular G-quadruplexes has been noted in a variety of human oncogenes, including c-Myc, Bcl-2, VEGF, Hif-1α, Ret, c-Kit, PDGF-A, KRAS, and c-Myb (reviewed in refs. 34 and 46). Although Sp1 has been found to be a prime factor that binds to G-quadruplex-forming elements,46 other transcriptional factors, such as hnRNP K and CNBP, that bind to single-stranded elements have also been described.32,33 In addition, proteins that recognize and bind to G-quadruplexes may also play an important role.47 As has been previously noted, “...transcription is inevitably coupled with dynamic perturbation of the double helix [and] the mechanisms [mechanosensor] discussed here may prove to be widespread if not universal.”26 This statement may also be equally applicable to transcriptional activation/silencing mechanisms involving G-quadruplexes and i-motifs, since sequences that can form these structures are found in high abundance in regions upstream of the transcription start sites in eukaryotic organisms.12
Implication for Design of Drugs That Target Transcriptional Control via Dynamically Induced Secondary DNA Structures
The insight provided by the model (Figure 8) for the mechanosensor mechanism and activation/silencing of the c-Myc that involves the NHE III1 and the FUSE provides a number of opportunities for small molecule–mediated external control of gene expression. As eluded to earlier, the globular structures of the G-quadruplex and i-motif determined by the primary base sequence of the NHE III1 afford a unique opportunity to target in this case a 31-base-pair sequence with small drug-like molecules. Evidence from several laboratories28,48-50 already demonstrates that targeting G-quadruplexes has effect on transcription of c-Myc, c-Kit, KRAS, and VEGF, but in the majority of cases selectivity has yet to be determined. The abundance of these putative G-quadruplex-forming units in eukaryotic promoters12,51 suggests that drug selectivity may be a problem, but the diversity of folding patterns and loop sizes offers opportunities to distinguish between these structures. Only sparse information is available on drug targeting of i-motif structures,52 and here the folding patterns may diverge considerably between single-stranded templates at acidic pH levels and negatively supercoiled induced structures (see before). This may be less of an issue for G-quadruplexes, but in the case of c-Myc there is a subtle difference between the G-quadruplex that predominates in a single-stranded state and that found in this study. Of particular interest is the possibility of drug targeting the complex structure that exists within a combined G-quadruplex/i-motif complex as shown in Figure 8 (A).
In addition to the NHE III1, the mechanosensor mechanism involving the FUSE/FBP/FIR system also provides a unique opportunity to modulate gene transcription. Indeed, in a collaborative effort between scientists at NCI and Abbott labs the targeting of the interface between the FUSE and FBP has been achieved with benzoylanthranilic acid–type molecules identified through SAR by NMR.53 Although this approach was not carried forward into cellular studies, it remains a viable complementary approach to that of targeting the NHE III1.
We believe the potential “widespread if not universal”26 molecular features of this transcriptionally induced negative superhelicity provide an enormous opportunity in drug discovery and development, perhaps equivalent to that of drug targeting of kinases. Quarfloxin, a first-in-class drug now in phase 2 clinical trials, is believed to exert its mechanism of action through targeting G-quadruplexes.54,55 Much remains to be learned in this area, and in particular, proof of the existence of these structures in the promoter elements of cells and their involvement in modulation of gene transcription are important objectives of this lab and others involved in this emerging area for drug discovery.
MATERIALS AND METHODS
Enzymes and Oligonucleotide DNA
T4 polymerase kinase, DNase I, and S1 nuclease were purchased from Promega. PAGE-purified oligonucleotides were obtained from Sigma Genosys.
Plasmid DNA
For in vitro footprinting of the c-Myc promoter region, we used the supercoiled form of the human c-Myc reporter Del4, which was kindly provided by Kenneth W. Kinzler (Johns Hopkins University, Baltimore, MD). The Del4 plasmid contains ∼850 bp of c-Myc sequence, including the two major promoters (P1 and P2) and the NHE III1 region.28 The mutant reporter plasmid Del4-DM was constructed by introducing point mutations into the specific guanine residues within the NHE III1 region of Del4 using a QuickChange mutagenesis kit (Stratagene) to destabilize the G-quadruplex-forming ability of this region.28
In Vitro Footprinting of the c-Myc Promoter Region with DNase I, S1 Nuclease, and Chemical Reagents
In vitro plasmid footprinting experiments were performed as described previously.16 In brief, a supercoiled form of the plasmids (1 μg of Del4 or Del4-DM) was incubated in 20 μL Tris-HCl buffer (20 mM, pH 7.6) without or with 100 mM KCl at 37 °C overnight and then treated with DNase I (0.5 U) or S1 nuclease (200 U) for 2 min. DNA was precipitated with ethanol and resuspended in double-distilled water after vacuum drying. For chemical footprinting, plasmid DNA was treated for 2 min with 0.5% DMS and 10 min with 10 mM KMnO4 or 1 mM molecular Br2 (KBr with KHSO5) as previously described.39 The reactions were then terminated by adding 50 μL of stop mix containing 0.6 M Na-acetate (pH 5.2), and unreacted chemicals were removed by ethanol precipitation. Following ethanol precipitation, the DNA pellet was dried and resuspended with 100 μL of double-distilled water, and the samples were heated at 90 °C for 30 min to induce strand cleavage. Following thermal treatments, the DNA samples were completely dried and resuspended with double-distilled water. To map cleavage sites on the plasmid DNA, linear amplification by PCR was performed using a Thermo Sequenase Cycle Sequencing kit (USB), following the manufacturer's instructions, with the 32P-labeled gene-specific primers GGGCCGGTGGGCGGAGATTAGCG and CGCGCGTAGTTAATTCATGCGGC to amplify the top and bottom strands of the plasmid DNA, respectively. PCR was carried out using cycling conditions consisting of an initial 4-min denaturation step at 94 °C, 1 min at 60 °C, and 1 min at 72 °C, for a total of 40 cycles. After primer extension, the samples were dried under vacuum, and the DNA pellet was resuspended in 15 μL of formamide dye and electrophoresed in a 8% urea-acrylamide gel for 8 h at 1400 V. Dideoxy sequencing reactions were carried out using the same labeled primer, according to the protocol provided by the manufacturer. The dried gel was exposed to a phosphorimager screen and quantified using ImageQuant software.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the National Institutes of Health (CA94166 and CA109069). We thank David Levens, Shankar Balasubramanian, Andrew Travers, and Danzhou Yang for insightful discussions. We are grateful to David Bishop for preparing, proofreading, and editing the final version of the manuscript and figures.
Abbreviations
- NHE III1
Nuclease Hypersensitive Element III1
- FUSE
Far Upstream Element
- FBP
FUSE Binding Protein
- FIR
FBP Interacting Repressor
- PPC
Pre-Promoter Escape Complex
- EC
Elongation Complex
- TFIIH
Transcription Factor II H
References
- 1.Marcu KB. Regulation of expression of the c-myc proto-oncogene. Bioessays. 1987;6:28–32. doi: 10.1002/bies.950060108. [DOI] [PubMed] [Google Scholar]
- 2.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 3.Pelengaris S, Khan M. The c-Myc oncoprotein as a treatment target in cancer and other disorders of cell growth. Expert Opin. Ther. Targets. 2003;7:623–642. doi: 10.1517/14728222.7.5.623. [DOI] [PubMed] [Google Scholar]
- 4.Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 5.Marcu KB, Patel AJ, Yang Y. Differential regulation of the c-Myc P1 and P2 promoters in the absence of functional tumor suppressors: implications for mechanisms of deregulated Myc transcription. Curr. Top. Microbiol. Immunol. 1997;224:47–56. doi: 10.1007/978-3-642-60801-8_4. [DOI] [PubMed] [Google Scholar]
- 6.Nowell P, Finan J, Dalla-Favera R, Gallo RC, ar-Rushdi A, Romanczuk H, Selden JR, Emanuel BS, Rovera G, Croce CM. Association of amplified oncogene c-myc with an abnormally banded chromosome 8 in a human leukaemia cell line. Nature. 1982;306:494–497. doi: 10.1038/306494a0. [DOI] [PubMed] [Google Scholar]
- 7.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 8.Dalla-Favera R, Westin E, Gelmann EP, Martinotti S, Bregni M, Wong-Staal F, Gallo RC. The human onc gene c-myc: structure, expression, and amplification in the human promyelocytic leukemia cell line HL-60. Haematol. Blood Transfus. 1983;28:247–254. doi: 10.1007/978-3-642-68761-7_47. [DOI] [PubMed] [Google Scholar]
- 9.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. U.S.A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. U.S.A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphey DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy JG, Heywood SM. A long polypyrimidine/polypurine tract induces an altered DNA conformation on the 3′ coding region of the adjacent myosin heavy chain gene. Nucleic Acids Res. 1987;15:8069–8085. doi: 10.1093/nar/15.19.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Lin XH, Qiu QQ, Deuel TF. Modulation of transcription of the platelet-derived growth factor A-chain gene by a promoter region sensitive to S1 nuclease. J. Biol. Chem. 1992;267:17022–17031. [PubMed] [Google Scholar]
- 16.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS protooncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 19.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benham CJ. Theoretical analysis of conformational equilibria in superhelical DNA. Ann. Rev. Biophys. Biophysical Chem. 1985;14:23–45. doi: 10.1146/annurev.bb.14.060185.000323. [DOI] [PubMed] [Google Scholar]
- 21.Williams DL, Kowalski D. Easily unwound DNA sequences and hairpin structures in the Epstein-Barr virus origin of plasmid replication. J. Virol. 1993;67:2707–2715. doi: 10.1128/jvi.67.5.2707-2715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JS, Ashley C, Hampel KJ, Bradley R, Scraba DG. A stable interaction between separated pyrimidine·purine tracts in circular DNA. J. Mol. Biol. 1995;1995;252:283–288. doi: 10.1006/jmbi.1995.0495. [DOI] [PubMed] [Google Scholar]
- 23.Ashley C, Lee JS. A triplex-mediated knot between separated polypurine–polypyrimidine tracts in circular DNA blocks transcription by Escherichia coli RNA polymerase. DNA Cell Biol. 2000;19:235–241. doi: 10.1089/104454900314500. [DOI] [PubMed] [Google Scholar]
- 24.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 27.DesJardins E, Hay N. Repeated CT elements bound by zinc finger proteins control the absolute and relative activities of the two principal human c-myc promoters. Mol. Cell. Biol. 1993;13:5710–5724. doi: 10.1128/mcb.13.9.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-Myc transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-Myc promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 31.Simonsson T, Pribylova M, Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 32.Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 33.Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, Dreyfuss G, Levens D. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 1993;268:18249–18258. [PubMed] [Google Scholar]
- 34.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siebenlist U, Henninghausen L, Battey J, Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 36.Weller P, Bark C, Janson L, Pettersson U. Transcription analysis of a human U4C gene: involvement of transcription factors novel to snRNA gene expression. Genes Dev. 1988;2:1389–1399. doi: 10.1101/gad.2.11.1389. [DOI] [PubMed] [Google Scholar]
- 37.Evans T, Efstratiadis A. Sequence-dependent S1 nuclease hypersensitivity of a heteronomous DNA duplex. J. Biol. Chem. 1986;261:14771–14780. [PubMed] [Google Scholar]
- 38.Łoziński T, Wierzchowski KL. Mg2+-modulated KMnO4 reactivity of thymines in the open transcription complex reflects variation in the negative electrostatic potential along the separated DNA strands. Footprinting of Escherichia coli RNA polymerase complex at the λPR promoter revisited. FEBS J. 2005;272:2838–2853. doi: 10.1111/j.1742-4658.2005.04705.x. [DOI] [PubMed] [Google Scholar]
- 39.Ross SA, Burrows CJ. Cytosine-specific chemical probing of DNA using bromide and monoperoxysulfate. Nucleic Acids Res. 1996;24:5062–5063. doi: 10.1093/nar/24.24.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saluz HP, Jost JP. Approaches to characterize protein-DNA interactions in vivo. Crit. Rev. Eukaryot. Gene Expr. 1993;3:1–29. [PubMed] [Google Scholar]
- 41.Johnston BH, Rich A. Chemical probes of DNA conformation: detection of ZDNA at nucleotide resolution. Cell. 1985;42:713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- 42.Guo K, Gokhale V, Hurley LH, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36:4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dabrowiak JC, Goodisman J, Ward B. Quantitative DNA footprinting. Methods Mol. Biol. 1997;90:23–42. doi: 10.1385/0-89603-447-X:23. [DOI] [PubMed] [Google Scholar]
- 44.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front. Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Kouzine F, Nie Z, Chung H-J, Elisha-Feil Z, Weber A, Zhao K, Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todd AK, Neidle S. The relationship of potential G-quadruplex sequences in cis-upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res. 2008;36:2700–2704. doi: 10.1093/nar/gkn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 48.Ou T-M, Lu Y-J, Zhang C, Huang Z-S, Wang X-D, Tan J-H, Chen Y, Ma K-L, Wong K-Y, Tang JC-O, Chan AS-C, Gu L-Q. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-Myc by quindoline derivatives. J. Med. Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 49.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: Small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D, Liu W-J, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 52.Fedoroff O. Yu., Rangan A, Chemeris VV, Hurley LH. Cationic porphyrins promote the formation of i-motif DNA and bind peripherally by a nonintercalative mechanism. Biochemistry. 2000;39:15083–15090. doi: 10.1021/bi001528j. [DOI] [PubMed] [Google Scholar]
- 53.Huth JR, Yu L, Collins I, Mack J, Mendoza R, Isaac B, Braddock DT, Muchmore SW, Comess KM, Fesik SW, Clore GM, Levens D, Hajduk PJ. NMR-driven discovery of benzoylanthranilic acid inhibitors of far upstream element binding protein binding to the human oncogene c-myc promoter. J. Med. Chem. 2004;47:4851–4857. doi: 10.1021/jm0497803. [DOI] [PubMed] [Google Scholar]
- 54.Bates P, Mergny J-L, Yang D. Quartets in G-major. The first international meeting on quadruplex DNA. EMBO Reports. 2007;8:1003–1010. doi: 10.1038/sj.embor.7401073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cylene Pharmaceuticals website: http://www.cylenepharma.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.