Abstract
The balance between the inductive signals and endogenous anti-apoptotic mechanisms determines whether or not programmed cell death occurs. The widely expressed inhibitor of apoptosis gene family includes three closely related mammalian proteins: c-IAP1, c-IAP2, and hILP. The anti-apoptotic properties of these proteins have been linked to caspase inhibition. Here we show that one member of this group, hILP, inhibits interleukin-1β-converting enzyme-induced apoptosis via a mechanism dependent on the selective activation of c-Jun N-terminal kinase 1. These data demonstrate that apoptosis can be inhibited by an endogenous cellular protein by a mechanism that requires the activation of a single member of the mitogen-activating protein kinase family.
Programmed cell death or apoptosis is a fundamental event in the developmental and homeostatic processes of all multicellular organisms (1, 2). Activation of programmed cell death is mediated by the caspase family of cysteine proteases (3, 4). The activation and activity of the caspases is, in turn, regulated by several families of both pro- and anti-apoptotic cellular proteins, including the Bcl-2 and inhibitor of apoptosis (IAP) families (5–11). Many stimuli that initiate apoptosis mediate their effects through the activation or inhibition of protein kinase signaling cascades including those involving the mitogen-activating protein kinase (MAPK) family (12–18). For this reason, considerable attention has been placed on the role of c-Jun N-terminal kinase (JNK), p38, or extracellular signal-regulated kinases (ERKs) in apoptosis. The role of JNK activation in apoptosis has recently been reviewed (19).
The IAP family (hILP, c-IAP1, and c-IAP2), represents a potentially important group of endogenous regulatory proteins that also may be connected to intracellular signaling cascades (6–10). c-IAP1 and c-IAP2, but not hILP, have been shown to bind to TRAF1 and TRAF2, two components of the tumor necrosis factor (TNF) signaling complex, that activates the MAPK pathways (7, 10). Therefore, we examined the possibility that the IAP proteins might modulate the activity of intracellular MAPKs.
MATERIALS AND METHODS
Transfection of Cultured Cells.
Cells were transfected using Lipofectamine reagents (GIBCO) for 4 hr and incubated for 18 hr before lysis. Lysis was performed for 30 min at 4°C with lysis buffer (25 mM Hepes, pH 7.6/1% Triton X-100/137 mM NaCl/3 mM β-glycerophosphate/3 mM EDTA/0.1 mM sodium orthovanadate/1 mM phenylmethylsulfonyl fluoride). Equivalent amounts of protein were immunoprecipitated as quantified by densitometry after Western blot analysis (data not shown). Endogenous JNK1 was immunoprecipitated from cell lysates using 20 μl of agarose-protein A (Pierce) preincubated with JNK1 antibody (Santa Cruz Biotechnology). Samples were incubated at 4°C for 2 hr and washed twice with lysis and kinase buffers before performing the kinase assay. Hemagglutinin-tagged proteins were immunoprecipitated using 20 μl of agarose-protein A (Pierce) preincubated with anti-hemagglutinin antibody (Boehringer Mannheim) and FLAG-tagged proteins with 20 μl agarose conjugated with M2-FLAG mAb (Kodak).
Kinase Assay.
In vitro kinase assays were carried out at 37°C for 20 min using immunoprecipitated JNK1, 10 μg of kinase substrate (ATF-2 or MBP), and 100 μM [γ-32P]ATP [10 μCi (1 Ci = 37 GBq)] in 20 μl of 5× kinase reaction buffer (100 mM Hepes, pH 7.6/100 mM MgCl2/125 mM β-glycerophosphate/0.5 mM sodium orthovanadate/10 mM DTT). Reactions were terminated by adding Laemmli sample buffer, products were separated on SDS/12% polyacrylamide gel and quantified by PhosphorImager (Molecular Dynamics). All protein kinases used in this work were first checked for their intrinsic capacity to be activated by known stimuli as UV light (50 J/m2 per 10 sec) for JNK and p38 and phorbol 12-myristate 13-acetate (0.1 mM) or H2O2 (0.1 mM) for ERK2 and ERK5/BMK1, respectively (data not shown). All gels shown are representative of data obtained from at least three independent experiments done in duplicate.
Plasmids.
cDNA carrying jnk1 and jnk1AF (jnk1-dn) were expressed from a pCMV5 vector (Invitrogen); p38, erk2, erk5/bmk1, mkk4, c-iap1, c-iap2, hilp, and the hilp deletion mutants from pcDNA3 (Invitrogen), interleukin 1β-converting enzyme (ICE) was expressed both from pcDNA3 or from pβactM10Z that contains pro-ICE fused to lacZ (20); mekk1, mekk1ΔK432M (mekk1-dn), and jnkkAA (mkk4-dn) from pSRα. The dominant negative JNKK (MKK4-dn) used in this study has been constructed by substitution of the two phosphorylation sites Ser-257 and Thr-261 to Ala (kindly provided by Z. Wu and M. Karin, University of California, San Diego). The capacity of the MKK4 mutant to act as a dominant negative was determined by the ability to block JNK1 activation in the presence of wild-type MEKK1. JNK1 (100 ng), MEKK1 (10 ng), and MKK4-dn (50, 200, and 500 ng) were cotransfected in 293 cells; 80% inhibition of MEKK1-induced JNK1 activation was detected in the presence of the highest concentration of MKK4-dn, whereas no inhibition was observed in the presence of empty vector (data not shown). MEKK1-dn activity was tested as follows: MEKK1 wild-type (10, 50 ng), MEKK1-dn (200 ng), and NF-κB-Luc reporter gene (p5xNF-κB-Luc, 5 μg) were transfected in 293 cells and activation of NF-κB was detected as calculated by luciferase activity. MEKK1 induced NF-κB activation was 90% inhibited in the presence of MEKK1-dn whereas no inhibition was detected when using empty vector (data not shown). JNK1 dominant negative mutant was constructed using site-directed mutagenesis by PCR (21). We found that the dominant negative JNK1 mutant (JNK1AF) was unable to phosphorylate ATF-2. In addition the effect of overexpression of JNK1AF on endogenous JNK1 activation in unstimulated or UV stimulated cells was tested as follows: JNK1AF (200 and 600 ng) was transfected, all endogenous JNK1 and transfected JNK1AF were immunoprecipitated from the lysates using saturating amount of antibody (30 μg JNK1 C-17, Santa Cruz Biotechnology) and the activity of JNK1 detected by kinase assay. The activity of endogenous JNK1 was 80% inhibited in the presence of JNK1AF as compared with positive control cells transfected with empty vector (data not shown). The expression of JNK1AF did not inhibit expression of Myc-tagged hILP in any of the in vivo and in vitro experiments as assessed by Western blot analysis using an anti-Myc antibody (c-Myc 9E10, Santa Cruz Biotechnology; data not shown). The hILP and deletion mutants were cloned in pcDNA3 vector as follows: Δ449–497 is a deletion construct encoding residues 1–449; Δ399–497 vector encodes residues 1–399 (kindly provided by R. Clem and J. M. Hardwich, Johns Hopkins School of Hygiene and Public Health, Baltimore); Δ449–497 was constructed by PCR-mediated fusion, using Pfu polymerase, of residues 1–342 to residues 445–497.
Detection of Apoptotic Cells.
Expression of ICE, JNK1, JNK1-dn and p38-dn was checked by Western blot on duplicate lysates of original transfections used for the apoptosis assays. These results showed consistent expression of all proteins and that cotransfection of multiple constructs did not interfere with the level of individual protein expression (data not shown). Apoptosis experiments were carried out using ICE expressed from pcDNA3 or from pβactM10Z; similar results were obtained with both constructs. Expression of ICE was detected using an antibody against β-galactosidase (β-gal), because the commercially available antibodies against ICE were not sensitive enough to detect expression. AnnexinV/fluorescein isothiocyanate (FITC) staining was performed as follows: 293 transfected cells were washed once in binding buffer (10 mM Hepes, pH 7.4/140 mM NaCl/2.5 mM CaCl2) and then stained with AnnexinV/FITC-propidium iodide (PI, BioWhittaker) that allows the differentiation between total number of dead cells (AnnexinV/FITC stained) and necrotic cells (PI stained). Cells that are stained only with AnnexinV/FITC but not with PI are apoptotic. Apoptotic cells expressing β-gal were detected as described (20).
RESULTS AND DISCUSSION
hILP Selectively Activates JNK1.
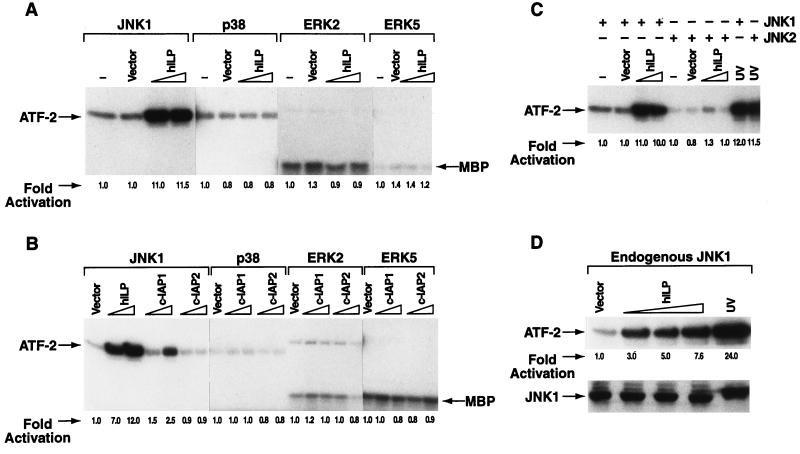
To test whether hILP, c-IAP1, or c-IAP2 could induce activation of the known MAPKs pathways (22, 15), COS-7, or 293 cells were transiently transfected with expression vectors encoding hilp, c-iap1, or c-iap2 together with vectors encoding epitope tagged jnk1, p38, erk2, or erk5/bmk1. MAPK proteins were immunoprecipitated and their activation measured in an in vitro kinase assay using either ATF-2 or MBP as substrate. Phosphorylation of ATF-2 was ≈10-fold higher in cells expressing both hILP and JNK1 than in the cells expressing JNK1 alone (Fig. 1A) suggesting that hILP could activate JNK1. Surprisingly, this activation was selective for JNK1 because none of the other MAPKs was activated by hILP. Thus hILP strongly activates JNK1 but not p38, ERK2, or ERK5/BMK1. c-IAP1 weakly activated JNK1 (2.5-fold) while no activation was detectable with c-IAP2 (Fig. 1B). Moreover, expression of c-IAP1 or c-IAP2 did not result in activation of any other MAPKs (Fig. 1B). Activation of each one of the MAPKs used in these studies was established using prototypic activators. In each case robust activation was observed with the in vitro kinase assay (data not shown).
Figure 1.
hILP selectively activates JNK1. (A) Effect of hILP expression on MAPK activation. COS-7 or 293 cells were cotransfected with FLAG-jnk1, -p38, -erk2, and -erk5/bmk1 (100 ng) in the absence or in the presence of increasing concentrations of Myc-tagged-hilp (100 and 600 ng). The amount of transfected cDNA was kept constant in each samples adding control pcDNA3 vector. In vitro kinase assay was performed using substrates ATF-2 for JNK1/p38 or myelin basic protein (MBP) for ERK2 and ERK5/BMK1. The fold-activation is indicated in each sample as quantitated by PhosphorImager with values normalized to the basal level of phosphorylation of each MAPK. Similar results were obtained in COS-7 or 293 cells for all the described kinase assays. (B) Expression of different IAP family members on MAPKs activation. Myc-tagged c-iap1 or c-iap2 cDNA (100, 600 ng) were cotransfected together with FLAG-jnk1, -p38, -erk2, and -erk5/bmk1 (100 ng) and activation was determined as described above. A sample transfected with jnk1 (100 ng) and hilp (100 and 600 ng) is also shown as control. (C) JNK2 isoform is not activated by hILP. Equal amounts of FLAG-jnk1 or HA-jnk2 (100 ng) were cotransfected together with hilp expression vector (100, 300, and 600 ng) and activation of JNK1 or JNK2 was determined. The results of samples containing 100 and 600 ng hILP are shown. Results of samples containing 300 ng of hILP are not shown because they behaved like samples containing 600 ng. JNK1 and JNK2 activation was also induced by UV stimulation as a positive control (last two lanes). (D) Activation of endogenous JNK1 by hILP. Cells were transfected with increasing amount of hilp (50, 200, and 600 ng) and endogenous JNK1 protein was immunoprecipitated using an antibody against JNK1. The Western blot analysis confirms that similar amounts of JNK1 proteins were immunoprecipitated and used in the kinase assay. The activity of the endogenous JNK1 protein after UV stimulation is also shown.
To determine whether hILP also activates JNK2—a closely related isoform of the JNK family—COS-7, and 293 cells were cotransfected with jnk2 and hilp expression vectors and assayed for kinase activation. hILP activated only the JNK1 isoform but not JNK2 (Fig. 1C). Many reports document the concurrent activation of JNK together with other members of the MAPK family (12, 23). At least one other example of selective preferential isoform activation within the JNK subfamily has recently been reported in TNF-α stimulated macrophages (24). However, the functional consequences of this preferential activation remains unknown. In contrast, treatment of cells with UV light caused >10-fold activation of both JNK1 and JNK2 (Fig. 1C).
We also asked whether hILP could activate endogenous JNK1. Endogenous JNK1 was immunoprecipitated from cells transfected with hilp and assayed for kinase activity. hILP was found to activate endogenous JNK1 in a dose-dependent manner (Fig. 1D).
Recent studies have shown that the activation of NF-κB can confer protection against TNF-induced apoptosis (25–27). However, we have failed to observe any up-regulation of NF-κB after ectopic expression of hILP (data not shown).
hILP Activates JNK1 Independent of the MEKK1/MKK4 Signaling Cascade.
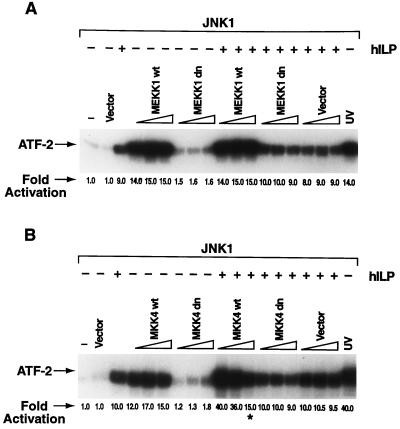
JNK1 is activated by another MAPK family member, MKK4, which in turn is activated by an upstream protein kinase, MEKK1 (22, 15). Dominant negative mutants of both MEKK1 and MKK4 [MEKK1-dn (28), and MKK4-dn] were used to determine whether hILP acted through MEKK1/MKK4 to activate JNK1. MEKK1 and MKK4 mutants were found to act as dominant negative proteins in control experiments (see Material and Methods). Cotransfection of hILP with MEKK1-dn in COS-7 or 293 cells did not significantly affect hILP-dependent JNK1 activation (Fig. 2A). Similar results were observed using MKK4-dn (Fig. 2B). These data suggest that hILP activates JNK1 by a mechanism that is independent of MEKK1 and MKK4.
Figure 2.
hILP activates JNK1 independent of the MEKK1/MKK4 signaling cascade. (A) Effects of MEKK1 dominant negative on hILP-dependent JNK1 activation. Increasing amounts (50, 200, and 700 ng) of dominant negative mekk1-dn or wild-type mekk1 (24), were cotransfected together with jnk1 (100 ng) in the presence or absence of hilp (100 ng). A kinase assay was performed on immunoprecipitated JNK1. (B) Effects of MKK4 dominant negative on hILP-dependent JNK1 activation. Wild-type mkk4 or mkk4-dn (50, 200, and 700 ng) were cotransfected together with jnk1 (100 ng) in the presence or absence of hilp (100 ng) and phosphorylation of ATF-2 determined. The asterisk indicates a ≈2-fold decrease in the JNK1 protein expression (data not shown), and therefore ATF-2 phosphorylation, due to the strong expression of MKK4.
The hILP RING Finger Domain Is Not Required for JNK1 Activation.
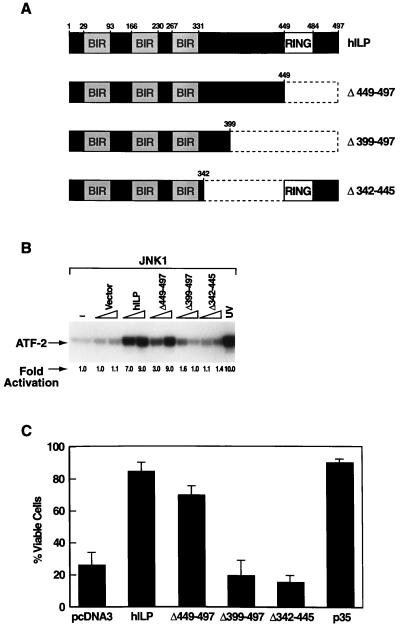
hILP contains three N-terminal baculovirus IAP repeats (BIRs) and a C-terminal zinc binding domain known as a RING finger (8). To identify domain(s) that are important for the activation of JNK1, deletion mutants of hILP (Fig. 3A) were analyzed for their capacity to activate JNK1 (Fig. 3B). The mutant lacking the RING finger domain (Δ449–497) was still able to activate JNK1 but required higher concentrations than the wild-type hILP. However mutants encoding only the BIR domains (Δ399–497), or lacking the region between the BIRs and the RING finger (Δ342–445) were unable to activate JNK1 (Fig. 3B). These results suggest that while the RING finger is dispensable, the BIRs and the region between the BIRs and the RING finger are necessary for the activation of JNK1.
Figure 3.
The hILP RING finger domain is not required for JNK1 activation. (A) Schematic representation of hILP deletion mutants. Mutant Δ449–497 is lacking the C-terminal RING finger domain; Δ399–497 encodes the BIR domains only; mutant Δ342–497 is lacking the region between the BIRs and the RING-finger. (B) JNK1 activation by hILP deletion mutants. Wild-type hilp or hilp deletion mutants (100 or 600 ng) were cotransfected into 293 or COS-7 cells together with jnk1 (100 ng). The ability of the deletion mutants to activate JNK1 was assayed. (C) Protective effects of hILP deletion mutants against ICE-induced apoptosis. 293 cells were transfected with ICE (500 ng) plus control vector or plus wild-type hilp or hilp deletion mutants (2 μg) and viability analysis was performed as previously described (8, 26).
Previous studies have shown that hILP protects cells from ICE-induced apoptosis (8, 10). The capacity of the hILP deletion mutants to protect 293 cells from ICE-induced death was analyzed. hILP mutant Δ449–497 was able to protect cells from death, although somehow less efficiently than wild-type hILP (compare 70% protection efficiency to 85% for wild-type hILP, Fig. 3C). However, hILP mutants Δ399–497 and Δ342–445, which did not activate JNK1, were not able to rescue cells from ICE-induced apoptosis (Fig. 3C). These data correlate well with the results of hILP deletion mutants to activate JNK1 in the kinase assay (Fig. 3B). Thus we have established a strong correlation between the JNK1 activation and protection from ICE-mediated death suggesting that JNK1 activation might be involved in the protective role of hILP in vivo. Our results slightly differ from data showing that the BIR domains of hILP are sufficient for protecting 293T cells from Bax-induced apoptosis (11), suggesting that more than one domain of hILP may be involved in protection against different apoptotic stimuli .
hILP Requires JNK1 for Protection Against ICE-Induced Apoptosis.
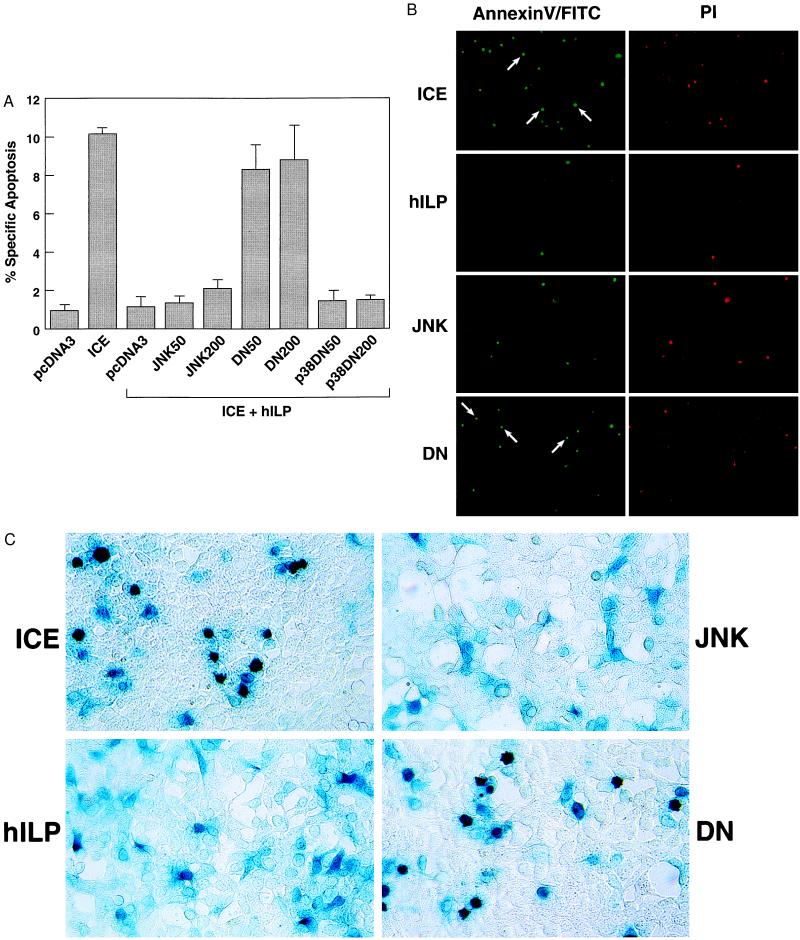
To further address the potential role of JNK1 activation in protection, a dominant negative mutant (JNK1-dn) was constructed with the dual phosphorylation sites, Thr-183 and Tyr-185, converted to Ala and Phe, respectively (21). 293 cells were transfected with ICE plus a control vector or plus hilp in the absence or presence of increasing concentrations of jnk1 or jnk1-dn mutant. A β-gal expressing plasmid was also transfected to allow quantitation of the transfection efficiency and cell morphology observation. The number of apoptotic cells was then determined by β-gal staining (20) or AnnexinV/FITC/PI staining. The presence of JNK-dn significantly reduced the ability of hILP to protect cells from apoptosis (Fig. 4; the number of apoptotic cells in this experiment is lower than in the previous experiment, Fig. 3C, because less ICE DNA was transfected to be able to cotransfect four different DNAs, ICE, hilp, jnk or jnk1-dn, and β-gal, and therefore titrate the effects of jnk1 or jnk1DN; see figure legends for more details). In contrast, transfection of a p38 dominant negative did not alter the effects of hILP (Fig. 4A). Furthermore expression of JNK1 or JNK1-dn in the absence of hILP did not rescue cells from ICE-induced apoptosis (data not shown). Thus, a functional JNK1 protein is required for the inhibition of ICE-mediated apoptosis by hILP. We also found that ICE by itself did not activate JNK1 and had no effect on hILP-mediated JNK1 activation (data not shown). The possibility that hILP could activate JNK1 by direct interaction was also investigated by coimmunoprecipitation. When JNK1 was immunoprecipitated no hILP was detected and the converse was true (data not shown), suggesting that other factors participate in this activation.
Figure 4.
hILP requires JNK1 for protection against ICE-induced apoptosis. (A) Effect of JNK1 wild-type and JNK1 dominant negative on ICE/hILP transfected cells. 293 cells were transfected with control vector pcDNA3 alone (50 ng; indicated as pcDNA3) or with plasmid encoding ICE (50 ng). Samples included in the bracket were transfected with ICE (in duplicate using pcDNA3/ICE or pβactM10Z, 50 ng) and hilp (200 ng) plus control vector pcDNA3 or one of the following: wild-type jnk1 (50 and 200 ng; JNK50 and JNK200); jnk1 dominant negative (50 and 200 ng; DN50 and DN200), p38 dominant negative (50 and 200 ng; p38DN50 and p38DN200). Parental pcDNA3 vector was added to normalize the amount of transfected DNA and a β-gal expressing plasmid to visualize transfected cells. % specific apoptosis indicates the number of ICE-induced apoptotic cells among the β-gal-positive (transfected cells). The number of apoptotic cells in this experiment is lower than in the previous experiment (Fig. 3C) because less ICE DNA was transfected to be able to cotransfect four different DNAs (ICE, hilp, jnk or jnk1-dn, and β-gal) and therefore to titrate the effects of jnk1 or jnk1-dn. Effects on cell viability were determined 24 hr after transfection with AnnexinV/FITC-PI staining. Data represent the mean between at least three experiments run in duplicate and scored blind. Dead cells were counted over five fields for each sample. (B) AnnexinV/FITC and PI staining of dead cells. Dead cells appear green with the AnnexinV/FITC staining whereas only necrotic cells are red due to the PI incorporation. The difference between cells counted with each staining represent the real number of apoptotic cells. Arrows indicate representative apoptotic cells. (C) 5-Bromo-4-chloro-3-indolyl β-d-galactoside staining of cells. cDNA expressing β-gal (50 ng) was transfected together with the different cDNA to allow quantification and morphological observation of the cells. Apoptotic cells appear to be smaller, rounder, and show condensed and misshapen nuclei compared with viable cells that are flat, well spread out, and with easily discernible nuclei.
JNK has been implicated on induction of programmed cell death in serum-deprived PC12 cells and in ceramide- or TNF-induced apoptosis in human monoblastic leukemic U937 cells (12, 17) but has been reported not to be sufficient for TNF-induced apoptosis of MCF7 and L929 cells (29, 30). Activation of the JNK substrate, c-Jun, has also been shown to promote apoptosis in sympathetic neurons and NIH 3T3 fibroblasts (16, 18). In addition, Fas-mediated JNK activation via the protein Daxx, leading to cell death in 293 and L929 cells, has been reported (31). Moreover, JNK activation has been shown recently to antagonize the anti-apoptotic action of Bcl-2 in N18TG neuroglioma cells (32). However, in many of these previous studies JNK activation is also accompanied by activation of other members of the MAPK family so that a specific role for JNK cannot be assigned. Our work supports the contention that selective activation of JNK1 is necessary for the anti-apoptotic activity of hILP. Other results suggesting an anti-apoptotic role for JNK can be found in studies showing that B cell survival after CD40 activation is accompanied by potent stimulation of JNK (13, 14). More recently, Sek1/MKK4 has been suggested to protect thymocytes from Fas- and CD3-mediated apoptosis (33).
The different activities of the JNK protein thus suggest a dual role of JNK in apoptosis that may be cell type specific and may depend on upstream signaling events including whether JNK1 is selectively activated to the exclusion of other MAPK family members.
In a recent report was shown that baculovirus IAPs, differently than p35, did not block active caspases-induced apoptosis but were able to inhibit their activation (34). In addition it has been suggested that the ability of hILP, c-IAP1, and c-IAP2, but not of neuronal apoptosis inhibitory protein (NAIP) to modulate programmed cell death, results from inhibition of the proteolytic activity of caspase-3 and -7 (11, 35). However, hILP was not able to inhibit the proteolytic activity of caspase-1 (ICE) by the same mechanism (11). All together these results suggest that alternative mechanisms for regulating caspases activity can be used to inhibit apoptosis. Here we provide data to support that hILP can also inhibit ICE-dependent apoptosis through a mechanism that involves the selective activation of JNK1. Our data provide a direct link between an endogenous IAP gene and selective activation of a member of the MAPK family. These findings suggest that approaches to modulate JNK1 may be useful in controlling apoptosis in a variety of different settings.
Acknowledgments
We thank Z. Wu and M. Karin for providing MKK4 and MEKK1 dominant negative expressing plasmids; R. Clem and J. M. Hardwick for providing hILP mutant encoding residues 1–399; Vladimir Kravchenko for testing the activity of MEKK1-dn; and C. Fearns, A. Ghetti, and J. Han for critical reading of the manuscript. This work was supported by Grants GM37696, AI15136, and Novartis Pharmaceuticals SFP1191 (M.G.S., B.W.M.R., and R.J.U); Grant P01DK49799 (C.B.T. and C.S.D.) from the National Institutes of Health. C.S.D. is a Special Fellow of the Leukemia Society of America.
ABBREVIATIONS
- IAP
inhibitor of apoptosis
- ICE
interleukin-1β-converting enzyme
- MAPK
mitogen-activated protein kinase
- JNK
c-Jun N-terminal kinase
- ERK
extracellular signal-regulated kinase
- TNF
tumor necrosis factor
- BIR
baculovirus IAP repeat
- β-gal
β-galactosidase
- FITC
fluorescein isothiocyanate
- PI
propidium iodide
References
- 1.White E. Gen Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 3.Chinnaiyan A M, Dixit V M. Curr Biol. 1996;6:555–562. doi: 10.1016/s0960-9822(02)00541-9. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson D W, Thornberry N A. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 5.Reed J C. Nature (London) 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 6.Clem R J, Duckett C S. Trends Cell Biol. 1997;7:337–339. doi: 10.1016/S0962-8924(97)01088-X. [DOI] [PubMed] [Google Scholar]
- 7.Rothe M, Pan M G, Henzel W J, Ayres T M, Goeddel D V. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 8.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J L, Gilfillan M C, Shiels H, Hardwick J M, Thompson C B. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 9.Liston P, Roy N, Tamai K, Lefebre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J-E, MacKenzie A, Korneluk R G. Nature (London) 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 10.Uren A G, Pakusch M, Hawkins C J, Puls K L, Vaux D L. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 12.Xia Z, Dickens M, Raingeraud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 13.Sakata N, Patel H R, Terada N, Aruffo A, Johnson G L, Gelfand E W. J Biol Chem. 1995;270:30823–30828. doi: 10.1074/jbc.270.51.30823. [DOI] [PubMed] [Google Scholar]
- 14.Berberich I, Shu G, Siebelt F, Woodgett J R, Kyriakis J M, Clark E A. EMBO J. 1996;15:92–101. [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson M J, Cobb M H. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 16.Ham J, Babij C, Whitfield J, Pfarr C M, Lallemand D, Yaniv M, Rubin L R. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 17.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, et al. Nature (London) 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 18.Bossy-Wetzel E, Bakiri L, Yaniv M. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cosulich S, Clarke P. Curr Biol. 1996;6:1586–1588. doi: 10.1016/s0960-9822(02)70779-3. [DOI] [PubMed] [Google Scholar]
- 21.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 20.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 22.Su B, Karin M. Curr Opin Immun. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 23.Raingeraud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, Davis R J. J Biol Chem. 1996;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 24.Chan E D, Winston B W, Jarpe M B, Wynes M W, Riches D W H. Proc Natl Acad Sci USA. 1997;94:13169–13174. doi: 10.1073/pnas.94.24.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 26.Wang C-Y, Mayo M W, Baldwin A S., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 27.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 28.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 30.Gardner A M, Johnson G L. J Biol Chem. 1996;271:14560–14566. doi: 10.1074/jbc.271.24.14560. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Kim I, Oh Y J, Lee K, Han P L, Choi E J. J Biol Chem. 1997;272:16725–16728. doi: 10.1074/jbc.272.27.16725. [DOI] [PubMed] [Google Scholar]
- 33.Nishina H, Fischer K D, Laszio R, Shahinian A, Hakem R, Rubie E, Bernstein A, Mak T W, Woodgett J R, Penninger J M. Nature (London) 1997;385:350–353. doi: 10.1038/385350a0. [DOI] [PubMed] [Google Scholar]
- 34.Seshagiri S, Miller L K. Proc Natl Acad Sci USA. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy N, Deveraux Q N, Takahashi R, Salvensen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]