Abstract
Crystallography of ribosomes, the universal cell nucleoprotein assemblies facilitating the translation of the genetic-code into proteins, met with severe problems owing to their large size, complex structure, inherent flexibility and high conformational variability. For the case of the small ribosomal subunit, which caused extreme difficulties, post crystallization treatment by minute amounts of a heteropolytungstate cluster allowed structure determination at atomic resolution. This cluster played a dual role in ribosomal crystallography: providing anomalous phasing power and dramatically increased the resolution, by stabilization of a selected functional conformation. Thus, four out of the fourteen clusters that bind to each of the crystallized small subunits are attached to a specific ribosomal protein in a fashion that may control a significant component of the subunit internal flexibility, by “gluing” symmetrical related subunits. Here we highlight basic issues in the relationship between metal ions and macromolecules and present common traits controlling in the interactions between polymetalates and various macromolecules, which may be extended towards the exploitation of polymetalates for therapeutical treatment.
Keywords: Ribosome, ribosomal functional flexibility, heteropolytungstates, crystal order, protein S2
Introduction
Metalloproteins play key roles in many aspects of life. Metal ions are also contributing significantly to the activity and conformational stability of ribozymes. This review highlights yet another aspect of the border between inorganic chemistry and biology: the crucial contribution of the interplay between mono and divalent ions as well as post crystallization treatment with metal clusters to ribosomal crystallography.
Ribosomes are giant ribonucleoprotein assemblies that translate the genetic code into proteins in all living cells. They are built of two subunits of unequal size that associate upon the initiation of protein biosynthesis to form a functional particle and dissociate once this process is terminated. Protein biosynthesis is performed cooperatively by the two ribosomal subunits. While elongation proceeds, the small subunit provides the decoding-center and controls translation fidelity, and the large one contains the site where the peptide bonds are being formed, called the peptidyltransferase-center (PTC), as well as the protein exit tunnel. mRNA carries the genetic code to the ribosome, and tRNA molecules bring the amino acids to the ribosome. The ribosome possesses three tRNA binding sites, called A-(aminoacyl), P-(peptidyl) and E-(exit) sites (Figure 1). The tRNA anticodon loops interact with the mRNA on the small subunit whereas the tRNA 3′ ends interact with the large subunit, where peptide bonds are being formed. Hence, in addition to the intersubunit bridges that are built of flexible components of both subunits, the tRNA molecules are the entities combining the two subunits within the active ribosome. The elongation cycle includes decoding, peptide bond formation, the detachment of the P-site tRNA from the growing polypeptide chain, the release of a deacylated tRNA molecule and the advancement of the mRNA together with the tRNA molecules attached to it from the A- to the P- and then to the E-site, all driven by the GTPase activity of two G-protein factors.
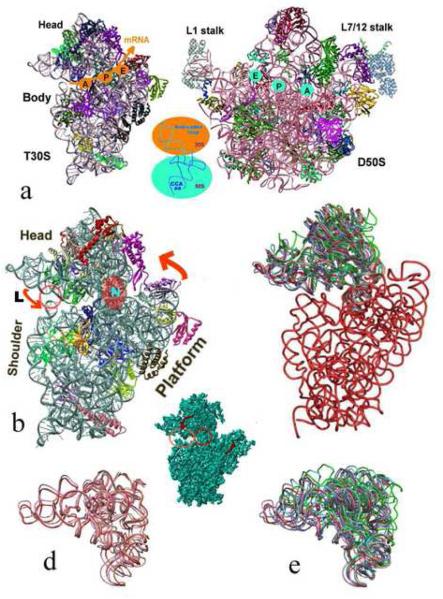
Figure 1.
Functional mobility of the ribosomal subunits:
a. The interface surfaces of the high resolution structures of the small (left) and the large (right) ribosomal subunits from Thermus thermophilus, T30S [2] and Deinococcus radiodurans, D50S [4], respectively, with their main structural regions. The approximate mRNA channel on the small subunit and the positions of the three tRNA sites on both subunits are shown, with a representative tRNA molecule placed between the two subunits, showing the regions interacting with each of the subunits.
b. An enlarged view of the small subunit indicating its functionally relevant motions (the two arrows), the position of the latch (indicated by L) , participating actively in mRNA attachment; and the single RNA chain called neck (shown as “N”) that seems to act as the structural element facilitating the “head” motions, which involved in opening/closing the latch. The pink circle indicates the position that closes and opens during the latch motion, for creating a pore for the incoming mRNA.
c-e. rRNA backbone representation of the small subunit. The body, shown in red, is almost identical in all known crystal structures, whereas various “head” folds have been detected and are shown in different colors. (Red: the tungstenated T30S structure [2]; pink and wheat: the two traceable folds among the ensemble of conformations existing in the T30S low resolution crystals; cyan and green: the two conformations existing in the crystal of the entire ribosome from E. coli, E70S [5]; grey and blue: the fold of functional complexes of the entire ribosome from Thermus thermophilus, T70S [9] and [10], respectively. The T30S non-tungstenated structure [3] is almost identical to that observed in [9], therefore is shown in grey).
d. Shows the assembly of structured observed in the low resolution crystal form of T30S.
Inserted is the 7A resolution structure of T30S [75] that shows clearly the structural domains and the possible motions (indicated by arrows). The left red circle indicates the position of the latch and the right circle is positioned on the neck.
Ribosomes from all kingdoms are composed of long RNA (rRNA) chains, accounting for 2/3 of their mass (apart from mitochondrial ribosome, where the ratio RNA/protein is somewhat different) and different proteins (r-proteins). The small and the large bacterial ribosomal subunits are of molecular weights of 0.85 and 1.45 Mega Dalton respectively. The small subunit (called 30S in prokaryotes) contains an rRNA chain (called 16S) of ~1500 nucleotides and 20-21 r-proteins, and the large one (called 50S in prokaryotes) is composed of two rRNA chains (23S and 5S RNA) of about 3000 nucleotides in total, and 31-35 r-proteins, depending on the source.
The recently determined crystal structures of ribosomal particles [1-5] and of their functional complexes [e.g. [6-10]) show that all ribosomes functions utilize primarily rRNA-mediated processes that may be assisted by r-proteins. They also indicate, mainly indirectly, that within the ribosome metal ions bind to both r-proteins and rRNA in a fashion similar albeit different to those seen in systems composed of either RNA or proteins.
Metal ions and the structure and function of RNA and protein enzymes
A large repertoire of RNA structural motifs that typically sequester metal ions appears in ribozymes, namely catalytic RNA molecules [11-13], where they contribute to folding pathways and tertiary structure maintenance (as reviewed in [14]). In addition, although some ribozymes can utilize alternative catalytic mechanisms [15,16], in many cases ribozymes catalysis depends on metal ions (e.g. [17]) functioning as cofactors that are explicitly implicated in the chemical mechanism of the catalysis [18]. Examples are RNA polymerization [19, 20], the aldol reaction[21], splicing [22], self-splicing, ligation, packing [23-27] and resolving ribozyme misfolding [28] and gene control utilizing riboswitches [29]. Nevertheless, even for RNA enzymes (e.g. group I and II introns) specific and nonspecific interactions [30] of a few or many [31] monovalent cations and magnesium ions are mainly essential for adopting and maintaining the proper compact tertiary structures (e.g. [32, 33]). Experimental and simulation results established that the stability of large RNA ribozymes is largely determined by a combination of counter ion charges [34, 35] that kinetic intermediates regulate proper RNA folding [36] and that the packing efficiency of condensed cations depends on their excluded volumes [37, 38].
Metal ions bound to protein-enzymes are often involved in enzymatic catalysis and form part of the active site. They are involved in a large range of activities, including catalysis, electron transfer, energy reorganization, oxygen and carbon dioxide transport, redox, polymerization and similar tasks. As such they play key roles in all aspects of life, yielding numerous studies in bioinorganic chemistry, including medicinal chemistry (as reviewed in [39]). Metal ion cofactors may appear either as isolated ions or can be coordinated with either nonprotein organic compounds (e.g. porphyrin in hemoproteins) or with protein side chains (e.g. iron-sulfur clusters) and contrary to the limited repertoire of cations bound to ribozymes (magnesium, potassium, sodium, ammonium, etc), an impressive variety of metal ions are bound to proteins. Some examples of metalloenzymes are nitrite reductase, cytochrome oxidase, cytochrome ba3, azurin and other small electron transfer proteins that utilize cupric ferrous or ferric ions, other proteins that utilize copper and iron, such as catalase, hemoglobin and myoglobin; alcohol dehydrogenase, carbonic anhydrase, DNA polymerase and matrix proteins that utilize zinc; glucose 6-phosphatase and hexokinase that unitize magnesium; arginase (manganese); urease (nickel); glutathione peroxidase (selenium); alcohol dehydrogenase, and anti-HIV-1 and antitumor proteins MAP30 that contain manganese and zinc. Tungsten ions are less frequent in metalloenzymes. They appear in transport proteins, or act as a cofactor of Fe4S4 cluster, or participate in redox reactions or contribute to shielding, mostly in hyperthermophilic enzymes [40-44].
Metals and ribosomal flexibility
Mono and bivalent metal ions that are required for proper ribosomal activity [45, 46], seem to maintain the active conformation rather than catalyze the ribosomal main enzymatic activity, namely peptide bond formation [47]. Specifically, ions are located in proximity to various functionally relevant regions, or shown to stabilize structurally sensitive regions, such as the intersubunit interface as well as the kink in the mRNA path that is placed at the boundary between the A and P sites, which prevents mRNA slippage [49]. Thus, the mono and divalent metal ions that are vital for ribosome function are actually stabilizing the functional conformations, rather than contributing significantly to the enzymatic activity.
Despite the stabilization by mono and divalent ions, the ribosomes exhibit significant flexibility obtained by small and large internal motions. Examples of global motion that could be implicated from the high resolution structures include (a) the L1 stalk swing that allows the release of the exiting tRNA [4], (b) the latch-like mechanism that facilitates mRNA attachment and progression, which results from a substantial platform-head concerted motion, in which the entire subunit “head” seems to modulate between an ensemble of conformations (Figure 1) [2, 4, 5, 9, 10, 48, 49], (c) a ratchet-like motion of the entire subunit [5] that can be associated with the elongation cycle, in accord with previous cryo electron microcopy observations [50], and (d) a conformational modulation facilitated partially by the non ribosomal elongation factor G [51].
The linkage between crystal quality and heteropolytungstate clusters
The high resolution structures that emerged recently verify that ribosomes are precisely engineered machines that utilize conformational variability for optimizing their functional efficiency, but interfere with obtaining well ordered crystals. Among all ribosomal particles, the small subunit is the less suitable for crystallographic analysis, owing to its inherent conformational dynamics. Indeed, contrary to the marked tendency of large subunits to crystallize, only one crystal form has so far been obtained from the small subunit [52]. For example the crystals obtained from 70S ribosomes that were assembled from purified subunits, were found to consist solely of 50S subunits [53] whereas the supernatant of the crystallization drop contained, instead of intact small subunits, its isolated proteins and fragmented 16S rRNA chain. The low stability of the small subunit appears to be the reason for the poor resolution (about 10 Å) of the early crystals of the small ribosomal subunits from Thermus thermophilus, T30S [54-55]. It also seems to account for the unsuitability of the small ribosomal subunit cryo-EM reconstructions for extracting initial phase sets, although similar studies that were performed successfully for large ribosomal subunits [56].
Neither of the ribosomal crystal types that diffract to molecular resolution was obtained solely from purified ribosomal particles. In all cases additives had to be used in various fashions. For example, minute amounts of Cd++ in the crystallizing droplet led to a significant gain in the internal order (i.e. from 6 to ~2.8 Å) of the crystals of the large ribosomal subunits from Haloarcula marismortui, H50S, which contain over 3 M of monovalent ions (K+, Na+) [57]. More striking is the improvement of the resolution of the crystals of T30S, from 7-9 to 3 Å, by post crystallization treatments by minute amounts (less than 10−2 micromolar) of a heteropolytungstate cluster (see below and in [2].
Heteropolytungstates are of exceptional stability over a wide range of pH and redox states and may be considered as nanoscale magnets or function as catalysts in aqueous solutions [58-69]. Their effective phasing resolution in the absence of preferred orientation is limited to 4-5 Å, even when sophisticated spherical averaging techniques are being used [70]. Nevertheless, their symmetrical compounds yielded derivatives of biological macromolecules that were found useful for phasing, particularily in systems possessing a high internal symmetry that could correlate with that of the clusters [71-72]. Example are [(W3O2(O2CCH3)6]2+, which has trigonal symmetry and binds to the three fold axis and (NaP5W30O110)−14 that possesses an internal five fold symmetry that coincided with the 5-fold axis of the crystals of riboflavin synthase [73].
In the absence of internal symmetry the heteropolytungstate clusters bind to the ribosomal particles in non-specific manners, hence they were initially useful for phasing at low resolution. Among them, (NH4)6(P2W18O62)14H2O (called here W18) was found suitable for validating results obtained by electron microscopy combined with molecular replacement searches [74-77], or by previous identification at low resolution of smaller heavy atom compounds (e.g. tetrakis(acetoxymercuri)-methane, called also TAMM) bound to exposed sulfhydryls for localizing functional regions (i.e. the mRNA 5′end) or selected ribosomal proteins (i.e. S11 and S13) [78-81]. Later reports of compounds that generated medium and low resolution phases in ribosomal crystals include Na16[(O3PCH2PO3)4W12O36]40H2O), (Cs7(P2W17O61Co(NC5H5))14H2O) [52], (phSn)4(AsW9O33)2] and Cs5(PW11O39[Rh2CH3COO2] [77], H4SiO4[12WO], Li10(P2W17O61) [82], ((NH4)6(P2W18O62)14H2O) and ((TMA)2Na2[Nb2W4O19]18H2O) [76].
Contrary to the non-specific or non-complete binding reported above, post crystallization treatment of T30S crystals by minute amounts of W18 caused a dramatic improved in the crystalline order, expressed by increase in resolution from the initial 7-9 Å to 3 Å [2, 77, 76], indicating firm and quantitative attachment to well defined locations. Indeed, all of the fourteen clusters that bind to a single T30S particle were detected in close proximity to ribosomal-proteins, making numerous interactions with various side chains (mainly positively charged) that are exposed on the proteins surfaces and/or positioned along flexible extended loops or terminus extensions (Figure 2). In several cases the clusters trapped flexible protein termini that were found to be disordered in the non-tungstenated crystals in a manner that consequently influenced the rigidity of the rather flexible 30S ribosomal subunit (Figures 2 and 3).
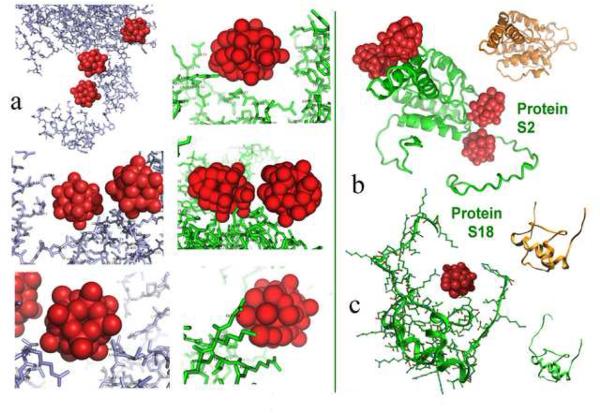
Figure 2.
(a) Typical heteropolytungstate interactions in both T30S and the LDL receptor extracellular domain. Note the similarity of the cluster “nests” created in the two systems. Also common is the high density of the tungsten clusters, which frequently appear very close to each other. (b) examples for conformational alterations induced by the cluster binding in two r-proteins: S2 and S18. For comparison the structures of the same proteins in the nontungstenated T30S [3] are also shown (in gold).
In all the heteropolytungstate clusters are shown as groups of 12 [83] or 18 [2] red balls, according to the number of the W atoms in the respective cluster 9 (Na3PW12O40 and (NH4)6(P2W18O62)14H2O). The LDL receptor extracellular domain is shown in metal blue. R-proteins S2 and S18 are shown in green in the tungstenated T30S [2] and in gold in the nontungstenated T30S [3]. Amino acid numbering for proteins of T30S is according to E. coli. In items showing highly dense regions, the numbering was removed, for clarity.
(b and c) show the structures of proteins S2 (b) and S18 (c) within the tungstenated [2] and non-tungstenated [3] T30S crystals in. For both the main chain is shown (in green and gold respectively). Note the marked difference in the conformation of the termini of protein S2. Also, note that all atoms of protein S18 are resolved in the tungstenated crystal, including those embracing the cluster, whereas over a dozen aminoacids are disordered in the nontungstenated crystal. The all-atom presentation is also shown, for highlighting typical “nest” architecture.
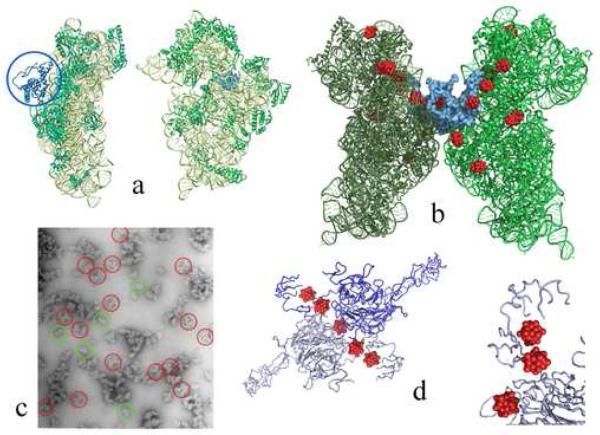
Figure 3.
The contacts holding together and rigidify the symmetry related pairs.
(a) Backbone representations of the front (right) and the side (left) views of T30S. The T30S particle is shown in green, except for protein S2 that is shown in blue.
(b) The pair of T30S (in green and dark green) related by the 2-fold rotation axis, with tungsten clusters (in red) bound to them. Protein S2, which is involved in the symmetrical interactions, is shown in a space-fill manner, in blue.
(c) An electron microscopy negatively stained image of the dissolved crystals. Examples for 30S pairs that maintain their crystallographic interactions are circles in red. The green circles indicate typical single particles.
(d) The symmetry relation within the crystals of LDL receptor extracellular domain after treatment with the heteropolytungstate cluster (red). In the bottom right, part of the structure is shown from a slightly different view, which represents the “nest” created by the protein for accommodating the cluster.
The post-crystallization treatment by minute amounts of W18 was performed under controlled heating, the common procedure for functional activation of ribosomal particles [45]. Nevertheless, the improvement in resolution was neither accompanied by alterations of the unit cell dimensions nor by crystal symmetry (a=b=407 Å, c=176 Å, P41212). However, data collected from the W18 treated crystals could not merge with data obtained from the native crystals, indicating that a major conformational rearrangement has occurred upon the W18 treatment, thus an apparent new crystal form [2] has been created. This indicates that although conformational changes are not routinely induced within crystals due to the limitation of the motion imposed by the crystal network, the T30S crystals not only tolerated but also benefited from the post-crystallization internal rearrangements.
All of the W18 clusters within the tungstenated crystals interact with ribosomal proteins, in positions that may significantly reduce the global mobility of the T30S particles within the crystal network. Among these, the interactions with protein S2 (Figures 2 and 3) were found crucial for the increase in resolution. In both native and the tungstenated T30S crystals, pairing of T30S particles positioned across the crystallographic 2-fold symmetry axis is a main feature of the crystallographic network. However, in contrast to the tungstenated crystals, in the native crystals the inter particle contacts that are formed between the two particles across the symmetry axis relating them, the “head” of the 30S particles (Figure 1) is still free to move [58].
Head motions have been shown to play an important role in ribosome function, as it was correlated with mRNA progression [4,5,9, 48,49]. Protein S2 is located on the subunit periphery, opposite to the RNA feature that seems to facilitate the head motions called the “neck” or helix H28 (Figure 3). The few interactions of protein S2 with this neck occur far from its flexible termini. Hence the four W18 clusters that bind to these termini are situated so that they cannot form any physical interaction with the neck or the head. However, the significant conformational changes (Figure 2) in these termini that were caused by the binding of the cluster, and their proximity to the 2-fold symmetry axis (Figure 3), fixed the flexible termini in an interwoven interaction network, which minimize the mobility of the entire vicinity. Thus, despite the large distance between the locations of the W18 clusters and the central feature acquiring head mobility, the extremely stable network of contacts around the crystallographic 2-fold symmetry axis limits head motions. Remarkably, this network of interactions remains in dissolved crystals, and therefore difficulties were encountered when attempting to fully dissolve the crystals and pairs of 30S particle could be seen by electron microscopy when inspecting the solution containing the dissolved crystals (Figure 3). Importantly, structural analysis indicated that the native crystals contain more than two different head conformations (Figure 1), whereas the tungstenated 30S particles are trapped at a specific conformation that was later found to mimic the conformation found in crystals of functionally active ribosomes [7, 9,10] and small subunits [3] (Figure 1). Likewise, one of the clusters bound to T30S fixes the conformation of the flexible termini of the r-protein S18 (Figure 2) in a fashion mimicking its involvement in the binding of the C-terminal domain of initiation factor IF3 [53].
Resolution increase was also obtained by treating crystals of LDL receptor extracellular domain with a somewhat smaller heteropolytungstate cluster, namely (Na3PW12O40) [83]. It appears that despite the significant differences between these two systems, namely a giant riboprotein complex vs. a single protein enzyme, both systems are utilizing comparable interactions in a similar mechanism, namely exploiting crystallographic symmetry for the trapping a specific conformation (Figures 2 and 3). In the case of the W18 tungstenated T30S, alongside the improvement of the internal order, individual W atoms could be resolved and therefore all bound 252 W atoms, the 868 oxygens and 28 phosphorous atoms in the 14 bound clusters could be efficiently used for phasing [2]. In contrast, in studies performed independently on T30S crystals obtained under the same conditions, the related compound that was used for phasing, Li10(P2W17O61), led to reduction rather than increase of the resolution [82].
An additional metal compound that yielded a similar increase in resolution of crystals of the small subunit is Os-hexamine chloride [82]. This compound has been also used for improving the order of other RNA crystals [25, 84], but contrary to the heteropolytungstates that bind to the surface and/or to flexible extension, loops and tails of proteins [2, 83], Os hexamine chloride interacts with RNA chains in a fashion that may increase their rigidity.
Conclusions
As an ion, tungsten does not exhibit exceptional affinity to proteins. In contrast, compounds containing negatively charged tungstate ions exhibit outstanding affinity to proteins, so that dense binding of a few clusters in close proximity is rather common (Figures 2 and 3). There is a preference of the tungstate clusters to bind to positively charged side chains (i. e. lysines and arginines), but interactions with alanines, glutamines and leucines were also detected (Figure 2). A marked tendency of long protein stretches to form multi-interactions “nests” that can embrace clusters has been observed in T30S as well as in the LDL receptor extracellular domain, which also packs as a dimer around a 2-fold symmetry axis and appears to be stabilized by the tungsten cluster [83] (Figure 3). As the binding of W18 to protein S18 created a similar “nest” for accommodating the heteropolytungstate cluster, this mode of interactions represents a common trait. Therefore it is conceivable that the polymetalates that serve as therapeutical agents (e.g. [85]) including providing antitumor activity [86] interfering with virus replication by inhibiting viral DNA and RNA polymerases [87,88] interact with their target proteins in a similar manner.
Finally, we would like to note that this study gives us a great pleasure, as it combines the determination of the high resolution structure of the ribosome with the work of one of our most distinguished mentors, the late F. Albert Cotton, who dedicated over 5 decades of his life to tungsten and similar metals [89, 90].
ACKNOWLEDGMENTS
Thanks are due to all members of the ribosome group at the Weizmann Institute for their constant assistance and to M. Pope for the heteropolytungstate clusters. Support was provided by the US National Inst. of Health (GM34360) and the Kimmelman Center for Macromolecular Assemblies. AY holds the Martin and Helen Kimmel Professorial Chair.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ban N, Nissen P, Hansen J, et al. Science. 2000;289:905. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 2.Schluenzen F, Tocilj A, Zarivach R, et al. Cell. 2000;102:615. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 3.Wimberly BT, Brodersen DE, Clemons WM, Jr., et al. Nature. 2000;407:327. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 4.Harms J, Schluenzen F, Zarivach R, et al. Cell. 2001;107:679. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 5.Schuwirth BS, Borovinskaya MA, Hau CW, et al. Science. 2005;310:827. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Hansen J, Ban N, et al. Science. 2000;289:920. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 7.Yusupov MM, Yusupova GZ, Baucom A, et al. Science. 2001;292:883. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 8.Bashan A, Agmon I, Zarivach R, et al. Mol Cell. 2003;11:91. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 9.Selmer M, Dunham CM, Murphy FV, Iv, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 10.Korostelev A, Trakhanov S, Laurberg M, et al. Cell. 2006;126:1065. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Hougland JL, Kravchuk AV, Herschlag D, et al. PLoS Biol. 2005;3:e277. doi: 10.1371/journal.pbio.0030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedor MJ, Williamson JR. Nat Rev Mol Cell Biol. 2005;6:399. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 13.Woodson SA. Curr Opin Chem Biol. 2005;9:104. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Pyle M. J Biol Inorg Chem. 2002;7:679. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 15.Rupert PB, Ferre-D'Amare AR. Nature. 2001;410:780. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 16.Hanna R, Doudna JA. Curr Opin Chem Biol. 2000;4:166. doi: 10.1016/s1367-5931(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 17.Leclerc F, Karplus M. J Phys Chem B. 2006;110:3395. doi: 10.1021/jp053835a. [DOI] [PubMed] [Google Scholar]
- 18.Lonnberg T, Lonnberg H. Curr Opin Chem Biol. 2005;9:665. doi: 10.1016/j.cbpa.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Cramer P, Bushnell DA, Kornberg RD. Science. 2001;292:1863. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 20.Gnatt AL, Cramer P, Fu J, et al. Science. 2001;292:1876. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 21.Fusz S, Eisenfuhr A, Srivatsan SG, et al. Chem Biol. 2005;12:941. doi: 10.1016/j.chembiol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Stahley MR, Strobel SA. Science. 2005;309:1587. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 23.Guo F, Gooding AR, Cech TR. Mol Cell. 2004;16:351. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Golden BL, Kim H, Chase E. Nat Struct Mol Biol. 2005;12:82. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 25.Cate JH, Gooding AR, Podell E, et al. Science. 1996;273:1678. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 26.Stahley MR, Strobel SA. Curr Opin Struct Biol. 2006;16:319. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Nahas MK, Wilson TJ, Hohng S, et al. Nat Struct Mol Biol. 2004;11:1107. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 28.Jiang YF, Xiao M, Yin P, et al. RNA. 2006;12:561. doi: 10.1261/rna.2188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serganov A, Polonskaia A, Phan AT, et al. Nature. 2006;441:1167. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travers KJ, Boyd N, Herschlag D. Rna. 2007;13:1205. doi: 10.1261/rna.566007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahley MR, Adams PL, Wang J, et al. J Mol Biol. 2007;372:89. doi: 10.1016/j.jmb.2007.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su LJ, Waldsich C, Pyle AM. Nucleic Acids Res. 2005;33:6674. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ke A, Ding F, Batchelor JD, et al. Structure. 2007;15:281. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Vicens Q, Cech TR. Trends Biochem Sci. 2006;31:41. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Kisseleva N, Khvorova A, Westhof E, et al. RNA. 2005;11:1. doi: 10.1261/rna.7127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldsich C, Pyle AM. J Mol Biol. 2008;375:572. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W, Lee JY, Nowotny M. Mol Cell. 2006;22:5. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Koculi E, Hyeon C, Thirumalai D, et al. J Am Chem Soc. 2007;129:2676. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farver O. Textbook of Drug Design and Discovery, Taylor and Francis Publisher. 2002:368. [Google Scholar]
- 40.Sugio T, Kuwano H, Hamago Y, et al. J Biosci Bioeng. 2004;97:378. doi: 10.1016/S1389-1723(04)70222-4. [DOI] [PubMed] [Google Scholar]
- 41.Chan MK, Mukund S, Kletzin A, et al. Science. 1995;267:1463. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- 42.Seiffert GB, Ullmann GM, Messerschmidt A, et al. Proc Natl Acad Sci U S A. 2007;104:3073. doi: 10.1073/pnas.0610407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy R, Adams MW. J Bacteriol. 2002;184:6952. doi: 10.1128/JB.184.24.6952-6956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bevers LE, Hagedoorn PL, Krijger GC, et al. J Bacteriol. 2006;188:6498. doi: 10.1128/JB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miskin R, Zamir A, Elson D. J Mol Biol. 1970;54:355. doi: 10.1016/0022-2836(70)90435-3. [DOI] [PubMed] [Google Scholar]
- 46.Bayfield MA, Dahlberg AE, Schulmeister U, et al. Proc Natl Acad Sci U S A. 2001;98:10096. doi: 10.1073/pnas.171319598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmeing TM, Huang KS, Kitchen DE, et al. Mol Cell. 2005;20:437. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Schuwirth BS, Day JM, Hau CW, et al. Nat Struct Mol Biol. 2006;13:879. doi: 10.1038/nsmb1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pioletti M, Schluenzen F, Harms J, et al. Embo J. 2001;20:1829. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank J. Chem Biol. 2000;7:R133. doi: 10.1016/s1074-5521(00)00127-7. [DOI] [PubMed] [Google Scholar]
- 51.VanLoock MS, Agrawal RK, Gabashvili IS, et al. J Mol Biol. 2000;304:507. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- 52.Yonath A, Harms J, Hansen HA, et al. Acta Crystallogr A. 1998;54:945. doi: 10.1107/s010876739800991x. [DOI] [PubMed] [Google Scholar]
- 53.Gluehmann M, Zarivach R, Bashan A, et al. Methods. 2001;25:292. doi: 10.1006/meth.2001.1241. [DOI] [PubMed] [Google Scholar]
- 54.Yonath A, Glotz C, Gewitz HS, et al. J Mol Biol. 1988;203:831. doi: 10.1016/0022-2836(88)90216-1. [DOI] [PubMed] [Google Scholar]
- 55.Trakhanov S, Yusupov M, Shirokov V, et al. J Mol Biol. 1989;209:327. doi: 10.1016/0022-2836(89)90282-9. [DOI] [PubMed] [Google Scholar]
- 56.Harms J, Tocilj A, Levin I, et al. Structure Fold Des. 1999;7:931. doi: 10.1016/s0969-2126(99)80120-8. [DOI] [PubMed] [Google Scholar]
- 57.von Bohlen K, Makowski I, Hansen HA, et al. J Mol Biol. 1991;222:11. doi: 10.1016/0022-2836(91)90730-t. [DOI] [PubMed] [Google Scholar]
- 58.Dawson B. Acta Crystallogr. 1953;6:113. [Google Scholar]
- 59.Pope MT, Papaconstantinou E. Inorg. Chem. 1967;6:1147. [Google Scholar]
- 60.D'Amour H. Acta Cryst. 1976;B32:729. [Google Scholar]
- 61.Brown GM, Noe Spirlet MR, Busing WR, et al. Acta. Crystallogr B. 1977;33:1038. [Google Scholar]
- 62.Contant R. Inorg. Synth. 1990;27:104. [Google Scholar]
- 63.Xin F, Pope MT. Organometallic. 1994;13:4881. [Google Scholar]
- 64.Wei X, Dickman MH, Pope MT. Inorg. Chem. 1997;36:130. [Google Scholar]
- 65.Zhang XY, O'Connor CJ, Jameson GB, et al. Inorg Chem. 1996;35:30. doi: 10.1021/ic950770p. [DOI] [PubMed] [Google Scholar]
- 66.Sazani G, Pope MT. Dalton Trans. 2004:1989. doi: 10.1039/b402421d. [DOI] [PubMed] [Google Scholar]
- 67.Muller A, Peters F, Pope MT, et al. Chem Rev. 1998;98:239. doi: 10.1021/cr9603946. [DOI] [PubMed] [Google Scholar]
- 68.Belai N, Sadakane M, Pope MT. J Am Chem Soc. 2001;123:2087. doi: 10.1021/ja005578n. [DOI] [PubMed] [Google Scholar]
- 69.Kogan V, Aizenshtat Z, Neumann R. New Journal of Chemistry. 2002;26:272. [Google Scholar]
- 70.Fu J, Gnatt AL, Bushnell DA, et al. Cell. 1999;98:799. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 71.Lowe J, Stock D, Jap B, et al. Science. 1995;268:533. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 72.Ladenstein R, Bacher A, Huber R. J. Mol. Biol. 1987;195:751. doi: 10.1016/0022-2836(87)90196-3. [DOI] [PubMed] [Google Scholar]
- 73.Alizadeh MH, Harmalker SP, Jeannin Y, et al. J. Am. Chem. Soc. 1985;107:2662. [Google Scholar]
- 74.Thygesen J, Weinstein S, Franceschi F, et al. Structure. 1996;4:513. doi: 10.1016/s0969-2126(96)00057-3. [DOI] [PubMed] [Google Scholar]
- 75.Janell D, Tocilj A, Kolln I, et al. In: Polyoxometalate Chemistry. Pope MT, editor. Kluwer Academic Publishers; 2001. p. 391. [Google Scholar]
- 76.Tocilj A, Schluenzen F, Janell D, et al. Proc Natl Acad Sci U S A. 1999;96:14252. doi: 10.1073/pnas.96.25.14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ban N, Nissen P, Hansen J, et al. Nature. 1999;400:841. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 78.Auerbach T, Pioletti M, Avila H, et al. J Biomol Struct Dyn. 2000;17:617. doi: 10.1080/07391102.2000.10506553. [DOI] [PubMed] [Google Scholar]
- 79.Bashan A, Agmon I, Zarivach R, et al. Cold Spring Harb Symp Quant Biol. 2001;66:43. doi: 10.1101/sqb.2001.66.43. [DOI] [PubMed] [Google Scholar]
- 80.Weinstein S, Jahn W, Glotz C, et al. J Struct Biol. 1999;127:141. doi: 10.1006/jsbi.1999.4135. [DOI] [PubMed] [Google Scholar]
- 81.Bartels H, Gluehmann M, Janell D, et al. Cell Mol Biol (Noisy-le-grand) 2000;46:871. [PubMed] [Google Scholar]
- 82.Clemons WM, Jr., May JL, Wimberly BT, et al. Nature. 1999;400:833. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 83.Rudenko G, Henry L, Henderson K, et al. Science. 2002;298:2353. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 84.Golden BL, Gooding AR, Podell ER, et al. Science. 1998;282:259. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 85.Jeffrey T, Hill CL, Judd DA. Chem. Res. 1998;327–357:98. [Google Scholar]
- 86.Wang X, Liu J, Li J, et al. J Inorg Biochem. 2003;279:94. doi: 10.1016/s0162-0134(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 87.Cherman JC, Sinoussi FC, Jasmin C. Biockem. Biophys. Res. Commun. 1975;1229:65. doi: 10.1016/s0006-291x(75)80361-5. [DOI] [PubMed] [Google Scholar]
- 88.Herve M, Sinoussi-Barre F, Chermann JC, et al. Biochem Biophys Res Commun. 1983;222:116. doi: 10.1016/0006-291x(83)90404-7. [DOI] [PubMed] [Google Scholar]
- 89.Cotton FA. Chemical Reviews. 1955:551. [Google Scholar]
- 90.Cotton FA, Donahue JP, Gruhn NE, Lichtenberger DL, Murillo CA, Timmons DJ, Van Dorn LO, Villagran D, Wang X. Inorg. Chem. 2006;45:201. doi: 10.1021/ic0515709. F. A. [DOI] [PubMed] [Google Scholar]