Abstract
We have identified five complementation groups of peroxisome biogenesis (peb) mutants in Saccharomyces cerevisiae by a positive selection procedure. Three of these contained morphologically recognizable peroxisomes, and two appeared to lack the organelle altogether. The packaging of peroxisomal proteins in these mutants has been analyzed with a new gentle cell fractionation procedure. It employs digitonin titration for the selective permeabilization of yeast plasma and intracellular membranes. Proteins were measured by enzymatic assay or by quantitative chemiluminescent immunoblotting. With this gentle fractionation method, it was demonstrated that two mutants are selectively defective in assembling proteins into peroxisomes. Peb1-1 packages catalase and acyl-CoA oxidase within peroxisomes but not thiolase. Peb5-1 packages thiolase and acyl-CoA oxidase within peroxisomes but not catalase. The data suggest that the peroxisome biogenesis machinery contains components that are specific for each of three classes of peroxisomal proteins, represented by catalase, thiolase, and acyl-CoA oxidase. In the two mutants lacking morphologically recognizable peroxisomes, peb2-1 and peb4-1, all three enzymes were mislocalized to the cytosol.
Full text
PDF

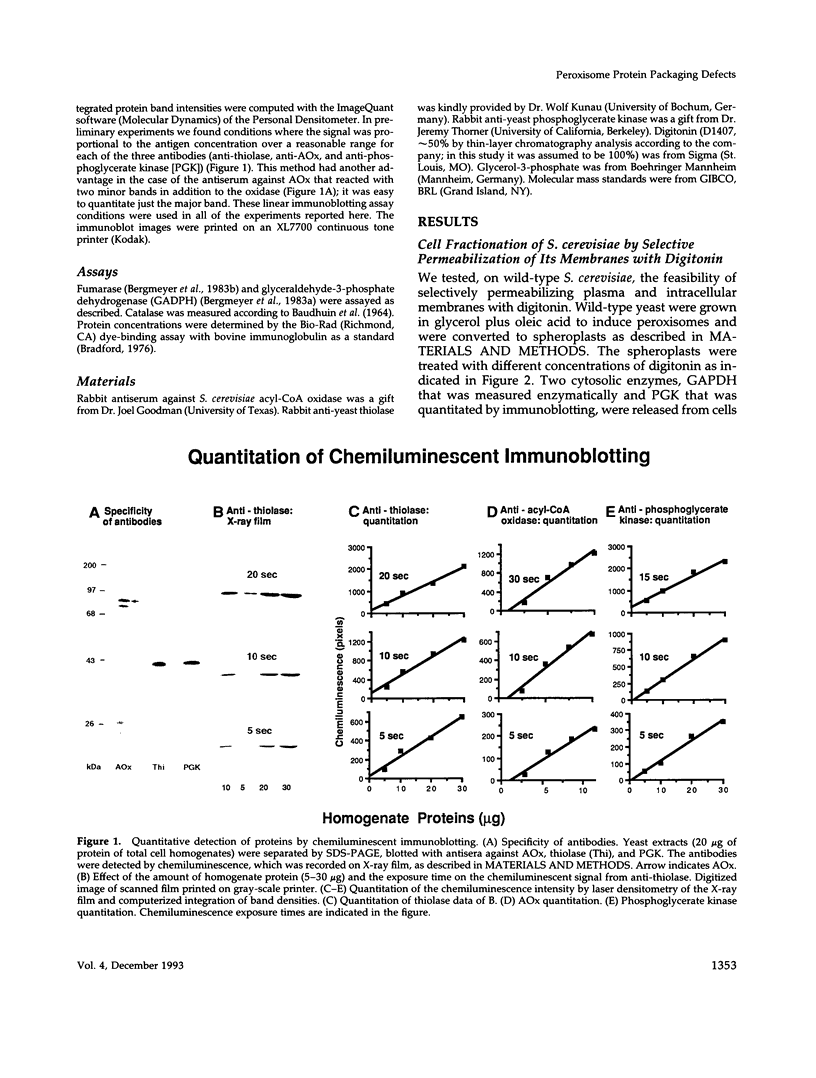
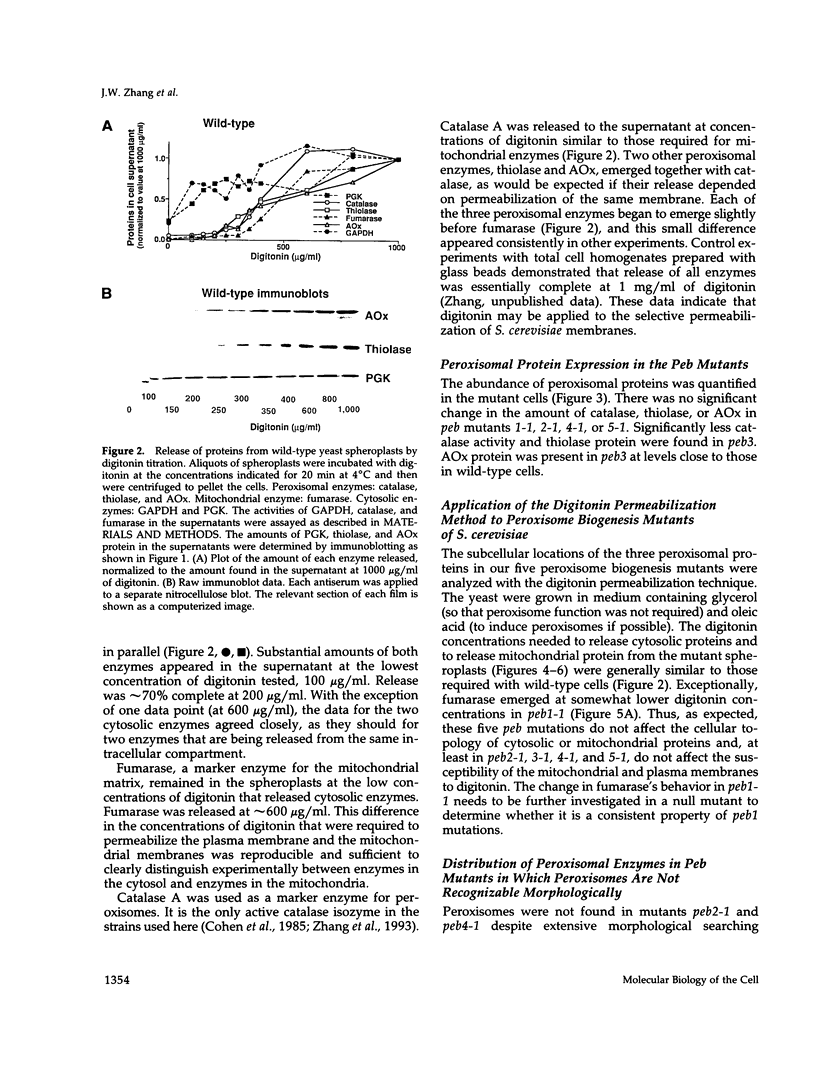
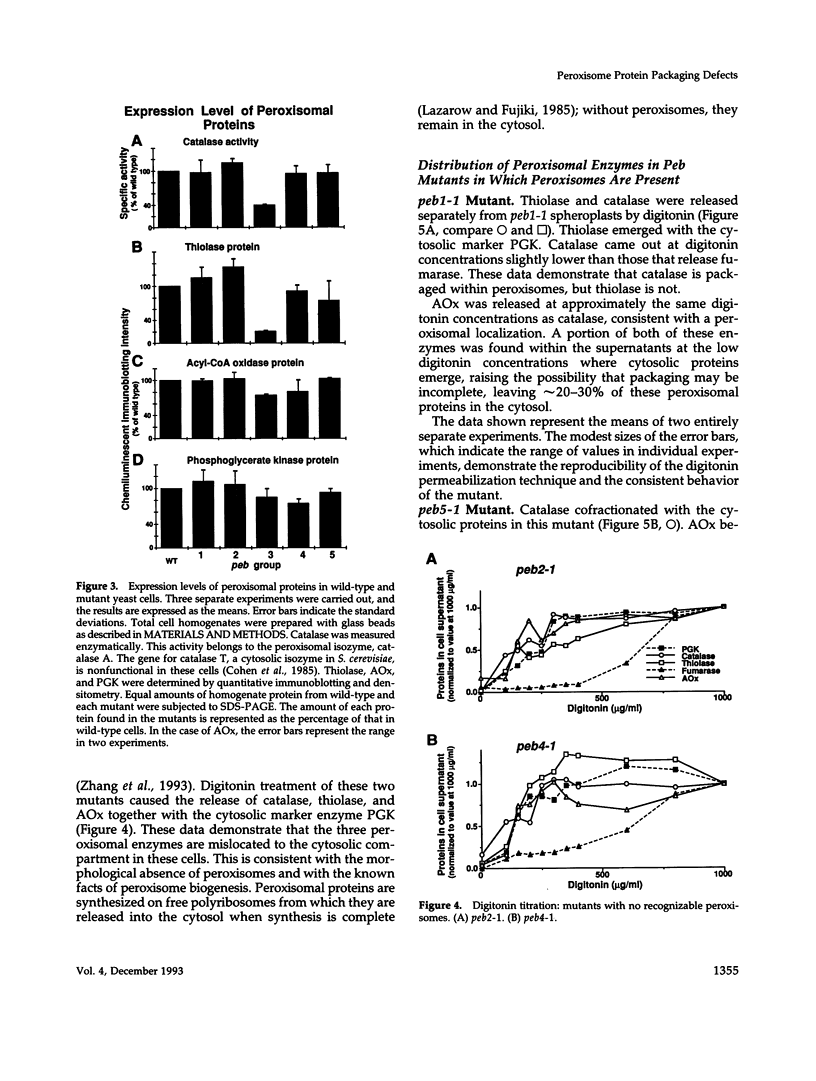



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Marr R. S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990 Sep;111(3):807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexson S. E., Fujiki Y., Shio H., Lazarow P. B. Partial disassembly of peroxisomes. J Cell Biol. 1985 Jul;101(1):294–304. doi: 10.1083/jcb.101.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudhuin P., Beaufay H., Rahman-Li Y., Sellinger O. Z., Wattiaux R., Jacques P., De Duve C. Tissue fractionation studies. 17. Intracellular distribution of monoamine oxidase, aspartate aminotransferase, alanine aminotransferase, D-amino acid oxidase and catalase in rat-liver tissue. Biochem J. 1964 Jul;92(1):179–184. doi: 10.1042/bj0920179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- CHANCE B. The reactions of catalase in the presence of the notatin system. Biochem J. 1950 Apr;46(4):387–402. doi: 10.1042/bj0460387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G., Fessl F., Traczyk A., Rytka J., Ruis H. Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol Gen Genet. 1985;200(1):74–79. doi: 10.1007/BF00383315. [DOI] [PubMed] [Google Scholar]
- Colbeau A., Nachbaur J., Vignais P. M. Enzymic characterization and lipid composition of rat liver subcellular membranes. Biochim Biophys Acta. 1971 Dec 3;249(2):462–492. doi: 10.1016/0005-2736(71)90123-4. [DOI] [PubMed] [Google Scholar]
- De Duve C. The separation and characterization of subcellular particles. Harvey Lect. 1965;59:49–87. [PubMed] [Google Scholar]
- Diaz R., Stahl P. D. Digitonin permeabilization procedures for the study of endosome acidification and function. Methods Cell Biol. 1989;31:25–43. doi: 10.1016/s0091-679x(08)61600-3. [DOI] [PubMed] [Google Scholar]
- Dmochowska A., Dignard D., Maleszka R., Thomas D. Y. Structure and transcriptional control of the Saccharomyces cerevisiae POX1 gene encoding acyl-coenzyme A oxidase. Gene. 1990 Apr 16;88(2):247–252. doi: 10.1016/0378-1119(90)90038-s. [DOI] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Schneider M., Howell S. H., Garrard L. J., Goodman J. M., Distel B., Tabak H., Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990 Jan;9(1):85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Krisans S., Keller G. A., Subramani S. Antibodies directed against the peroxisomal targeting signal of firefly luciferase recognize multiple mammalian peroxisomal proteins. J Cell Biol. 1990 Jan;110(1):27–34. doi: 10.1083/jcb.110.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F., Langeder A., Raupachova J., Binder M., Hartig A. Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. J Cell Biol. 1993 Feb;120(3):665–673. doi: 10.1083/jcb.120.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Longley R. P., Rose A. H., Knights B. A. Composition of the protoplast membrane from Saccharomyces cerevisiae. Biochem J. 1968 Jul;108(3):401–412. doi: 10.1042/bj1080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon M. T., Veenhuis M., Trapp S. B., Goodman J. M. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J Bacteriol. 1990 Oct;172(10):5816–5827. doi: 10.1128/jb.172.10.5816-5827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T., Tsukamoto T., Hata S., Yokota S., Miura S., Fujiki Y., Hijikata M., Miyazawa S., Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991 Dec 31;181(3):947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Rattray J. B., Schibeci A., Kidby D. K. Lipids of yeasts. Bacteriol Rev. 1975 Sep;39(3):197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small G. M., Szabo L. J., Lazarow P. B. Acyl-CoA oxidase contains two targeting sequences each of which can mediate protein import into peroxisomes. EMBO J. 1988 Apr;7(4):1167–1173. doi: 10.1002/j.1460-2075.1988.tb02927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991 Nov;10(11):3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer R., Shio H., Han Y. S., Cohen G., Lazarow P. B. Peroxisomes in Saccharomyces cerevisiae: immunofluorescence analysis and import of catalase A into isolated peroxisomes. Mol Cell Biol. 1991 Jan;11(1):510–522. doi: 10.1128/mcb.11.1.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leij I., Van den Berg M., Boot R., Franse M., Distel B., Tabak H. F. Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J Cell Biol. 1992 Oct;119(1):153–162. doi: 10.1083/jcb.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R. J., Kos M., Roest B., Meijer A. J., Schrakamp G., Heymans H. S., Tegelaers W. H., van den Bosch H., Schutgens R. B., Tager J. M. Activity of peroxisomal enzymes and intracellular distribution of catalase in Zellweger syndrome. Biochem Biophys Res Commun. 1984 Sep 28;123(3):1054–1061. doi: 10.1016/s0006-291x(84)80240-5. [DOI] [PubMed] [Google Scholar]
- Zhang J. W., Han Y., Lazarow P. B. Novel peroxisome clustering mutants and peroxisome biogenesis mutants of Saccharomyces cerevisiae. J Cell Biol. 1993 Dec;123(5):1133–1147. doi: 10.1083/jcb.123.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]