Abstract
We have developed a strategy for preparing tethered lipid bilayer membrane patches on solid surfaces by DNA hybridization. In this way, the tethered membrane patch is held at a controllable distance from the surface by varying the length of the DNA used. Two basic strategies are described. In the first, single-stranded DNA strands are immobilized by click chemistry to a silica surface, whose remaining surface is passivated to prevent direct assembly of a solid supported bilayer. Then giant unilamellar vesicles (GUVs) displaying the antisense strand, using a DNA-lipid conjugate developed in earlier work (Chan, Lengerich et al. 2008), are allowed to tether, spread and rupture to form tethered bilayer patches. In the second, a supported lipid bilayer displaying DNA using the DNA-lipid conjugate is first assembled on the surface. Then GUVs displaying the antisense strand are allowed to tether, spread and rupture to form tethered bilayer patches. The essential difference between these methods is that the tethering hybrid DNA is immobile in the first, while it is mobile in the second. Both strategies are successful; however, with mobile DNA hybrids as tethers, the patches are unstable, while in the first strategy stable patches can be formed. In the case of mobile tethers, if different length DNA hybrids are present, lateral segregation by length occurs and can be visualized by fluorescence interference contrast microscopy making this an interesting model for interactions that occur in cell junctions. In both cases, lipid mobility is high and there is a negligible immobile fraction. Thus, these architectures offer a flexible platform for the assembly of lipid bilayers at a well-defined distance from a solid support.
Keywords: Tethered lipid membrane, surface-immobilized DNA, Giant unilamellar vesicles
Supported lipid bilayers (SLBs) have been widely used as a model for cell membranes (Sackmann 1996; Chan and Boxer 2007) and to investigate membrane components including proteins in a simpler context apart from the complex cellular environment. SLBs are assembled by Langmuir-Blodgett techniques or spontaneous fusion of unilamellar vesicles on carefully prepared surfaces, usually hydrophilic solid supports, such as glass (Seu, Pandey et al. 2007), silica, mica (Richter, Berat et al. 2006), or TiO2 (Rossetti, Bally et al. 2005). Although SLBs have the advantages of simple formation, easy handling and are well-suited for investigation by a suite of surface sensitive methods due to their planar geometry, the close proximity of the lower leaflet to the solid support often leads to unfavorable interactions with integral membrane proteins. Recognizing this limitation, many groups have described methods to separate the membrane from the solid support including polymer cushioned bilayers (Wagner and Tamm 2000; Merzlyakov, Li et al. 2006; Renner, Osaki et al. 2008), polymer tethered membranes (Naumann, Prucker et al. 2002; Purrucker, Fortig et al. 2004; Koper 2007) and tethered vesicles (Yoshina-Ishii and Boxer 2003; Stadler, Falconnet et al. 2004; Yoshina-Ishii, Miller et al. 2005).
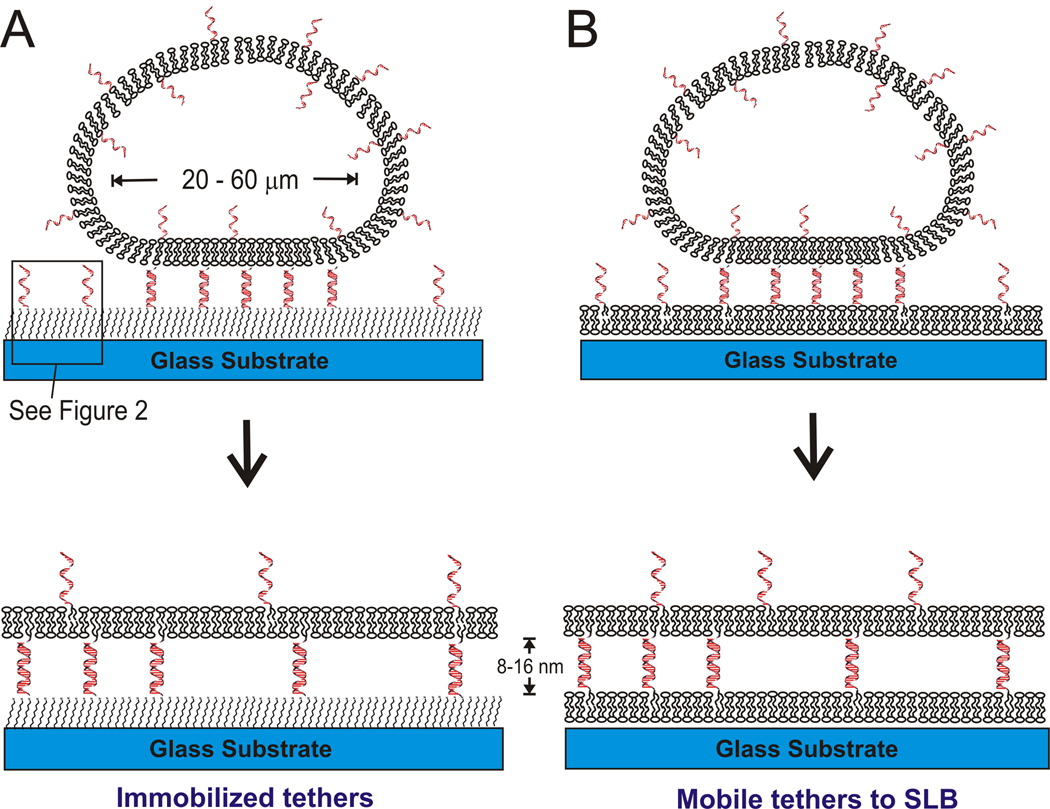
In this paper, we describe two approaches for tethering lipid membranes using DNA linkages to separate the membrane from surface interactions. In one strategy (Figure 1A and Figure 2), DNA tethers are coupled directly to the substrate by using click chemistry (Kolb, Finn et al. 2001) to graft alkyne-functionalized DNA onto a glass or silica substrate which is silanized by azide-terminated alkyl silane. Giant unilamellar vesicles (GUVs, diameter of 10 – 60µm) displaying complementary DNA-functionalized lipids bind to the DNA functionalized surface via hybridization, then rupture and spread to form planar tethered bilayer patches. In an alternative strategy (Figure 1B), DNA-conjugated lipids are incorporated into an SLB, which allows the DNA tethers to remain mobile. Then GUVs displaying the antisense DNA are bound by hybridization and ruptured to form bilayer patches. In both approaches, the length of the DNA and hence distance of the tethered bilayer from the surface can be controlled at will and, as far as we know, this is a unique feature. The first system produces stable patches, while in the second interesting dynamic behavior is observed, in part because the tethers are mobile.
Figure 1.
schematic diagram of DNA-tethered membrane formation by GUV rupture onto (A) self-assembled alkylsilane monolayers, see Figure 2 for details; and (B) supported lipid bilayers. The GUV membrane contains DNA-linked lipids, which mediate vesicle binding and rupture. In (A), the glass substrate is functionalized with an azide-terminated alkyl-siloxane monolayer to which complementary DNAs are immobilized via the click reaction (see Figure 2). The distance from the substrate is controllable by adjusting the DNA length, about 8 nm for 24mer and 16 nm for 48mer DNA. In (B), a supported lipid bilayer presenting DNA is first formed on the glass substrate, and then exposed to GUVs presenting the complementary DNA to form a DNA-tethered membrane. In both cases, tethered membrane patches whose area is approximately that of the original GUVs are formed.
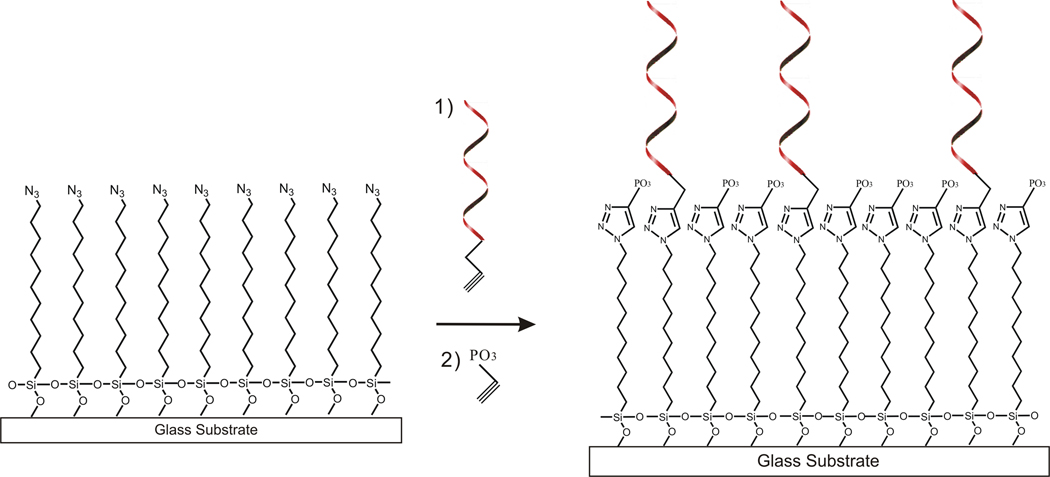
Figure 2.
The general synthetic scheme to make the immobilized DNA surface (c.f. Fig. 1A). 5' alkyne modified DNA (Alkyne-C6-5' DNA) is clicked to the azide-modified glass surface in step 1 at approximately 1% coverage, and the remaining free azides are then clicked with ethynyl phosphonic acid in step 2.
Materials and Methods
Formation of supported bilayers
Small unilamellar vesicles (SUV) were prepared by vesicle extrusion through 100nm polycarbonate membranes (Whatman) as described previously (Yoshina-Ishii and Boxer 2009). They were composed of egg phosphatidylcholine (EggPC, Avanti Polar Lipids, Alabaster, AL), 1 mol% Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-DHPE, Molecular Probes, Eugene, OR) or 2 mol% of Oregon Green® 488 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (OG-DHPE, Molecular Probes), and DNA-lipids conjugates (Chan, Lengerich et al. 2008) anchored at the 5' end and with either 24- or 48-mer poly T as the sequence. DNA-lipid conjugates were dissolved in a 1:1 mixture of acetonitrile and water. An aliquot of this solution was mixed with 40 µl of 10mM phosphate buffered saline (PBS) without salt to achieve the desired average DNA mol%. Then, 10µl of extruded SUV suspension (10mg/ml) was added and incubated overnight at 4°C. In the case of two different lengths of DNA tether, an equimolar mixture of 5'-lipid-24 mer poly T and 5'-lipid-TAACTACAGAATTTATACTATCCCGGGTCACAGCAGAGAAACAAGATA −3' (a 48 mer) was used.
Supported lipid bilayers were formed by vesicle fusion by exposing a glass cover slip to the above SUVs that were diluted to 0.2 mg/ml in 10 mM PBS, pH 7.2, containing 100 mM sodium chloride, for 10 min. The glass cover slips (VWR) were cleaned by soaking in 90°C 7x ICN detergent (diluted 1:6 in double distilled water, ICN Pharmaceuticals, Costa Mesa, CA), rinsed extensively in double distilled water, and baked at 400 °C for 4 h. The rupture of GUVs rarely occurred on the SLBs containing less than 0.1 mol% DNA-lipid conjugates, while most GUVs ruptured above 0.5 mol% DNA-lipid conjugates. It is hard to make SLBs containing more than 0.5 mol% DNA-lipid, because SLB formation is interfered probably due to high negative charge of DNAs. These membranes were observed with a Nikon TE300 inverted epifluorescence microscope using a 40X oil immersion objective (N.A. = 1.2).
Protein micro-patterned lipid bilayer surfaces were prepared by the method described previously (Kung, Kam et al. 2000). 100 µm square bovine serum albumin (BSA) barriers were transferred from polydimethylsiloxane (PDMS, Dow Corning, Midland, MI) elastomer stamps containing a microscale pattern. The PDMS stamps were formed by curing on silicon wafers patterned by standard photolithographic techniques. Then an SUV suspension was added to form SLBs separated by BSA square grids.
Preparation and characterization of surfaces displaying immobilized DNA
11-azidoundecyl-1-trimethoxysilane was prepared from 11-bromoundecyl-1-trimethoxysilane (Gelest) according to a published procedure (Fu and Yu 2001). Azidosilane monolayers were vapor-deposited in a glass vacuum desiccator (Jencons part no. 08–625A) fitted with a Teflon stopcock. 100 µL of neat azidosilane was injected onto filter paper in the bottom of the desiccator, and 0.5g of MgSO4·7H2O was placed in a foil boat in the bottom of the desiccator as a water source for the silanization reaction. The detergent cleaned glass slides were oxygen plasma cleaned for 5 min. prior to deposition and placed in a slide rack supported above the azidosilane liquid. The desiccator was then evacuated on a Schlenk line to ∼300 mTorr with a liquid nitrogen-trapped mechanical pump. The Teflon valve on the desiccator was then closed, and the chamber was placed in a 110°C preheated oven for 48h. After deposition, the surfaces were rinsed with toluene and isopropanol, and then dried in a stream of nitrogen.
Azide-modified glass surfaces were exposed for 2h (Figure 2) to aqueous solutions containing: 100 µM CuSO4, 100 µM tris(triazolylmethyl)amine (TTMA) Cu ligand, 1 mM sodium ascorbate, and 5 µM alkynyl oligonucleotides. These are 5' alkynyl modified polyT oligonucleotides with a six carbon linker (Alkynyl-C6-polyT), either 24 or 48 mer matching in length. Following this, the surfaces were rinsed with water and exposed to a second click solution containing: 100 µM CuSO4, 100µM TTMA Culigand, 1 mM sodium ascorbate, and 1 mM ethynyl phosphonic acid for 1h, to passivate unreacted azide. The clicked surfaces were then rinsed with 3X saline–sodium citrate (SSC) buffered solution, 150 mM HEPES/0.1%SDS/1XSSC, and finally 150 mM HEPES/150mM NaCl in water to remove nonspecifically adsorbed oligonucleotides.
The thickness change accompanying the formation of azidosilane-monolayer was monitored by single-wavelength ellipsometry using Si wafers with ∼ 260 nm thick SiO2 films instead of glass slides. The Si chips (0.5 × 0.5 cm) were detergent cleaned, baked and silanized as described above, and the exact thickness of the SiO2 layer and azidosilane-monolayer from the Si mirror was measured before and after silanization. The auto-nulling-four-zone analysis of the ellipsometer (Nanofilm Technologie, Goettingen, Germany) was used to generate delta and psi values, with 532 nm light and 54 ° incidence angle. The results were fit to a model of Si/SiO2/monolayer with parameters of nSi = 4.1562, κSi = 0.0419, nSiO2 = 1.4605 and nmonolayer = 1.4578 (Ali, Bessueille et al. 2008). The root-mean-square (RMS) surface roughness change was measured by atomic force microscopy (AFM), performed in contact mode at 1 Hz scan rate in air using a Nanoscope III (Digital Instruments Inc., Santa Barbara, CA), using silicon cantilevers. RMS roughness, defined as the average height (z) taken from the mean data plane, was calculated for a 1 × 1 µm area using the instrument software.
The density of DNA molecules on the substrate is roughly estimated by the fluorescence intensity from complementary Cy5 labeled DNA. DNA immobilized substrates were prepared as described above using solutions with several different alkynyl-DNA (24mer polyT) concentrations (0, 1, 1.5, 2.5, and 5 nmol/mL), followed by passivating with ethynyl phosphonic acid and rinsing steps. The dependence on incubation time was also tested, but because there was no significant difference after 2hr, every reaction was incubated for 2hr. 5µM of 5'-Cy5-modified 24 mer polyA oligonucleotides in buffered solution (pH 7.2) containing 0.1M sodium phosphate and 0.1M NaCl were introduced onto the substrates and incubated for 1hr. Nonspecifically adsorbed oligonucleotides were removed using the same rinsing procedure as for DNA immobilization. After Cy5 labeled complementary DNA was bound, the fluorescence intensity was determined immediately with a Nikon TE300 (Nikon USA, New York USA) inverted microscope using a 100X oil immersion objective illuminating with a high-pressure mercury lamp and an excitation filter transmitting 590–650 nm and emission filter of 663–738nm. The fluorescence signal was detected with a cooled 16-bit CCD camera (Roper-scientific, Tucson, AZ), and the data were processed with Metamorph imaging software. To correlate the average fluorescence intensity with a surface density, a calibration measurement was performed. For this purpose, solid-supported membranes of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC;Avanti Polar Lipid) with 0, 0.26, 0.78, and 1.3 pmoles/cm2 (equivalent to an SLB containing 0.5 mol% DNA) of 5' poly T DNA-lipid conjugates were prepared and measured in the exactly same way as described for the immobilized DNA surfaces. A comparison of fluorescence intensities per identical area provides the number of active DNAs on the substrate. It is expected that some inaccessible DNA is present on the SLB substrate. Although DNA-lipid conjugate is added to the outer leaflet of the SUV, during formation, both the inner and outer surface of the SUV may end up facing the glass support. While DNA may be repelled from the surface and largely end up on the upper leaflet, where it can bind the fluorescently labeled antisense strand, the precise distribution is not known, and so the amount of DNA-lipid conjugate added to the SUVs represents an upper limit to the amount available, and amounts are compared with this limit.
Giant unilamellar vesicle preparation and tethered membrane formation
GUVs ranging in size from 10–60 µm were prepared as described previously (Longo and Ly 2007) with minor modifications. Phospholipid compositions in GUVs are egg PC with 1 mol% TR-DHPE for visualization and 1 mol% of DNA-lipid conjugates with either 24- or 48-mer poly A as the sequence. In the case of two different lengths of DNA tethers, equimolar mixture of 5'-lipid-24-mer poly A and 5'-lipid-TATCTTGTTTCTCTGCTGTGACCCGGGATAGTATAAATTCTGTAGTTA −3' (a 48 mer) was used. When the DNA-lipid conjugates were added to the lipid mixture before electroformation, 1µl of 0.3 mM DNA-lipid conjugate aqueous solution was mixed with 50µl of 0.5 mg/ml lipid mixture dissolved in 2:1 choloform:methanol, then this was coated on Pt electrodes. When DNA-lipid conjugates were added after the GUV preparation, a desired amount of the DNA-lipid conjugate stock solution was mixed with the GUV suspension in 0.5 M sucrose solution to achieve approximately 1 mol% DNA-lipid and incubated overnight at 4°C, as described above for SUVs. The GUVs were stored at 4°C and used within 2 days.
DNA-tethered lipid membrane patch formation from the GUVs was slightly different for the two approaches. The DNA-immobilized glass cover slips (Figure 1A) were covered by a CoverWell perfusion chamber gasket (9 mm diameter, 0.5 mm thickness, Molecular Probes), and filled with 40 µl of 10 mM PBS containing 240 mM sodium chloride for osmotic balance with the 0.5 M sucrose solution inside the GUVs. 5µl of GUV suspension (0.2µg/ml) in 0.5 M sucrose was added through the hole of the gasket. The GUVs settled down and made contact to the substrate surface due to the dense sucrose solution inside. As DNA hybridization progressed, spontaneous rupture of GUVs to form tethered membrane patches was monitored by epifluorescence and confocal microscopy. After about 20 min, excess vesicles were removed by rinsing with PBS buffer. Because of the flow stress of the washing buffer, some flattened GUVs (see below) were ruptured. The rupture of GUVs can also be induced by osmotic shock by exchanging the buffer to 10mM PBS with 50mM NaCl, though this did not work for all flattened GUVs. In the case of the mobile DNA substrate (Figure 1B), the same amount of GUV suspension was pipetted directly above the SLB which was kept in 10 mM PBS buffer with 240 mM NaCl under the well of the gasket. All SLBs under the tethered patches contain 2 mol% of OG dye, not seen in the figures because TR filters were used. The tethering process was similar, though the stability of the resulting patches was very different as discussed below.
Fluorescence interference contrast (FLIC) microscopy
Variable incidence angle FLIC (VIA-FLIC) data provide information on the distance between the solid support and the tethered bilayer patch (Ajo-Franklin, Ganesan et al. 2005). Three systems were examined: (i) patches tethered using polyA / T 24-mer DNA oligonucleotides to supported bilayers assembled on SiO2 films ∼ 230, ∼ 260 and ∼ 330 nm thick on Si wafers; (ii) bilayer patches tethered with a mixture of non-repeating 48-mer and polyA / T 24-mer DNA oligonucleotides to supported bilayers on SiO2 films of thickness ∼ 260 nm on Si wafers; and (iii) tethered bilayer patches hybridized with polyA / T 48-mer DNA covalently attached to the solid support formed on SiO2 films of thickness ∼ 260 nm and ∼ 330 nm on Si wafers. VIA-FLIC data were fit in Matlab, largely using the previously described model (Ajo-Franklin, Ganesan et al. 2005). Briefly, the hydrophobic region of lipid bilayers was defined to be 3.78 nm thick, and Texas Red was modeled as equally distributed between two layers, each 0.1 Å from the top and bottom bilayer/water interfaces, respectively. The absorption transition dipole orientation was allowed to vary initially before being fixed at 60° relative to bilayer normal. This value is quite different from the expected value near 90° (Ajo-Franklin, Kam et al. 2001). This difference may be because the DNA-tethered lipid membranes are more prone to fluctuations than a bilayer directly on a solid support, although deviations from expected values have been reported for other dyes in supported bilayers (Sund, Swanson et al. 1999). Transmission data for the microscope filters and absorption and emission spectra for the dyes were provided by the manufacturers and used to characterize the wavelength dependence of excitation and emission in the model. Thicknesses of the SiO2 spacer layer separating the lipids from the Si mirror were determined by ellipsometry. In the model, two free parameters were used for the fit – a distance parameter and a scaling factor. For tethered membranes hybridized with DNA covalently linked to the solid support, the water layer thickness d0 beneath the bilayer was the variable distance parameter. For tethered membranes attached to supported bilayers, d0 was set to 1.4 nm, and the 3.78 nm thickness of the lower (supported) bilayer was also included in the model. The Fresnel reflection and transmission coefficients at the interfaces of these two layers were included by matrix algebra in the 'bulk' substrate beneath the layer of interest, extending the approach of Lambacher and Fromherz (Lambacher and Fromherz 2002). The variable distance parameter in this case was defined as dsep, the thickness of the water layer between the lower (supported) bilayer and the tethered membrane patch.
Confocal microscopy and fluorescence recovery after photobleaching (FRAP) analysis
Confocal imaging was performed using a Leica TCS SP2 AOBS laser scanning confocal microscope. For imaging GUVs and tethered membrane patches, the specimens were formed on coverslips as described above. TR was excited with a 594 nm HeNe laser line set at 20–30% of maximum laser output (4 mW) and imaged with a 600 to 640 nm band-pass filter using a 100X Plan-Apochromat oil immersion objective (NA1.4) at a definition of 1024 × 1024 pixels with the pinhole set at 1 airy disk. A series of optical sections with a z-spacing of 122 nm were taken and collected using the equipped Leica confocal software. 3D reconstruction of Z-stack images were processed by Volocity (Improvision, Lexington, MA) visualization software.
An automated FRAP module in the Leica confocal software was used for diffusion coefficient measurement by spot photobleaching. DNA-tethered membrane patches, SLBs formed from GUVs, and SLBs formed by small vesicle fusion were measured within the same hour to minimize atmospheric temperature differences. The same GUVs were used to form both tethered patches and SLBs. Three samples were examined for each lipid bilayer and at least 5 spots on each sample were measured. Each FRAP experiment started with 6 pre-bleaching image scans using a 100X oil immersion objective (NA 1.4) with excitation from a 594 nm HeNe laser line set at 25% of maximum laser output (4 mW) and detection with a 600 to 640 nm band-pass filter at a definition of 256 × 256 pixels with the pinhole set at 1 airy disk, followed by bleaching of a 3 ∼ 5 µm circular spot in the center of a round 40 ∼ 60 µm diameter tethered membrane patch for 2.08 seconds, using a 488 nm line of high-powered argon laser (100% of 200mW full output). Post-bleaching image acquisition was immediately started to monitor fluorescence recovery, at the same setting as pre-bleaching. Time intervals of each image were adjusted from 208 ms to 300 ms to collect enough data points while avoiding further photo-bleaching during recovery.
Diffusion coefficients were estimated following the approach of Jonsson (Jonsson, Jonsson et al. 2008) with simplifications, a modification of the traditional integral method of Axelrod (Axelrod, Koppel et al. 1976). After the background intensity, defined as the average intensity of the regions that is devoid of lipid membrane was subtracted, the average fluorescence intensity, B(t), inside a circle of radius R centered on the bleached region and average intensity U(t) of TR labeled lipid membrane regions far apart from bleached spot were obtained. Then, FRAP recovery curves were normalized as the intensity ratio of bleached region/unbleached region (Phair and Misteli 2000), to correct the loss of fluorescence caused by imaging.
Circular averaging of the images around the center of the bleached spot was performed to reduce the noise of the intensity profile. Fluorescence intensities of each pixel, I(x, y, t), the same distance r from the center was averaged to yield I(r, t). Under the assumption that bleaching profiles generated by the scanning beam can be described by a Gaussian function (Braga, Desterro et al. 2004; Jonsson, Jonsson et al. 2008), because of diffusion during bleaching, the initial intensity profile after bleaching, I(r, 0) can be fitted to:
where I0 is the intensity before bleaching, and K and w are determined by fitting (Fig. 7B). The solution of Fick’s second law for this initial expression can be shown to be:
where R is the radius of a circle centered on the bleached spot, β is the immobile fraction, and the diffusion coefficient D is calculated from τ = w2/4D. Parameters τ and β are obtained from the fit of this equation to the normalized FRAP recovery curves (Fig. 7C). All fittings procedures were performed by using nonlinear regression in MATLAB.
Figure 7.
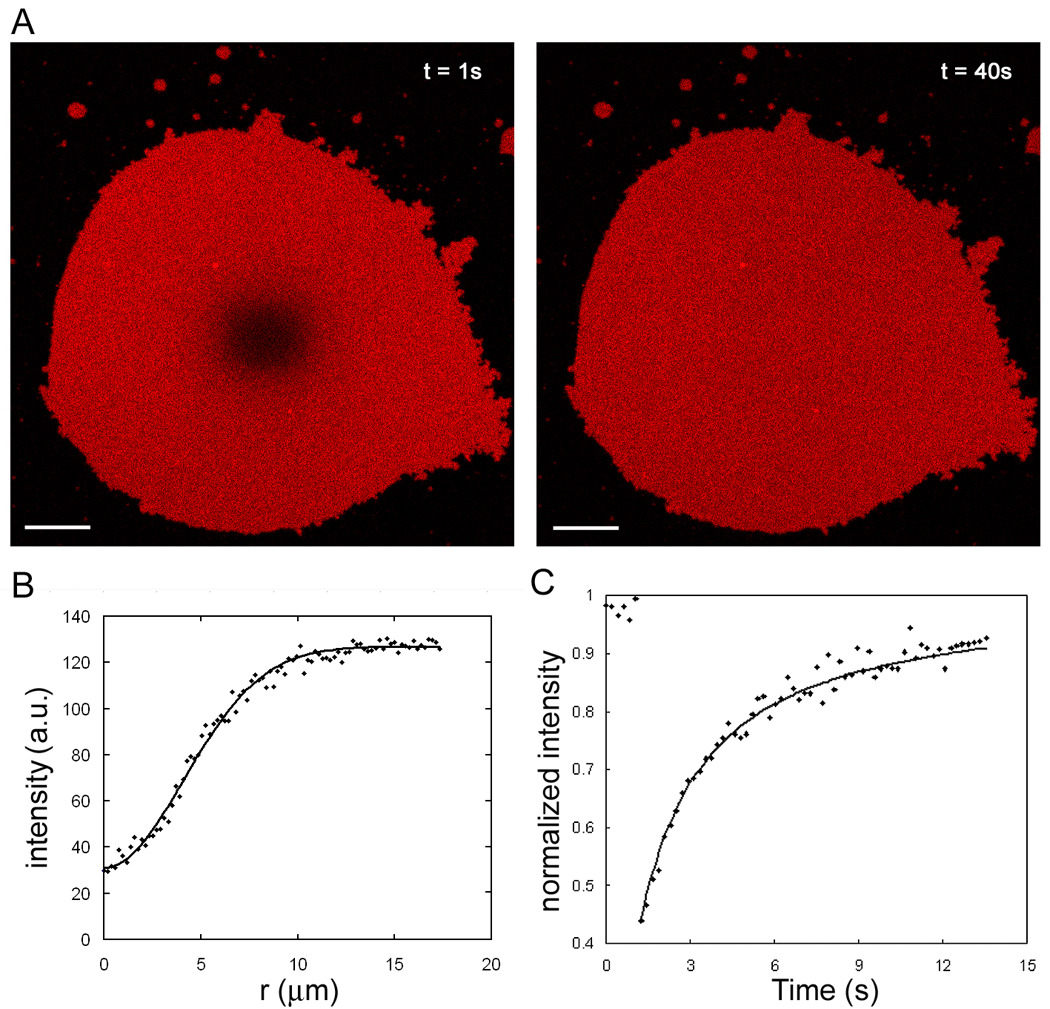
FRAP analysis of lipid mobility in tethered membrane patches. (A) The images show a representative example of recovery after photobleaching of 4 µm bleached spot (left) in a fluorescently labeled DNA-tethered membrane patch. This is on the immobilized DNA tethers as Figure 1A. Scale bar is 5 µm. (B) Circular averaged radial profile (dots) from the center of bleached spot is generated from the initial intensity profile after bleaching, I(r, t = 0), to reduce noise. K and w values (see Materials and Methods) from a fit to a Gaussian function (line) are used for plotting graph C. (C) Representative normalized fluorescence intensity recovery curve (dots) of bleached spot is plotted. This is fitted to obtain diffusion coefficient and mobile fraction (line). The first 6 data points are intensities before bleaching.
In most previous reports describing lateral diffusion coefficient comparisons, SLBs were made by Langmuir-Blodget transfer or small vesicle fusion methods. However, tethered lipid bilayers are small roughly circular patches less than 100µm in diameter; likewise SLBs made by GUVs rupture on glass substrates form circular supported lipid bilayer patches of the same dimensions as tethered membrane patches. Therefore, the results of SLBs formed by GUV rupture as well as continuous SLBs were tested and compared.
Results and Discussion
Fabrication and Characterization of the DNA grafted surface
We have designed two kinds of DNA-presenting surfaces onto which lipid membranes can be tethered. One surface has covalently bound DNA tethers (Figure 1A), and the other is a DNA-presenting supported lipid membrane (Figure 1B). In the latter case, the glass substrate is covered by a lipid membrane, which prevents tethered vesicles, GUVs or the membrane patches reported here from interacting with the glass surface. Furthermore, the DNA-lipid conjugates are free to diffuse laterally in both the supported lipid membrane and the tethered patch, and thus the hybridized tethers should also diffuse; this is a major difference between the architectures in Figures 1 A and B.
For the covalently linked DNA system, phosphonic acid termination prevents the exposed hydrophobic azide monolayer from interacting nonspecifically with the lipid membrane, maintaining the overall stability of the tethered membrane. Thickness changes of the surfaces during preparation of the DNA bound substrate were analyzed by ellipsometry and AFM was used to analyze the topography. These thickness measurements were done on flat Si wafers with SiO2 films of thickness ∼ 260 nm. The average thickness change measured by ellipsometry after azidosilane-monolayer assembly is 15–20Å, consistent with the expected chemical structure. RMS surface. roughness measured by AFM of the Si/SiO2 wafer (< 2 Å) increased to ∼ 1nm after the DNA immobilization. This slight roughness increase is normal for silanized surfaces (Popat, Sharma et al. 2003), and possibly reflects imperfect coating of the alkyl silane. Although this small roughness does not hinder the formation and stability of the tethered membrane and is small compared with the tether length (Naumann, Schiller et al. 2003), it may be an important variable to consider in some high sensitivity applications.
The density of DNA coupled to the azidosilane-covered surface was estimated by fluorescence as described in the Materials and Methods. Cy5-labeled complementary DNA was hybridized to DNA on the surface, and the fluorescence intensity was converted to a number density per area by comparing the intensity with that of SLBs presenting a known mol fraction of DNA (Ajo-Franklin, Yoshina-Ishii et al. 2005). Because the fluorescence is from a small fraction of a monolayer, it is very weak, and the experimental errors are quite large. Fixing the incubation time at 2 hours, the DNA coverage increases when a higher concentration of alkynyl-oligonucleotides is used (Fig. 3), and varies from about 1.2 pmol/cm2 to 2.6 pmol/cm2 (equivalent to an SLB displaying ∼1 mol% DNA), when 5mM alkynyl-oligonucleotides was used. Approximately 700 pmol/cm2 of azide groups are present on the surface based on the number of SiOH groups/cm2 (Wasserman, Tao et al. 1989). Although only a small fraction of surface azides underwent the click reaction with alkynyl-DNA, higher densities of immobilized DNA density were not pursued because this range of DNA coverage was sufficient to allow stable tethering of membranes. Reproducible spontaneous GUV rupture is observed when 5 µM alkynyl-DNA was incubated for 2hr; therefore, the DNA immobilized substrates in the rest of this study were prepared using this set of conditions.
Figure 3.
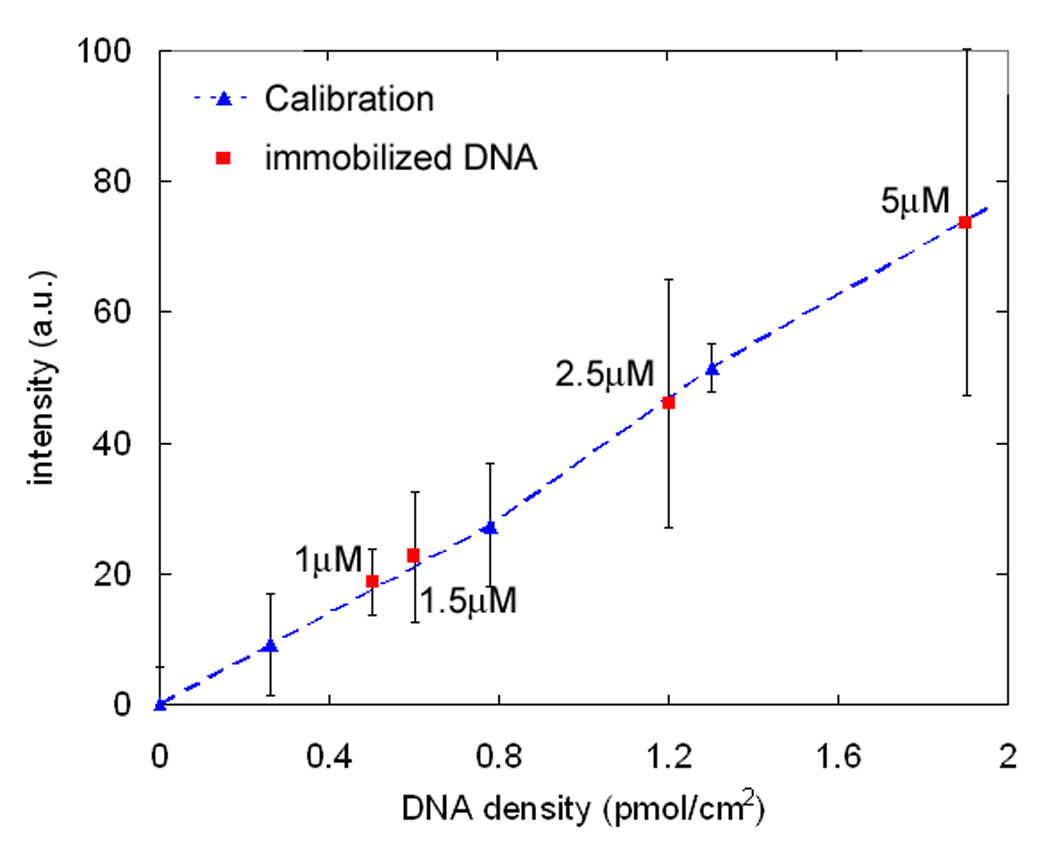
Estimation of click-immobilized DNA density. DNA-immobilized substrates are prepared with the click reaction for 2 hr using different concentrations of alkynyl-oligonucleotides. Cy5 labeled complementary DNA was then hybridized and its fluorescence intensity, reflecting the immobilized DNA density, was measured. Fluorescence intensities are calibrated with supported lipid bilayers displaying known amounts of DNA-lipid conjugates (blue triangles and dotted line, see text). The apparent surface density of immobilized DNA incubated with labeled alkynyl-oligonucleotides concentration is then estimated (red squares).
Tethered lipid membrane patch formation by GUV rupture
Tethered membrane patches were formed by the introduction, binding, and subsequent rupture of GUVs presenting complementary DNA onto the two types of surface described above. DNA-lipid conjugates were inserted into the GUVs by two different methods. First, DNA-lipid conjugates were included in the lipid mixtures before electrode coating and GUV electroformation, so the mol% of DNAs in the GUV should be determined by the composition of lipid mixture. Because the inner and outer leaflets of GUVs formed by electroformation are expected to be identical, DNAs are displayed on both the inside and outside of the GUVs, and DNA are consquently displayed on the top surface of the tethered patch in the final assembly; this can be used to tether vesicles on the upper surface of the patch or for building another layer (data not shown). Applications of these patches as platforms for more complex assemblies and processes will be discussed in subsequent work. Second, DNA-lipid conjugates were mixed with already-formed GUVs, so DNA-lipids should insert only on the outside of the GUVs, as is the case when DNA-lipids are added to SUVs (Yoshina-Ishii, Chan et al. 2006), and therefore, the top leaflet of the tethered bilayer, which was the inner leaflet of a GUV, will not display DNA. Consistent with this, vesicles displaying the antisense strand could not be tethered to this upper surface (data not shown). Because the DNA displayed on the tethered bilayer plays no role in the experiments described below, the first DNA insertion method was mostly used.
GUV rupture to form tethered membrane patches was indistinguishable whether mobile or immobile DNA tethers were used; however, the tethered bilayers with mobile tethers are much less stable as discussed in the next section. This can be visualized by fluorescence imaging, either in projection by epifluorescence or by confocal microscopy, from which a 3D image can be reconstructed (Figure 4). Because tethering and rupture are typically quite rapid and usually faster than the time required to obtain a full confocal image, the epifluorescence images on the left side of Figure 4 are for a single GUV undergoing binding and rupture, while the confocal images on the right side are representative examples of GUVs and patches at comparable stages. Initially the GUVs retain their spherical shape; as more of the DNA on the GUV binds to the surface, they undergo deformation and start to flatten as the contact area expands from the initial point of contact, giving a hemispherical shape as seen in reconstructed images from confocal microscopy (Fig. 4 B). Because there is a structure above the focal plane, the epifluorescence microscopy images show a characteristic halo around the circular contact area (Fig. 4 A). As the binding progresses further, the halo disappears, indicating that the GUV has become almost flat (Fig. 4 C and D). At this point, a hole, which appears as a darker area in Figure 4E, forms and expands while concurrently the total area of the flattened GUV also expands. This appears to initiate the rupture process and is likely driven by the tension caused by multiple DNA binding. The upper part of the flattened GUVs ruptures and spreads over the substrate, allowing more DNA hybridization. This spread of the upper membrane is seen as the expansion of an area with roughly half the fluorescence intensity per unit area than the original flattened GUV, because images of the flattened GUV contain intensity from two bilayers whereas the resulting tethered patch is a single bilayer. After rupture, the bright regions (two bilayers) and dim regions (one bilayer) are in the same focal plane (roughly 122 nm thick), thus their height difference is not resolvable with confocal microscopy (Fig. 4 F). The initial hole forms at a random location in the upper membrane, which causes an asymmetrical expansion of the upper membrane due to the irregular structure of the ruptured membrane. From these observations, the outer leaflet of the GUV becomes the bottom leaflet of the tethered membrane and the inner leaflet of the GUV becomes the top leaflet. For some GUVs (about 20∼40% of the population), this process does not occur spontaneously and is arrested as a stable hemispherical shape as in Fig. 4 C; about half of these can be ruptured by osmotic shock. This osmotic shock induced tethered lipid bilayer formation with GUVs was reported by Kunding and Stamou (Kunding and Stamou 2006); however, the details of the formation process were not described.
Figure 4.
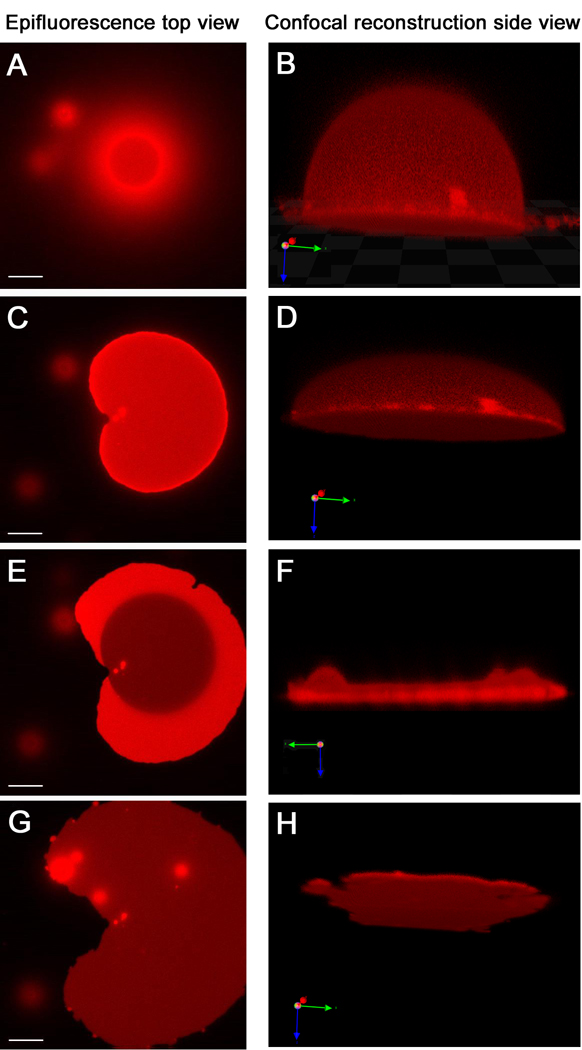
Giant vesicle to tethered membrane patch transformation observed by fluorescence microscopy. These images are of stable tethered membranes with immobile tethers (Fig 1A); those with mobile tethers are very similar. The epifluorescence microscopy images, left side, and confocal microscopy images in the corresponding state, right side, are displayed in parallel. While F is exhibited parallel to the surface, B, D and H are 20 degree tilted to show a better view of their 3-dimensional shape. (A and B) When the GUV starts to bind via DNA hybridization, only the bottom part of vesicle is in contact with the surface and shows a ring shape. Meanwhile, the upper part of the vesicle out of the focal plane appears as a cloudy halo around the ring. (C and D) Further binding of DNA from the vesicle to the surface flattens the vesicle asymmetrically. (E and F) Eventually, the upper membrane ruptures. Afterwards, a single bilayer remains (dark area) with some transient double bilayer (bright regions). (G and H) After all of the upper bilayer spreads, a homogeneous tethered membrane is formed. While the epifluorescence microscopy images are taken from the same GUV in about 5 minutes, the confocal microscopy images take much longer to collect and are of different GUVs captured at comparable times (see text). Scale bar of epifluorescence microscopy is 15µm.
Tethered planar membrane patch formation by GUV rupture is driven by DNA hybridization. Although occasional rupture of GUVs on SLBs without linkers has been reported (Wong and Groves 2002; Parthasarathy and Groves 2004), we do not observe rupture of GUVs without DNA. The time for the rupture process from GUV introduction to bilayer patch formation is 5∼15 minutes for spontaneous rupture and tends to be longer for low DNA surface density. This was examined crudely: the threshold DNA surface density to induce spontaneous rupture of a small population of bound GUVs is about 0.3 pmol/cm2, and almost all GUVs undergo rupture above 1 pmol/cm2. To prove that the tethered membrane is actually anchored by a DNA hybrid linkage, the salt-containing buffer was exchanged with deionized water to disrupt DNA hybridization which led to immediate loss of the membrane patch from the surface. This also shows that the solution in the thin gap (∼ 8 nm for the 24-mer hybrid and ∼ 16 nm for the 48-mer hybrid, see below) between the bilayer patch and substrate surface is readily water-permeable so that salt ions are washed out immediately. Water accessibility was also checked by a cobalt quenching experiment (Saurel, Cezanne et al. 1998). Co2+ completely quenches fluorescence from Texas red labeled lipid, and in the case of solid supported lipid membranes, 54% of the fluorescence intensity is immediately quenched because Co2+ in bulk solution cannot access the fluorescence dye in the bottom leaflet close to the substrate (Ajo-Franklin, Yoshina-Ishii et al. 2005). By contrast, the DNA-tethered bilayer patches containing TR-DHPE exhibited no fluorescence when they are observed within 10 seconds after Co2+ addition, so both sides of the tethered bilayer patch are fully accessible to the quencher.
Instability of DNA-tethered membrane patches on SLB
Bilayer patches deposited on DNA-presenting SLBs behave quite differently from those on covalently anchored DNA. Since the SLB is itself a passivating surface, no further surface modification is necessary as in the case of the unreacted azides in the SAM. Hybridized DNA tethers are expected to diffuse within the lipid membranes much like other lipid molecules, though their diffusion may be somewhat slower than for a DNA-lipid conjugate alone (Ajo-Franklin, Yoshina-Ishii et al. 2005) because both ends are anchored in the viscous hydrophobic parts of two bilayers. This avoids the limitation of obstructed lateral diffusion caused by immobile tethers (Deverall, Gindl et al. 2005). Most DNA-tethered lipid membrane patches on immobilized DNA surfaces remain largely unchanged over the course of a day; by contrast, most tethered bilayer patches on SLBs with mobile DNA are unstable and disappear over the course of several hours. Typically, two third of the patches disintegrate within 30 min, while one third remain up to 3 hours with no obvious environmental or size variable accounting for this difference. In some cases, the patches disappear from the interior of the patch, so this is likely not an effect that is dominated by the patch edge.
In order to gain some insight into the mechanism by which SLB-tethered membrane patches disappear, patterned SLBs displaying DNA were prepared using 100 µm square grids of BSA which confines the fluid SLB. As shown in Figure 5, when the patch disappears, red fluorescence appears in the underlying bilayer, which was prepared without a TR-labeled lipid. By using a patterned SLB, the fluorescent lipids are confined and can be readily visualized - the lipids rapidly diffuse away and are harder to visualize without this confinement. Two limiting models can be used to interpret this observation. First, all of the lipids in the tethered patch are transferred into the underlying SLB. However, it seems improbable that the SLB lateral density can be increased so much to absorb the large amount of lipids from the tethered patch, which is 10∼20% of the size of the confined SLB. Second, small (sub-diffraction limited) broken tethered bilayer fragments from the original patch could be scattered to the rest of the confined area. These evenly distributed tethered bilayers labeled with TR on a confined square SLB would also be imaged as the emergence of a dim red fluorescence over the entire confined region. The mobility of the DNA tethers makes either mechanism possible, while it is impossible with immobile tethers. Although we do not exclude the possibility that some of the tethered patches detach into the bulk solution, no evidence for such a process has been observed.
figure 5.
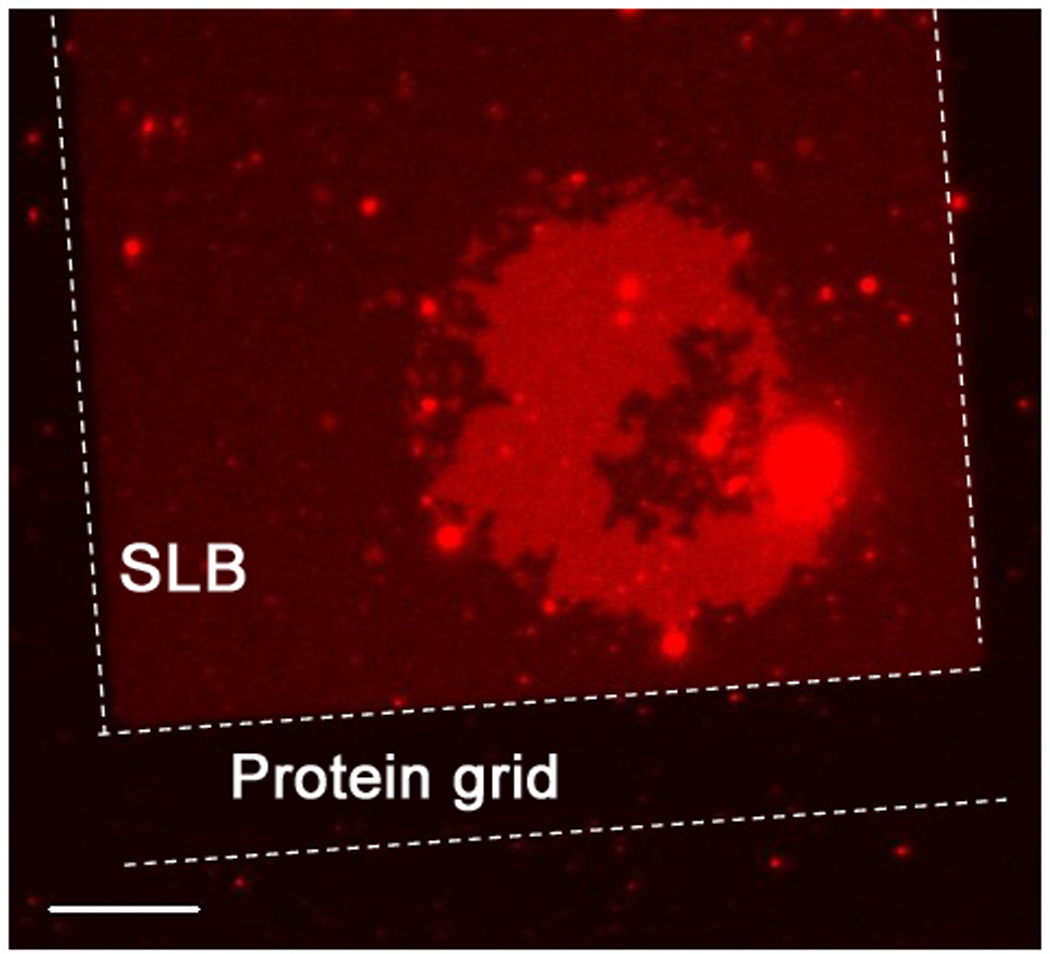
A fluorescently labeled tethered bilayer patch (bright red) with mobile DNA tethers on an unlabeled supported bilayer in the process of disintegrating. The supported bilayers are confined by a protein grid outlined with dotted lines, to observe the build-up of weak red fluorescence from the lipid materials that were part of the unstable tethered patch. This shows that the broken part of the tethered bilayer remains mostly on the SLB. One side of the rectangular protein grid is 100 µm and the scale bar is 15 µm.
Based on these observations, a tethered patch was created which covers an entire rectangular region. This will not have space so that small pieces of tethered bilayers can dissociate from the original tethered patch. Tethered patches created this way were more stable, supporting the suggestion that the tethered bilayer fragmentation caused by mobile DNA tethers limits the integrity of tethered patches on SLBs, though such patches tethered to patterned SLBs are not as stable as the tethered patches with immobile tethers. Because of the mobile DNA tethers and the existence of the SLB, this instability appears to be an intrinsic problem with this architecture. With some relatively stable populations for up to hours, some measurements still can be performed. However, the use of tethered lipid bilayers with mobile tethers is inevitably limited for some applications. The dynamic nature of this system mimics that of biological membrane-to-membrane junctions, and thus will be useful to investigate surface protein interaction or membrane behavior. Variable length DNA tethers are an example of this and are briefly discussed below.
Measurement of the distance between tethered membrane patches and the surface
VIA-FLIC was used to determine the distance between the substrate and the tethered membrane. Changing the incidence angle of excitation light creates predictable changes in the intensity of light perpendicular to the mirror surface due to interference, and the fluorescence intensity changes can be precisely related to the distance of the fluorophores from the reflective underlying surface (Ajo-Franklin, Ganesan et al. 2005). For interferometry measurements, tethered lipid bilayer patches were assembled on atomically flat Si chips with thermally grown SiO2 surfaces of fixed thicknesses. Three kinds of tethered bilayers were analyzed and modeled: tethered bilayer patches with mobile and immobile DNA tethers, and mobile tethers with two different lengths of tethers. Due to the instability of tethered patches with mobile tethers, only limited data could be obtained.
Because the DNA linking the lipid membrane and substrate acts as a spacer, the gap should vary depending on the length of DNA used. Tethered bilayer patches linked by 24-mer mobile DNA tethers to a SLB give a bilayer-bilayer separation distance of 6 ± 1 nm (see Materials and Methods), which is in range for the expected linker length of ∼8 nm. Analysis of tethered bilayer patches linked to a solid support by 48-mer immobile DNA tethers give a substrate-bilayer separation distance of 14 ± 6 nm, again in the range expected for this longer hybrid. In both cases, the fluorescence intensity across the patch was very homogeneous indicating that the patch is parallel to the support. These results show that the distance from the substrate is controllable on the nanometer scale, a unique feature of this tethering strategy since any reasonable DNA length can be prepared. Considering that the size of most cytosolic domains of transmembrane proteins does not exceed 5 nm, the gap of DNA-tethered lipid bilayers can provide enough space to prevent contact between transmembrane proteins and substrates.
An interesting situation arises when different length DNA-lipid conjugates are displayed on both surfaces as illustrated in Figure 6A. In this case, the behavior is expected to be quite different for immobile vs. mobile tethers. A fluorescence image of a bilayer patch tethered to a supported bilayer by only a 24mer DNA tether formed on a reflective Si/SiO2 wafer is shown in Figure 6 B, left. The fluorescence intensity observed across the patch is nearly constant, reflecting the well-defined distance of the tethered patch from the SLB, and, in turn, its well-defined distance from the reflecting substrate. If the tethered patches were to undulate and hence have inhomogeneous height profiles, this would be visible as variations in fluorescence intensity. At least on the lateral length scale resolvable by fluorescence microscopy (∼ hundreds of nm), there is no evidence of such fluorescence variation, indicating that the tethered bilayers with constant length DNA linkers are fairly flat (Chan and Boxer 2007). By contrast, when GUVs that have an equimolar mixture of two different lengths of DNAs were used to form DNA-tethered membrane patches to an SLB displaying both lengths of antisense DNA on an SiO2/Si surface, two distinct levels of fluorescence intensity were observed in micrometer scale domains (Figure 6 B, right). This indicates qualitatively that the tethered patch is not uniformly flat. Also, the observation of micrometer-sized domains suggests that the DNA tethers, initially presumed to be randomly distributed, must spontaneously reorganize and segregate by length upon hybridization. VIA-FLIC analysis was performed to quantify these height differences on the nanometer scale. The regions exhibit different intensity variations as a function of the angle of incidence, i.e. different VIA-FLIC behavior (Figure 6 C). The intensity differences arise from height differences, presumably caused by different lengths of hybrid DNA supporting each domain. When the two levels were fit separately (Figure 6 C), bilayer-bilayer separation distances were obtained: 10 ± 1 and 16 ± 1 nm for the dimmer and brighter regions, respectively. This is consistent with the length of 24mer and 48mer DNA supports. The measured distance of the 24mer DNA is longer than the value obtained for the 24mer only. This may be because a repeating sequence (poly A and T) was used for 24mer. The repeating sequence could be hybridized flexibly, especially adjacent to higher 48mer tethers. It is also possible that small number of 48mer tethers exists in 24mer rich domains and this makes the gap larger because the segregation can be incomplete. When tethered patches with two lengths of DNA linkers were formed on the DNA immobilized substrate which displays randomly distributed two lengths of the complementary DNAs, segregated height domains were not observed. Consequently, the fluorescence variations built by the height difference between individual DNA tether lengths on the small lateral length scale (about 70 nm2 for each DNA tethers) are not resolvable, and the microscopy image looks homogeneous.
Figure 6.
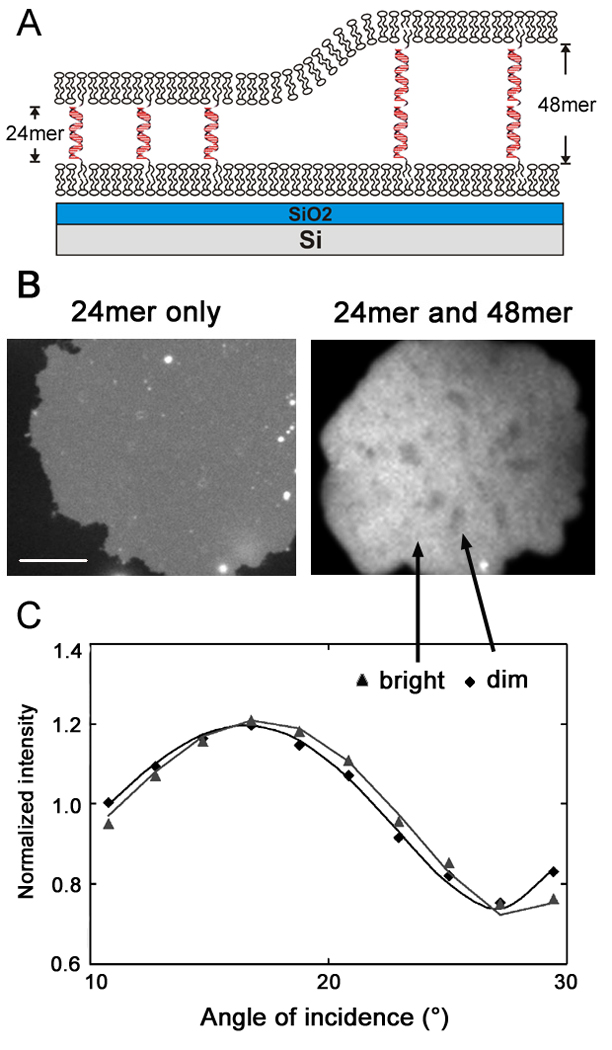
DNA tether segregation by different lengths of DNA hybridization on a mobile SLB (c.f Fig. 1B). (A) Schematic illustration of segregation involving two lengths of DNA hybrids between the mobile membrane surfaces. The substrate is 260 nm of SiO2 of grown on atomically flat Si to allow for quantitative analysis by VIA-FLIC. (B) Fluorescence image of Texas red labeled tethered bilayer patches. When only 24-mer DNA tethers were used (left, poly A/poly T), the intensity is homogeneous across the membrane. With mixed 24-mer (poly A/poly T) and 48-mer (fully overlapping sequence) tethers, bright and dark domains were observed that indicate regions where the DNA hybrids have segregated by length. Left scale bar is 15 µm, and right image is 10 µm wide. (C) The fluorescence signal as a function of angle of incidence for selected bright and dark regions of the 24/48-mer tethered patch. The intensity counts are normalized by dividing by the mean counts recorded per image for each region, so that the shape of each curve can be more easily compared. The lines show the fits of the VIA-FLIC model to the data: for the dim region, the separation distance is fit as 10.0 nm, for the bright region, it is fit as 16.9 nm.
This phenomenon is similar in concept to the work of Parthasarathy and Groves, who observe lipid membrane bound protein reorganization in intermembrane junctions. In their case, the domains were distinguished by dense and sparse regions of membrane-bound IgG (Parthasarathy and Groves 2004), rather than by binding events between membranes. The mechanism discussed in this study is expected to be similar to the one observed with tethered patches, i.e. membrane mechanical forces drive rearrangement and segregation of DNA linkers with different heights. The segregated patterns are formed randomly and do not change over tens of minutes, because coarsening or movement of domains requires local rearrangement of different lengths of DNA hybrid tethers across domains. This segregation experiment demonstrates that there is little resistance to the mobility of DNA tethers because weak membrane mechanical forces were sufficient to drive the rearrangement. The low resistance of the mobility is also indirect evidence that DNA tethers neither attract or repel each other, and moreover do not adhere or interact with lipid membranes, which could cause drag. Many variations on this concept are readily achieved with DNA hybrids as models for membrane junctions, and this will be described in future publications.
Diffusion coefficient of Lipids in DNA-tethered membrane patches
Diffusion coefficients and mobile fractions were measured by FRAP experiments to demonstrate lateral continuity and measure fluid dynamic properties of DNA-tethered lipid bilayers as well as measure diffusion coefficients of lipid molecules. Figure 7 shows the spot-bleaching recovery region of a tethered membrane patch and the normalized fluorescence intensity recovery curve. Diffusion coefficients and mobile fractions of DNA-tethered membrane patches, SLBs formed by GUVs, and SLBs are summarized in Table 1. The values (3.4 µm2s−1 ) of SLB diffusion coefficients are somewhat higher than most reports (Wagner and Tamm 2000; Naumann, Prucker et al. 2002; Renner, Osaki et al. 2008), which were between 0.5 and 2.5 µm2s−1 for SLBs composed of synthetic PCs on glass prepared in a similar process. This difference is possibly because these investigators used the classical approach of Axelrod or Soumpasis (Axelrod, Koppel et al. 1976; Soumpasis 1983), which can cause significant underestimation of diffusion coefficients, especially for highly mobile molecules (Braga, Desterro et al. 2004). The diffusion coefficients measured for SLB patches on glass from GUV rupture (3.2 µm2s−1) and from vesicle fusion (3.4 µm2s−1 ) are almost the same, as discussed above. Diffusion coefficients for lipids in patches with mobile tethers (4.8 µm2s−1 ) are somewhat smaller than with immobilized DNA tethers (6.5 µm2s−1), so there is no evidence for hindered diffusion due to immobile tethers. Relatively stable patches with mobile tethers had to be chosen due to the instability described above, and it is quite possible that this selected population could have biased properties, e.g. tethered patches with lower diffusion may be more stable. In contrast to most reports of tethered or polymer-supported lipid bilayers, almost no immobile fraction is observed for the DNA tethered membrane patches, suggesting that this is a particularly good system for minimizing interactions with the substrate.
Table 1.
Diffusion coefficients and mobile fractions determined by FRAP for lipids in DNA-tethered bilayer pathces and solid supported bilayers
| Diffusion coefficient [µm2s−1] | Mobile fraction [%] | |
|---|---|---|
| DNA-tethered patch on immobilized tether | 6.5 ± 1.0 | 0.99 ± 0.01 |
| DNA-tethered patch on mobile tether | 4.8 ± 0.2 | 0.97 ± 0.03 |
| SLB formed by GUVs | 3.2 ± 0.4 | 0.93 ±0.02 |
| SLB formed by vesicle fusion | 3.4 ± 0.2 | 0.97 ±0.03 |
Diffusion coefficients of the tethered membrane patches are 1.5–2.0 times higher than SLBs without DNA tethers. This result corresponds to the case of free-standing bilayers in GUVs, whose diffusion coefficients are 2 times larger than SLBs (7.8 µm2s−1 versus 3.1 µm2s−1 on glass) (Przybylo, Sykora et al. 2006). Thus the DNA spacers provide an environment that is more similar to free-standing bilayers than solid-supported bilayers.
In summary, two novel architectures have been developed based on DNA lipid conjugates that we originally developed for tethering and fusing vesicles (Yoshina-Ishii and Boxer 2003; Chan, van Lengerich et al. 2009). Stable membrane patches are obtained when the DNA displayed on the surface is covalently fixed on the surface, while unstable patches are obtained when the DNA is displayed on a fluid SLB. Thus far, we have not observed coalescence of the patches to form a continuous tethered bilayer in either architecture. The stable patches should be useful for displaying membrane proteins in a planar format that retains all the advantages of the planar geometry, yet keeps the proteins away from the solid support. The unstable patches exhibit interesting dynamics when multiple lengths of DNA hybrids are present and this can only occur because the lipid anchors on both surfaces are mobile.
Acknowledgements
This work was supported by National Institutes of Health grant GM069630, the NSF Biophysics Program, and by the MRSEC Program of the NSF under award DMR-0213618 (CPIMA). P.V.G. was supported by a Benchmark Stanford Graduate fellowship and Center for Probing the Nanoscale Graduate Prize fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Ajo-Franklin CM, Ganesan PV, et al. Variable incidence angle fluorescence interference contrast microscopy for z-imaging single objects. Biophys J. 2005;89(4):2759–2769. doi: 10.1529/biophysj.105.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajo-Franklin CM, Kam L, et al. High refractive index substrates for fluorescence microscopy of biological interfaces with high z contrast. Proc Natl Acad Sci U S A. 2001;98(24):13643–13648. doi: 10.1073/pnas.241208698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajo-Franklin CM, Yoshina-Ishii C, et al. Probing the structure of supported membranes and tethered oligonucleotides by fluorescence interference contrast microscopy. Langmuir. 2005;21(11):4976–4983. doi: 10.1021/la0468388. [DOI] [PubMed] [Google Scholar]
- Ali MB, Bessueille F, et al. Use of ultra-thin organic silane films for the improvement of gold adhesion to the silicon dioxide wafers for (bio)sensor applications. Materials Science and Engineering C. 2008;28:628–632. [Google Scholar]
- Axelrod D, Koppel DE, et al. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16(9):1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga J, Desterro JM, et al. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Mol Biol Cell. 2004;15(10):4749–4760. doi: 10.1091/mbc.E04-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-H, Lengerich MBv, et al. Lipid-anchored DNA mediates vesicle fusion as observed by lipid and content mixing. Biointerphases. 2008;3(2):FA17–FA21. doi: 10.1116/1.2889062. [DOI] [PubMed] [Google Scholar]
- Chan YH, Boxer SG. Model membrane systems and their applications. Curr Opin Chem Biol. 2007;11(6):581–587. doi: 10.1016/j.cbpa.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YH, van Lengerich B, et al. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci U S A. 2009;106(4):979–984. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverall MA, Gindl E, et al. Membrane lateral mobility obstructed by polymer-tethered lipids studied at the single molecule level. Biophys J. 2005;88(3):1875–1886. doi: 10.1529/biophysj.104.050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YS, Yu SJ. Immobilization of Homogeneous Palladium(II) Complex Catalysts on Novel Polysiloxanes with Controllable Solubility: Important Implications for the Study of Heterogeneous Catalysis on Silica Surfaces This work was supported in part by the National Science Council of Taiwan. The authors are grateful to the NSC South Precision Instrument Center at National Cheng Kung University for performing the solution (29)Si NMR experiments. Angew Chem Int Ed Engl. 2001;40(2):437–440. [PubMed] [Google Scholar]
- Jonsson P, Jonsson MP, et al. A method improving the accuracy of fluorescence recovery after photobleaching analysis. Biophys J. 2008;95(11):5334–5348. doi: 10.1529/biophysj.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, et al. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Koper I. Insulating tethered bilayer lipid membranes to study membrane proteins. Mol Biosyst. 2007;3(10):651–657. doi: 10.1039/b707168j. [DOI] [PubMed] [Google Scholar]
- Kunding A, Stamou D. Subnanometer actuation of a tethered lipid bilayer monitored with fluorescence resonance energy transfer. J Am Chem Soc. 2006;128(35):11328–11329. doi: 10.1021/ja062545k. [DOI] [PubMed] [Google Scholar]
- Kung LA, Kam L, et al. Patterning Hybrid Surfaces of Proteins and Supported Lipid Bilayers. Langmuir. 2000;16:6773–6776. [Google Scholar]
- Lambacher A, Fromherz P. Luminescence of dye molecules on oxidized silicon and fluorescence interference contrast microscopy of biomembranes. Journal of the Optical Society of America B. 2002;19(6):1435–1453. [Google Scholar]
- Longo ML, Ly HV. Micropipet aspiration for measuring elastic properties of lipid bilayers. Methods Mol Biol. 2007;400:421–437. doi: 10.1007/978-1-59745-519-0_28. [DOI] [PubMed] [Google Scholar]
- Merzlyakov M, Li E, et al. Surface-supported bilayers with transmembrane proteins: role of the polymer cushion revisited. Langmuir. 2006;22(24):10145–10151. doi: 10.1021/la061976d. [DOI] [PubMed] [Google Scholar]
- Naumann CA, Prucker O, et al. The polymer-supported phospholipid bilayer: tethering as a new approach to substrate-membrane stabilization. Biomacromolecules. 2002;3(1):27–35. doi: 10.1021/bm0100211. [DOI] [PubMed] [Google Scholar]
- Naumann R, Schiller SM, et al. Tethered Lipid Bilayers on Ultraflat Gold Surfaces. Langmuir. 2003;19:5435–5443. [Google Scholar]
- Parthasarathy R, Groves JT. Protein patterns at lipid bilayer junctions. Proc Natl Acad Sci U S A. 2004;101(35):12798–12803. doi: 10.1073/pnas.0402168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404(6778):604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Popat CK, Sharma S, et al. AFM analysis of organic silane thin films for bioMEMS applications. SURFACE AND INTERFACE ANALYSIS. 2003;35:205–215. [Google Scholar]
- Przybylo M, Sykora J, et al. Lipid diffusion in giant unilamellar vesicles is more than 2 times faster than in supported phospholipid bilayers under identical conditions. Langmuir. 2006;22(22):9096–9099. doi: 10.1021/la061934p. [DOI] [PubMed] [Google Scholar]
- Purrucker O, Fortig A, et al. Supported membranes with well-defined polymer tethers--incorporation of cell receptors. Chemphyschem. 2004;5(3):327–335. doi: 10.1002/cphc.200300863. [DOI] [PubMed] [Google Scholar]
- Renner L, Osaki T, et al. Supported lipid bilayers on spacious and pH-responsive polymer cushions with varied hydrophilicity. J Phys Chem B. 2008;112(20):6373–6378. doi: 10.1021/jp711479f. [DOI] [PubMed] [Google Scholar]
- Richter RP, Berat R, et al. Formation of solid-supported lipid bilayers: an integrated view. Langmuir. 2006;22(8):3497–3505. doi: 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- Rossetti FF, Bally M, et al. Interactions between titanium dioxide and phosphatidyl serine-containing liposomes: formation and patterning of supported phospholipid bilayers on the surface of a medically relevant material. Langmuir. 2005;21(14):6443–6450. doi: 10.1021/la0509100. [DOI] [PubMed] [Google Scholar]
- Sackmann E. Supported membranes: scientific and practical applications. Science. 1996;271(5245):43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- Saurel O, Cezanne L, et al. Influence of annexin V on the structure and dynamics of phosphatidylcholine/phosphatidylserine bilayers: a fluorescence and NMR study. Biochemistry. 1998;37(5):1403–1410. doi: 10.1021/bi971484n. [DOI] [PubMed] [Google Scholar]
- Seu KJ, Pandey AP, et al. Effect of surface treatment on diffusion and domain formation in supported lipid bilayers. Biophys J. 2007;92(7):2445–2450. doi: 10.1529/biophysj.106.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumpasis MD. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys J. 1983;41(1):95–97. doi: 10.1016/S0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B, Falconnet D, et al. Micropatterning of DNA-tagged vesicles. Langmuir. 2004;20(26):11348–11354. doi: 10.1021/la0482305. [DOI] [PubMed] [Google Scholar]
- Sund SE, Swanson JA, et al. Cell membrane orientation visualized by polarized total internal reflection fluorescence. Biophys J. 1999;77(4):2266–2283. doi: 10.1016/S0006-3495(99)77066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner ML, Tamm LK. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophys J. 2000;79(3):1400–1414. doi: 10.1016/S0006-3495(00)76392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman SR, Y.-Tao T, et al. Structure and Reactivity of Alkylsiloxane Monolayers Formed by Reaction of Alkyltrichlorosilanes on Silicon Substrates. Langmuir. 1989;5(4):1074–1087. [Google Scholar]
- Wong AP, Groves JT. Molecular topography imaging by intermembrane fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2002;99(22):14147–14152. doi: 10.1073/pnas.212392599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina-Ishii C, Boxer SG. Arrays of mobile tethered vesicles on supported lipid bilayers. J Am Chem Soc. 2003;125(13):3696–3697. doi: 10.1021/ja029783+. [DOI] [PubMed] [Google Scholar]
- Yoshina-Ishii C, Chan YH, et al. Diffusive dynamics of vesicles tethered to a fluid supported bilayer by single-particle tracking. Langmuir. 2006;22(13):5682–5689. doi: 10.1021/la0534219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina-Ishii C, Miller GP, et al. General method for modification of liposomes for encoded assembly on supported bilayers. J Am Chem Soc. 2005;127(5):1356–1357. doi: 10.1021/ja043299k. [DOI] [PubMed] [Google Scholar]