Abstract
Intrinsically unstructured domains occur in one-third of all proteins and are characterized by conformational flexibility, protease sensitivity, and the occurrence of multiple phosphorylation. They provide large interfaces for diverse protein-protein interactions. Peptidylglycine α-amidating monooxygenase (PAM), an enzyme essential for neuropeptide biosynthesis, is a secretory granule membrane protein. As one of the few proteins spanning the granule membrane, PAM is a candidate to relay information about the status of the granule pool and conditions in the granule lumen. Here, we show that the PAM cytosolic domain is unstructured. Mass spectroscopy and two-dimensional gel electrophoresis demonstrated phosphorylation at 10–12 sites in the cytosolic domain. Stimulation of exocytosis resulted in coupled phosphorylation and dephosphorylation of specific sites and in the endoproteolytic release of a soluble, proteasome-sensitive cytosolic domain fragment. Analysis of granule-rich tissues, such as pituitary and heart, showed that a similar fragment was generated endogenously and translocated to the nucleus. This multiply phosphorylated unstructured domain may act as a signaling molecule that relays information from secretory granules to both cytosol and nucleus.
One-third to one-half of all eukaryotic proteins include a disordered region more than 50 amino acids long (1). These intrinsically unstructured domains are characterized by low hydrophobicity and high net charge, features that contribute to their flexibility, low secondary structure, and protease sensitivity (2). Unstructured domains are common in proteins that play key roles in complex pathways like cell cycle regulation, endocytic trafficking, and control of transcription (2, 3). Their ability to mediate several low affinity interactions with specific interactors makes them well suited to participation in multistep processes.
Intrinsically unstructured domains are often sites of multiple phosphorylation (2). The presence of an extended unstructured domain may provide a larger interface for protein-protein interactions, with phosphorylation at multiple sites contributing to increased local order, facilitating cooperativity, and driving specific intermolecular interactions (4). Identification of phosphorylation sites in the unstructured domains of several proteins and mutational analyses have confirmed their functional significance (5–7). Bioinformatic analyses reveal an increase in their prevalence with organism complexity and an association with alternative splicing (8).
Regulated exocytosis of secretory granules is crucial for maintaining neuronal homeostasis. Very little is known about the feedback mechanisms that signal the status of the secretory granule pool and regulate recycling. This process involves even greater complexity in neurons where the site of release is distant from the cell body. Peptidylglycine α-amidating monooxygenase (PAM),2 a type I integral membrane protein that catalyzes one of the final steps in the biosynthesis of neuropeptides, is composed of two independently folded catalytic domains, a single transmembrane domain and a highly conserved 86-amino acid cytosolic domain whose sequence suggested that it might lack structure. Removal of this cytosolic domain limited the access of PAM to secretory granules and eliminated endocytosis (9, 10). Radiolabeling studies demonstrated that phosphorylation of PAM-1 was restricted to its cytosolic domain (11), and trafficking roles were identified for phosphorylation at two specific residues, Ser937 and Ser949 (12, 13).
C-terminal amidation is the final reaction in the generation of many bioactive peptides, and PAM is one of the small number of secretory granule proteins that span the granule membrane and has been shown to be recycled and reused in secretory granules (14, 15). Biochemical studies demonstrated that PAM was sensitive to the pH changes that occur as granules mature (16), and analysis of a cell line in which expression of PAM-1 could be induced suggested an important role for PAM-1 in signaling from the granule lumen to the cytosol (17, 18). Cytosolic PAM-CD interactors include Kalirin and Trio, Rho GDP/GTP exchange factors that affect cytoskeletal organization (19, 20), and P-CIP2 (KIS, Uhmk1), a protein kinase that binds to and phosphorylates PAM-CD at Ser949, a site known to affect its endocytic trafficking (19).
We set out to explore the hypothesis that this key region of PAM was involved in relaying information from the secretory granule lumen. We demonstrate that PAM-CD achieves this by being intrinsically unstructured and acting as a display site for multiple phosphorylation. Mass spectroscopy was used to identify additional sites of phosphorylation, and two-dimensional gel electrophoresis was used to demonstrate regulated multiple site phosphorylation in pituitary cells. A newly developed antibody to the C terminus of PAM was used to identify a soluble fragment of PAM-CD (sf-CD). Formed in response to stimulated secretion, sf-CD was identified in nuclei isolated from both heart atrium and pituitary. Production of sf-CD was limited in cells expressing phosphomimetic mutants of PAM. This study demonstrates that the unstructured domain of PAM acts as a signaling module by utilizing multisite phosphorylation, proteolysis, and nuclear translocation.
EXPERIMENTAL PROCEDURES
Purification of PAM Cytosolic Domain
Recombinant PAM-CD was purified from Escherichia coli as described (21), with an additional step of purification by reverse phase chromatography on a C-18 μBondapak column (Waters) equilibrated with 0.1% trifluoroacetic acid and eluted with a gradient to 40% acetonitrile, 0.1% trifluoroacetic acid over 60 min; purity as assessed by SDS-PAGE was >99%; MALDI-TOF analysis revealed that the purified protein terminated at Ser961. PAM-CD-3P and PAM-CD-5P (residues 896–976) with phosphomimetic Ser to Asp and Thr to Glu mutations at positions 932, 937, and 945 (3P) and also at 946 and 949 (5P) were expressed as glutathione S-transferase fusion proteins in pGEX-6P2 (Amersham Biosciences). Purification was accomplished using glutathione-Sepharose 4B resin equilibrated with 50 mm Tris-HCl, 150 mm NaCl, pH 7.4, and phenylmethylsulfonyl fluoride (0.3 mg/ml). The bound glutathione S-transferase fusion proteins were cleaved with PreScission protease (200 μg of protein, 8 units of enzyme, 24 h, 4 °C in 50 mm NaH2PO4, 150 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA). When expressed as a glutathione S-transferase-PAM-CD fusion protein, purification yielded intact PAM-CD.
Circular Dichroism
Circular dichroism spectra were recorded at 20 °C using a Jasco J-715 spectropolarimeter (Jasco, Easton, MD). Purified PAM-CD* (20 μm) in 1 mm sodium phosphate, pH 7.4, was used. Spectra were the average of 10 scans acquired using a scan rate of 20 nm/min and a response time of 8 s. The background signal of the buffer was subtracted from the spectra. Protein concentration was determined by measuring absorbance at 280 nm.
Cell Culture
Stably transfected AtT-20 cells expressing PAM-1 (22), PAM-1/899 (10), PAM-1/TS/DD (12), PAM-1/S937D (13), or Myc-TMD-CD (23) were grown in Dulbecco's modified Eagle's medium/F-12 with 25 mm Hepes, 10% NuSerum, 10% fetal bovine serum, 0.5 mg/ml G418, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were passaged weekly and maintained for less than 20 passages, since changes in phosphorylation pattern and a loss of stimulated secretion were observed in cells maintained in culture for longer times.
Anterior pituitary cultures were prepared from ex-pregnant rats as described (15). Plastic dishes (12-well) were coated with protamine and NuSerum. Dissociated cells (1 pituitary/well) were plated in Dulbecco's modified Eagle's medium/F-12 (Invitrogen or Mediatech) containing 25 mm HEPES, 10% NuSerum, 10% fetal bovine serum, penicillin/streptomycin, and 10 μm cytosine arabinoside. The next day, the serum-containing medium was replaced with complete serum-free medium (Dulbecco's modified Eagle's medium/F-12 (1:1), 25 mm HEPES, pH 7.4, penicillin/streptomycin, insulin/transferrin/selenium, 1 mg/ml fatty acid-free bovine serum albumin) and cytosine arabinoside (10 μm) and maintained in this medium until harvesting. Cells were maintained for a total of 3 days in vitro.
Stimulation of Secretion and Preparation of Extracts
PAM-1 AtT-20 cells and anterior pituitary cultures were rinsed with complete serum-free medium prior to stimulation with complete serum-free medium containing 1 μm phorbol myristate acetate along with 12.5 nm calyculin A (Sigma) for 1 h and, when indicated, 2 μm MG132 (Sigma) for 3 h. Myc-TMD-CD cells were also treated with 0.5 mm cyclic 8-bromo-AMP for 1 h. Cells were extracted with 20 mm NaTES, 10 mm mannitol (TM; pH 7.4), 50 mm NaF, 5 mm EDTA at 4 °C containing protease inhibitors (0.3 mg/ml phenylmethylsulfonyl fluoride, 50 μg/ml lima bean trypsin inhibitor, 2 μg/ml leupeptin, 16 μg/ml benzamidine, and 2 μg/ml pepstatin), 200 μm sodium orthovanadate, and 12.5 nm calyculin A. Extracts were passed 10 times through a 25-gauge needle and centrifuged at 1000 × g for 5 min to remove cell debris and nuclei. Membranes were prepared by centrifugation at 430,000 × g for 15 min. Pellets were resuspended in TM with 1% Triton X-100 (Pierce SurfActs), pH 7.4 (TMT), allowed to tumble at 4 °C for 30 min, and clarified by centrifugation at 10,000 × g for 10 min. Protein concentrations were determined using the bicinchoninic acid assay with bovine serum albumin as the standard (Pierce), and samples were subjected to SDS-PAGE and Western blot analysis (15). Where indicated, membrane protein extracts (150 μg) were treated with calf intestinal alkaline phosphatase (CIP; 30 units; New England BioLabs) at 37 °C for 20 min in 0.5% TMT, 10 mm MgCl2 with protease inhibitors. Immunoprecipitated PAM-1 prepared from AtT-20 extracts received a second addition of CIP and an additional 20-min incubation.
Two-dimensional PAGE
Solubilized membrane proteins were precipitated with two volumes of 100% ethanol at −20 °C overnight. The protein pellet obtained by centrifugation at 10,000 × g for 30 min at 4 °C was resuspended in 10 μl of 2% ASB-14 in TM (Sigma) and sonicated. Rehydration buffer (7 m urea, 2 m thiourea, 2% ASB-14, 0.5% ampholytes 4–7 (Invitrogen), 100 mm dithiothreitol, 0.001% bromphenol blue) was added and mixed for 1 h at room temperature. The sample was loaded into Zoom IPGRunner cassettes (Invitrogen), and an ImmobilineTM Dry Strip pH 4–7 isoelectric focusing gel (IPG) (Amersham Biosciences) was placed into the well. Rehydration of IPG strips was carried out overnight, after which the strips were transferred to IPGPhor coffins. Isoelectric focusing was carried out at 20 °C using an IPGPhor electrophoresis unit (Amersham Biosciences) as follows: 200 V, 30 min; 500 V, 30 min; 1000 V, 30 min; ramp to 4000 V over 30 min; 4000 V for 12,000 V-h). After focusing, the IPG strip was incubated for 15 min with 1% dithiothreitol in equilibration buffer (6 m urea, 50 mm Tris, pH 8.8, 30% glycerol, 2% SDS), followed by a 15-min alkylation with 2% iodoacetamide in equilibration buffer. IPG strips were then placed into the IPG wells of 4–20% Zoom gels (Invitrogen) and sealed in place with 0.5% agarose with bromphenol blue to be fractionated.
Western Blot Analysis
After fractionation by SDS-PAGE, proteins were electroblotted to polyvinylidene difluoride membranes (Millipore). Antigen-antibody complexes were visualized using horseradish peroxidase-conjugated secondary antibody and Super Signal West Pico chemiluminescence substrate (Pierce). To reprobe with a different antibody, membranes were stripped by incubation at 50 °C for 30 min in 62.5 mm Tris-HCl, pH 6.7, 2% SDS containing 0.1 m 2-mercaptoethanol. PAM-1 was visualized using a rabbit polyclonal antibody (JH629) to Exon A (rPAM-1-(394–498)); this antibody recognizes both PHM and PAL produced from PAM-1 (11). Rabbit polyclonal antisera to phospho-Ser949 (JH 2541) (12) and phospho-Ser937 (JH1922) (13) were used to detect PAM-CD phosphorylated at these residues. A polyclonal antibody (C-stop) to the final 12 residues of PAM was raised by immunizing rabbits with the Cys-extended peptide (Cys-Y965SAPLPKPAPSS976) conjugated to keyhole limpet hemocyanin (Covance); following ammonium sulfate precipitation and dialysis, C-stop antibody was affinity-purified using AffiGel 10 (Bio-Rad) resin conjugated to peptide antigen. Actin was immunoblotted with a monoclonal antibody (JLA20) from the Developmental Studies Hybridoma Bank (University of Iowa). Polyclonal antibody to histone H3 (Cell Signaling Technology, Inc.) was used to identify nuclear fractions. Immunoblots of two-dimensional gels were analyzed using ImageJ software (National Institutes of Health). Where indicated, membranes were cut above the 25 kDa marker; the two pieces were incubated separately with the same stock of C-stop and second antibody to eliminate any possibility of competition for limited amounts of antibody and to allow the use of different exposure times.
Immunoprecipitation and Mass Spectrometry
Membrane extracts prepared from AtT-20 cell lines overexpressing PAM-1 or Myc-TMD-CD were immunoprecipitated with Exon A or C-stop polyclonal antibody using Protein A beads (200 μg of membrane protein). Bound proteins were eluted by boiling in Laemmli sample buffer and separated on 4–20% Tris-glycine gels (Invitrogen). Gels were stained with Coomassie Brilliant Blue R250 or GelCode Blue (Pierce), and the band of interest was excised for mass spectrometric analysis. Gel pieces were processed for in situ enzymatic digestion with endoproteinase Lys-C or chymotrypsin (24), followed by separation into flow-through and titanium dioxide-bound fractions. TiO2 Top Tips (Glygen Corp.) were washed with 40 μl of 100% acetonitrile, followed by 0.2 m sodium phosphate, pH 7.0, and then by 0.5% trifluoroacetic acid, 50% acetonitrile. The acidified digest was spun onto the TopTip (1000 rpm for 1 min and then 3000 rpm for 2 min) prior to a 0.5% trifluoroacetic acid, 50% acetonitrile (40 μl) wash to generate the flow-through fraction. Phosphopeptides were eluted using 3 × 30 μl of 28% NH4OH. Both fractions were dried prior to dissolving in 3 μl of 70% formic acid and dilution with 7 μl of 0.1% trifluoroacetic acid. Liquid chromatography-MS/MS analysis (5 μl injected) was performed on a Thermo Scientific LTQ-Orbitrap XL equipped with a Waters nanoAcquity (25). MS/MS and MS3 spectra were obtained utilizing neutral loss (32.7 triply, 49 doubly, and 98 singly charged). Spectra were analyzed using the Mascot algorithm (26) along with manual verification of the sequence.
Isolation and Analysis of Nuclei
Cardiomyocyte and pituitary nuclei were purified from tissue taken from adult female Sprague-Dawley rats (27). Tissue rinsed with L-15 medium was suspended in 10–15 volumes of MA buffer (10 mm Tris-HCl, pH 7.4, 2 mm MgCl2, 0.25 m sucrose) with phenylmethylsulfonyl fluoride and protease inhibitors. Tissue was homogenized for 10–12 s with a Polytron homogenizer at setting 4 (homogenate), followed by centrifugation at 1000 × g for 10 min. The pellet was resuspended in 10 volumes of MA buffer, further homogenized with a Teflon homogenizer (6 strokes), and filtered through a nylon membrane (pore size 70 μm); the supernatant (S1) was retained for analysis. The filtrate was centrifuged at 1000 × g for 10 min, and the pellet was resuspended in MA buffer containing 0.5% Triton X-100; the supernatant (S2) was retained for analysis. After centrifugation as above, the pellet was resuspended in 800 μl of MB buffer (10 mm Tris-HCl, pH 7.4, 2 mm MgCl2, 2.2 m sucrose) with phenylmethylsulfonyl fluoride and protease inhibitor mixture. Resuspended nuclei (gradient input) were layered onto a 1.4-ml layer of 2.4 m sucrose in MB buffer and centrifuged for 2 h at 86,000 × gavg in a TLS55 rotor in a Beckman TL-100 ultracentrifuge. All but 200 μl of buffer covering the white pellet at the bottom of the tube was removed; the remainder was diluted with 4 volumes of MA buffer so that nuclei could be pelleted by centrifugation at 2000 × g for 20 min. The supernatant was discarded, and the white nuclear pellet was resuspended in 40 μl of MA buffer. Freshly isolated nuclei were fixed in 3.7% (w/v) formaldehyde in PBS (50 mm sodium phosphate, 150 mm NaCl, pH 7.4) for 30 min at 4 °C, and aliquots were placed onto polylysine-coated coverslips as described (28). After rinsing with PBS, nuclei were permeabilized with 0.075% Triton X-100 in PBS for 20 min and blocked with 2 mg/ml bovine serum albumin in PBS for 1 h at room temperature. Fixed nuclei were incubated with affinity-purified C-stop antibody (1:200) diluted in blocking buffer overnight. Specificity was confirmed by preblocking the antibody with antigenic peptide (10 μg/ml) for 1 h at room temperature prior to immunostaining. Following extensive rinsing with PBS, nuclei were incubated in Cy3-conjugated second antibody (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature in the dark. TOPRO3 (1:500 in PBS for 30 min) was used to identify nuclei. Coverslips were mounted on slides using Prolong Gold anti-fade reagent (Molecular Probes). Confocal microscopy was performed in the Center for Cell Analysis and Modeling using a Zeiss LSM510 microscope (Thornwood, NY).
RESULTS
PAM-CD Is Unstructured, with Multiple Predicted Phosphorylation Sites
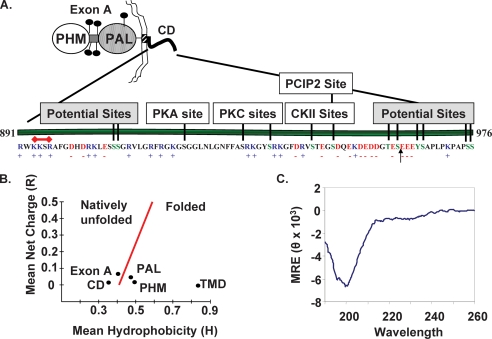
The cytosolic domain of PAM, although not as well conserved as its catalytic cores, is more highly conserved than its signal sequence or transmembrane domain (29). Basic residues predominate near the transmembrane domain, with the K893KSR sequence meeting the criteria for a monopartite nuclear localization signal (30); acidic residues are prevalent near the C terminus (Fig. 1A). Of the 15 Ser/Thr residues in rat PAM-CD, Disphos (Disorder-enhanced Phosphorylation Predictor) (available on the World Wide Web) identifies 11 as likely phosphorylation sites (probability score of >0.5) (Fig. 1A). The five sites previously identified by in vitro phosphorylation of recombinant PAM-CD using purified protein kinase C, protein kinase A, casein kinase 2, and P-CIP2 bring the total number of sites to be considered to 13 (11, 31).
FIGURE 1.
PAM-CD is unstructured and has many predicted phosphorylation sites. A, the independently folded PHM and PAL catalytic domains of PAM-1 are separated by an O-glycosylated linker region (exon A) and followed by a transmembrane helix that terminates with a stop-transfer signal (R891WKK) that overlaps a putative nuclear localization signal (K893KSR; red bar) that connects to a cytosolic domain terminating with Ser976. An arrow indicates the C terminus of PAM-CD* purified from bacterial lysates and subjected to MALDI-TOF analysis. Phosphorylation sites predicted by Disphos (green letters) or demonstrated experimentally using purified protein kinases are shown. B, the mean net charge and mean hydrophobicity of PHM (rPAM-1-(42–356)), PAL (rPAM-1-(498–820)), exon A (rPAM-1-(393–497)), PAM-TMD (rPAM-1-(867–890)), and PAM-CD (rPAM-1-(891–976)) were calculated and plotted according to Uversky et al. (31). C, secondary structure of purified recombinant PAM-CD was determined by circular dichroism spectroscopy at 20 °C at pH 7.4 The mean residue ellipticity (MRE) is in degrees·cm2·dmol−1. The far-UV spectrum has a single negative peak at 200 nm, a pattern characteristic of a random coil.
Mean hydrophobicity and mean net charge can be used to predict regions expected to have a natively unfolded structure (32). When applied to PAM, the catalytic cores (PHM and PAL), along with its single transmembrane domain, fell into the folded category (Fig. 1B). In contrast, both PAM-CD and the region between PHM and PAL (Exon A) fell into the natively unfolded category. IUPRED, a Web server that predicts intrinsically unstructured regions in proteins, also predicts PAM-CD to be intrinsically unstructured (33). Recombinant PAM-CD purified from bacterial lysates was used to test this prediction. Consistent with the protease sensitivity expected of natively unfolded proteins, MALDI-TOF analysis indicated that PAM-CD purified from bacteria expressing rat PAM-1-(898–976) terminated at Ser961. Antibody directed to the C terminus of PAM (-Ser976) confirmed the absence of this epitope, and this C-terminally truncated fragment of PAM-CD is here referred to as PAM-CD*. The secondary structure of purified PAM-CD* was analyzed by circular dichroism spectroscopy (Fig. 1C). The spectrum obtained, with its negative peak at 200 nm, indicated that PAM-CD* was largely a random coil at pH 7.4. NMR studies of purified 15N-labeled PAM-CD were consistent with this conclusion (data not shown).
PAM-CD Is Subject to Basal and Secretagogue-stimulated Phosphorylation in AtT-20 Cells and Primary Pituitary Cells
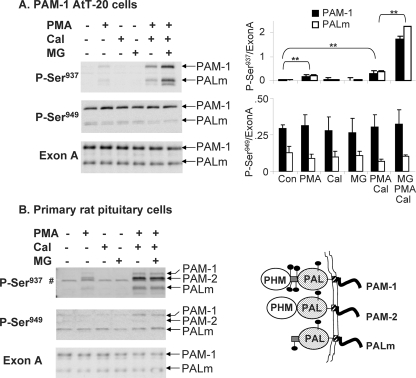
Antisera specific for the protein kinase C site in PAM-CD, Ser(P)937, and for the P-CIP2 or Casein Kinase 2 site, Ser(P)949, were used to compare PAM phosphorylation in AtT-20 corticotrope tumor cells and primary pituitary cells. In both systems, stimulated exocytosis evoked by treatment with the secretagogue phorbol myristate acetate (phorbol ester) was necessary before phosphorylation of Ser937 was detectable (Fig. 2, A and B). Although calyculin A alone was without effect, inclusion of this inhibitor of protein phosphatases 1 and 2A in phorbol ester-stimulated cultures increased levels of Ser(P)937 in PAM-1 AtT-20 cells and in primary cultures. In primary pituitary cells, Ser(P)937 was detected in PAM-1, PAM-2 (an alternatively spliced isoform that lacks exon A), and PALm (Fig. 2B, right). Although inclusion of a proteasome inhibitor (MG132) increased Ser(P)937 levels in AtT-20 cells, it did not do so in pituitary cells.
FIGURE 2.
Basal and stimulated phosphorylation of PAM-CD. PAM-1 AtT-20 cells (A) and primary cultures of rat anterior pituitary cells (B) were exposed to phorbol ester (PMA) (1 μm for 1 h), calyculin A (Cal) (12.5 nm for 1 h), MG132 (MG) (2 μm for 3 h) alone or in combination, as indicated, before extraction and Western blot analysis using antibody to Ser(P)937, Ser(P)949, or exon A. Data for PAM-1 AtT-20 cells from two separate experiments and three separate gels were quantified by taking the ratio (arbitrary units) of the Ser(P)937 or Ser(P)949 signals to the exon A signal for PAM-1 or PALm (right); *, p < 0.02 by independent t test. In rat pituitary cells, PAM-2, which cannot be detected by the exon A antibody, was not well recognized by the Ser(P)949 antibody; #, a nonspecific band. Pituitary cultures were analyzed two times with similar results, but use of animals of variable ages and hormonal status precluded averaging the data. A schematic comparing PAM-1 to PAM-2 and PALm is shown in the bottom right.
When the same samples were examined using antibody specific for Ser(P)949, a site phosphorylated by P-CIP2 and by casein kinase 2, basal phosphorylation was apparent in both AtT-20 cells and primary pituitary cells (Fig. 2). Neither stimulation with phorbol ester nor the addition of calyculin A had any effect on the level of phosphorylation at this site. Although PAM-2 is more prevalent than PAM-1 in rat pituitary, it was barely detected by the Ser(P)949-specific antibody, revealing the occurrence of isoform-specific phosphorylation. This simple analysis of factors affecting phosphorylation at these two sites revealed rapid turnover and site-specific regulation, validating the use of stably transfected AtT-20 cells as a model for pituitary endocrine cells.
Mass Spectrometric Identification of Phosphorylation Sites
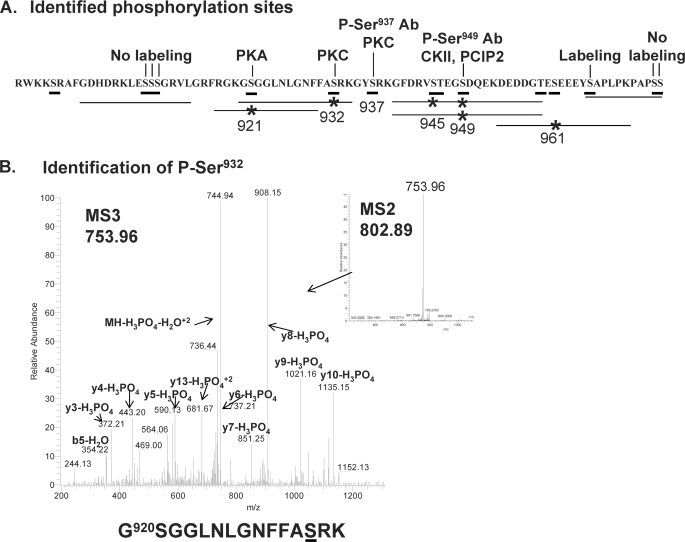
To identify phosphorylation sites in PAM, membranes were prepared from AtT-20 cell lines stably expressing PAM-1 or its Myc-tagged transmembrane/cytosolic domain (Myc-TMD-CD). To facilitate identification of phosphorylation sites, cells were pretreated with phorbol ester and calyculin A before membrane preparation. The solubilized membrane proteins were immunoprecipitated using antibody to exon A or the C terminus of PAM-1. Following SDS-PAGE, Coomassie-stained bands corresponding to PAM-1 or Myc-TMD-CD were excised for analysis. Peptides prepared using endoproteinase Lys-C or chymotrypsin were separated into a phosphopeptide-enriched fraction by binding to titanium dioxide beads. Both the bound and flow-through fractions were subjected to liquid chromatography-MS/MS analysis on an LTQ Orbitrap. Phosphopeptides identified by MS/MS and verified manually are indicated by lines, with asterisks indicating phosphorylation sites (Fig. 3); the spectrum identifying Ser(P)932 is shown, and the remaining spectra are provided in the supplemental material. Five phosphorylation sites, Ser921, Ser932, Ser945, Ser949, and Ser961, were identified in this way; a doubly phosphorylated peptide containing both Ser(P)945 and Ser(P)949 was also identified. Despite the use of multiple different enzymes, no peptides containing Ser937 or Ser(P)937 were identified by mass spectrometry. Since phosphorylation at this site was demonstrated using a Ser(P)937-specific antibody, it is included in Fig. 3. Previous radiolabeling experiments using 32PO42−-containing medium to label truncated PAM-1 proteins expressed in AtT-20 cells suggested phosphorylation of Ser966, which was not identified by mass spectrometry. Taken together, we can conclude that at least 6 Ser residues in PAM-CD are phosphorylated.
FIGURE 3.
Summary of identified phosphorylation sites. A, the horizontal lines drawn below the sequence of PAM-CD identify the phosphopeptides whose sequences were verified; *, the presence of phosphorylated Ser or Thr. Phosphorylation sites identified by other means are indicated above the line. B, the inset contains the MS2 spectrum of the 802.892+ phosphopeptide precursor ion of PAM-1-(920–934); the LTQ Orbitrap MS3 spectrum is of the 753.962+ neutral loss ion showing phosphorylation of Ser932; this peptide was isolated from an endopeptidase Lys-C digest following TiO2 enrichment. Other spectra are included in the supplemental material.
PAM-CD Contributes to the Acidic Isoelectric Point of PAM-1
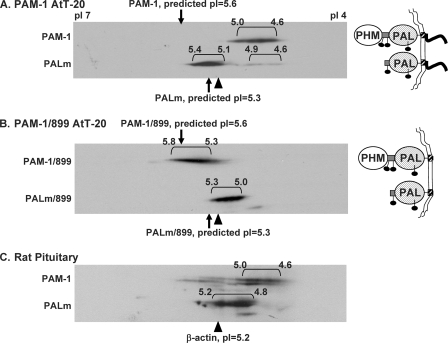
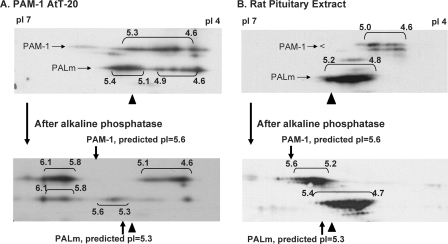
Two-dimensional gel electrophoresis was selected as a means of evaluating the phosphorylation state of PAM-1. Membrane proteins solubilized from PAM-1 AtT-20 cells were resolved on pH 4–7 linear Immobiline strips, and proteins were then separated by SDS-PAGE. Following transfer to a membrane, PAM-1 and PALm were visualized using antibody to exon A (Fig. 4A). A heterogeneous array of spots was observed for both PAM-1 and PALm. Isoelectric point calculations were based on a pI of 5.25 for β-actin, which was readily identified by Coomassie staining and verified by immunoblotting. Based solely on amino acid composition, PAM-1 and PALm should focus at pH 5.6 and 5.3, respectively. Predicted isoelectric points with a varying number of phosphates were calculated using ScanSite (available on the World Wide Web). PAM-1, with an array of spots extending from pH 4.6 to 5.0, was more acidic than expected. PALm was distributed among two heterogeneous groups, the major one focusing near the predicted pI (pI 5.1–5.4).
FIGURE 4.
Luminal domains of PAM-1 contribute to charge heterogeneity. Lysates (75 μg of protein) prepared from basal AtT-20 lines expressing PAM-1 (A) or PAM-1/899 (B) and solubilized membrane proteins from rat anterior pituitary (150 μg) (C) were separated by two-dimensional gel electrophoresis. Samples were focused on pH 4–7 linear pI strips to resolve charge variants and then separated based on molecular weight by SDS-PAGE. PAM-1 proteins were visualized using antibody to exon A. Isoelectric points were assigned using β-actin (pI 5.2) and the edges of the strips (pI 7.0 and pI 4.0) as reference points. The images shown are representative of five similar analyses of PAM-1 cells and four similar analyses of rat pituitary.
To assess the contribution of luminal domain modifications, such as N- and O-glycosylation, to the observed isoelectric point, PAM-1/899 was analyzed (Fig. 4B); this protein terminates just after its transmembrane domain, and its predicted isoelectric point is 5.6. PAM-1/899 yielded an array of species focusing from pH 5.3 to 5.8. Although differences in trafficking mean that the oligosaccharides attached to PAM-1 and PAM-1/899 need not be identical, the cytosolic domain of PAM-1 is thought to account for most of the 0.7-pH unit reduction in its isoelectric point. Cleavage of PHM from PAM-1/899 yields PALm/899, which focused at approximately the same pI as PALm. Although its luminal domain clearly makes PALm/899 more heterogeneous and more acidic than predicted, the absence of the cytosolic domain had little effect.
To verify that AtT-20 PAM-1 reflected the behavior of endogenous PAM-1, membrane extracts from rat anterior pituitary were subjected to isoelectric focusing (Fig. 4C). In addition to mature PAM-1, what is thought to be an immature form (34) was observed at a slightly lower molecular weight. As in AtT-20 cells, pituitary PAM-1 consistently focused from pH 4.6 to 5.0, demonstrating that it was substantially more acidic than predicted based on its amino acid composition. PALm in pituitary extracts focused from pH 4.8 to 5.2, mimicking the behavior of AtT-20 PALm. With a predicted pI of 5.3, PALm did not appear to be modified as extensively as PAM-1. We next explored the hypothesis that multiple phosphorylation of its cytosolic domain contributed to the increased acidity of PAM-1.
Multisite Phosphorylation of PAM-1
To determine the contribution of phosphorylation to its isoelectric point, we evaluated the effect of alkaline phosphatase on the isoelectric point of AtT-20 PAM-1 (Fig. 5A). Since phosphatase treatment of solubilized AtT-20 membranes was ineffective, PAM-1 was enriched by immunoprecipitation before digestion with alkaline phosphatase. Alkaline phosphatase treatment shifted the pI of most of the PAM-1 from pH 4.6–5.3 to pH 5.8–6.1. PALm shifted from an isoelectric point of pH 5.1–5.4 to an isoelectric point of pH 5.8–6.1 following phosphatase treatment. Despite multiple additions of phosphatase, a substantial amount of acidic PAM-1 and PALm remained. The different focusing patterns observed for PAM-1 and PALm in Figs. 4A and 5A reflect changes that occur with time in culture and differences in sample preparation (immunoprecipitates in Fig. 5A versus lysates in Fig. 4A).
FIGURE 5.
PAM-1 is multiply phosphorylated. A, extracts of basal PAM-1 AtT-20 cells were immunoprecipitated with polyclonal exon A antibody and analyzed before (top) and after (bottom) phosphatase treatment. Analysis of immunoprecipitates versus lysates may have contributed to the different pattern observed here and in Fig. 4A. B, solubilized membrane extracts (250 μg of protein) prepared from rat anterior pituitary were analyzed before (top) and after (bottom) phosphatase treatment. Samples were analyzed as described in the legend to Fig. 4. Phosphatase-treated PAM-1 showed a marked increase in pI, suggesting that multiple phosphates contributed to its low pI.
Pituitary extracts were also treated with alkaline phosphatase (Fig. 5B). The pI of pituitary PAM-1 shifted from pH 4.6–5.0 to pH 5.2–5.9; only a small shift in the pI of PALm was observed. The charge heterogeneity that remained following phosphatase treatment may reflect the presence of charged oligosaccharides, as observed in PAM-1/899. Based on calculated pI values, a shift of 0.4 pH units corresponds to the removal of 10–12 phosphate moieties from PAM-1. The fact that almost all of the PAM-1 focused at a substantially more basic position following phosphatase treatment indicates that almost all of it was multiply phosphorylated. PALm was less extensively phosphorylated than PAM-1.
Many of the Highly Charged Variants of PAM-1 Contain Ser(P)937 and Ser(P)949
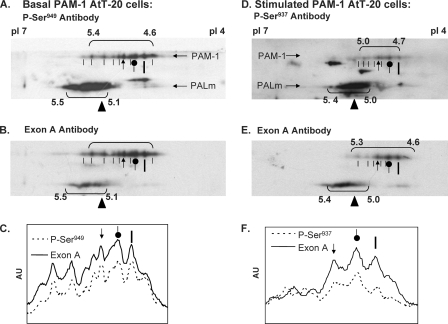
Under basal conditions, PAM-1 is recognized by antibody to Ser(P)949 but not by antibody to Ser(P)937. A comparison of the charge variants of PAM-1 recognized by the Ser(P)949 antibody and then by the Exon A antibody revealed an almost identical pattern, suggesting extensive basal phosphorylation at this site (Fig. 6, A and B). Extracts prepared from cells stimulated with phorbol ester in the presence of calyculin A were fractionated in a similar manner and visualized using antibody to Ser(P)937 followed by antibody to Exon A (Fig. 6, D and E).
FIGURE 6.
Identification of Ser(P)937 and Ser(P)949 in AtT-20 PAM-1. PAM-1 AtT-20 cells harvested under basal conditions (A–C) or after stimulation with phorbol ester plus calyculin A (D–F) were subjected to two-dimensional PAGE as in Fig. 4. Phosphospecific antibodies were applied first; Ser(P)949-specific antibody was used to analyze the basal sample (A), and Ser(P)937 antibody was used to analyze the stimulated sample (D). Blots were then stripped and exposed to exon A antibody to identify total PAM-1 (B and E). Horizontal line scans through the collection of PAM-1 spots identified by each antibody are shown; orientation marks from A and D are repeated to facilitate spot identification. Ser(P)937 was more prevalent among the less acidic variants of PAM-1. Similar results were obtained for independent samples. AU, arbitrary units.
A comparison of the basal and stimulated patterns observed using the exon A antibody revealed a shift of PAM-1 into more acidic forms; little change was observed in PALm (Fig. 6, B and E). Profile scans across the immunoblots are shown to facilitate comparison between the exon A and phosphorylation site-specific patterns (Fig. 6, C and F). The most acidic form of PAM-1 (pI 4.6) was barely detected by the Ser(P)937 antibody, whereas the less acidic forms (pI 5.0–5.3) were recognized. These differences may reflect antibody specificity or a hierarchy among phosphorylation sites. Although PALm is recognized by the Ser(P)937 and Ser(P)949 antibodies, individual charge variants were not well resolved.
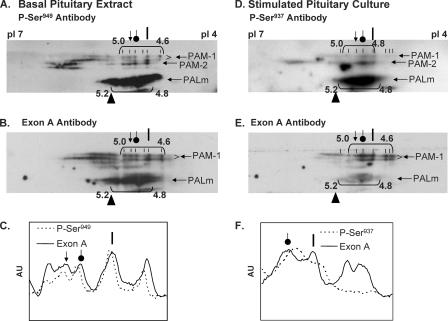
Phosphorylation of PAM-1 at Ser949 and at Ser937 Is Similar in AtT-20 Cells and Primary Pituitary Cells
The phosphorylation of PAM in primary cultures of rat anterior pituitary was examined in a similar manner. In membrane extracts prepared from tissue, PAM-1 yielded an array of charge variants focusing from pH 4.6 to 5.0 (Fig. 7, A and B). The Ser(P)949 antibody recognized each of the more acidic PAM-1 variants (pI < 5.0). Primary cultures treated with phorbol ester and calyculin A were analyzed using antibodies to Ser(P)937, followed by exon A antibody (Fig. 7, D and E). As in AtT-20 cells, secretagogue treatment shifted most of the PAM-1 into the more acidic variants (pI 4.6–5.0). The most negatively charged forms of PAM-1 (pI 4.6) were enriched following secretagogue treatment, but, as observed in AtT-20 PAM-1 cells, these variants were not detected by the Ser(P)937 antibody (Fig. 7F). This shift in isoelectric point suggests that phosphorylation at Ser937 is coupled to dephosphorylation at other sites. The combinatorial use of multiple phosphorylation sites allows unstructured domains to convey a great deal of information to interactor proteins.
FIGURE 7.
Identification of Ser(P)937 and Ser(P)949 in pituitary PAM-1. Rat anterior pituitaries (A–C) and pituitary cultures stimulated with phorbol ester plus calyculin A (D–F) were analyzed as in Fig. 6. The most acidic forms of PAM-1 were all recognized by the Ser(P)949 antibody. Ser(P)937 was detected only after stimulation, and the most acidic forms of PAM-1 were not phosphorylated at this site, demonstrating specificity in the combinatorial use of phosphorylation sites. The images shown are representative of two similar analyses. AU, arbitrary units.
A Soluble, Proteasome-sensitive PAM-CD Fragment Is Generated from PAM-1
Unstructured domains are typically protease-sensitive (2). To search for fragments of PAM-CD, we produced an antibody (C-stop) specific for a peptide encompassing its 12 C-terminal residues. To facilitate the identification of products, we compared total lysates prepared from AtT-20 cells expressing PAM-1 or Myc-TMD-CD. Myc-TMD-CD retains the PAM signal sequence and replaces both catalytic domains with the Myc epitope. Myc-TMD-CD traffics through secretory granules and the endocytic compartment much like PAM-1 (23).
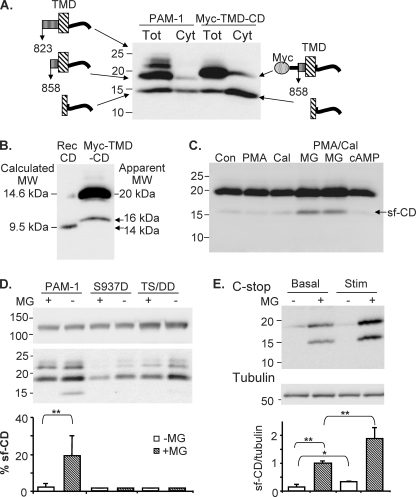
PAM-1 lysates contained C-stop fragments of 22, 19, and 16 kDa (Fig. 8A); cleavage of PAM-1 immediately after PAL (at Lys-Lys822) or closer to the transmembrane domain would generate products of this size. Myc-TMD-CD lysates contained a major 20-kDa protein recognized by antibodies to Myc (not shown) and C-stop (Fig. 8A); as in PAM-1 lysates, a 16-kDa protein was detected by the C-stop antibody. Based on its apparent mass, we hypothesized that the 16-kDa fragment lacked a transmembrane domain. To test this hypothesis, cytosolic fractions prepared from PAM-1 and Myc-TMD-CD cells were analyzed (Fig. 8A). The cytosolic fraction contained the 16 kDa band, here referred to as sf-CD.
FIGURE 8.
Identification of a soluble fragment generated from the cytosolic domain of PAM. A, cytosol (Cyt) and total lysate (Tot) prepared from MG132-treated PAM-1 and Myc-TMD-CD cells were analyzed using the C-stop antibody. B, purified recombinant PAM-CD-5P (calculated molecular mass 9.5 kDa) and total lysate prepared from MG132-treated Myc-TMD-CD cells were analyzed on 16% acrylamide gels. C, Myc-TMD-CD cells were treated with phorbol ester (PMA) (1 μm, 1 h), calyculin A (12.5 nm, 1 h), and MG132 (2 μm, 3 h) alone or in combination, as indicated, or cyclic 8-bromo-AMP (0.5 mm, 1 h) before extraction, separated on a 4–20% acrylamide gel, and analyzed by Western blot with C-stop antibody. D, PAM-1, PAM-1/S937D (S937D), and PAM-1/T946D,S949D (TS/DD) cells harvested under control conditions or after treatment with MG132 for 4.5 h were analyzed using the C-stop antibody; the top and bottom parts of the blot are shown. For both mutant lines, duplicate samples were analyzed; for PAM-1, n = 5; for each sample, sf-CD was expressed as a percentage of intact 120-kDa PAM. E, PAM-1 AtT-20 cells were treated with BaCl2 (2 mm) and phorbol ester (1 μm), using a paradigm known to deplete cells of hormone (15) in the presence or absence of MG132 (2 μm). Control cells kept in the presence or absence of MG132 were also examined. Lysates (20 μg of protein) were separated on a 4–20% acrylamide gel, and blots were probed with C-stop and tubulin antibodies. A representative gel is shown. Data were quantified by normalizing the signal for sf-CD to that for tubulin and setting the average for the basal/+MG samples in each of three experiments to 1.0 (bottom); *, p < 0.05; **, p < 0.02.
Attempts to sequence sf-CD by Edman degradation following immunoprecipitation from cytosol using the C-stop antibody were unsuccessful; since an adequate amount of protein appeared to be present, its N terminus may be blocked. To estimate the site of cleavage, we compared the behavior of intact recombinant PAM-CD (calculated molecular mass 9.5 kDa after cleavage from glutathione S-transferase) and Myc-TMD-CD lysates on stiffer gels (Fig. 8B). sf-CD generated from PAM-1 or from Myc-TMD-CD was about 1 kDa larger than recombinant PAM-CD, suggesting that the cleavage generating this soluble fragment occurred in the transmembrane domain.
We next sought to identify factors leading to the accumulation of sf-CD. Myc-TMD-CD cells were treated with various secretagogues, a phosphatase inhibitor, and a proteasome inhibitor, and levels of sf-CD were assessed (Fig. 8C). In the presence of MG132, levels of both intact Myc-TMD-CD and sf-CD increased substantially, suggesting a role for proteasomal degradation. Exposure to secretagogue for 30 min did not produce an increase in levels of sf-CD, even in the presence of MG132.
sf-CD Levels Are Decreased by Phosphorylation and Increased by Prolonged Stimulation
We next asked whether phosphorylation played a role in generation of sf-CD by using AtT-20 cell lines expressing PAM-1 with phosphomimetic mutations (Asp) at the PKC site, Ser937 (PAM-1/S937D) (13), or the P-CIP2 sites, Thr946 and Ser949 (PAM-1/TS/DD) (12) (Fig. 8D). These mutant PAM-1 proteins traverse the early parts of the endocytic pathway like PAM-1 but exhibit defects in the later parts of the endocytic recycling pathway and fail to return to the trans-Golgi network region as efficiently as PAM-1 (12, 13). Even in the presence of MG132, very little sf-CD could be detected in PAM-1/S937D or PAM-1/TS/DD lysates.
We next exposed AtT-20 cells to sequential applications of BaCl2 and phorbol ester so that they would continue to secrete endogenous peptide hormone over a long period of time, leading to secretory granule depletion (15). When AtT-20 PAM-1 cells were exposed to this stimulation paradigm in the absence or in the presence of MG132, an increased amount of sf-CD accumulated (Fig. 8E). Taken together, our data indicate that CD phosphorylation affects the ability of secretagogue to stimulate cleavage of PAM and release of a protease-sensitive soluble fragment, sf-CD, into the cytosol.
sf-CD Is Localized to the Nucleus
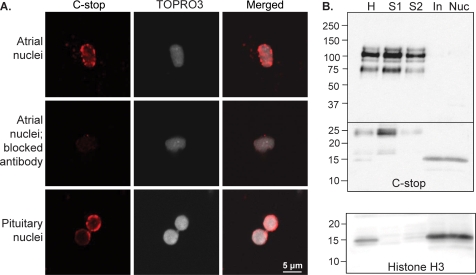
Expression of PAM-1 is known to alter cytoskeletal organization and regulated secretion (17). To determine whether sf-CD might play a role in signaling from the regulated secretory pathway to the nucleus, we isolated nuclei from rat atrium and pituitary. Atrial nuclei were visualized using the C-stop antibody and TOPRO3 (Fig. 9A). Staining for PAM-CD was speckled and was concentrated along the nuclear membrane; only a subset (∼5–10%) of the nuclei prepared from rat atrium were stained. Specificity was established by eliminating the signal following preincubation of the C-stop antibody with antigen. C-stop staining was also detected in nuclei isolated from rat pituitary (Fig. 9A); signal was again concentrated along the nuclear membrane, and levels varied in different nuclei.
FIGURE 9.
Localization of sf-CD to the nucleus. A, nuclei isolated from rat atrium and pituitary were visualized with C-stop antibody and TOPRO3; merged images are shown on the right. Preincubation with the antigenic peptide (Blocked Antibody) eliminated the C-stop signal. Punctate staining for PAM was concentrated along the nuclear membrane. B, equal amounts of protein (20 μg) from the preparation of rat pituitary nuclei were subjected to Western blot using C-stop antibody: homogenate (H), supernatant fractions (S1 and S2), gradient input (In), nuclear pellet (Nuc). Nuclei were enriched in sf-CD; the membrane was cut, but both parts were processed using the same solutions and exposure times. C, the identity of the nuclear fractions (12 μg) was verified by blotting for histone H3.
To determine whether sf-CD was responsible for the staining observed, nuclei purified from pituitary were subjected to Western blot analysis using the C-stop antibody (Fig. 9B). Homogenate, supernatants, gradient input, and nuclear pellets were examined. sf-CD was enriched in nuclear pellets prepared from the pituitary. The identity of the nuclear fraction was verified by the presence of histone 3 (Fig. 9B, bottom). Similar results were obtained when nuclei were purified from adult rat atrium (data not shown). Identification of endogenous sf-CD in the nucleus suggests that it plays a role in signaling pathways integrating control of the regulated secretory pathway with other aspects of cellular metabolism.
DISCUSSION
The PAM Gene Acquires Unstructured Domains
Unstructured regions in proteins can be predicted based on sequence and their prevalence increases with organism complexity (8, 35). Neuropeptides are synthesized in the same way in Hydra, creatures with a primitive nerve net, and in humans. PAM, one of the few enzymes unique to the synthesis of neuropeptides, has two highly conserved catalytic domains that function sequentially to produce amidated peptides from glycine-extended precursors. In Hydra and Drosophila, these two enzymes are encoded by separate genes. In vertebrates, a single gene encodes both enzymes and also encodes two putative unstructured domains; one is a linker region separating the catalytic cores, and the other is PAM-CD. As is typical of many putative unstructured domains, both regions are subject to alternative splicing (8).
The protease sensitivity of the two putative unstructured domains in PAM was apparent in early studies that defined its catalytic cores (36). As for many other Type I integral membrane proteins, its cytosolic domain plays a critical role in PAM trafficking through the biosynthetic and endocytic pathways. Later studies revealed a role for PAM-1 in relaying information about the status of the secretory granule lumen to cytosolic interactors, resulting in alterations in cytoskeletal organization and secretagogue responsiveness (18). We show here that the cytosolic domain of PAM is unstructured and that two features of unstructured domains, multiple sites for phosphorylation and protease sensitivity, play key roles in PAM function. Thirteen of the 15 Ser/Thr residues in PAM-CD are predicted phosphorylation sites, and our data indicate that much of the PAM-1 in AtT-20 cells and pituitary is phosphorylated at 10–12 sites.
PAM-CD Interacts with Multiple Proteins in a Regulated, Phosphorylation State-dependent Manner
Roles for multisite phosphorylation have been elucidated in proteins involved in cell cycle regulation, transcription, and signaling. Multiple phosphorylation of the intracellular loops and C-terminal tail of some G protein-coupled receptors is known to regulate arrestin recruitment and receptor internalization (37). Yeast Sic1, which regulates the cell cycle, is phosphorylated and degraded when its threshold number of six sites is phosphorylated (5). Yeast Rad9 is phosphorylated in response to DNA damage; phosphorylation of six sites in Rad9 allows Rad53 to bind and activate the checkpoint pathway. Mutation of any single site does not inhibit binding, and restoring single phosphorylation sites in an all Ala mutant does not restore wild type binding (38). Multiple phosphorylation of the unstructured N-terminal domain of transcription cofactor PC4 causes a gradual masking of its Lys-rich region and regulates its exposed surface for DNA interaction (6). Several proteins with intrinsically unstructured domains are modulated by phosphorylation (Tau, p27, stathmin, α-synuclein, and cystic fibrosis transmembrane conductance regulator) (39); CREB, for example, activates transcription of target genes upon its phosphorylation state-dependent interaction with CBP (40).
Analysis of the biosynthetic and endocytic trafficking of PAM-1 mutated to Asp or Ala at Ser937 and at Ser949 revealed roles for phosphorylation and dephosphorylation at each site (Fig. 10). Under basal conditions, almost all of the charge variants of PAM-1 were phosphorylated on Ser949 but not on Ser937. Following stimulation with phorbol myristate acetate, Ser937 was phosphorylated. The use of phosphatase inhibitors demonstrated that phosphorylation at this site is normally short lived. PAM-1 in which Thr946 and Ser949 were both mutated to Asp was cleaved to yield soluble PHM more efficiently than wild type protein but failed to traverse the endocytic pathway normally. Quantitative electron microscopic studies demonstrated that this phosphomimetic mutant was unable to enter the intraluminal vesicles of multivesicular bodies,3 and we showed here that levels of sf-CD were greatly reduced (Fig. 10). PAM-1 with these same residues mutated to Ala rapidly entered the intraluminal vesicles, generated large amounts of soluble 100-kDa PAM, and returned to secretory granules more efficiently than wild type PAM-1. Taken together, our data suggest that sf-CD is generated in the late endocytic pathway (Fig. 10).
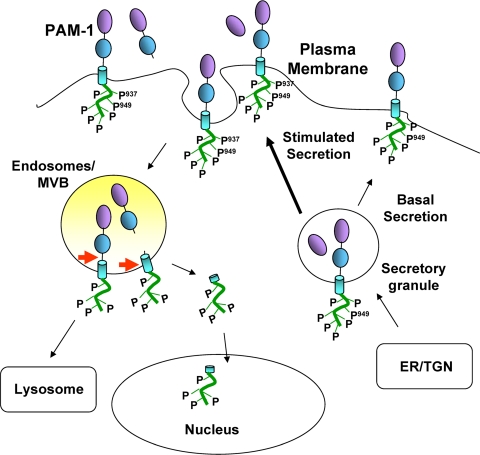
FIGURE 10.
Model. Under basal conditions, the unstructured PAM cytosolic domain is phosphorylated at multiple sites, including Ser949. Secretagogue stimulation results in the transient phosphorylation of Ser937 and dephosphorylation of other residues. The early steps of endocytosis are not affected by PAM phosphorylation, but dephosphorylation of multiple sites is necessary for generation of sf-CD, a soluble fragment that can enter the nucleus. Levels of sf-CD increase following prolonged stimulation of secretion and may relay information about the status of the secretory granule pool to the nucleus.
Unstructured domains have been noted in many proteins involved in endocytic trafficking and may provide a means of mediating the many specific low affinity interactions needed to control trafficking (41). P-CIP2, a protein kinase identified through its ability to bind PAM-CD, interacts with PAM-CD at Lys919, Leu926, and Phe929-Phe930 and phosphorylates Ser949, sites that extend over a 30-amino acid region (18). As for other unstructured domains, the few hydrophobic residues in PAM-CD play an important role in complex formation. The large surface area available for intermolecular contacts presumably provides specificity; whether PAM-CD undergoes induced folding upon interaction is not yet clear. The directionality necessary for trafficking may be driven by cooperative events that reflect multisite phosphorylation and dephosphorylation. In addition to P-CIP2, PAM-CD is known to interact with the spectrin repeat regions of two Rho GDP/GTP exchange factors, Kalirin and Trio, and with P-CIP1/RASSF9, a member of the RAS association domain-containing protein family (42).
A Soluble Fragment of PAM-CD Is Generated in Response to Secretagogue
PAM-1 is subject to endoproteolytic cleavages that produce soluble PHM and PAL, which are both released in response to secretagogue. Although these cleavages yield a cell-associated TMD-CD fragment, levels of this membrane protein are very low. Since PAM-CD is protease-sensitive, we explored the possibility that TMD-CD might accumulate in the presence of a proteasome inhibitor. Using an antibody to the C terminus of PAM-CD, both a membrane-associated fragment and a smaller cytosolic fragment were found. Although we have not yet identified the cleavage site, comparison with recombinant CD suggests that cleavage occurs in the transmembrane domain. Endogenous PAM-CD fragments of similar mass were identified in both pituitary and atrium, sites at which PAM is highly expressed. Phosphomimetic PAM-1 at Ser937 and Ser949 that have a defect in recycling in the late endocytic pathway are poor in generating sf-CD compared with wild type PAM-1. Therefore, dephosphorylation of PAM-1 at these sites is necessary for the release of sf-CD (Fig. 10).
Although an increase in sf-CD was not apparent after a single exposure to secretagogue, more intense stimulation of secretion through sequential exposure of AtT-20 cells to phorbol ester and BaCl2 in the presence of MG132 resulted in the accumulation of sf-CD (Fig. 10). ICA512, another Type I secretory granule membrane protein, undergoes μ-calpain-catalyzed cleavage following secretagogue-mediated entry of Ca2+ into insulin-secreting β-cells, triggering both changes in gene transcription and granule trafficking (43).
Endogenous sf-CD Is Localized to the Nucleus
As one of the final enzymes in the pathway leading to neuropeptides, PAM is well positioned to provide feedback about the status of the regulated secretory pathway. sf-CD could perform such a function. To determine whether endogenous sf-CD entered the nucleus, we turned to tissues expressing high levels of PAM. Nuclei isolated from rat pituitary were enriched in 16-kDa sf-CD (Fig. 10). Although sf-CD is small enough to pass through nuclear pores, its putative nuclear localization signal (KKSR) may play a role. Immunostaining revealed the presence of cross-reactive material in a subset of the nuclei isolated from pituitary and atria, with strong punctate staining along the nuclear membrane. When injected into the cytosol of AtT-20 cells or neurons, fluorescently tagged recombinant PAM-CD accumulated in the nucleus.4 Its ability to do so is sensitive to its phosphorylation state; recombinant PAM-CD with phosphomimetic mutations did not accumulate to the same extent as wild type PAM-CD. The interactions of PAM-CD with nuclear proteins may depend on features of its extended unstructured domain different from those that are important for interactions with cytosolic proteins.
Several proteins are known to produce cleaved signaling molecules that relay information to the nucleus. Soluble fragments generated from ICA512 (44), SREBP (45), Notch (46), and ATF6 (47) are larger and do not appear to be similar to sf-CD. Like PAM, other Type I integral membrane proteins with short cytosolic domains, which are often predicted to be unstructured (48), may yield soluble fragments with a role in signaling to the nucleus. Microarray data comparing gene expression in AtT-20 cells before and after expression of PAM-1 was induced identified a set of genes whose expression was altered.4 PAM-derived sf-CD may act as the messenger altering expression of this set of genes in response to signals generated in the lumen of the regulated secretory pathway in neurons, endocrine cells, and atrial myocytes.
Supplementary Material
Acknowledgments
We thank Dr. Mark Maciejewski for help with biophysical measurements, Dr. Erol Gulcicek for titanium enrichment of phosphopeptides, Drs. Steven E. Pfeiffer and Rashmi Bansal for use of the two-dimensional gel electrophoresis apparatus, and Darlene D'Amato for keeping the laboratory running.
This work was supported, in whole or in part, by National Institutes of Health, NIDDK, Grant DK32949; by NIDA Neuroproteomics Center Grant P30 DA018343; and by instrumentation obtained with funds from National Institutes of Health NCRR Clinical and Translational Science Award UL1 RR024139 and Shared Instrumentation Grant 1S10RR024617.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
N. Bäck, C. Rajagopal, R. E. Mains, and B. A. Eipper, manuscript in preparation.
V. P. Francone, M. F. Ifrim, C. Rajagopal, Y. Wang, J. H. Carson, R. E. Mains, and B. A. Eipper, manuscript in preparation.
- PAM
- peptidylglycine α-amidating monooxygenase
- PAM-CD
- PAM cytosolic domain
- sf-CD
- soluble fragment of cytosolic domain
- Myc-TMD-CD
- Myc-tagged transmembrane/cytosolic domain
- PHM
- peptidylglycine α-hydroxylating monooxygenase
- PAL
- peptidyl α-hydroxyglycine α-amidating lyase
- PALm
- PAL attached to the transmembrane/cytosolic domain
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- CIP
- calf intestinal alkaline phosphatase
- IPG
- ImmobilineTM Dry Strip pH 4–7 isoelectric focusing gel
- MS
- mass spectrometry
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. (2001) J. Mol. Graph. Model. 19, 26–59 [DOI] [PubMed] [Google Scholar]
- 2.Tompa P. (2002) Trends Biochem. Sci. 27, 527–533 [DOI] [PubMed] [Google Scholar]
- 3.Kalthoff C., Alves J., Urbanke C., Knorr R., Ungewickell E. J. (2002) J. Biol. Chem. 277, 8209–8216 [DOI] [PubMed] [Google Scholar]
- 4.Lenz P., Swain P. S. (2006) Curr. Biol. 16, 2150–2155 [DOI] [PubMed] [Google Scholar]
- 5.Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 6.Jonker H. R., Wechselberger R. W., Pinkse M., Kaptein R., Folkers G. E. (2006) FEBS J. 273, 1430–1444 [DOI] [PubMed] [Google Scholar]
- 7.Hsu J. M., Lee Y. C., Yu C. T., Huang C. Y. (2004) J. Biol. Chem. 279, 32592–32602 [DOI] [PubMed] [Google Scholar]
- 8.Dunker A. K., Silman I., Uversky V. N., Sussman J. L. (2008) Curr. Opin. Struct. Biol. 18, 756–764 [DOI] [PubMed] [Google Scholar]
- 9.Tausk F. A., Milgram S. L., Mains R. E., Eipper B. A. (1992) Mol. Endocrinol. 6, 2185–2196 [DOI] [PubMed] [Google Scholar]
- 10.Milgram S. L., Mains R. E., Eipper B. A. (1993) J. Cell Biol. 121, 23–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun H. Y., Milgram S. L., Keutmann H. T., Eipper B. A. (1995) J. Biol. Chem. 270, 30075–30083 [DOI] [PubMed] [Google Scholar]
- 12.Steveson T. C., Zhao G. C., Keutmann H. T., Mains R. E., Eipper B. A. (2001) J. Biol. Chem. 276, 40326–40337 [DOI] [PubMed] [Google Scholar]
- 13.Steveson T. C., Keutmann H. T., Mains R. E., Eipper B. A. (1999) J. Biol. Chem. 274, 21128–21138 [DOI] [PubMed] [Google Scholar]
- 14.Wasmeier C., Hutton J. C. (2001) J. Biol. Chem. 276, 31919–31928 [DOI] [PubMed] [Google Scholar]
- 15.Ferraro F., Eipper B. A., Mains R. E. (2005) J. Biol. Chem. 280, 25424–25435 [DOI] [PubMed] [Google Scholar]
- 16.Bell-Parikh L. C., Eipper B. A., Mains R. E. (2001) J. Biol. Chem. 276, 29854–29863 [DOI] [PubMed] [Google Scholar]
- 17.Ciccotosto G. D., Schiller M. R., Eipper B. A., Mains R. E. (1999) J. Cell Biol. 144, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alam M. R., Steveson T. C., Johnson R. C., Bäck N., Abraham B., Mains R. E., Eipper B. A. (2001) Mol. Biol. Cell 12, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alam M. R., Caldwell B. D., Johnson R. C., Darlington D. N., Mains R. E., Eipper B. A. (1996) J. Biol. Chem. 271, 28636–28640 [DOI] [PubMed] [Google Scholar]
- 20.Mains R. E., Alam M. R., Johnson R. C., Darlington D. N., Bäck N., Hand T. A., Eipper B. A. (1999) J. Biol. Chem. 274, 2929–2937 [DOI] [PubMed] [Google Scholar]
- 21.Yun H. Y., Johnson R. C., Mains R. E., Eipper B. A. (1993) Arch. Biochem. Biophys. 301, 77–84 [DOI] [PubMed] [Google Scholar]
- 22.Milgram S. L., Johnson R. C., Mains R. E. (1992) J. Cell Biol. 117, 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Meskini R., Galano G. J., Marx R., Mains R. E., Eipper B. A. (2001) J. Biol. Chem. 276, 3384–3393 [DOI] [PubMed] [Google Scholar]
- 24.Stone K. L., Williams K. R. (2004) Curr. Protocols Protein Sci. 11.3. [DOI] [PubMed] [Google Scholar]
- 25.Stone K. L., Crawford M., McMurray W., Williams N., Williams K. R. (2007) Methods Mol. Biol. 386, 57–77 [DOI] [PubMed] [Google Scholar]
- 26.Hirosawa M., Hoshida M., Ishikawa M., Toya T. (1993) Comput. Appl. Biosci. 9, 161–167 [DOI] [PubMed] [Google Scholar]
- 27.Boheler K. R., Chassagne C., Martin X., Wisnewsky C., Schwartz K. (1992) J. Biol. Chem. 267, 12979–12985 [PubMed] [Google Scholar]
- 28.Yao I., Iida J., Nishimura W., Hata Y. (2002) J. Neurosci. 22, 5354–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eipper B. A., Stoffers D. A., Mains R. E. (1992) Annu. Rev. Neurosci. 15, 57–85 [DOI] [PubMed] [Google Scholar]
- 30.Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldwell B. D., Darlington D. N., Penzes P., Johnson R. C., Eipper B. A., Mains R. E. (1999) J. Biol. Chem. 274, 34646–34656 [DOI] [PubMed] [Google Scholar]
- 32.Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 33.Dosztányi Z., Csizmok V., Tompa P., Simon I. (2005) Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 34.Maltese J. Y., Eipper B. A. (1993) Endocrinology 133, 2579–2587 [DOI] [PubMed] [Google Scholar]
- 35.Dyson H. J., Wright P. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 36.Husten E. J., Tausk F. A., Keutmann H. T., Eipper B. A. (1993) J. Biol. Chem. 268, 9709–9717 [PubMed] [Google Scholar]
- 37.Tobin A. B., Butcher A. J., Kong K. C. (2008) Trends Pharmacol. Sci. 29, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz M. F., Duong J. K., Sun Z., Morrow J. S., Pradhan D., Stern D. F. (2002) Mol. Cell 9, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 39.Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradoviæ Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 40.Radhakrishnan I., Pérez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., Wright P. E. (1997) Cell 91, 741–752 [DOI] [PubMed] [Google Scholar]
- 41.Dafforn T. R., Smith C. J. I. (2004) EMBO Rep. 5, 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherwood V., Manbodh R., Sheppard C., Chalmers A. D. (2008) Mol. Biol. Cell 19, 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ort T., Voronov S., Guo J., Zawalich K., Froehner S. C., Zawalich W., Solimena M. (2001) EMBO J. 20, 4013–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trajkovski M., Mziaut H., Altkrüger A., Ouwendijk J., Knoch K. P., Müller S., Solimena M. (2004) J. Cell Biol. 167, 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengoechea-Alonso M. T., Ericsson J. (2007) Curr. Opin. Cell. Biol. 19, 215–222 [DOI] [PubMed] [Google Scholar]
- 46.Bray S. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 47.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J. Y., Yang M. Q., Dunker A. K., Deng Y., Huang X. (2008) BMC Genomics 9, Suppl. 1, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.