Abstract
Guanylyl cyclases (GCs) and adenylyl cyclases (ACs) have fundamental roles in a wide range of cellular processes. Whereas GCs use GTP as a substrate to form cGMP, ACs catalyze the analogous conversion of ATP to cAMP. Previously, a model based on the structure of adenylate cyclase was used to predict the structure of the nucleotide-binding pocket of a membrane guanylyl cyclase, RetGC-1. Based on this model, we replaced specific amino acids in the guanine-binding pocket of GC with their counterparts from AC. A change of two amino acids, E925K together with C995D, is sufficient to completely alter the nucleotide specificity from GTP to ATP. These experiments strongly validate the AC-derived RetGC-1 structural model and functionally confirm the role of these residues in nucleotide discrimination.
Guanylyl cyclases (GCs) and adenylyl cyclases (ACs) have fundamental roles in activating kinases and regulating ion channels in a wide range of cells. Whereas the GCs use GTP as a substrate to form cGMP, ACs catalyze the stereochemically analogous conversion of ATP to cAMP (1–4). Because cGMP and cAMP have distinct biological roles, rigid selectivity of the cyclases for GTP or ATP, but not both, is imperative to their cellular function. Little is known, however, about the molecular determinants of substrate discrimination in these enzymes. Mutations have been isolated in AC of R. mellioti that abolish or reduce NTP selectivity as well as reduce catalytic activity (5), but there is little other genetic or biochemical data on this subject.
Crystal structures of several nucleotide-binding proteins complexed with their substrates or analogs have yielded fruitful information concerning the determinants of nucleotide specificity. The crystal structure of the bacterial cAMP-binding catabolite gene activator protein (CAP) complexed with cAMP (6) provided a basis for mutagenesis of specificity determinants in the cyclic nucleotide-gated channels (7). This same crystal structure was also used as a starting reference for altering a cAMP-binding site of cAPK (8).
Although no structure for a GC has been determined, the crystal structure of a C2 domain homodimer of type II AC (AC2) was solved recently (9). Homology modeling of this structure was used to predict the structure of the catalytic domain of a representative GC, bovine RetGC-1 (10). RetGC-1 plays a key role in vertebrate phototransduction and is a member of the family of membrane guanylyl cyclases, which includes the natriuretic peptide receptors GC-A and GC-B (for review see ref. 11). Activation of RetGC-1 occurs intracellularly through two calcium-binding proteins, GCAP-1 and GCAP-2, at calcium concentrations below ≈500 nM (12–15).
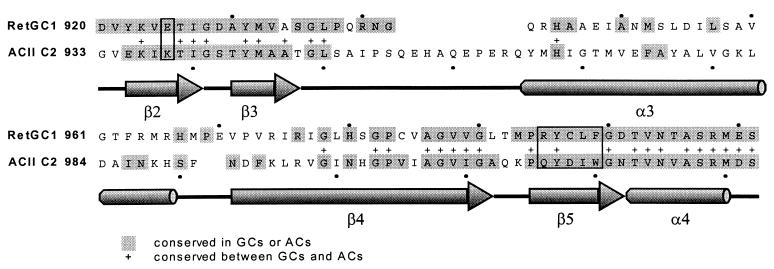
Modeling was used to dock GTP and ATP in best-fit conformations within the RetGC-1 model and AC C2 homodimer (10). These models suggested that several residues absolutely conserved in GC or AC but different between the two (boxed in Fig. 1) form a hydrogen bond network with the purine base of the nucleotide. This observation was recently reported independently by Tesmer et al. (16) based on their crystal structure of a VC1/IIC2 AC heterodimer complexed with an adenosine analogue.
Figure 1.
Sequence alignment of RetGC-1 and ACII C2 in catalytic domain. Amino acid residues conserved in all GCs or all ACs are denoted in gray. Residues conserved between the two types of cyclases are indicated with a “+”. Boxed are amino acids that approach the purine rings in the AC and GC models.
Structures derived by homology modeling and docking are tentative in the absence of corroborating experiments. We thus devised a mutagenesis strategy to test the veracity of the AC-derived RetGC-1 model and docked substrate. We replaced specific amino acids in the modeled guanine-binding pocket of RetGC-1 with their counterparts from AC. Substitution of two amino acids from AC changed the specificity of RetGC-1 from GTP to ATP while retaining GCAP-sensitive activation. Here we report a mutational and kinetic analysis of this substrate specificity change in RetGC-1.
MATERIALS AND METHODS
Modeling.
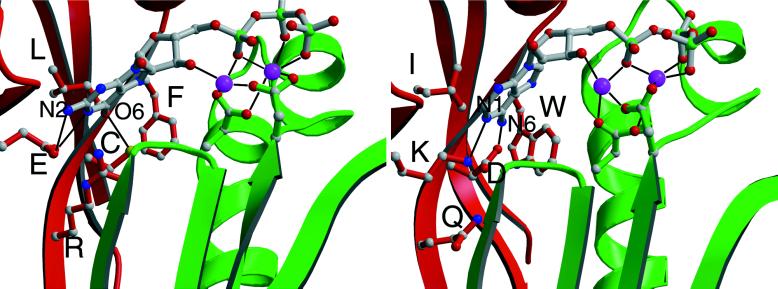
The starting point for the GTP complex was the protein model constructed previously (10), which was not altered. Two GTP molecules were docked into the same general location as they were previously. The conformation of the guanosine was modified to anti, as found for the adenosine moiety of the P-site inhibitor (16). Hydrogen-bonding constraints were incorporated between the guanine N1 and N2 and E925. A catalytic Mg2+ ion was incorporated by analogy to DNA polymerase (17). Fig. 2 was generated by using the program raster3d (18).
Figure 2.
Models of five residues in specificity-determining portion of active site of RetGC-1 (Left) and ACI (Right).
Mutagenesis and Expression in HEK293 Cells.
Signal overlap extension PCR (19) was used to introduce desired point mutations into human RetGC-1. A 750-bp fragment containing the mutation flanked by AatII and NdeI (for the E925K mutant) or NdeI and XbaI (for the mutations from 995 to 999) was subcloned into pBluescript (Stratagene) containing the full-length RetGC-1 cDNA in which the wild-type sequence between these sites had been removed. Clones were sequenced in the replaced region, and the entire RetGC-1 cDNA (3.6 kb) then was excised with HindIII/XbaI and subcloned into the eukaryotic expression plasmid pRC-CMV (Invitrogen). For the quadruple mutation, E/QYDIW, we used signal overlap extension PCR with E/QYDLF as a template. To make the mutants K/QYDIW, K/QYDLF, and K/RYDLF, we subcloned the 750-bp NdeI/XbaI fragment of E/QYDIW, E/QYDLF, and E/RYDLF into the vector pBluescript-RetGC-1 E925K, which had been cut at NdeI/XbaI. The 3.6-kb full-length GC-1 sequence containing the mutations was cut at HindIII/XbaI in pBluescript, excised, and subcloned into pRC-CMV.
Constructs were transiently transfected (calcium phosphate method) into human embryonic kidney (HEK293) cells. Cells were harvested after 48 hr essentially as described previously (20), except that instead of homogenizing, four strokes of a 26-gauge needle were used to lyse cells.
Western Blotting.
Equal amounts of total membrane protein were electrophoresed on a 7.5% SDS/PAGE gel and transferred to nitrocellulose membranes. Western blots were performed by using the polyclonal antibody CAT-Ab corresponding to M747-S1052 of RetGC-1. Preparation and characterization of this antibody is described elsewhere (R. P. Laura and J.B.H., unpublished data). The control lane labeled pRC-CMV contained membranes transfected with the plasmid pRC-CMV with no insert.
GC and AC Assays.
Transiently transfected membranes containing equal amounts of total protein were resuspended in GC buffer (100 mM KCl/50 mM Mops/7 mM 2-mercaptoethanol/10 mM MgCl/8 mM NaCl/1 mM EGTA). Measurement of guanylyl cyclase activity was carried out at 30°C for the indicated times essentially as described previously (20), except that ATP was omitted in the experiments shown in Fig. 3a. Stimulated reactions contained GCAP-2, which was prepared as described by Olshevskaya et al. (21). Measurement of adenylyl cyclase activity was the same as for GC activity, except ATP and cAMP (or radioactive forms) were substituted for GTP and cGMP. All experiments shown were repeated several times with similar results.
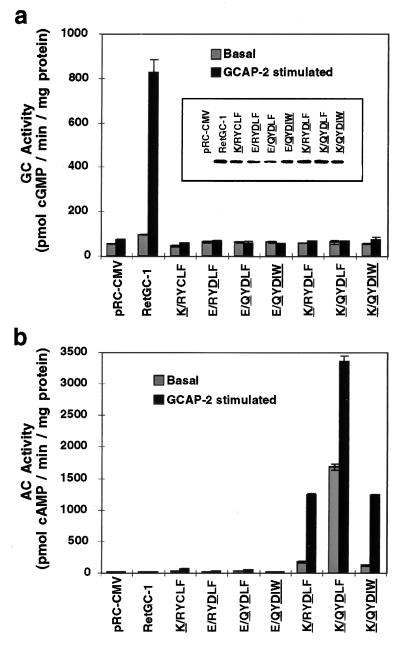
Figure 3.
GC and AC activities of wild-type and mutant RetGC-1 constructs. Membranes from transient transfections showing equivalent levels of expression of constructs (a Inset) were assayed for GC activity for 12 min (a) or AC activity for 30 min (b). Stimulated activity was measured in the presence of 4.1 μM GCAP-2. The changed residues in the mutant constructs are indicated in boldface and are underlined.
RESULTS
Mutant Design.
This study focused on five conserved residues that were predicted to form part of the guanine-binding site of RetGC-1: Glu-925, Arg-995, Cys-997, Leu-998, and Phe-999 (Fig. 2 Left). These five residues were sequentially changed to their AC counterparts, which are highly conserved among ACs but not conserved between ACs and GCs (Fig. 1). Residues thought to interact with the ribose and triphosphate of GTP and with the catalytic Mg2+ are conserved in both ACs and GCs, and were not altered. Of the five residues, the most prominent potential specificity determinant is Cys-997, which interacts with the O6 of guanine in the model. AC replaces this residue with an Asp that accepts a hydrogen bond from the adenine N6. The presence of the Asp in AC is expected to abolish guanine binding by an unfavorable electrostatic repulsion between the carboxylic acid side chain and the guanine O6. Glu-925 and Arg-995 are within salt bridge distance of each other, and both are near the edge of the purine ring containing the 1, 2, and 6 positions. In addition to stabilizing each other’s conformation, they approach three potential hydrogen bond partners on guanine: an acceptor at O6 and donors at N1 and N2. These residues are replaced by a Lys and a Gln, respectively, in AC. The AC Lys was predicted to interact with the hydrogen bond acceptor N1 on adenine and might discriminate against the protonated N1 and N2 of guanine. The AC Gln does not have a clear role in purine binding, but the Arg → Gln replacement serves to remove the guanine-binding Arg from the scene. The GC Leu and Phe do not appear to be directly involved in specific interactions; however, their invariance in GC and their spatial proximity to specific interactions led us to replace these by their AC counterparts, Ile and Trp.
Generation and Expression of Mutants.
We transfected wild-type and mutant proteins into HEK293 cells and demonstrated expression by using immunoblot analysis. We used an antibody to the RetGC-1 catalytic domain, CAT-Ab, which recognizes a single band of approximately 114 kDa corresponding to full-length RetGC-1. Wild-type and mutant proteins showed equivalent levels of expression in immunoblots (Fig. 3a Inset). Because we generated mutations in two regions of the GC-1 sequence, one within the β2–β3 loop of the RetGC-1 model (E925), and the others within the β4–β5 loop (R995, C997, L998, and F999), we refer to the wild-type RetGC-1 sequence as either “RetGC-1” or “E/RYCLF” and emphasize the mutated residues that correspond to the prototypical AC sequence in boldface and by underlining, e.g., K/QYDIW.
Measurement of GC and AC Activities.
We used equal amounts of total membrane protein to measure basal and stimulated GC and AC activities of expressed mutant and wild-type proteins. As a stimulator, we used a subsaturating amount of the calcium-binding protein GCAP-2, one of two isoforms in the retina that stimulate RetGC only at low calcium concentrations (15). Typically, we include ATP in our GC assays because RetGC-1 rapidly becomes insensitive to GCAP when incubated at physiological temperatures in the absence of ATP. Evidence suggests that the stabilizing effect of ATP is mediated through the kinase homology domain of RetGC-1 (22). We omitted ATP in the GC assay shown in Fig. 3a because we thought the presence of ATP could mask GTP binding if one of the mutants had a greater affinity for ATP than GTP. Thus, the GC activity of wild-type RetGC-1 we show here is less than we observe with ATP.
Fig. 3 shows that wild-type RetGC-1 (E/RYCLF) has significant GCAP-stimulated GC activity but no observable AC activity above basal levels. Neither GC nor AC activities could be detected with the single mutants E925K (K/RYCLF) and C997D (E/RYDLF), the double mutant R995Q;C997D (E/QYDLF), or the quadruple mutant R995Q; C997D; L998I; F999W (E/QYDIW), even at GTP or ATP concentrations of 10 mM (data not shown). In contrast, the K/QYDIW quintuple mutant had lost the ability to generate cGMP but exhibited high levels of AC activity when stimulated by GCAP-2.
We defined the minimal substitutions necessary for this change in specificity by generating the double and triple mutants K/RYDLF and K/QYDLF. K/RYDLF exhibits AC activity similar to the K/QYDIW mutant and likewise has no GC activity. This shows that only two amino acid substitutions, E925K with C997D, are required to completely change the specificity of RetGC-1 from GTP to ATP. The mutant K/QYDLF also has AC but no GC activity, but intriguingly has a 10-fold higher basal activity than the other specificity mutants and a 2-fold higher GCAP-stimulated activity. When we measured the half-saturation (K1/2) values for stimulation by GCAP-2, we found the K/QYDLF mutant is significantly more sensitive to GCAP-2. The average K1/2 [GCAP-2] value for K/QYDLF calculated from two separate experiments was 0.75 ± 1.5 mM, compared with 4.0 ± 1.8 mM for K/QYDIW and 2.9 ± 1.7 mM for K/QYDLF, or 5.4 ± 2.3 for wild-type RetGC-1 (20).
The high basal activity of the K/QYDLF mutant is significant but puzzling. K/QYDLF differs from the others only in that it has the combination of Gln from AC together with Leu and Phe from GC. We are not sure why the added presence of the Gln (or removal of the Arg) in this context causes heightened basal activity, because the R995Q change in the presence of the Ile and Trp (K/QYDIW) does not show enhanced activity.
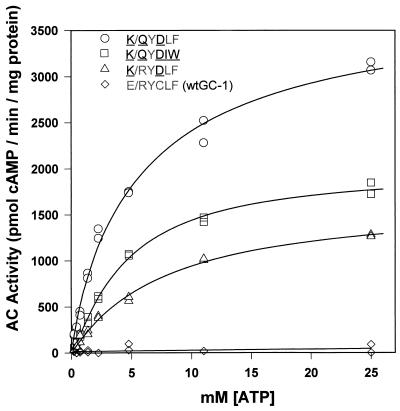
Measurement of Km.
To evaluate the catalytic abilities of the altered specificity mutants, we determined the Michaelis constants (Km) for ATP in the presence of GCAP-2 (Fig. 4). The Km values for ATP of K/QYDIW, K/RYDLF, and K/QYDLF were calculated to be between 3 and 8 mM, a 5- to 10-fold increase over wild-type RetGC-1 (Km [GTP] = 0.75 mM) (12). The Km [ATP] of AC I, in comparison, is 30–60 μM in the absence of the activator forskolin and approximately 10-fold higher with forskolin (23).
Figure 4.
Determination of Michaelis constants (Km) of altered specificity mutants. The activities of K/QYDIW, K/QYDLF, and K/RYDLF with increasing amounts of ATP were measured in the presence of 4.1 μM GCAP-2. A curve was fit to each plot by using the Hill equation v = [ATP]n Vmax/Kmn + [ATP]n. Km [ATP] values derived from the curve fit were 5.9 ± 1.6 mM for K/QYDLF, 4.5 ± 0.35 for K/QYDIW, and 7.7 ± 1.5 for K/RYDLF. In an independent set of experiments (not shown), we obtained comparable but slightly lower Km values of 3.6, 3.9, and 3.3 mM [ATP]. The Hill constants obtained from the curve fit were 0.89 ± 0.07, 1.19 ± 0.06, and 1.03 ± 0.07 for the respective mutants, which is similar to the noncooperative behavior we observe with wild-type RetGC-1.
DISCUSSION
The successful engineering of a highly active ATP-specific guanylyl cyclase with only two targeted mutations unambiguously confirms the roles of these residues in generating substrate specificity and strongly validates the AC-derived RetGC-1 structural model. The specificity change requires the presence of both mutations. The single mutations E925K or C997D completely knock out GC activity but alone are insufficient to confer AC activity. The single changes in AC I of K923A and D1000A, the counterparts to RetGC-1 E925 and C997, also decrease or abolish activity (23).
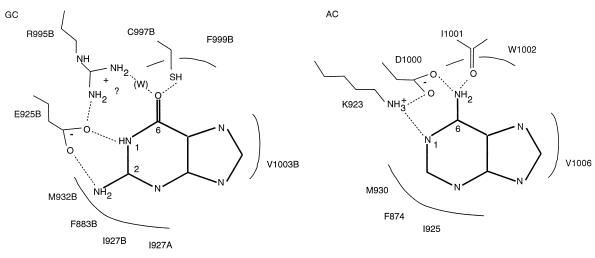
Based on the modeled RetGC-1 structure, we predicted that the introduction of Asp at 997 would be important for ATP binding and antagonistic to GTP binding. The role of the amino acid at position 925 in the RetGC-1 model was less clear. In the GC and AC models, this position approached the purine but did not directly hydrogen-bond to it. After we completed this study, the structure of adenylyl cyclase complexed with an adenosine analogue inhibitor was reported and showed a direct interaction between the adenine N1 and a Lys corresponding to E925 of RetGC-1 (16). On the basis of this structure, Tesmer et al. (16) proposed that the same two amino acid replacements described here would be responsible for the differences in specificity between GCs and ACs. This structure and our mutational data are consistent with a direct interaction between E925 and the N1 and N2 of the guanine ring (Fig. 5).
Figure 5.
Hypothesis for purine selectivity by guanylyl and adenylyl cyclases. Adapted from Liu et al. (10) and revised following Tesmer et al. (16) and a key role for the E925K mutant. The “A” or “B” after the RetGC-1 amino acid designation represents the polypeptide chain of the dimer.
A direct interaction between E925 and the guanine ring would be analogous to the finding by Zagotta and coworkers that an Asp interacting with the N1 and N2 of the cGMP guanine is essential for the cGMP-selective activation of rod cyclic-nucleotide-gated ion channels (7). These authors note that carboxylate interactions with guanine N1 and N2 are a common feature of high-affinity guanine nucleotide-binding proteins. This scheme of molecular recognition is widespread but not completely general. Corbin and coworkers (8) engineered a high-affinity cGMP-binding site in the regulatory subunit of protein kinase A by a single Ala → Thr replacement that created an interaction between the introduced OH and the guanine N2.
With the exception of the hyperactive mutant K/QYDLF, the specific activities of the altered specificity mutants were comparable with that of wild-type RetGC-1. That two of these mutants retain normal fold activation by GCAP shows that these changes are localized to the purine-binding pocket and have not altered the mechanism by which GCAP activates RetGC. The third AC-active mutant with higher basal activity has a reduced fold activation by GCAP-2 and lower K1/2 [GCAP-2] and could provide clues to the mechanism of activation. A similar high basal activity in a guanylyl cyclase was observed in a catalytic domain mutant of the atrial natriuretic peptide (ANP) receptor GC-A, but this protein was not ANP-responsive (24). The high basal activity of K/QYDLF could reflect a subtle change, possibly affecting the interface between the two GC monomers.
The measured Km [ATP] of the ATP-specific mutants were increased relative to the Km [GTP] of wild-type RetGC-1. Because these were steady-state measurements, the increase could be because of a decreased affinity for ATP or an increased rate of cyclization or product release. Further experiments to directly measure substrate binding would differentiate between these possibilities. At present, however, we cannot functionally isolate RetGC-1 from membranes, and measurement of substrate-binding affinities from membrane preparations would be unfeasible.
It is often difficult to engineer complete specificity changes in enzymes while at the same time maintaining wild-type levels of activity (25). Our successful conversion of a GC into an AC based on the RetGC-1 structural model provides strong validation for the veracity of the AC-derived structure. In doing so, it clarifies the key differences between adenylyl and guanylyl cyclases while increasing our confidence in the similarity of active-site details. Further studies will be necessary to compare and contrast the mechanisms of regulation in the two enzymes.
Acknowledgments
These studies were supported by National Institutes of Health Grant EY06641 to J.B.H.
ABBREVIATIONS
- GC
guanylyl cyclase
- AC
adenylyl cyclase
- CAP
catabolite gene activator protein
- GCAP
guanylyl cyclase activator protein
Note Added in Proof
The Km measurements shown in Fig. 4 were generated using 10 mM MgCl2. Because the substrate for the cyclase reaction may be a complex of Mg2+ with ATP, we were concerned that Mg2+ may have been limiting in those experiments at the highest ATP concentrations. To address this possibility, we repeated the experiment using 50 mM MgCl2 and obtained Km values of 14.0 ± 3.4 for K/QYDLF, 23.8 ± 10.1 for K/RYDLF, and 24.6 ± 7.9 for K/QYDIW.
References
- 1.Eckstein F, Romaniuk P J, Heideman W, Storm D R. J Biol Chem. 1981;256:9118–9120. [PubMed] [Google Scholar]
- 2.Gerlt J A, Coderre J A, Wolin M S. J Biol Chem. 1980;255:331–334. [PubMed] [Google Scholar]
- 3.Senter P D, Eckstein F. J Biol Chem. 1983;258:6741–6745. [PubMed] [Google Scholar]
- 4.Koch K-W, Eckstein F, Stryer L. J Biol Chem. 1990;265:9659–9663. [PubMed] [Google Scholar]
- 5.Beuve A, Danchin A. J Mol Biol. 1992;225:933–938. doi: 10.1016/0022-2836(92)90093-y. [DOI] [PubMed] [Google Scholar]
- 6.Weber I T, Steitz T A. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 7.Varnum M D, Black K D, Zagotta W N. Neuron. 1995;15:619–625. doi: 10.1016/0896-6273(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 8.Shabb J B, Ng L, Corbin J D. J Biol Chem. 1990;265:16031–16034. [PubMed] [Google Scholar]
- 9.Zhang G, Liu Y, Ruoho A E, Hurley J H. Nature (London) 1997;386:247–253. doi: 10.1038/386247a0. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Ruoho A E, Rao V D, Hurley J H. Proc Natl Acad Sci USA. 1997;94:13414–13419. doi: 10.1073/pnas.94.25.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garbers D L, Lowe D G. J Biol Chem. 1994;269:30741–30744. [PubMed] [Google Scholar]
- 12.Dizhoor A M, Lowe D G, Olshevskaya E V, Laura R P, Hurley J B. Neuron. 1994;12:1345–1352. doi: 10.1016/0896-6273(94)90449-9. [DOI] [PubMed] [Google Scholar]
- 13.Gorczyca W A, Gray-Keller M P, Detwiler P B, Palczewski K. Proc Natl Acad Sci USA. 1994;91:4014–4018. doi: 10.1073/pnas.91.9.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palczewski K, Subbaraya I, Gorczyca W A, Helekar B S, Ruiz C C, Ohguro H, Huang J, Zhao X, Crabb J W, Johnson R S, et al. Neuron. 1994;13:395–404. doi: 10.1016/0896-6273(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 15.Dizhoor A M, Olshevskaya E V, Henzel W J, Wong S C, Stults J T, Ankoudinova I, Hurley J B. J Biol Chem. 1995;270:25200–25206. doi: 10.1074/jbc.270.42.25200. [DOI] [PubMed] [Google Scholar]
- 16.Tesmer J J G, Sunahara R K, Gilman A G, Sprang S R. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 17.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 18.Merritt E A, Murphy E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 19.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Laura R P, Dizhoor A M, Hurley J B. J Biol Chem. 1996;271:11646–11651. doi: 10.1074/jbc.271.20.11646. [DOI] [PubMed] [Google Scholar]
- 21.Olshevskaya E V, Hughes R E, Hurley J B, Dizhoor A M. J Biol Chem. 1997;272:14327–14333. doi: 10.1074/jbc.272.22.14327. [DOI] [PubMed] [Google Scholar]
- 22.Tucker C L, Laura R P, Hurley J B. Biochemistry. 1997;36:11995–12000. doi: 10.1021/bi971212k. [DOI] [PubMed] [Google Scholar]
- 23.Tang W-J, Stanzel M, Gilman A G. Biochemistry. 1995;34:14563–14572. doi: 10.1021/bi00044a035. [DOI] [PubMed] [Google Scholar]
- 24.Wedel B J, Foster D C, Miller D E, Garbers D L. Proc Natl Acad Sci USA. 1997;94:459–462. doi: 10.1073/pnas.94.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedstrom L. Curr Opin Struct Biol. 1994;4:608–611. [Google Scholar]