Summary
Dimethylallyltransferase (DMATase) transfers a five-carbon isoprenoid moiety from dimethylallyl pyrophosphate (DMAPP) to the amino group of adenosine at position 37 of certain tRNAs. Reported here are the crystal structures of Pseudomonas aeruginosa DMATase alone and in complex with pyrophosphate at 1.9 angstrom resolution. Surprisingly, the enzyme possesses a central channel spanning the entire width of the enzyme. Both the accepting substrate tRNA and the donating substrate DMAPP appear to enter the channel from opposite sides in an ordered sequence, with tRNA first and DMAPP second. And the RNA modification reaction occurs in the middle of the channel once the two substrates have met. The structure of DMATase is homologous to a class of small soluble kinases involved in biosynthesis of nucleotide precursors for nucleic acids, indicating its possibly evolutionary origin. Furthermore, specific recognition of the pyrophosphate by a conserved loop in DMATase, similar to the P-loop commonly seen in diverse nucleotide-binding proteins, demonstrates that DMATase is structurally and mechanistically distinct from farnesyltransferase, another family of prenyltransferases involved in protein modification.
Keywords: structural biology, RNA modification, enzyme mechanism, protein channel, tRNA
Introduction
Among the 107 modified nucleotides discovered in RNA to date 1; 2, several are hypermodified requiring more than one-step enzymatic reactions to achieve their final products. These hypermodified nucleotides are usually found near the tRNA anticodon, either at the wobble position (position 34) or at the 3' adjacent to the anticodon. The biological roles of these hypermodifications in tRNA are believed to enhance specific codon:anticodon recognition between mRNA and tRNA, a critical step that ensures high fidelity and specificity of protein translation by the ribosome 3; 4.
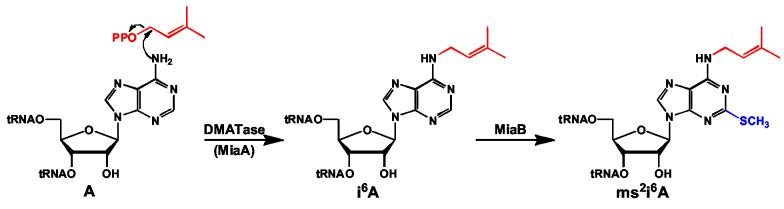
DMATase (named MiaA in E. coli) converts an adenosine (A) to an N6-isopentenyladenosine (i6A) at position 37 of all tRNAs that read codons beginning with uridine 5; 6. i6A is further converted to 2-methylthio-N6-isopentenyladenosine (ms2i6A) by a bifunctional enzyme named MiaB in E. coli 7; 8 (Fig. 1). DMATase belongs to a large family of enzymes called prenyltransferases that catalyze the alkylation of electron-rich acceptors by the hydrophobic moiety of allylic isoprenoid pyrophosphate 9; 10. Within this family, farnesyltransferase (FTase) is the enzyme most relevant to the study reported here. Both DMATase and FTase are engaged in modifications of macromolecules (tRNAs for DMATase and proteins for FTase) and both utilize the allylic isoprenoid pyrophosphates as their carbon sources for the reactions. FTase is a well-studied Zn2+-dependent enzyme that is involved in the modification of various, important signaling proteins 11; 12. Structural studies indicate that FTase is an all α-helical protein 13. Recognition of the pyrophosphate moiety is achieved through direct interactions with side chains of some conserved amino acids 14. Several lines of evidence suggested that DMATase functions differently from FTase. First, unlike FTase, DMATase is Mg2+ dependent. Moreover, DMATase binds acceptor substrate tRNA first and allylic substrate DMAPP second 15. To investigate the mechanism by which DMATase functions, we determined crystal structures of DMATase from P. aeruginosa alone and in complex with pyrophosphate. We find that DMATase has a fold different from FTase, and that the fold most closely resembles that of some kinases. Further, we find that recognition of the pyrophosphate moiety of the substrate is achieved via a GxTxxGK(T/S) motif similar to that found in a wide variety of NTP binding proteins 16; 17; 18. Based on structures and biochemical experiments 15; 19; 20, we here propose a mechanism.
Figure 1.
The chemical reaction carried out by DMATase. The dimethylallyl moiety in DMAPP is transferred to the amino group of A37 in certain tRNAs, resulting in the formation of i6A, which is further modified by a bifunctional enzyme MiaB in E. coli to produce ms2i6A.
Results and Discussion
Structure of P. aeruginosa DMATase
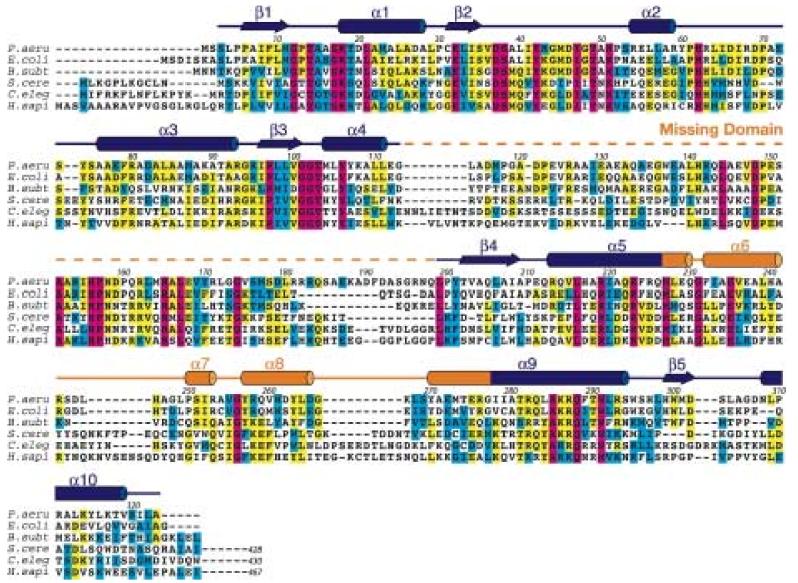
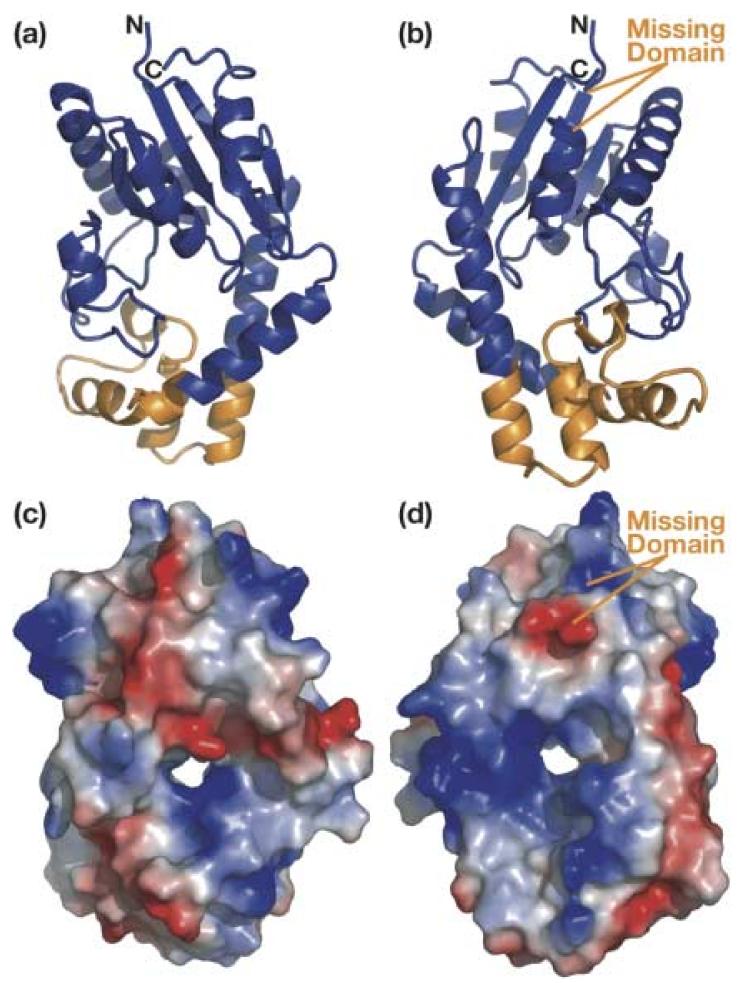
The crystal structure of DMATase from P. aeruginosa was determined using multiple wavelength anomalous dispersion (MAD) phasing, and the structure of the DMATase alone has been refined to 1.9 Å resolution (R/Rfree = 21.1/23.0%; see Table 1). Several additional data sets have also been collected, from crystals grown in the presence of DMAPP or DMASPP (dimethylallyl S-thiopyrophosphate), or from crystals that were soaked with DMAPP or DMASPP. Thus, two additional structures (DMATase in complex with a pyrophosphate, an Mg2+ ion, and a Tris molecule; and DMATase in complex with a pyrophosphate and an Mg2+ ion) were also refined (Table 1). These three structures are essentially identical except for the presence of ligands in the latter two. Electron density map did not show residues 114–198 (Fig. 2, dashed line in orange). Inspection of crystal packing indicates that there is sufficient room in the crystal to accommodate the missing domain (Supporting information, Fig. S1). Furthermore, SDS gel analysis of dissolved crystals reveals that DMATase within the crystal is intact (Supporting information, Fig. S2), indicating that the missing domain (residues 114-198) is structurally disordered in our crystals and that is why it is not observed. As discussed in later section, the missing domain is likely to be involved in tRNA substrate binding, and it is possible that it is ordered only in the presence of tRNA substrate. The structure of the remaining protein (residues 3-113 and 199-323) consists of ten α helices and five β strands (Fig. 2). The five β strands form a parallel β-sheet, and six of the ten helices flank both sides of the β-sheet [Fig. 3(a), 3(b)]. They constitute the top portion of the structure, which is structurally homologous to a class of kinases involved in biosynthesis of precursors for nucleic acids 21; 22 [Fig. 3(a), 3(b), colored blue; Fig. 4]. Three additional helices (α6, α7, and α8) form the bottom portion of the structure, which is unique to DMATase [Fig. 3(a), 3(b), colored orange]. The missing domain is located between α4 and β4 [Fig. 3(b); Fig. 2]. A search through the Protein Data Bank (PDB) did not yield any deposited structure with sequence homology to the missing domain.
Table 1.
Statistics of data collection and refinementa
| Crystals | SeMet DMATase |
DMATase | DMATase + PP + Tris |
DMATase + PP | ||
|---|---|---|---|---|---|---|
| Inflection | Peak | Remote | ||||
| Space group | P41 | |||||
| Unit cell | 76.42, 76.42, 57.26 | 76.1, 76.1, 58.3 | 76.2, 76.2, 57.8 | 76.3, 76.3, 57.5 | ||
| 90.00, 90.00, 90.00 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | |||
| Resolution (Å) | 50.0 – 2.2 | 50.0 – 2.2 | 50.0 – 2.2 | 50.0 – 1.9 | 50.0 – 1.9 | 50.0 – 2.2 |
| Wavelength (Å) | 0.9794 | 0.9793 | 0.9747 | 0.9793 | 0.9793 | 0.9793 |
| Unique reflections | 16,957 | 16,912 | 16,853 | 25,837 | 25,178 | 16,969 |
| Redundancy | 12.2 | 12.4 | 12.1 | 6.5 | 6.3 | 5.0 |
| Completeness (%) | 100.0 (99.9) |
100. 0 (99.9) |
99.3 (96.2) |
98.1 (92.3) | 96.0 (88.2) | 99.9 (99.4) |
| Average I/σ (I)b | 10.2 (8.0) | 10.9 (9.8) | 9.1 (6.4) | 15.4 (5.8) | 15.5 (5.3) | 14.3 (7.5) |
| Rsymc (%) | 8.8 (25.8) | 8.4 (22.4) | 7.9 (27.8) | 7.3 (28.7) | 6.5 (28.8) | 5.5 (22.9) |
| Refinement | ||||||
| Resolution (Å) | 50.0 – 1.9 | 50.0 – 1.9 | 50.0 – 2.2 | |||
| Reflections (free) | 21,738 (1,876) | 20,885 (1,768) | 14,484 (1,242) | |||
| Rcrystald (Rfreee) (%) | 21.1 (23.0) | 20.9 (23.5) | 22.0 (25.0) | |||
| r.m.s.d. bonds (Å) | 0.0049 | 0.0060 | 0.0056 | |||
| r.m.s.d. angles (°) | 1.15 | 1.29 | 1.15 | |||
Mean figure-of-merit (FOM) for phasing = 0.53 from SOLVE and 0.76 after RESOLVE.
I/σ (I) is the mean reflection intensity/estimated error.
, where I is the intensity of an individual reflection and <I> is the average intensity over symmetry equivalents.
, where Fo and Fc are the observed and calculated structure factor amplitudes.
Rfree is equivalent to Rcrystal but calculated for a randomly chosen set of reflections that were omitted from the refinement process.
Figure 2.
Conservation of DMATase from bacteria to human. The sequence of P. aeruginosa DMATase is compared with representatives of other prokaryotic (E. coli and B. subtilis) and eukaryotic (S. cerevisiae, C. elegans and human) DMATases. Residue number over the alignment corresponds to P. aeruginosa DMATase. The conserved residues are boxed in color, with completely conserved residues in magenta, identical residues in yellow and similar residues in cyan. The secondary structure of DMATase is depicted schematically above the primary sequence, with α-helices highlighted as cylinders and β-sheets as arrows. The putative RNA binding domain disordered in our structure is in dashed line. The eukaryotic DMATase contains an additional C-terminal domain of roughly 100 amino acids (not shown).
Figure 3.
Structure of P. aeruginosa DMATase. (a) Ribbon representation of the structure from the DMAPP binding side. The top portion of the protein that is homologous to the kinase is in blue and the bottom portion that is unique to DMATase is in orange. (b) Ribbon representation of the structure from the tRNA binding side [180° vertical rotation of (a)]. An additional putative RNA binding domain that is disordered in our structure is labeled in orange. (c) & (d) Surface representation of the structure with the same orientation as in (a) and (b), respectively. The positively charged surface is in blue and the negatively charged is in red.
Figure 4.
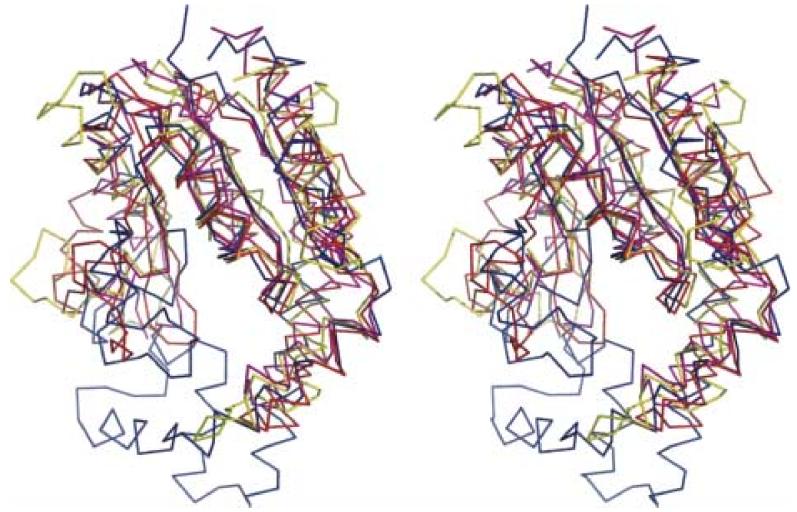
Stereoview of Cα superposition of the DMATase structure with the three top scores from Dali structural homology search. DMATase is in blue, S. cerevisiae GMP kinase (PDB entry 1GKY, Z core = 12.1, rmsd = 2.9 Å) is in red, human UMP/CMP kinase (PDB entry 1TEV, Z core = 11.8, rmsd = 3.3 Å) is in magenta, and L. major CMP kinase (PDB entry 1Y63, Z core = 10.9, rmsd = 3.2 Å) is in yellow.
As expected, the structure of the portion homologous to the kinase [Fig. 3(a), 3(b), colored blue] forms a cleft that contains the conserved P-loop, the site of ATP binding in kinases. Addition of the structural component of the bottom portion, however, closes the cleft and results in formation of a channel [Fig. 3(c), 3(d)]. As discussed in detail later, this channel is where the substrates meet and the RNA modification reaction occurs.
Dali structural homology search 23 reveals that the top three scores are all small (less than 200 amino acids) kinases that catalyze phosphorylation of NMP (N = G, C and U) at the expense of ATP (PDB entry 1GKY, 1TEV, 1Y63) 21; 22. These enzymes play a critical role in the pathway that supplies nucleotide precursors for DNA and RNA synthesis. With the exception of the conserved P-loop close to the N-terminus, DMATase does not show significant sequence similarity to these three enzymes. Additional differences between DMATase and these kinases arise from two sources. One is the bottom portion of the DMATase structure that connects α5 and α9 [Fig. 3(a), 3(b), colored orange]; this portion coincides with a short loop in kinases (Fig. 4). The other is the missing domain that connects α4 and β4 [Fig. 3(b)], which, again, is present as a short loop in kinases.
Recognition of Pyrophosphate in DMAPP by DMATase
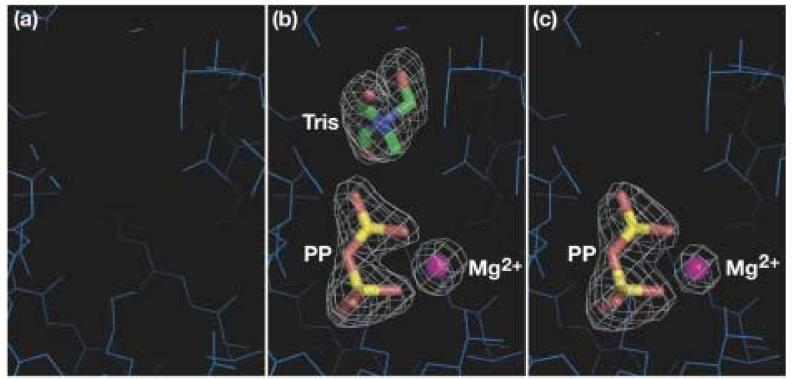
Incorporation of DMAPP or DMASPP into the channel of DMATase was unsuccessful despite extensive efforts. Crystallization of DMATase with its substrate or inhibitor was carried out in the presence of 2.5 mM DMAPP or DMASPP in Tris buffer. However, instead of a DMAPP or DMASPP, a pyrophosphate, an Mg2+ ion, and a Tris molecule were found in the channel [Fig. 5(b)]. Interestingly, binding of Tris by DMATase requires the presence of the pyrophosphate because no Tris was found in the channel when DMATase was crystallized alone [Fig. 5(a)]. In order to eliminate a remote possibility that the binding of the combined pyrophosphate and Tris is somehow preferred by DMATase to that of DMAPP or DMASPP, we have carried out soaking experiments. We first soaked the crystals of DMATase (crystallized in the absence of any ligand) in HEPES buffer to eliminate Tris. We then soaked the Tris-free crystals with 2.5 mM DMAPP or DMASPP for three hours. In this case, a pyrophosphate and an Mg2+ ion, but not Tris, were bound in the channel [Fig. 5(c)]. Spectroscopic analysis of the remaining DMAPP and DMASPP showed the vast majority of the compounds were intact, indicating that the source of the pyrophosphate bound in the channel of DMATase was likely from the residual pyrophosphate impurity in the purchased compounds. Therefore, the logical explanation of our failure to incorporate DMAPP or DMASPP into the channel of DMATase is that DMATase is incapable of binding DMAPP in the absence of a tRNA substrate. This is consistent with the binding assay carried out by Moore and Poulter 15.
Figure 5.
No DMAPP found in the channel in the absence of tRNA substrate. Close-up view of the structure of the DMATase channel (a) crystallized alone; (b) crystallized in the presence of DMAPP or DMASPP; (c) crystallized alone but soaked with HEPES buffer followed by DMAPP or DMASPP soaking. DMATase is represented by thin, blue line. Pyrophosphate and Tris are shown in stick representation and the Mg2+ ion is in small sphere. Atoms for the ligands are colored individually, with carbon in green, nitrogen in blue, oxygen in red, phosphate in yellow, and magnesium ion in magenta. The contour of the simulated annealing omit electron maps in (b) and (c) is 5.0σ.
Likely Binding of tRNA Substrate by DMATase
Our structural analysis, together with mutational studies carried out by Soderberg and Poulter 20, shed light on the possible mechanism of tRNA binding by DMATase. tRNA most likely approaches DMATase from the opposite side of the channel relative to where the pyrophosphate binds because this side contains many positively charged residues [shown in blue, Fig. 3(d)], which are likely to complement the negatively charged tRNA substrate. Furthermore, the missing domain, which we presume to be the RNA binding domain, is also located on this side [Fig. 3(d)]. This is consistent with the mutational studies carried out by Soderberg and Poulter, where they reported that most mutations affecting the Km of tRNA were found either on this side of DMATase or in the missing domain 20.
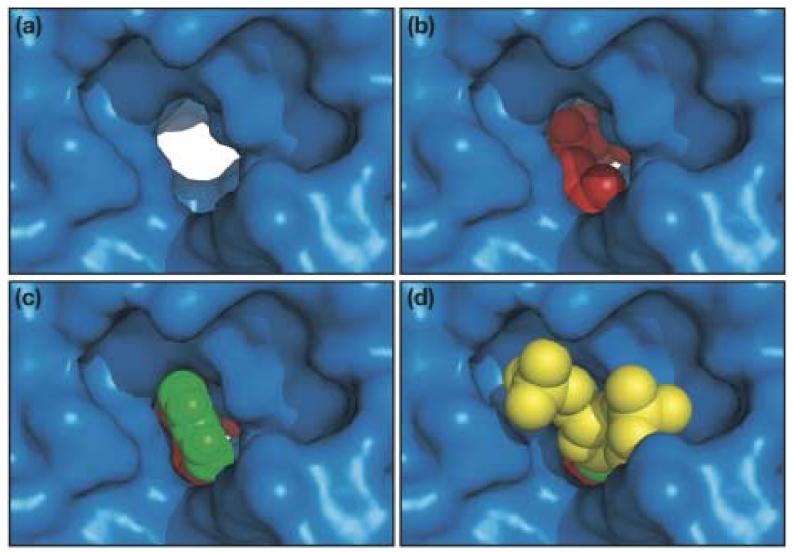
We constructed a docking model in which the targeted nucleotide A37 was manually docked onto the structure of DMATase from this side of the channel. Docking the entire tRNA, or even the anticodon stem-loop, was not possible because of the unstructured nature of the missing domain. The base of A37 fits very well into the channel [Fig. 6(c)]. Our model shows that the base of A37 is sandwiched between the side chains of the conserved L284 and S38 residues. The amino group of A37, where the modification occurs, is in hydrogen bonding distance with the side chain of the conserved D37, a likely general base for the reaction. The strictly conserved Q288 is also in hydrogen bonding distance with the amino group and the N1 of A37, which makes Q288 to be the possible recognition element of A37. The ribose of A37 and the attached phosphate groups also fit nicely within the surrounding platform near the entrance of the channel [Fig. 6(d)].
Figure 6.
Close-up view of the channel from the RNA binding side with (a) nothing bound; (b) modeled DMAPP, an Mg2+ ion, and three water molecules coordinated to the Mg2+ ion; (c) everything in (b) and the modeled base of A37; (d) everything in (c) and the modeled ribose and 5'- and 3'-phosphates of A37. DMATase is depicted in a surface representation and is in the same orientation as in Fig. 3(d). The molecules in the channel are in spheres, with DMAPP and water molecules in red, Mg2+ in magenta, base A37 in green, and the ribose and phosphates of A37 in yellow.
We also modeled in the DMA moiety, together with the bound pyrophosphate, resulting in a model of a DMAPP bound to DMATase [Fig. 6(b)]. We found that one of the terminal methyl groups of the modeled DMAPP fit tightly between the side chains of two conserved hydrophobic residues, M227 and V255. Since DMATase is incapable of binding DMAPP without the tRNA substrate, the association of the tRNA with DMATase, especially through flipping the base of A37 into the channel, must cause a subtle but important conformational change in the DMA binding pocket to allow DMAPP to be accommodated. Therefore, although the approximate location of DMA moiety is likely to be as shown in Fig. 6(b), the detailed interactions of DMA moiety with the surrounding amino acids in DMATase should be interpreted with caution.
Proposed Mechanism of DMATase-Catalyzed tRNA Modification Reaction
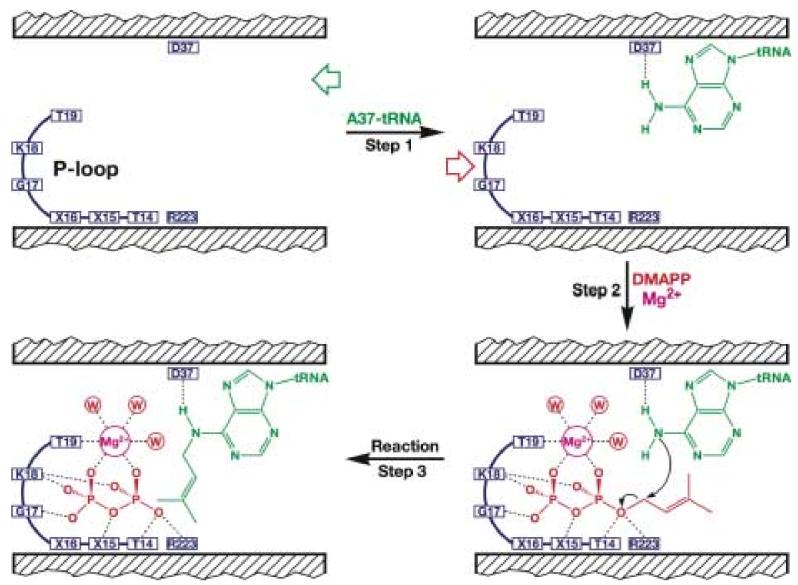
In docking A37 and the DMA moiety, we have been guided by our structure and biochemical studies by Poulter and coworkers. If our model is correct, it predicts a likely mechanism of substrate binding and catalysis carried out by DMATase as follows. First, the acceptor substrate tRNA approaches DMATase from the end of the channel opposite where the pyrophosphate binds. Association of tRNA with DMATase, achieved through interaction with the positively charged amino acids on the surface of DMATase and in the missing domain, allows the base of A37 to enter the channel through a base-flipping mechanism commonly seen in other DNA and RNA modification enzymes 24; 25; 26 (Fig. 7, Step 1). The conserved D37 residue forms a hydrogen bond with the amino group of A37. The equivalent D37A mutation in the E. coli enzyme resulted in 20-fold reduction of the enzymatic activity 20. Second, subtle but important conformational changes in DMATase caused by tRNA binding allow the donating substrate DMAPP to enter the opposite end of the channel (Fig. 7, Step 2). Recognition of the pyrophosphate moiety in DMAPP is achieved by interacting with the conserved P-loop in DMATase, as well as coordinating with an Mg2+ ion. Particularly relevant are the hydrogen bonding interactions between the side chains of the conserved T14 and R223 residues and the bridging oxygen in DMAPP (Fig. 7). The equivalent T14A and R223A mutations in the E. coli enzyme resulted in 600- and 25-fold reduction of the enzymatic activity, respectively 20. Third, activation of the carbon atom that is directly attached to the pyrophosphate by T14 and R223 allows the carbon atom to accept the electron-rich amino group of A37 (Fig. 7, Step 3). Thus, the possible roles of the side chains of T14 and R223 are to stabilize the leaving pyrophosphate group, and to activate the carbon atom directly connected to the pyrophosphate during the reaction. On the other hand, D37 is likely to act as the general base by abstracting a proton from the amino group in A37.
Figure 7.
Proposed mechanism of DMATase-catalyzed reaction. The tRNA substrate binds to the right side of channel and the base of A37 flips into the channel (Step 1). DMAPP enters the channel from the left side and is stabilized by interacting with the P-loop as well as by coordinating with an Mg2+ ion (Step 2). Nucleophilic attack of the amino group in A37 on the DMA moiety of DMAPP results in the DMA moiety to be transferred from DMAPP to the A37 of tRNA, made possible by activation of the carbon atom directly linked to the pyrophosphate in DMAPP by the side chains of T14 and R223, as well as deprotonation of the amino group in A37 with the side chain of D37 (Step 3).
Possibly Evolutionary Origin of DMATase
Dali search indicates that the top portion of DMATase is structurally homologous to a class of small soluble kinases that are involved in biosynthesis of nucleotide precursors for nucleic acids (Fig. 4). Furthermore, the recognition of pyrophosphate moiety of DMAPP is achieved by a conserved GxTxxGK(T/S) motif, similar to the P-loop in kinases as well as other nucleotide binding proteins. Therefore, it is not hard to envision that DMATase could have evolved from an ancient kinase-like protein with only two insertions. One insertion is in the loop between α5 and α9 of the progenitor protein. This insertion would have closed the ATP binding site in the progenitor protein, resulting in formation of the channel in DMATase. The other insertion is in the loop between α4 and β4. This insertion would have created a domain between α4 and β4, which is missing in our structure but is presumably required for the binding of tRNA substrate. Sequence alignment of DMATase from diverse organisms appears to support the second insertion, as the amino acid sequences immediately after α4, and immediately before β4, do not align well (existence of gaps) (Fig. 2).
Despite structural similarity of DMATase to kinases based on Dali search (Fig. 4), DMATase does not show significant sequence similarity to these enzymes except for the conserved N-terminus, where the P-loop of kinases is located. Therefore, the only function remaining from the evolutionary precursor protein appears to be the recognition of the di- or triphosphate in ATP by the conserved P-loop, which is now employed by DMATase for the recognition of the pyrophosphate moiety in DMAPP. Interestingly, the P-loop in DMATase also appears to have evolved because it is not exactly the same P-loop seen in kinases. The conserved sequence of the P-loop in the kinases is GxxxxGK(T/S) 21; 22. This is also the conserved sequence of P-loop in ATPases 16 as well as G-proteins 17; 18, indicating that the recognition of di-phosphate in NTP has been conserved during evolution of the diverse NTP-binding proteins. The P-loop in DMATase, however, has an additional conservation, a threonine (T) at the third amino acid of the standard P-loop in NTP-binding proteins [GxTxxGK(T/S) instead of GxxxxGK(T/S), Fig. 2]. This is the amino acid (T14 in P. aeruginosa DMATase) that plays the most important role in catalysis (Fig. 7).
In summary, the structural studies of DMATase presented here reveal that DMATase possesses a central channel spanning the width of the enzyme where the tRNA modification reaction occurs. This is the first reported RNA or DNA modifying enzyme that utilizes a channel to carry out its reaction. In addition, DMATase sequentially binds the acceptor tRNA substrate at one entrance of the channel and then the donating substrate DMAPP at the opposite entrance. The presence of this central channel and the ordered sequential binding of substrates are two unique features of DMATase when compared with other known prenyltransferases or other transferases involved in RNA and DNA modification. Furthermore, the recognition of the pyrophosphate moiety in DMAPP is achieved by a P-loop, indicating it is mechanistically distinct, in terms of substrate binding, from other prenyltransferases.
Materials and Methods
Cloning, Overexpression and Purification of P. aeruginosa DMATase
The open reading frame (ORF) of the putative DMATase from P. aeruginosa was identified from a BLAST search of the NCBI database based on sequence homology to the published E. coli miaA gene 6. The gene was amplified from the P. aeruginosa genomic DNA (purchased from ATCC, GeneID 15600138) and cloned into a pLM-1 vector between EcoRI and HindIII restriction sites 27.
Recombinant DMATase-pLM-1 vector was transformed into BL21(DE3) E. coli expression cells. A 10 mL overnight culture was used to inoculate 2 L of LB supplemented with 50 μg/mL ampicillin. Cells were cultured at 25 °C until OD600nm reached 0.6 and then induced with 1 mM IPTG for 12 hours. Cells were harvested by centrifugation at 5,000 × g for 20 minutes.
Purification of DMATase using a FPLC system was conducted as follows: cell pellet was suspended in DEAE buffer A (50 mM Tris-HCl, pH 8.5, 1 mM EDTA, 1 mM PMSF) and lysed using a French press. Cell lysate was centrifuged, and the supernatant was passed through a preparatory DEAE anion exchange column. The protein was eluted with DEAE buffer B (same as DEAE buffer A except 1 M NaCl). Fractions containing DMATase were pooled and injected into a HiLoad hydrophobic column equilibrated with HiLoad buffer A (20 mM Tris-HCl, pH 7.8, 1.0 M (NH4)2SO4, 1 mM DTT). Proteins were eluted with HiLoad buffer B [same as HiLoad buffer A except no (NH4)2SO4]. Fractions containing DMATase were pooled, concentrated, and washed with DEAE buffer A. The sample was then applied to a MonoQ anion exchange column and the same DEAE buffers were used to elute proteins. Finally, fractions containing DMATase from MonoQ were purified by a Superdex 200 size exclusion column using a buffer containing 20 mM HEPES (pH 7.0), 200 mM NaCl, and 1 mM DTT. The enzymatic activity of the purified DMATase was confirmed by an in vitro assay using the in vitro transcribed E. coli tRNATyr and DMAPP (purchased from Echelon Biosciences Inc., Salt Lake City, Utah) as substrates.
Selenomethionine labeled protein was prepared using the following protocol: 5 mL of overnight culture was used to inoculate 2 liters of minimal media supplemented with ampicillin and thiamine at final concentrations of 100 mg/L and 50 mg/L, respectively. The cell culture was grown at 25 °C to OD600nm of 0.6 and 40 mL of methionine suppression buffer (comprised of 200 mg of L-Lys-HCl, L-Threonine and L-Phenylalanine, and 100 mg of L-Leucine, L-Isoleucine and L-Valine in water) was added. After another 30 minutes of shaking at 25 °C, the cell culture was induced with 0.5 mM IPTG and 50 mg/L of selenomethionine was added. Incubation at 25 °C was continued for another 18 hours and the cells were harvested. The selenomethionine labeled protein was purified analogous to the native protein.
Crystallization, Data Collection, and Structural Determination
Crystals of DMATase alone were obtained at both 4 °C and 18 °C using the hanging drop vapor diffusion method by mixing DMATase (6 mg/mL) with equal volume of a well solution containing 18 % PEG6000 (w/v), 0.1 M NaCl, 0.1 M Tris-HCl, pH 8.5. Crystals of selenomethionine-labeled protein were obtained under similar conditions. Diamond-shaped crystals were harvested after four weeks of growth. In order to carry out data collection at a low temperature, the crystals were step-wise soaked in a cryo-protecting solution containing all the components of the well solution supplemented with 5-to-20 % glycerol. The cryo-protected crystals were then mounted in a nylon loop and flash-frozen in liquid nitrogen. Both the native and three-wavelength anomalous dispersion (MAD) data were collected at the 22-ID beamline at the Advanced Photon Source (APS). Data were reduced with Denzo and Scalepack 28. For phase determination, the resolution range from 30 to 2.2 Å was chosen. Nine of the eleven expected sites (assuming DMATase acts as a monomer in an asymmetric unit) were located using program Solve 29. Phases were improved with solvent flattening and ∼85 % of the model was automatically built using program Resolve 30. Several rounds of manual building using program O 31, followed by refinement with program CNS 32, resulted in a final model with an R-factor of 21.1 % (Rfree = 23.0 %). Final refinement statistics are given in Table I.
Crystals of DMATase in complex with a pyrophosphate and a Tris molecule were grown analogously to DMATase alone, with the exception that 2.5 mM of DMAPP or DMASPP was present in the crystallization drop. An attempt to soak DMAPP or DMASPP into the channel of DMATase was conducted using crystals of DMATase alone. The crystals were first soaked with a solution containing 18 % PEG6000 (w/v), 0.1 M NaCl, 0.1 M HEPES, pH 8.0, in order to eliminate the Tris molecules within the crystals. The crystals were then soaked in the same solution in the presence of 2.5 mM DMAPP or DMASPP for 3 hours. The crystals were harvested similarly to DMATase alone, with the exception that DMAPP or DMASPP was present in the cryo-protecting solution. Structures were refined using the structure of DMATase alone as the starting model (Table I).
Construction of a Docking Model
The nucleotide A37 and its 3'-linked phosphate group from the structure of yeast tRNAiMet (PDB entry 1YFG) were employed for a docking model. The base A37 was first manually docked into the channel from the RNA binding side using program PyMOL 33. The ribose was then rotated through the glycosylic bond that links to the base to make it fit the surrounding structure better. Finally, the position of the phosphate group of A37 is adjusted through the rotation of the O5'-C5' bond. The Dimethylallyl (DMA) group was also manually modeled into the channel of DMATase approximately where the Tris was bound. The oxygen atom in the bound pyrophosphate that bridges PP and DMA in DMAPP was identified based on its interaction with the side chains of the conserved T14 and R223.
Supplementary Material
Acknowledgements
We thank the staffs of beamline 22ID at APS (J. Chrzas, Z. Jin) for their support during data collection, W. van de donk and R. Gumport for critical reading of the manuscript. This work was supported by the NIH (grant CA90954 to R.H.H.).
Abbreviations used
- DMATase
Dimethylallyltransferase
- DMAPP
dimethylallyl pyrophosphate
- DMASPP
dimethylallyl S-thiopyrophosphate
- FTase
farnesyltransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–96. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crain PF, Rozenski J, McCloskey JA. The RNA Modification Database. 2006 on World Wide Web http://medlib.med.utah.edu/RNAmods/
- 3.Curran JF. Modified nucleosides in translation. In: Grosjean H, Benne B, editors. Modification and Editing of RNA. ASM Press; Washington, D.C.: 1998. pp. 493–516. [Google Scholar]
- 4.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–73. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum N, Gefter ML. Delta 2 -isopentenylpyrophosphate: transfer ribonucleic acid 2 -isopentenyltransferase from Escherichia coli. Purification and properties of the enzyme. J. Biol. Chem. 1972;247:5675–80. [PubMed] [Google Scholar]
- 6.Caillet J, Droogmans L. Molecular cloning of the Escherichia coli miaA gene involved in the formation of delta 2-isopentenyl adenosine in tRNA. J. Bacteriol. 1988;170:4147–52. doi: 10.1128/jb.170.9.4147-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierrel F, Bjork GR, Fontecave M, Atta M. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 2002;277:13367–70. doi: 10.1074/jbc.C100609200. [DOI] [PubMed] [Google Scholar]
- 8.Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 2004;279:47555–63. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 9.Poulter CD, Rilling HC. Prenyl transferases and isomerase. In: Spurgeon SL, editor. Biosynthesis of Isoprenoid Compounds. Vol. 1. John Wiley & Sons; New York: 1981. pp. 161–224. [Google Scholar]
- 10.Liang PH, Ko TP, Wang AH. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002;269:3339–54. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 11.Glomset JA, Farnsworth CC. Role of protein modification reactions in programming interactions between ras-related GTPases and cell membranes. Annu. Rev. Cell Biol. 1994;10:181–205. doi: 10.1146/annurev.cb.10.110194.001145. [DOI] [PubMed] [Google Scholar]
- 12.Schafer WR, Rine J. Protein prenylation: genes, enzymes, targets, and functions. Annu. Rev. Genet. 1992;26:209–37. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- 13.Park HW, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science. 1997;275:1800–4. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 14.Long SB, Casey PJ, Beese LS. Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate. Biochemistry. 1998;37:9612–8. doi: 10.1021/bi980708e. [DOI] [PubMed] [Google Scholar]
- 15.Moore JA, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: a binding mechanism for recombinant enzyme. Biochemistry. 1997;36:604–14. doi: 10.1021/bi962225l. [DOI] [PubMed] [Google Scholar]
- 16.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–8. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 17.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–45. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 18.Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–32. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 19.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry. 2000;39:6546–53. doi: 10.1021/bi992775u. [DOI] [PubMed] [Google Scholar]
- 20.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: site-directed mutagenesis of highly conserved residues. Biochemistry. 2001;40:1734–40. doi: 10.1021/bi002149t. [DOI] [PubMed] [Google Scholar]
- 21.Stehle T, Schulz GE. Refined structure of the complex between guanylate kinase and its substrate GMP at 2.0 A resolution. J. Mol. Biol. 1992;224:1127–41. doi: 10.1016/0022-2836(92)90474-x. [DOI] [PubMed] [Google Scholar]
- 22.Segura-Pena D, Sekulic N, Ort S, Konrad M, Lavie A. Substrate-induced conformational changes in human UMP/CMP kinase. J. Biol. Chem. 2004;279:33882–9. doi: 10.1074/jbc.M401989200. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Protein structure comparison by allignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 24.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–69. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 25.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, Liu X, Huang RH. Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate. Nat. Struct. Biol. 2003;10:781–8. doi: 10.1038/nsb976. [DOI] [PubMed] [Google Scholar]
- 27.MacFerrin KD, Chen L, Terranova MP, Schreiber SL, Verdine GL. Overproduction of proteins using the expression-cassette polymerase chain reaction. Methods Enzymol. 1993;217:79–102. doi: 10.1016/0076-6879(93)17057-c. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Methods in Enzymology. Vol. 277. Academic Press; San Diego, CA: 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta. Cryst. 1999;D55:849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC. Reciprocal-space solvent flattening. Acta Cryst. 1999;D55:1863–71. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. The PyMOL Molecular Graphic System. 2002 on World Wide Web http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.