Abstract
Terminally misfolded or unassembled proteins in the early secretory pathway are degraded by a ubiquitin and proteasome-dependent process known as ER-associated degradation (ERAD). How substrates of this pathway are recognized within the ER and delivered to the cytoplasmic ubiquitin conjugating machinery is unknown. We report here that OS-9 and XTP3-B/erlectin are ER-resident glycoproteins that bind to ERAD substrates and, via the SEL1L adaptor, to the ER membrane-embedded ubiquitin ligase Hrd1. Both proteins contain conserved mannose 6-phosphate receptor homology (MRH) domains which are required for interaction with SEL1L, but not with substrate. OS-9 associates with the ER chaperone GRP94 which, together with Hrd1 and SEL1L, is required for the degradation of an ERAD substrate, mutant α1-antitrypsin. These data suggest that XTP3-B and OS-9 are components of distinct, partially redundant quality control surveillance pathways that coordinate protein folding with membrane dislocation and ubiquitin conjugation in mammalian cells.
Keywords: ER-associated degradation (ERAD), OS-9, GRP94, XTP3-B, SEL1L, Hrd1
Folding of nascent polypeptide chains in the endoplasmic reticulum (ER) is monitored by a quality control process that ensures that only correctly folded and assembled proteins are deployed1. Terminally misfolded or unassembled polypeptides are destroyed by a process known as ER-associated degradation (ERAD)2-4. Because ERAD substrates initially are wholly or partially situated within the ER lumen, they must first be dislocated across the ER membrane in order to be degraded. Covalent ubiquitin conjugation is required for tight coupling of substrate dislocation and targeting to the 26S proteasome for destruction5-7. Multi-protein complexes containing the yeast8-11 and mammalian12, 13 ER ubiquitin ligases have been identified and proposed as nexuses for the recognition, dislocation and ubiquitination aspects of ERAD.
The ability to distinguish native proteins from non-native states and folding intermediates in the ER lumen has been proposed to exploit a “glycan code” in which progressive trimming of terminal mannose residues on Man9GlcNAc2 Asn-linked glycans by mannosidases serves as a molecular timer that specifies the folding history of the substrate protein14. An important clue to understanding how this code is deciphered by the quality control apparatus was the discovery of EDEM (Htm1p in yeast), an α-mannosidase-like, ER-resident protein proposed to function as a mannose-specific lectin that captures glycoproteins locked in futile cycles of folding and unfolding15, 16. EDEM and Htm1p are required for degradation of some, but not all misfolded glycoproteins.
Recently, a second lectin, Yos9p, was identified in yeast and proposed to function as a crucial bridge linking mannose recognition to the ubiquitin conjugation system9, 17-19. Yos9p has an N-terminal signal sequence and a C-terminal HDEL peptide that are respectively required to deliver Yos9p to, and retain it in the ER lumen (Fig. 1a). Deletion of Yos9p abrogates the degradation of the glycosylated but not the non-glycosylated forms of the lumenal ERAD substrate CPY*17, 18, 20. Yos9p is able to discriminate mutant (CPY*) from wild-type CPY and appears to bind selectively to substrates that contain Man8GlcNAc2 or Man5GlcNAc2 glycans19. Yos9p is recruited into a protein complex containing the Hrd1p ubiquitin ligase and the lumenal chaperone Kar2p through association with Hrd3p, a lumenally-exposed transmembrane tetratricopeptide repeat (TPR)-containing protein known to associate with Hrd1p8, 9, 21. Thus, Yos9p is a central component of a lumenal quality control surveillance system that connects substrate recognition to protein degradation.
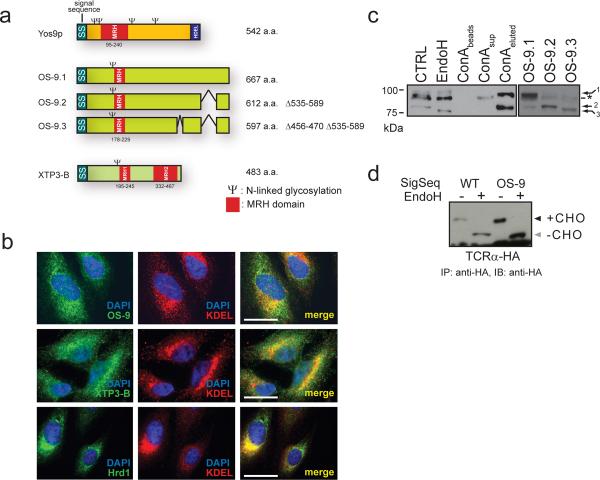
Figure 1. The lectins OS-9 and XTP3-B are ER resident proteins.
a. Domain architecture for XTP3-B, OS-9 and Yos9p. Conserved MRH domains (red) and potential N-linked glycosylation sites (Ψ) are indicated. b. Colocalization of endogenous OS-9, XTP3-B and Hrd1 (green) with the ER-resident proteins (anti-KDEL, red) and nuclei (DAPI, blue) by immunofluorescence in HeLa cells. Antibody specificity is presented in Supplemental Information (Fig. S1) c. Separation of OS-9 isoforms by SDS-PAGE and detected by anti-OS-9. 1% Triton X-100 lysates from HEK293 cells untreated (lane 1), + EndoH (lane 2), concanavalin A-Sepharose (ConA) fractions (lanes 3-5) and transiently expressing OS-9.1, 2 & 3 (lanes 6-8). Non-specific background band is indicated by (*). Full scans are presented in Supplemental Information (Fig. S5). d. Transient expression of TCRα-HA in HEK293 cells with endogenous (left) or OS-9 (right) signal sequence. Samples were immunoprecipitated with anti-HA, treated -/+ EndoH, separated by SDS-PAGE and immunoblots probed with anti-HA.
The human genome contains two genes, XTP3-B (XTP3 transactivated protein B; Erlectin22) and OS-9 (Fig. 1a) which share ~15% identity with Yos9p and ~23% identity with each other. Despite low overall homology among the three proteins, the presence of predicted N-terminal signal sequences, MRH domains and N-linked glycosylation sites suggests that XTP3-B and/or OS-9 may serve a function similar to Yos9p. XTP3-B was previously identified based on its ability to interact with kremen 2 (Krm2), a plasma membrane coreceptor for Dickkopf (Dkk) in the Wnt/β-catenin signaling pathway22. XTP3-B has two MRH domains and, despite lacking a C-terminal KDEL sequence, is localized in the ER lumen22. Binding to Krm2 in vitro requires the C-terminal MRH domain and is abolished by Krm2 deglycosylation, consistent with the conclusion that XTP3-B is a lectin.
OS-9 was originally identified in a screen for genes that are upregulated in osteosarcoma23 and myeloid leukemia24. Fragments of OS-9 have also been independently identified in a number of yeast 2-hybrid screens using cytoplasmic proteins as bait25-27. The interaction of OS-9 with proteins or domains located within the cytoplasm are surprising, as all three alternatively spliced isoforms are predicted to contain a canonical N-terminal signal sequence, an MRH domain and an N-linked glycan24 (Fig. 1a).
Here we show that OS-9 and XTP3-B are both ER-resident lectins that bind to ERAD substrates and to the membrane-embedded Hrd1-SEL1L ubiquitin ligase complex. Our data suggest that these lectins form an ERAD nexus that coordinates substrate recognition in the ER lumen with ubiquitin conjugation in the cytoplasm.
RESULTS
OS-9 and XTP3-B/Erlectin are ER resident proteins
Endogenous XTP3-B and OS-9 in HEK293 cells exhibited a prominent perinuclear reticular pattern of expression with extensive overlap with immunofluorescence from a anti-KDEL antibody, similar to the pattern exhibited by Hrd1 (Fig. 1b). Endogenous OS-9 in HEK293 cells migrated as two predominant electrophoretic species corresponding to OS-9.1 and OS-9.2 (Fig. 1c). We were unable to detect OS-9.3, consistent with the previous finding indicating that OS-9.1 and OS-9.2 mRNA is far more abundant24. Digestion with endoglycosidase H increased the mobilities of bands corresponding to OS-9.1 and OS-9.2, supporting their likely identities as ER-resident glycoproteins. OS-9.1 and OS-9.2 bound to concanavalin A (ConA), a lectin that selectively binds to high-mannose N-linked oligosaccharides, and were specifically eluted by methyl α-D-mannopyranoside. Together with the finding that the N-terminal 34 amino acids from OS-9 can functionally replace the signal sequence of an unrelated type I membrane protein (TCR-α; Fig. 1d), we conclude that isoforms 1 and 2 of endogenous OS-9, like XTP3-B, are bona-fide ER-resident glycoproteins, consistent with a potential role in quality control surveillance in the ER lumen.
OS-9 and XTP3-B interact with the Hrd1-SEL1L ubiquitin ligase
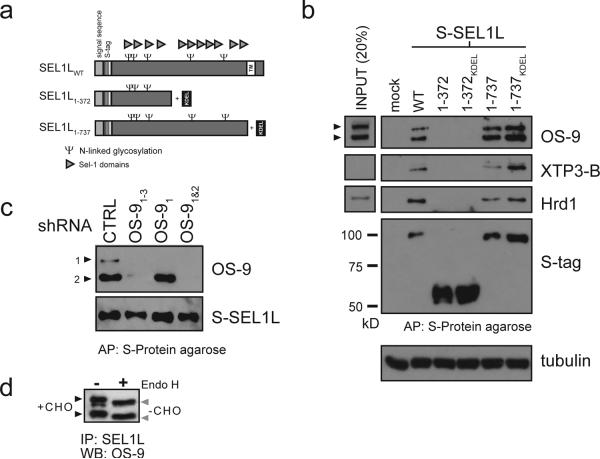
SEL1L is a component of an ER multiprotein complex implicated in the process of recognition and/or dislocation of misfolded proteins12, 28. Like its yeast ortholog, Hrd3p, mammalian SEL1L is a type I transmembrane glycoprotein with the bulk of the protein, composed of twelve copies of the short tetratricopeptide-like “Sel1” repeats29, exposed to the ER lumen (Fig. 2a). Previous studies have demonstrated that SEL1L interacts with the transmembrane ERAD components Hrd1, Derlin1 and Derlin2 as well as the cytoplasmic protein VCP/p9712. Full length S-tagged SEL1L (SEL1LWT) coprecipitated endogenous XTP3-B as well as the two OS-9 isoforms together with Hrd1, suggesting that a multiprotein complex containing orthologs of Hrd1p-Hrd3p-Yos9p is conserved in mammalian cells (Fig. 2b). Deletion of the C-terminal portion of SEL1L containing eight Sel1 repeats (SEL1L1-372) abolished all of these interactions, establishing that an intact lumenal domain is required for complex formation. Deletion of the C-terminal transmembrane domain (SEL1L1-737) decreased but did not abolish capture of Hrd1, OS-9 and XTP3-B (Fig. 2b), similar to findings observed for Hrd3p interactions with Hrd1p and Yos9p21. This reduced association is probably due to secretion of SEL1L1-737 as the mutant protein could be readily detected in the media (data not shown). Wild-type levels of interaction were restored when we appended a KDEL retrieval signal to the SEL1L1-737 construct. Thus, the transmembrane domain of SEL1L is required for its retention in the ER, but not for its interaction with Hrd1, XTP3-B or OS-9. The identities of the two OS-9 isoforms pulled-down by SEL1L were confirmed in transfected cells expressing S-tagged SEL1L with isoform-specific short-hairpin RNAs (shRNAs, Fig. 2c). Finally, the sensitivity of both SEL1L-bound OS-9 isoforms to EndoH digestion (Fig. 2d) supports the conclusion that these protein complexes are present within the ER lumen.
Figure 2. The lumenal domain of SEL1L scaffolds Hrd1, OS-9 and XTP3-B.
a. Schematic diagram of full length and truncated SEL1L. Potential N-linked glycosylation sites (Ψ) and Sel-1 domains (triangle) are indicated. b. S-tagged versions of full length (WT), truncated (1-372, 1-737) and KDEL-amended (1-372KDEL, 1-737KDEL) SEL1L expressed in HEK293 cells. From normalized protein amounts, coprecipitation of indicated proteins (OS-9, XTP3-B and Hrd1) with representative input controls (20% of starting crude lysate) were assessed by immunoblot with the designated antibodies. c. Coexpression of S-SEL1LWT with shRNA targeting the indicated OS-9 isoforms and affinity purification as in Fig. 2b. Immunoblots were probed for S-tag and OS-9. shRNA specificity is also presented in Supplemental Information (Fig. S2, S3) d. Pulldowns of SEL1LWT treated -/+ EndoH and probed for OS-9.
OS-9 and SEL1L are required for degradation of some ERAD substrates
Both XTP3-B and OS-9 were co-immunoprecipitated with the null Hong Kong variant of α1-antitrypsin (NHK), a well-characterized ERAD substrate16, 30, 31 (Fig. 3a). XTP3-B also bound to RI332, a mutant of ribophorin I and, like NHK, a lumenal ERAD substrate32, but not the T-cell receptor α subunit (TCRα), an unstable type I transmembrane protein33, 34 (Fig. 3b).
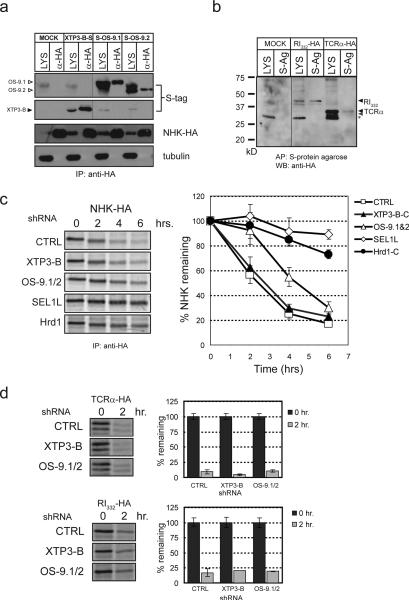
Figure 3. OS-9 and XTP3-B are required for ERAD.
a. Coexpression of S-tagged XTP3-B and OS-9.1/2 with NHK-HA. Triton X-100 lysates (LYS, 20%) and anti-HA immunoprecipitates (α-HA) were separated by SDS-PAGE and immunoblots are shown for the S-tag (top), NHK (middle) and tubulin (bottom). Hairline indicates where western blot exposures were joined. Full scans are presented in Supplemental Information (Fig. S5). b. Coexpression of S-tagged XTP3-B with TCRα-HA and RI332-HA. Crude lysates (20%) and material coprecipitated by S-protein agarose were probed with anti-HA. Non-specific background band is indicated by (*). c. Representative pulse-chase assay of NHK coexpressed with shRNA (CTRL/GFP, XTP3-B-C, OS-9.1&2, Hrd1-C and SEL1L, left). Bands were quantified by phosphorimager and expressed as the percent of the value at time = 0. Composite data for degradation time courses of NHK coexpressed with shRNA against CTRL (open square), XTP3-B (closed triangle), OS-9.1/2 (open triangle), SEL1L (open diamond) and Hrd1 (closed circle) (right). Data shown represent the mean and S.E.M. from at least 5 individual experiments. The efficacy and specificity of each shRNA are presented in Supplemental Information (Fig. S2, S3) d. Representative pulse-chase assays for the HA-tagged ERAD substrates TCRα and RI332 coexpressed with shRNAs targeting XTP3-B and OS- 9.1/2 used in 3c (left). Bar graph (right) represents composite pulse-chase data from at least 3 individual experiments quantified as in 3c. Mean and S.E.M. for 0 and 2 hr. time points are shown.
Knockdown of endogenous SEL1L or Hrd1 dramatically stabilized NHK, establishing that this ubiquitin ligase complex is essential for NHK degradation (Fig. 3c, S3). While knockdown of XTP3-B had no measurable effect on the kinetics or extent of NHK degradation, we found that depletion of OS-9 significantly impaired NHK clearance (Fig. 3c, S3). Knockdown of both XTP3-B and OS-9 together did not enhance the effect observed with OS-9 shRNA alone, suggesting that XTP3-B does not functionally compensate for the absence of OS-9 (data not shown). By contrast, no appreciable stabilization of TCRα or RI332 was observed following depletion of OS-9 or XTP3-B (Fig. 3d). Thus, the MRH-domain containing proteins XTP3-B and OS-9 interact with and are functionally required for distinct sets of ERAD substrates.
SEL1L links XTP3-B and OS-9 to Hrd1
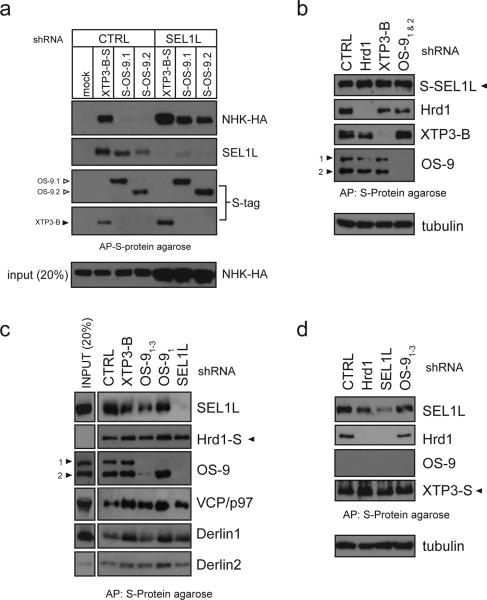
We used affinity capture combined with shRNA knockdown of various components in order to define the organization of OS-9, SEL1L and Hrd1 into functional complexes. Although S-tagged XTP3-B and OS-9 are both able to bind to NHK, we observed vastly greater levels of substrate bound at steady-state to the former compared with the latter (Fig. 4a, CTRL). We propose that this difference in the amount of NHK bound to the two lectins reflects more efficient transfer of the substrate to downstream degradation machinery from OS-9, which is required for NHK degradation than from XTP3-B, which is dispensable. This is supported by the observation that SEL1L knockdown drastically increased the total amount of NHK as well as the amount of NHK bound to OS-9 (Fig. 4a, SEL1L). Consistently, S-tagged SEL1L bound robustly to endogenous XTP3-B and OS-9 as well as to Hrd1; none of these interactions is affected by knockdown of XTP3-B, OS-9 or Hrd1 (Fig. 4b). By contrast, SEL1L depletion abrogated binding of S-tagged Hrd1 to OS-9 (Fig. 4c), and S-tagged XTP3-B to Hrd1 (Fig. 4d) without affecting the other Hrd1 and XTP3-B interactions. These data establish that SEL1L mediates the interaction between the ERAD ubiquitin ligase Hrd1, and the two lectins, OS-9 and XTP3-B and places them functionally upstream of the Derlin-VCP/p97 dislocation complex.
Figure 4. OS-9/XTP3-B interaction with Hrd1 is mediated through SEL1L.
a. Coexpression of NHK-HA, CTRL or SEL1L shRNA and S-tagged XTP3-B, OS-9.1 and OS-9.2 in HEK293 cells. Complexes were affinity purified by S-protein agarose from 1% TritonX-100 lysates. No S-tagged protein was expressed in CTRL lane. Immunoblots are shown for NHK (top), SEL1L (middle) and S-tag (bottom). A lysate input of 20% of the IP is probed for NHK (anti-HA, very bottom). b. Coexpression of S-tagged SEL1L with shRNA (CTRL/GFP, Hrd1-C, XTP3-B-C and OS-91&2) in HEK293 cells. Complexes were affinity purified from 1% CHAPS lysates by S-protein agarose. Loading was normalized to approximately equal amounts of S-SEL1L. c. Hrd1-S coexpressed with indicated shRNA and processed as in Fig. 4b. Input lysate representing 20% of the starting material for each affinity purification is shown on left. d. Coexpression of S-tagged XTP3-B and indicated shRNA (described above). In all cases, samples were separated by SDS-PAGE and western blots probed with the indicated antibodies.
OS-9 and XTP3-B interact with ER chaperones
To characterize the protein complexes associated with mammalian Yos9p orthologs, detergent-soluble proteins associated with affinity-purified S-tagged XTP3-B and OS-9 were identified by “shotgun” tandem LC-MS/MS analysis (Table 1). S-tagged lectins were captured from whole cells and from a rough microsome fraction enriched in ER (Supplemental Information, Fig. S4). Both lectins bound strongly to SEL1L, confirming the interactions we observed from functional depletion and pull-downs, and to GRP78/BiP, the lumenal Hsp70 class chaperone. Strikingly, the most prominent interaction for OS-9 was with GRP94, the ER-resident Hsp90 paralog.
Table 1.
LC-MS/MS analyses of complexes affinity purified with S-tagged ER lectins.
| OS-9.2 RM | OS-9.2 WC | XTP3-B RM | XTP3-B WC | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name |
Function/Role | Accession | #Scans Unique Peptides % Covarage Rank Order |
#Scans Unique Peptides % Covarage Rank Order |
#Scans Unique Peptides % Covarage Rank Order |
#Scans Unique Peptides % Coverage Rank Order |
||||||||||||
| GRP94 | ER chaperone | P14625 | 83 | 41 | 67% | 1 | 151 | 52 | 86% | 1 | NA | 8 | 5 | 8% | 12 | |||
| BiP | ER chaperone | P11021 | 65 | 29 | 58% | 2 | 125 | 37 | 75% | 3 | 22 | 15 | 31% | 2 | 70 | 28 | 59% | 1 |
| SEL1L | ER component | Q9UBV2 | 23 | 12 | 30% | 4 | 23 | 12 | 32% | 11 | 26 | 13 | 35% | 1 | 52 | 21 | 55% | 2 |
| OS-9 | ER lectin | Q13438 | 54 | 21 | 52% | 3 | 146 | 37 | 98% | 2 | NA | NA | ||||||
| Ribophorin I | N-linked glycosylation | P04843 | 7 | 6 | 13% | 5 | 37 | 19 | 42% | 5 | NA | NA | ||||||
| XTP3-B | ER lectin | Q96DZ1 | NA | NA | 13 | 8 | 3 | 20 | 8 | 3 | ||||||||
Abbreviations: RM (rough microsome fraction), WC (whole cell lysate).
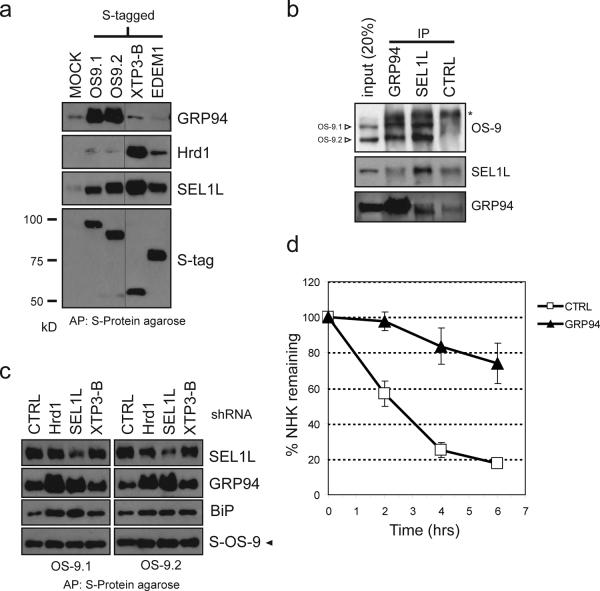
We used immunoblot analysis to confirm the interaction of GRP94 with ER quality control components. S-tagged OS-9, but not EDEM or XTP3-B robustly bound to endogenous GRP94, confirming the interaction detected by mass spectrometry (Fig. 5a). Moreover, endogenous OS-9 (both isoforms) was clearly detected in immunoprecipitates of endogenous GRP94, confirming that the lectin-chaperone interaction is not an artifact of overexpression of S-tagged protein (Fig. 5b). Although both GRP94 and SEL1L both bind to OS-9, neither protein precipitated the other, indicating that they must form distinct complexes with OS-9. The interaction between S-tagged OS-9 and GRP94 was not disrupted by knockdown of SEL1L, Hrd1 or XTP3-B further supporting our conclusion that the interaction between GRP94 does not depend on the other components of the ERAD complex (Fig. 5c). In fact, the amount of BiP and GRP94 captured with OS-9 was substantially elevated when SEL1L or Hrd1 levels were reduced. Together, these data suggest that OS-9 partitions between a Hrd1-SEL1L complex and one containing substrate (Fig. 4a) and GRP94. Finally, knockdown of GRP94 strongly stabilized NHK (Fig. 5d), establishing a requirement for this ER chaperone in degrading at least one well-established ERAD substrate.
Figure 5. XTP3-B and OS-9 interact with ER quality control components.
a. S-tagged versions of OS-9.1 & 2, XTP3-B and EDEM1 were expressed in HEK293 cells, lysed in 1% CHAPS and affinity purified by S-protein agarose. Samples were separated by SDS-PAGE and westerns blots performed with antibodies for Hrd1, SEL1L, GRP94 and S-tag to detect coprecipitating proteins. Hairline indicates where western blot exposures were joined. Full scans are presented in Supplemental Information (Fig. S5). b. Immunprecipitations from 1% TritonX-100 detergent lysate of HEK293 cells with anti-GRP94, anti-SEL1L and a control antibody (anti-HA). Western blots were probed with antibodies for OS-9, GRP94 and SEL1L. c. Coexpression of S-tagged OS- 9.1 and OS-9.2 with indicated shRNA (described in Fig. 4b) in HEK293 cells with affinity purification as in Fig. 5a. Western blots were probed with antibodies against S-tag, GRP94, BiP and SEL1L. d. Composite data of pulse-chase assays for NHK coexpressed with a CTRL (open square) or GRP94 (closed triangle) shRNA. Data are presented as in Fig. 3c. Mean and S.E.M. were determined from 4 individual experiments.
Role of N-glycans in XTP3-B and OS-9 function
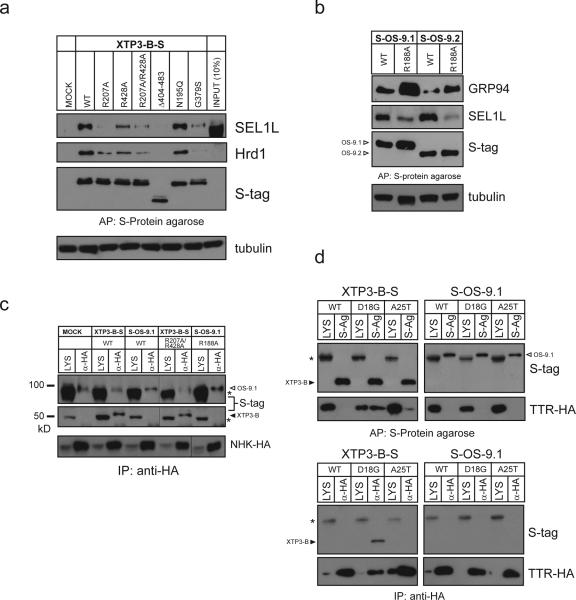
The presence of MRH domains in XTP3-B and OS-9 suggests that binding to N-glycans may contribute to their ability to recognize other components of the lumenal quality control surveillance apparatus or, as has been suggested for Yos9p, substrates17, 19. We found that the interaction of XTP3-B with SEL1L and Hrd1 was strongly impaired by disrupting the putative sugar-binding pockets in both the first (R207A) and second (R428A), as well as a both (R207A/R428A) MRH domains (Fig. 6a). SEL1L binding was completely abolished by deleting the C-terminal 79 amino acids containing one of the two MRH domains, as assessed by immunoblot analysis (Fig. 6a) or mass spectrometry (data not shown). Binding to the ubiquitin ligase complex was decreased by G379S, a mutation previously reported to interfere with the interaction between XTP3-B/Erlectin and Krm222 but not by elimination of XTP3-B's sole N-linked glycan (N195Q). As with XTP3-B, MRH domain mutants (R188A) of either OS-9.1 or OS-9.2 exhibited a reduced amount of coprecipitated SEL1L (Fig. 6b). This reduced binding to SEL1L was accompanied by an increase in captured GRP94, as observed for knockdown of SEL1L or Hrd1 (Fig. 5c). These findings raise the possibility that OS-9's MRH domain is required for binding to SEL1L— a requirement that could be either direct (i.e., OS-9 binds to SEL1L) or indirect (i.e., OS-9 binds to SEL1L via substrate).
Figure 6. Dependence of N-glycan recognition for XTP3-B/OS-9 interaction with ERAD components and substrate.
a. Transient expression of S-tagged wild-type, mutant (R207A, R428A, R207A/R428A, N195Q, G379S) and a C-terminal truncation (Δ404-483) of XTP3-B in HEK293 cells, processed as in Fig. 4d and probed for Hrd1, SEL1L and S-tag by western blot. b. Expression of S-tagged wild type and MRH mutant (R188A) of OS-9.1. Immunoblots were performed with antibodies against S-tag, SEL1L and GRP94. c. NHK-HA coexpression with MRH mutants of XTP3-B (R207A/R428A) and OS-9.1 (R188A). Complexes were brought down with anti-HA and immunoblots probed with anti-S-tag (top) and anti-HA (bottom). Hairline indicates where western blot exposures were joined. Full scans are presented in Supplemental Information (Fig. S5). d. HA-tagged transthyretin (TTR-HA) wild-type (WT), D18G and A25T were coexpressed in HEK293 cells with S-tagged XTP3-B and OS-9.1. Complexes were affinity purified by S-protein agarose (top) or anti-HA (bottom) from cells lysed in 1% TritonX-100. Immunoblots were performed with anti-S-tag and anti-HA (TTR). Non-specific background bands are indicated by (*).
To assess the role of sugar recognition in ERAD substrate binding we evaluated the ability of S-tagged XTP3-B or OS-9 harboring mutant MRH domains to bind NHK (Fig. 6c). The amount of NHK captured by these mutants, however, was similar to that bound to the wild-type proteins, suggesting that, like Yos9p, the MRH domains of XTP3-B and OS-9 are dispensable for substrate binding. To determine whether substrate N-glycans are required, we coexpressed either XTP3-B or OS-9.1 with transthyretin (TTR), a non-glycosylated serum protein normally secreted by the liver. Mutations that destabilize the wild-type TTR homotetramer cause the protein to form amyloid deposits that underlie dominantly inherited familial TTR amyloidoses35. Wild-type TTR was not captured to an appreciable extent by XTP3-B or OS-9.1 (Fig. 6d, top), consistent with the fact that this protein is efficiently folded in transfected cells36. By contrast, D18G and A25T, two amyloidogenic mutants previously shown to be ERAD substrates36, were detected in pull-downs of S-tagged XTP3-B but not S-tagged OS-9.1. This result was confirmed in the reverse experiment in which HA-tagged D18G captured XTP3-B but not OS-9 (Fig. 6d, bottom). The amount of TTR coprecipitated with XTP3-B correlated with the extent to which these TTR variants interact with BiP and inversely with the efficiency with which they are secreted36. Therefore, XTP3-B, but not OS-9, is capable of selectively binding to folding-defective lumenal proteins lacking N-glycans.
DISCUSSION
In this paper we describe two MRH-domain containing ER resident proteins in mammalian cells that bind selectively to misfolded proteins and interact with the membrane-embedded ubiquitin ligase Hrd1 via SEL1L. Although these proteins share features with the Yos9p-Hrd1-Hrd3 system described in yeast, our data reveal several key differences between the yeast and mammalian systems. Chief among these is the unexpected role for GRP94, a protein which, despite its similarity to cytoplasmic Hsp90, has hitherto not been shown to have a direct role in ER quality control or ERAD. Our data implicate GRP94 in the role of regulating the assembly dynamics of protein complexes which hand lumenal substrates off to the ubiquitination and dislocation apparatus.
A model of how the lumenal surveillance mechanism might coordinate with ubiquitination machinery in the mammalian ER is presented in Figure. 7. Terminal mannoses of core N-glycans on proteins engaged in futile cycling in the ER lumen, or perhaps cycling between the ER and the cis Golgi, are progressively trimmed by mannosidases to a configuration compatible with binding to OS-9 or XTP3-B. How substrates partition between these two lectins is presently unclear, but our evidence suggests that the interaction with XTP3-B is unproductive for the substrates examined in the present work. Since mutation of the MRH domains does not affect substrate capture, and since XTP3-B is able to discriminate between native and misfolded non-glycosylated TTR, we conclude that substrate interaction must involve additional, as yet undetermined structural features displayed by misfolded proteins. Our data suggest that instead of mediating substrate interaction, the MRH domains of OS-9 and XTP3-B may contribute to interaction with glycans on other ER-resident components of the ERAD/quality control apparatus (e.g. SEL1L). This conclusion is supported by the observation that the amount of SEL1L and Hrd1 coprecipitated with either lectin was strongly diminished by mutation of the MRH domains.
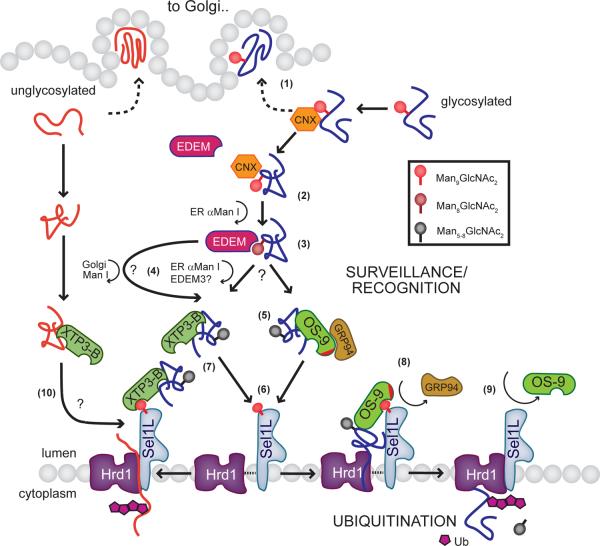
Figure 7. Model for coordinating lumenal surveillance with ubiquitination in mammalian ERAD.
Nascent glycoproteins interact with the ER-resident chaperone calnexin (CNX) where they are either folded correctly and are released for subsequent trafficking to the Golgi apparatus (1) or in the case of misfolded proteins, re-enter the calnexin cycle in an attempt to facilitate their maturation. If folding remains unproductive, the substrate becomes the target for demannosylation by ER α-mannosidase I to a Man8GlcNac2 form (2). The trimmed mannose structure is recognized by EDEM and displaced from the CNX cycle (3). Further demannosylation to a Man5 or Man6 form occurs by the actions of either ER resident mannosidases (α-mannosidase I or possibly EDEM3), or in the case of cycling between the ER and the Golgi apparatus, Golgi Mannosidase I49 (4). By analogy to Yos9p, these lower mannose structures may be part of the signal recognized by XTP3-B and OS-9 (5). OS-9 scaffolds the ER chaperones GRP94 and BiP which may also help to prevent aggregation or facilitate unfolding of the substrate. OS-9/XTP3-B complexes bound to substrate subsequently interact with the lumenal domain of SEL1L (potentially through their MRH domains) which may facilitate the release of substrate in the case of OS-9 (6). XTP3-B forms a stable ternary complex with SEL1L and Hrd1 (7). For OS-9, binding to SEL1L displaces GRP94 and the substrate is transferred to the Hrd1-SEL1L complex (8). The final step is release of OS-9 and dislocation of the substrate by the dislocation apparatus and subsequent ubiquitination in the cytoplasm (9). In an alternative pathway, misfolded, non-glycosylated proteins may also enter this pathway, perhaps through interactions with XTP3-B (10).
One intriguing possibility, suggested by the present work, is that substrate recognition may be facilitated or mediated by molecular chaperones such as GRP94 and BiP, which associate with OS-9. GRP94 is the ER lumenal member of the Hsp90 class of chaperones. Unlike other molecular chaperones, cytoplasmic Hsp90 appears to be required for folding only a limited range of substrates, most notably the steroid hormone receptors and other proteins involved in signal transduction37. Physiological substrates for GRP94 have not been identified, although a role in the assembly of toll-like receptors has been suggested38, 39. Despite being an abundant protein in the cytoplasm, Hsp90 appears to function in different cellular pathways through its ability to interact with distinct sets of co-chaperones which regulate its interactions with substrates40. We speculate that, by analogy to the role of Hsp90 in assembling cytoplasmic receptor complexes, GRP94 may control the transfer of OS-9 (together with bound ligand) to the membrane ubiquitin ligase/dislocation complex. Although we observed a strong GRP94 signal in OS-9 pulldowns, we failed to detect this chaperone coprecipitating with SEL1L, despite the presence of abundant OS-9. Thus, like Hsp90, which dissociates from steroid hormone receptors upon hormone binding, GRP94 binding to OS-9 appears to be mutually exclusive with that of SEL1L. Further study will be needed to address the relative contribution of GRP94 to ERAD substrate recognition versus regulating the assembly and disassembly of the Hrd1-SEL1L-OS-9 ERAD complex.
A notable difference between Yos9p and its mammalian counterparts is the absence of a KDEL or other potential ER retention sequence from the mammalian proteins. Without a canonical retrieval sequence, XTP3-B/OS-9 retention may be conferred by association with known ER resident proteins, as suggested for EDEM1 and EDEM214. Retention of OS-9 in the ER might be accomplished by virtue of its interaction with the ER membrane through SEL1L or indirectly, through binding to GRP94/BiP. The secretion of overexpressed forms of XTP3-B22 and OS-9 (data not shown) suggests that the available pool of retaining molecules is limited.
Hrd1 and SEL1L, like their yeast counterparts, are present in a 1:1 stoichiometric complex in the ER membrane12, 41. Interestingly though, shotgun mass spectrometry analyses of microsomal fractions from a variety of mouse tissues found a significantly greater number of SEL1L scans (5-7 fold) as compared to Hrd142, suggesting that not all SEL1L may be associated with Hrd1. We find that the interaction between Hrd1 and SEL1L is labile and sensitive to detergents (Supplemental Information, Fig. S1). This differs from the interaction between the yeast orthologs Hrd1p-Hrd3p, where the stoichiometric complex is captured quite readily41. While S-tagged Hrd1 coprecipitates both SEL1L and OS-9 (Fig. 4a), S-tagged OS-9 efficiently captures SEL1L but not Hrd1 (Fig. 5a). By contrast, the XTP3-B-Hrd1-SEL1L complex can be coprecipitated with similar stoichiometries irrespective of the capture reagent. This suggests that SEL1L may interact sequentially with OS-9 and Hrd1, perhaps because the Hrd1-SEL1L interaction is destabilized by SEL1L's interaction with OS-9. Moreover, the dramatic enhancement of NHK association with OS-9 following SEL1L knockdown suggests that SEL1L binding to these lectins may facilitate the transfer of substrate to Hrd1. Our data showing that SEL1L's interactions with OS-9 and Hrd1 do not require the SEL1L transmembrane domain is consistent with those observed between Yos9p and Hrd1p with Hrd3p9, 21, 41. Moreover, the Hrd3p truncation41 as well as the original Sel-1 from C.elegans29 are both functional, despite the lack of a transmembrane domain. It is not unreasonable to envision that the Hrd1-SEL1L interaction, mediated through the lumenal domains of both proteins, could be modulated by the binding of lumenal proteins like OS-9.
Although XTP3-B and OS-9 share the ability to bind to a broad range of glycosylated and non-glycosylated proteins, knockdown experiments reveal that neither lectin is essential for this process. While further study is needed to clarify the details of this complex pathway, the data reported here contribute to our understanding of the mechanisms by which misfolded proteins are recognized and sorted for destruction in the mammalian ER.
METHODS
Plasmids and constructs
The following constructs were provided: Hrd1 (Dr. Vincent Chau), SEL1L (Dr. Ida Biunno), V5-His6-OS-9 (Dr. Gregg Semenza) and transthyretin (TTR: wild-type, D18G, A25T; Dr. Jeffery Kelly). XTP3-B, OS-9.2 and EDEM1 were cloned from full length ESTs (Open Biosystems). XTP3-B and EDEM1 were amended with a C-terminal S-tag (KETAAAKFERQHMDS) and cloned into pcDNA3.1(-). The N-terminal S-tag for SEL1L and OS-9.1 & 2 was constructed from the signal sequence from bovine preprolactin followed by the S-tag and in frame fusion without endogenous signal sequences. All point mutations were made by Quikchange mutagenesis (Stratagene). The degradation substrates α1-anti-trypsin NHK variant, NHKQQQ, TTR, TCRα, and RI332 were HA tagged at the C-terminus and subcloned into pcDNA3.1(-).
Short-hairpin RNAs (shRNAs)
Target sequences for shRNA mediated knockdown (Supplemental Information, Table S1) were identified from the literature or generated using the siRNA Selection Program at the Whitehead Institute43, online repository (RNAi Codex, http://katahdin.cshl.org:9331/rnai/repository/scripts/newmain.pl) or a web-based design tool (Ambion, http://www.ambion.com/techlib/misc/psilencer_converter.html), with each based on the rules of Tuschl44. All shRNAs were cloned into the pSUPERSTAR expression vector45.
Cell culture and transfections
HEK293s/HeLa S3 cells were maintained in DMEM + 10% fetal bovine serum (FBS). Cells were transfected by either Fugene-6 (Roche) or the calcium-phosphate coprecipitation technique. Stable, clonal cell lines were selected by resistance to neomycin (1mg/ml) and limiting dilution.
Immuno-/affinity- purification and immunoblotting
All cells were harvested manually in PBS, and lysed in buffer46 containing either 1% Triton X-100 or 1% CHAPS and supplemented with protease inhibitor cocktail (Roche). Lysates were spun twice, first at 1000 × g and the supernatant respun at 20,000 × g. After preclearing, S-tagged proteins were affinity purified from cell lysates by S-protein agarose (Novagen). Samples were washed thrice in lysis buffer without detergent, resuspended in loading buffer + 5mM DTT and treated with EndoH (NEB) where specified. Samples were run on uniform or gradient SDS-PAGE gels and transferred to PVDF membrane for immunoblotting.
Antibodies
The following antibodies were used for experiments: anti-KDEL (Assay Designs), anti-Hrd1 (Abgent, kind gift of E. Wiertz), anti-SEL1L (Axxora, kind gift of H. Ploegh), anti-OS-9 (Abcam, Proteintech), anti-GRP94 (kind gift of C. Nicchitta, Assay Designs), anti-VCP/p97 (Novus Biologicals), anti-HA (12CA5 and HA.11), anti-α-tubulin (Abcam), anti-Hsp90 (BD Biosciences), anti-F1-ATPase (Santa Cruz Biotechnologies), anti-Derlin-1 and anti-Derlin-2 (kind gift of Y. Ye). Rabbit polyclonal antibodies against a peptide (a.a. 357-376) of human XTP3-B were generated and affinity purified.
Metabolic labeling and pulse-chase assays
Confluent plates of transfected HEK293 cells were incubated in methionine/cysteine (Met/Cys)-free DME for 10 min. Cells were metabolically pulse-labeled by supplementing met/cys-free DME with 35S-Met/Cys (NEN EXPRE 35S35S: 50 μCi/6 cm. dish) for 15 min. Rinsing cells twice with ice-cold phosphate buffered saline (PBS) supplemented with 5mM Met/Cys terminated labeling. Labeled cells were gently harvested, pelleted at 1,000 r.p.m. for 10 sec. and lysed according to conditions above. Incorporation of 35S-Met/Cys was normalized using previously described methods47. HA-tagged proteins were immunopurified by anti-HA 12CA5 coupled Protein A Sepharose and separated by SDS-PAGE. Dried gels were exposed and quantified by phosphoimaging (Molecular Devices).
Mass spectrometry
Rough microsomes were prepared from HEK293s stably expressing S-tagged proteins according to the protocol of Stephens et al.48. Starting whole cell and rough microsome fractions were then lysed in 1% TritonX-100 and affinity purified by S-protein agarose. Bead-bound complexes were washed thrice in lysis buffer46, followed by 5 times in 50 mM ammonium bicarbonate, pH 8. Bound proteins were eluted by overnight treatment with Rapigest (Waters Inc.), followed by trypsin digestion (Promega) prior to injection into the mass spectrometer. Samples were analyzed on a system consisting of a CTC-PAL autosampler (Leap Technologies), a capillary gradient HPLC pump (Agilent Model 1100) and a linear-ion-trap mass spectrometer (Model LTQ, ThermoFisher Scientific). Solvents used for the chromatography were 0.1% aqueous formic acid (Solvent A) and 0.1% formic acid in acetonitrile (Solvent B). The chromatographic separations were performed on a 0.32 mm × 15 cm C18 reversed-phase column (MicroTech Scientific) at 8 microliters/min flow rate with a gradient of 100% Solvent A to 40% Solvent B over 104 minutes. Column eluant was electrosprayed directly into the source of the mass spectrometer. The mass spectrometer was operated in a data-dependent MS/MS mode in which the instrument cycled between full MS scans (m/z 300-2000) and intervening MS/MS scans on the ten most intense ions occurring in the MS scan. The acquired MS/MS spectra were searched using the Mascot protein database search program (Matrix Science) against a full database of human protein sequences to which the set of S-tagged proteins sequences and S-protein sequence were added.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Weissman, I. Biunno, V. Chau, C. Nicchitta, H. Ploegh, G. Semenza, J. Kelly and E. Wiertz for reagents and technical advice, members of the Kopito lab for helpful discussion, and S. Duttler and J. Olzmann for critical reading of the manuscript. This work was supported by grants from NIDDK, NIGMS, and the Cystic Fibrosis Foundation. JCC was supported by a postdoctoral fellowship from Cystic Fibrosis Research, Inc.
REFERENCES
- 1.Hammond C, Helenius A. Quality control in the secretory pathway. Current Opinion in Cell Biology. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 2.Lippincott-Schwartz J, Bonifacino JS, Yuan LC, Klausner RD. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- 3.McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 6.Shamu CE, Flierman D, Ploegh HL, Rapoport TA, Chau V. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol Biol Cell. 2001;12:2546–2555. doi: 10.1091/mbc.12.8.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarosch E, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nature Cell Biology. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 9.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 10.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 11.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 12.Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulze A, et al. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299(5611):1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 16.Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299(5611):1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 17.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 18.Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Buschhorn BA, Kostova Z, Medicherla B, Wolf DH. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 2004;577:422–426. doi: 10.1016/j.febslet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 21.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 22.Cruciat CM, Hassler C, Niehrs C. The MRH protein Erlectin is a member of the endoplasmic reticulum synexpression group and functions in N-glycan recognition. J Biol Chem. 2006;281:12986–12993. doi: 10.1074/jbc.M511872200. [DOI] [PubMed] [Google Scholar]
- 23.Su YA, Hutter CM, Trent JM, Meltzer PS. Complete sequence analysis of a gene (OS-9) ubiquitously expressed in human tissues and amplified in sarcomas. Mol Carcinog. 1996;15:270–275. doi: 10.1002/(SICI)1098-2744(199604)15:4<270::AID-MC4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 24.Kimura Y, Nakazawa M, Yamada M. Cloning and characterization of three isoforms of OS-9 cDNA and expression of the OS-9 gene in various human tumor cell lines. J Biochem (Tokyo) 1998;123:876–882. doi: 10.1093/oxfordjournals.jbchem.a022019. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama T, Yaoi T, Kuwajima G, Yoshie O, Sakata T. Ca2(+)- dependent interaction of N-copine, a member of the two C2 domain protein family, with OS-9, the product of a gene frequently amplified in osteosarcoma. FEBS Lett. 1999;453:77–80. doi: 10.1016/s0014-5793(99)00700-0. [DOI] [PubMed] [Google Scholar]
- 26.Litovchick L, Friedmann E, Shaltiel S. A selective interaction between OS-9 and the carboxyl-terminal tail of meprin beta. J Biol Chem. 2002;277:34413–34423. doi: 10.1074/jbc.M203986200. [DOI] [PubMed] [Google Scholar]
- 27.Baek JH, et al. OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1alpha. Mol Cell. 2005;17:503–512. doi: 10.1016/j.molcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant B, Greenwald I. The Caenorhabditis elegans sel-1 gene, a negative regulator of lin-12 and glp-1, encodes a predicted extracellular protein. Genetics. 1996;143:237–247. doi: 10.1093/genetics/143.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirao K, et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein ERAD and mannose trimming. J Biol Chem. 2006 doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- 31.Oda Y, et al. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Virgilio M, Weninger H, Ivessa NE. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- 33.Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 34.Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 35.Johnson SM, et al. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 36.Sekijima Y, et al. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, et al. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 40.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 41.Gardner RG, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. The Journal of cell biology. 2000;151(1):69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kislinger T, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 43.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 45.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 46.Christianson JC, Green WN. Regulation of nicotinic receptor expression by the ubiquitin-proteasome system. EMBO J. 2004;23:4156–4165. doi: 10.1038/sj.emboj.7600436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269(41):25710–25718. [PubMed] [Google Scholar]
- 48.Stephens S, Dodd R, Lerner R, Pyhtila B, Nicchitta C. Analysis of mRNA Partitioning Between the Cytosol and Endoplasmic Reticulum Compartments of Mammalian Cells. In: Wilusz J, editor. Methods in Molecular Biology. Vol. 419. Humana Press; Totowa, NJ: (in press)) [DOI] [PubMed] [Google Scholar]
- 49.Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A. Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem Biophys Res Commun. 2007;362:626–632. doi: 10.1016/j.bbrc.2007.08.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.