Abstract
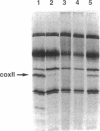
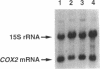
The ability to replace wild-type mitochondrial DNA sequences in yeast with in vitro-generated mutations has been exploited to study the mechanism by which the nuclearly encoded PET111 protein specifically activates translation of the mitochondrially coded COX2 mRNA. We have generated three mutations in vitro that alter the COX2 mRNA 5'-untranslated leader (UTL) and introduced them into the mitochondrial genome, replacing the wild-type sequence. None of the mutations significantly affected the steady-state level of COX2 mRNA. Deletion of a single base at position -24 (relative to the translation initiation codon) in the 5'-UTL (cox2-11) reduced COX2 mRNA translation and respiratory growth, whereas insertion of four bases in place of the deleted base (cox2-12) and deletion of bases -30 to -2 (cox2-13) completely blocked both. Six spontaneous nuclear mutations were selected as suppressors of the single-base 5'-UTL deletion, cox2-11. One of these mapped to PET111 and was shown to be a missense mutation that changed residue 652 from Ala to Thr. This suppressor, PET111-20, failed to suppress the 29-base deletion, cox2-13, but very weakly suppressed the insertion mutation, cox2-12. PET111-20 also enhanced translation of a partially functional COX2 mRNA with a wild-type 5'-UTL but a mutant initiation codon. Although overexpression of the wild-type PET111 protein caused weak suppression of the single-base deletion, cox2-11, the PET111-20 suppressor mutation did not function simply by increasing the level of the protein. These results demonstrate an intimate functional interaction between the translational activator protein and the mRNA 5'-UTL and suggest that they may interact directly.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman S. H., Gatti D. L., Gellefors P., Douglas M. G., Tzagoloff A. ATP13, a nuclear gene of Saccharomyces cerevisiae essential for the expression of subunit 9 of the mitochondrial ATPase. FEBS Lett. 1991 Jan 28;278(2):234–238. doi: 10.1016/0014-5793(91)80124-l. [DOI] [PubMed] [Google Scholar]
- Anziano P. Q., Butow R. A. Splicing-defective mutants of the yeast mitochondrial COXI gene can be corrected by transformation with a hybrid maturase gene. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5592–5596. doi: 10.1073/pnas.88.13.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M., Grivell L. A. Genetic approaches to the study of mitochondrial biogenesis in yeast. Antonie Van Leeuwenhoek. 1992 Aug;62(1-2):131–153. doi: 10.1007/BF00584467. [DOI] [PubMed] [Google Scholar]
- Bordonné R., Dirheimer G., Martin R. P. Expression of the oxi1 and maturase-related RF1 genes in yeast mitochondria. Curr Genet. 1988 Mar;13(3):227–233. doi: 10.1007/BF00387768. [DOI] [PubMed] [Google Scholar]
- Cabral F., Schatz G. Identification of cytochrome c oxidase subunits in nuclear yeast mutants lacking the functional enzyme. J Biol Chem. 1978 Jun 25;253(12):4396–4401. [PubMed] [Google Scholar]
- Cabral F., Solioz M., Rudin Y., Schatz G., Clavilier L., Slonimski P. P. Identification of the structural gene for yeast cytochrome c oxidase subunit II on mitochondrial DNA. J Biol Chem. 1978 Jan 10;253(1):297–304. [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G., Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence of subunit 2 of yeast cytochrome oxidase. J Biol Chem. 1979 Sep 25;254(18):9324–9330. [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986 Nov;6(11):3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Specific translational activation by nuclear gene products occurs in the 5' untranslated leader of a yeast mitochondrial mRNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Suppression of a defect in the 5' untranslated leader of mitochondrial COX3 mRNA by a mutation affecting an mRNA-specific translational activator protein. Mol Cell Biol. 1993 Aug;13(8):4806–4813. doi: 10.1128/mcb.13.8.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Seaver E. C., Fox T. D. The PET54 gene of Saccharomyces cerevisiae: characterization of a nuclear gene encoding a mitochondrial translational activator and subcellular localization of its product. Genetics. 1989 Jun;122(2):297–305. doi: 10.1093/genetics/122.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P. J., Papadopoulou B., Grivell L. A. In-vitro translation of mitochondrial mRNAs by yeast mitochondrial ribosomes is hampered by the lack of start-codon recognition. Curr Genet. 1993 Jan;23(1):22–27. doi: 10.1007/BF00336745. [DOI] [PubMed] [Google Scholar]
- Dekker P. J., Papadopoulou B., Grivell L. A. Properties of an abundant RNA-binding protein in yeast mitochondria. Biochimie. 1991 Dec;73(12):1487–1492. doi: 10.1016/0300-9084(91)90182-z. [DOI] [PubMed] [Google Scholar]
- Dekker P. J., Stuurman J., van Oosterum K., Grivell L. A. Determinants for binding of a 40 kDa protein to the leaders of yeast mitochondrial mRNAs. Nucleic Acids Res. 1992 Jun 11;20(11):2647–2655. doi: 10.1093/nar/20.11.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow N. D., Michaels G. S., Montoya J., Attardi G., O'Brien T. W. Mechanism of mRNA binding to bovine mitochondrial ribosomes. J Biol Chem. 1989 May 15;264(14):8328–8338. [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Douglas M. G., Butow R. A. Variant forms of mitochondrial translation products in yeast: evidence for location of determinants on mitochondrial DNA. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1083–1086. doi: 10.1073/pnas.73.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992 Jul;13(1):18–20. [PubMed] [Google Scholar]
- Folley L. S., Fox T. D. Site-directed mutagenesis of a Saccharomyces cerevisiae mitochondrial translation initiation codon. Genetics. 1991 Nov;129(3):659–668. doi: 10.1093/genetics/129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Folley L. S., Mulero J. J., McMullin T. W., Thorsness P. E., Hedin L. O., Costanzo M. C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Genetic and physical analysis of the mitochondrial gene for subunit II of yeast cytochrome c oxidase. J Mol Biol. 1979 May 5;130(1):63–82. doi: 10.1016/0022-2836(79)90552-7. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Sanford J. C., McMullin T. W. Plasmids can stably transform yeast mitochondria lacking endogenous mtDNA. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7288–7292. doi: 10.1073/pnas.85.19.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Staempfli S. Suppressor of yeast mitochondrial ochre mutations that maps in or near the 15S ribosomal RNA gene of mtDNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1583–1587. doi: 10.1073/pnas.79.5.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Haffter P., Fox T. D. Suppression of carboxy-terminal truncations of the yeast mitochondrial mRNA-specific translational activator PET122 by mutations in two new genes, MRP17 and PET127. Mol Gen Genet. 1992 Oct;235(1):64–73. doi: 10.1007/BF00286182. [DOI] [PubMed] [Google Scholar]
- Haffter P., McMullin T. W., Fox T. D. A genetic link between an mRNA-specific translational activator and the translation system in yeast mitochondria. Genetics. 1990 Jul;125(3):495–503. doi: 10.1093/genetics/125.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., McMullin T. W., Fox T. D. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991 Feb;127(2):319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. M., Clark-Walker G. D. Nucleotide sequence of the cytochrome oxidase subunit 2 and val-tRNA genes and surrounding sequences from Kluyveromyces lactis K8 mitochondrial DNA. Yeast. 1990 Sep-Oct;6(5):403–410. doi: 10.1002/yea.320060505. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Johnston S. A., Anziano P. Q., Shark K., Sanford J. C., Butow R. A. Mitochondrial transformation in yeast by bombardment with microprojectiles. Science. 1988 Jun 10;240(4858):1538–1541. doi: 10.1126/science.2836954. [DOI] [PubMed] [Google Scholar]
- Liao H. X., Spremulli L. L. Effects of length and mRNA secondary structure on the interaction of bovine mitochondrial ribosomes with messenger RNA. J Biol Chem. 1990 Jul 15;265(20):11761–11765. [PubMed] [Google Scholar]
- Liao H. X., Spremulli L. L. Interaction of bovine mitochondrial ribosomes with messenger RNA. J Biol Chem. 1989 May 5;264(13):7518–7522. [PubMed] [Google Scholar]
- McMullin T. W., Fox T. D. COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J Biol Chem. 1993 Jun 5;268(16):11737–11741. [PubMed] [Google Scholar]
- McMullin T. W., Haffter P., Fox T. D. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990 Sep;10(9):4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Le Corre D., Panvert M., Blanquet S., Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis U., Körte A., Rödel G. Association of cytochrome b translational activator proteins with the mitochondrial membrane: implications for cytochrome b expression in yeast. Mol Gen Genet. 1991 Nov;230(1-2):177–185. doi: 10.1007/BF00290666. [DOI] [PubMed] [Google Scholar]
- Mittelmeier T. M., Dieckmann C. L. In vivo analysis of sequences necessary for CBP1-dependent accumulation of cytochrome b transcripts in yeast mitochondria. Mol Cell Biol. 1993 Jul;13(7):4203–4213. doi: 10.1128/mcb.13.7.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero J. J., Fox T. D. PET111 acts in the 5'-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993 Mar;133(3):509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K., Schnier J. A temperature-sensitive mutant in the gene rplX for ribosomal protein L24 and its suppression by spontaneous mutations in a 23S rRNA gene of Escherichia coli. EMBO J. 1986 Jun;5(6):1373–1376. doi: 10.1002/j.1460-2075.1986.tb04369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmen J. D., Kloeckener-Gruissem B., McEwen J. E. Molecular cloning and nucleotide sequence of the nuclear PET122 gene required for expression of the mitochondrial COX3 gene in S. cerevisiae. Nucleic Acids Res. 1988 Nov 25;16(22):10783–10802. doi: 10.1093/nar/16.22.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou B., Dekker P., Blom J., Grivell L. A. A 40 kd protein binds specifically to the 5'-untranslated regions of yeast mitochondrial mRNAs. EMBO J. 1990 Dec;9(12):4135–4143. doi: 10.1002/j.1460-2075.1990.tb07636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Ely K. R. Control of translational repression by protein-protein interactions. Nucleic Acids Res. 1992 Apr 11;20(7):1649–1655. doi: 10.1093/nar/20.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutre C. G., Fox T. D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987 Apr;115(4):637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratje E., Mannhaupt G., Michaelis G., Beyreuther K. A nuclear mutation prevents processing of a mitochondrially encoded membrane protein in Saccharomyces cerevisiae. EMBO J. 1983;2(7):1049–1054. doi: 10.1002/j.1460-2075.1983.tb01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D. Post-transcriptional steps in the expression of chloroplast genes. Annu Rev Cell Biol. 1992;8:1–28. doi: 10.1146/annurev.cb.08.110192.000245. [DOI] [PubMed] [Google Scholar]
- Rogers D., Bussey H. Fidelity of conjugation in Saccharomyces cerevisiae. Mol Gen Genet. 1978 Jun 14;162(2):173–182. doi: 10.1007/BF00267874. [DOI] [PubMed] [Google Scholar]
- Rödel G., Fox T. D. The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5' untranslated leader. Mol Gen Genet. 1987 Jan;206(1):45–50. doi: 10.1007/BF00326534. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Kindle K. L., Stern D. B. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of beta-glucuronidase translational fusions. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevarino K. A., Poyton R. O. Mitochondrial membrane biogenesis: identification of a precursor to yeast cytochrome c oxidase subunit II, an integral polypeptide. Proc Natl Acad Sci U S A. 1980 Jan;77(1):142–146. doi: 10.1073/pnas.77.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K. W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991 May;5(5):773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Shen Z. H., Fox T. D. Substitution of an invariant nucleotide at the base of the highly conserved '530-loop' of 15S rRNA causes suppression of yeast mitochondrial ochre mutations. Nucleic Acids Res. 1989 Jun 26;17(12):4535–4539. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub J. M., Maliga P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 1993 Feb;12(2):601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick C. A., Fox T. D. Saccharomyces cerevisiae positive regulatory gene PET111 encodes a mitochondrial protein that is translated from an mRNA with a long 5' leader. Mol Cell Biol. 1987 Aug;7(8):2728–2734. doi: 10.1128/mcb.7.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993 May;134(1):21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. D., Jefferson L. M. Elevated mitochondrial RNA in a Chinese hamster mutant deficient in the mitochondrially encoded subunits of NADH dehydrogenase and cytochrome c oxidase. J Biol Chem. 1990 Nov 5;265(31):18852–18859. [PubMed] [Google Scholar]
- Yaffe M. P. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]