Abstract
S-adenosyl-l-methionine (AdoMet) dependent methyltransferases (MTases) are involved in biosynthesis, signal transduction, protein repair, chromatin regulation and gene silencing. Five different structural folds (I–V) have been described that bind AdoMet and catalyze methyltransfer to diverse substrates, although the great majority of known MTases have the Class I fold. Even within a particular MTase class the amino-acid sequence similarity can be as low as 10%. Thus, the structural and catalytic requirements for methyltransfer from AdoMet appear to be remarkably flexible.
‘There are many paths to the top of the mountain, but the view is always the same.’…Chinese proverb
[(1996) The Columbia World of Quotations, New York Columbia University Press]
Following ATP, S-adenosyl-l-methionine (AdoMet) is the second most widely used enzyme substrate [1]. The majority of AdoMet-dependent reactions involve methyltransfer, leaving the product S-adenosyl-l-homocysteine (AdoHcy). The huge preference for AdoMet over other methyl donors, such as folate, reflects favorable energetics resulting from the charged methylsulfonium center: the ΔG° for (AdoMet + Hcy → AdoHcy + Met) is −17 kcal mol−1 – over double that for (ATP → ADP + Pi) [1]. Methylation substrates range in size from arsenite to DNA and proteins, and the atomic targets can be carbon, oxygen, nitrogen, sulfur or even halides [2,3].
The first structure of an AdoMet-dependent methyltransferase (MTase), determined in 1993, was for the DNA C5-cytosine MTase M.HhaI [4]. For several years thereafter, a variety of additional MTases, with a wide range of different substrates, were found to share the same basic structure. More recently however, AdoMet-dependent methylation has been found to be the target of functional convergence that is catalyzed by enzymes with remarkably distinct structures. The Protein Data Bank (PDB) currently includes >100 structures for 50 distinct AdoMet-dependent MTases from 31 different classes of enzymes as defined by the Enzyme Classification (EC) system (Table 1; for a more extensive list see Ref. [5]).
Table 1.
Known structures of AdoMet-dependent methyltransferases
| EC Classification | Name | PDB code |
|---|---|---|
| Class I | ||
| 2.1.1.2 | Guanidinoacetate N-MTase | 1KHH |
| 2.1.1.5 | Betaine homocysteine MTase | 1LT7 |
| 2.1.1.6 | Catechol O-MTase | 1VID, 1JR4 |
| 2.1.1.8 | Histamine N-MTase | 1JQD, 1JQE |
| 2.1.1.20 | Glycine N-MTase | 1BHJ, 1D2H, 1D2G, 1D2H, 1XVA, 1D2C |
| 2.1.1.28 | Phenylethanolamine N-MTase | 1HNN |
| 2.1.1.37 | DNA (cytosine) C5-MTase | 1G55 |
| 2.1.1.43 | Dot1p-like histone-lysine N-MTase | 1NW3 |
| 2.1.1.46 | Isoflavone O4′-MTase | 1FP2 |
| 2.1.1.48 | rRNA (adenine) N6-MTase | 1QAM, 1QAN, 1QAO, 1QAQ, 1YUB |
| 2.1.1.63 | O6-Alkylguanine-DNA MTase | 1EH6, 1SFE |
| 2.1.1.68 | Caffeate O-MTase | 1KYW, 1KYZ, 1KNS, 1L0U |
| 2.1.1.72 | Adenine-specific DNA N6-MTase | 1G38, 1ADM, 1AQI, 1AQJ, 2ADM, 1EG2, 1G60, 2DPM |
| 2.1.1.73 | Cytosine-specific DNA C5-MTase | 10MH, 1DCT, 1FJX, 1HMY, 1MHT, 2HMY, 3MHT, 4MHT, 5MHT, 6MHT, 7MHT, 8MHT, 9MHT, 1KHC |
| 2.1.1.77 | Protein-l-isoaspartate O-MTase | 1D15, 1L1N, 1JG1, 1JG2, 1JG3, 1JG4, 1KR5 |
| 2.1.1.79 | Cyclopropane-fatty-acyl-phospholipid synthase | 1KP9, 1KPG, 1KPH, 1L1E |
| 2.1.1.80 | Protein-glutamate O-MTase | 1AF7, 1BC5 |
| 2.1.1.113 | Cytosine-N4-DNA-MTase | 1BOO |
| 2.1.1.125 | Protein Arginine N-MTase | 1F3L, 7G6Q, 1OR8, 1ORH, 1ORI |
| 2.1.1.132 | Precorrin-6y C-MTase | 1KXZ |
| 2.7.7.19 | Polynucleotide adenylyltransferase | 1VPT, 3MAG, 1B42, 1BKY, 1EAM, 1EQA, 1JSZ, 1JTE, 1JTF, 1P39, 1V39, 1VP3, 1VP9, 1VPT, 2VP3, 3MCT, 4DCG, 1AV6 |
| 2.1.1.? | Monoethanolamine MTase | 1L2Q |
| 2.1.1.? | Isoliquiritigenin (chalcone) O2′-MTase | 1FP1 |
| 2.1.1.? | Protein-glutamine N5-MTase | 1NV8 |
| 2.1.1.? | FtsJ RNA MTase | 1EIZ |
| 2.1.1.? | Hypothetical MTase | 1IM8, 1I9G, 1IXK, 1M6Y, 1DUS |
| Class II | ||
| 2.1.1.13 | Reactivation domain of MetH | 1MSK |
| Class III | ||
| 2.1.1.133 | Precorrin-4 C11-Mtase | 1CBF, 2CBF |
| Class IV | ||
| 2.1.1.? | rRNA 2′O-MTase | 1GZ0 |
| Class V | ||
| 2.1.1.43 | Histone-lysine N-MTase | 1ML9, 1MT6, 1H3I, 1MUF, 1N6A, 1N6C, 1O9S, 1MVX, 1MVH |
| 2.1.1.127 | [Ribulose-bisphosphate-carboxylase]-lysine N MTase | 1MLV |
The purpose of this review is to compare and contrast the five known structurally distinct families of AdoMet-dependent MTases (Classes I–V). The phenomenon of enzymes from distinct structural families catalyzing the same reaction, termed enzyme analogy, has been noted for several decades [6]. Many enzymes exhibit ‘catalytic promiscuity’ resulting in a pluripotency that can be shaped by mutation and selection [7,8]. This can lead to a given protein structure playing several distinct catalytic roles [9], but also results in distinct protein structures playing a common catalytic role. Perhaps such flexibility is particularly easy where highly exergonic reactions are involved. As ATP is the only enzyme substrate more widely used than AdoMet, it seems logical that the current champion for greatest number of analogous families is the ATP-dependent protein phosphoryltransferases (protein kinases), with seven known structurally distinct families [10]. Nonetheless, this degree of analogy appears to be quite rare, and the five known families of AdoMet-dependent MTases also provide an impressive example of functional convergence.
In the beginning, a MTase was a MTase was a MTase
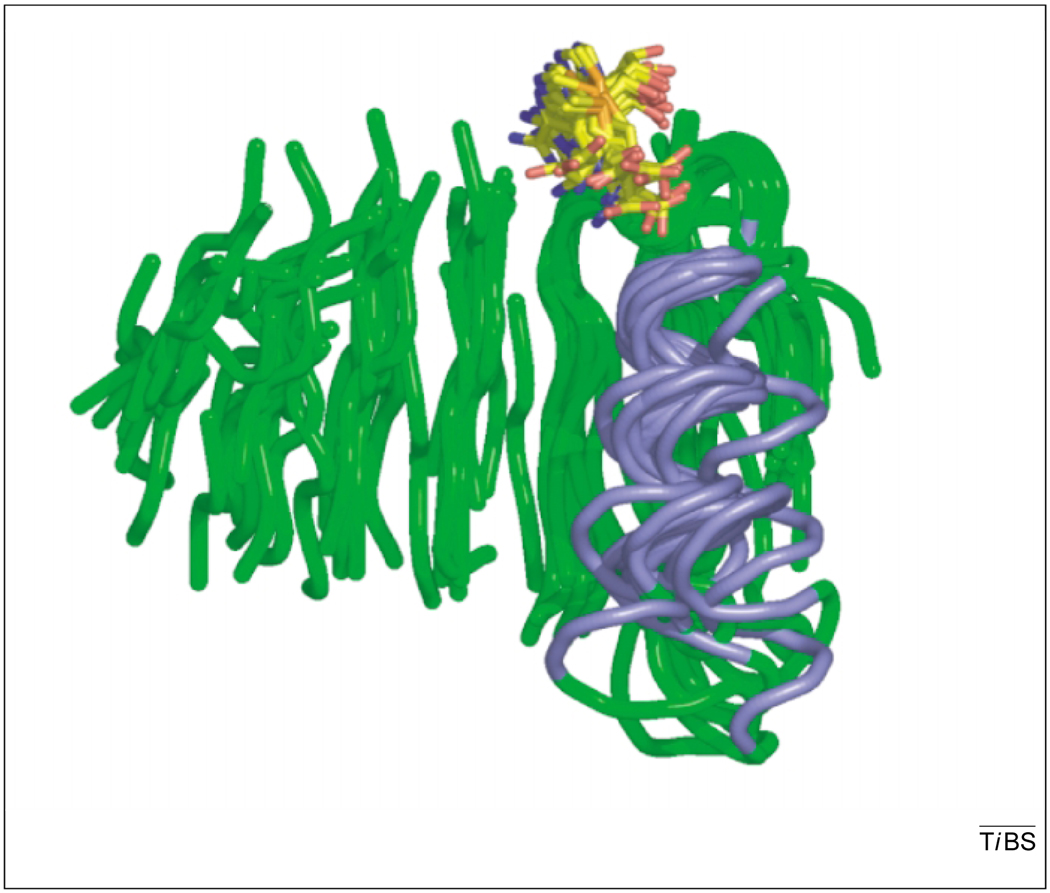
Starting with the M.HhaI DNA-MTase structure in 1993 [4], a continuing string of structures for AdoMet-dependent MTases have been reported. These structures are remarkably similar, comprising a seven-stranded β sheet with a central topological switch-point and a characteristic reversed β hairpin at the carboxyl end of the sheet (6 ↑ 7 ↓ 5 ↑ 4 ↑ 1 ↑ 2 ↑ 3 ↑; Fig. 1a). This sheet is flanked by α helices to form a doubly wound open αβα sandwich, and is henceforth referred to as the Class I MTase structure (Fig. 1a). The first β strand typically ends in a GxGxG (or at least GxG) motif – the hallmark of a nucleotide-binding site – bending sharply underneath the AdoMet to initiate the first α helix. The only other strongly conserved position is an acidic residue at the end of β2 that forms hydrogen bonds to both hydroxyls of the AdoMet ribose [11]. The spatial conservation of this class of MTase is so pronounced that superimposing the cores (150 amino acids on average) of twenty MTases, gives root-mean-square deviations for the Cα atoms of just 3.6 ± 0.5Å when 190 pair-wise comparisons are averaged. Figure 2 displays the alignment of 14 unique structures displaying their β sheet and first α helix. Several groups have noted that, with the exception of strand seven, this structure is quite similar to that of the NAD(P)-binding Rossmann-fold domains [11]. In both groups of proteins, the central topological switch-point results in a deep cleft in which the AdoMet or NAD(P) binds.
Fig. 1.
The tertiary structures of the five classes of S-adenosyl-l-methionine (AdoMet)-dependent methyltransferases (MTases). In each case a representative tertiary structure (left) and topology diagram (right) is shown. (a) Class I: tertiary structures have been determined for >33 family members, most containing a seven-stranded β sheet flanked by α helices (enzyme:PDB code, M.HhaI:6MHT). (b) Class II: the reactivation domain of methionine synthase contains a series of long β strands and binds to AdoMet in a shallow groove on the surface of the domain (MetH:1MSK). (c) Class III: the bilobal structure of CbiF contains an AdoMet-binding site between the two αβα domains, and a groove in the N-terminal domain is proposed to be the active-site cleft (CbiF:1CBF). (d) Class IV: the SPOUT family of RNA MTases contains a novel knot structure (magenta) at the C terminus that contributes to the AdoHcy-binding site (YibK:1MXI). (Helices that are not conserved between family members are shown in pale pink in the topology diagram.) (e) Class V: the SET-domain containing histone-lysine N-MTase family is formed by the combination of three small β sheets. The AdoMet-binding site is formed on a shallow groove of the protein and the substrate active site is proximal to the C-terminal tail, which is also involved in a knot-like structure (Set7/9:1O9S). Figures were generated using PYMOL (http://www.pymol.org).
Fig. 2.
The strong structural similarity within the Class I methyltransferases (MTases) can been seen in the alignment of 14 structures via their β strands. For clarity, only the peptide backbone of the catalytic core containing the main β sheet (green) and the first α helix (blue) is shown. The cofactor (AdoMet or AdoHcy) is shown in stick model with carbon (yellow), oxygen (red), nitrogen (blue) and sulfur (orange). Structures include protein isoaspartate O-MTase (PDB code: 1I1N), protein arginine N-MTase (1F3L), cyclopropane-fatty-acyl-phospholipid synthase (1KPH), catechol O-MTase (1VID), M.TaqI (2ADM), M.HhaI (6MHT), CheR (1AF7), FtsJ (1EIZ), isoliquiritigenin O-MTase (1FP1), ErmC′ (1QAN), VP39 (1VPT), PrmC (1NV8), mj0882 (1DUS), hypothetical rv2118c (1I9G).
Although highly conserved, the Class I family contains several members with variant structural features [5]. Some Class I MTases are homodimeric or even tetrameric [12], but most are monomers. One of the smallest MTases – catechol O-MTase [13] – consists of just the consensus structural core, whereas most MTases contain auxiliary domains that are inserted throughout the MTase fold and appear to play roles in substrate recognition. Strands six and seven are reversed in the primary sequence of protein isoaspartyl MTase [14], and are absent from protein arginine (R) MTase (PRMT) [15]. Finally, circular permutation of the overall topology has been proven for some DNA MTases [16,17] and is predicted for some RNA MTases [18]. Together, these differences suggest structural flexibility, but it (incorrectly) appeared for a time that AdoMet-dependent MTases were variations on a single basic theme.
Many of the known Class I MTases act on DNA to regulate gene expression, to repair mutations or to protect against bacterial restriction enzymes. Initially, it was a mystery as to how MTases acted on nucleotides that are held inside the DNA duplex by base pairing and stacking –seemingly inaccessible to the active site of an enzyme. The answer came from the M.HhaI DNA C5-MTase complexed with a synthetic DNA duplex [19]. In a process termed ‘base flipping’, the enzyme simply rotates the target DNA base,~180° on its flanking phosphodiester bonds such that the base projects into the catalytic pocket [20]. This strategy helps explain the tremendous diversity of substrates accommodated by MTases sharing the Class I structure.
A lesson in cobalamin
As early as 1996, there was a hint that not all AdoMet-dependent MTases would follow the same structural theme. The Escherichia coli cobalamin (vitamin B12)-dependent methionine synthase, MetH, generates methionine from homocysteine, transferring a methyl group from a folate derivative to the bound cobalamin and thence to homocysteine. Periodically, the B12 cobalt is oxidized to a dead-end form, and reactivation requires reductive methylation using AdoMet, flavodoxin and an additional structural domain. The MetH reactivation domain (Class II MTase) – dominated by a long, central, antiparallel β-sheet flanked by groups of helices at either end (Fig. 1b) – looks nothing like the Class I MTases either in overall architecture or interactions with AdoMet [21]. AdoMet is bound in an extended conformation to a shallow groove along the edges of the β strands, forming hydrogen bonds to a conserved RxxxGY motif [21]. Large conformational changes are required to position the reactivation domain near the cobalamin substrate within the main catalytic domain [21,22]. However interesting from a biochemical standpoint, the flavodoxin requirement and subordinate role of this reactivation domain led many to underestimate its relevance to understanding independent MTases.
Two years later the MTase field was again surprised, this time by the homodimeric structure of CbiF – a MTase that acts on ring carbons of the large, planar precorrin substrates during cobalamin biosynthesis [23]. The active site in this structural family (Class III MTases) is tucked into a cleft between two αβα domains, each containing five strands and four helices (Fig. 1c). A GxGxG motif occurs at the C-terminal end of the first β strand, similar to the Class I enzymes, but surprisingly does not contact AdoMet (at least in the absence of the precorrin substrate). Instead, the AdoMet is tightly folded, and binds between the two domains. Based on sequence similarity, diphthine synthase – which acts on a protein His residue – is also predicted to adopt the Class III structure, which might indicate a broader substrate range for this class of MTase than was initially appreciated.
Knotty new MTase structures SET to SPOUT
This past year has provided two more disparate MTase structures. The SPOUT family of RNA MTases provides the only known cases of Class IV structure [24–26]. These enzymes are unique in three ways: (1) they include a six-stranded parallel β sheet flanked by seven α helices, of which the first three strands form half of a Rossman fold (Fig. 1d); (2) their active site is located near the subunit interface of a homodimer, and might encompass residues extending from both subunits; and (3) the topology of the structure is such that a significant portion of the C terminus is tucked back into the structure in a ‘knot’. This rare substructure is formed by the last ~30 residues, including the last α helix, and contains several catalytic residues that confirm its structural importance [25,26]. The structure of a SpoU homolog, Haemophilus influenzae (HI0766) YibK, has been determined in the presence of AdoHcy, and reveals the AdoHcy to be bound above strands four to six in a bent conformation [27] (Fig. 1d).
The most recently described structural family of AdoMet-dependent MTases is the SET-domain proteins [28–33]. Amino-acid sequence comparison [34] suggests that there are hundreds of these proteins, and several have been shown to methylate lysines in the flexible tails of histones or in Rubisco (ribulose-1,5-bisphosphate carboxylase) [35]. These Class V MTases contain a series of eight curved β strands forming three small sheets (Fig. 1e), with the C terminus tucked underneath a surface loop forming a knot-like structure similar to the Class IV MTases, but constructed on a totally different topology [28–33]. AdoHcy bound to the SET domain is kinked in a manner similar to that of the Class III CbiF-bound AdoHcy, and binds on a concave surface of the enzyme near an invariant tyrosine residue that has been implicated in the catalytic reaction. Flanking the SET domain are diverse sequences termed the pre- and post-SET regions, which are often essential for MTase activity and might participate in substrate recognition and specificity.
Conformations of AdoMet and AdoHcy
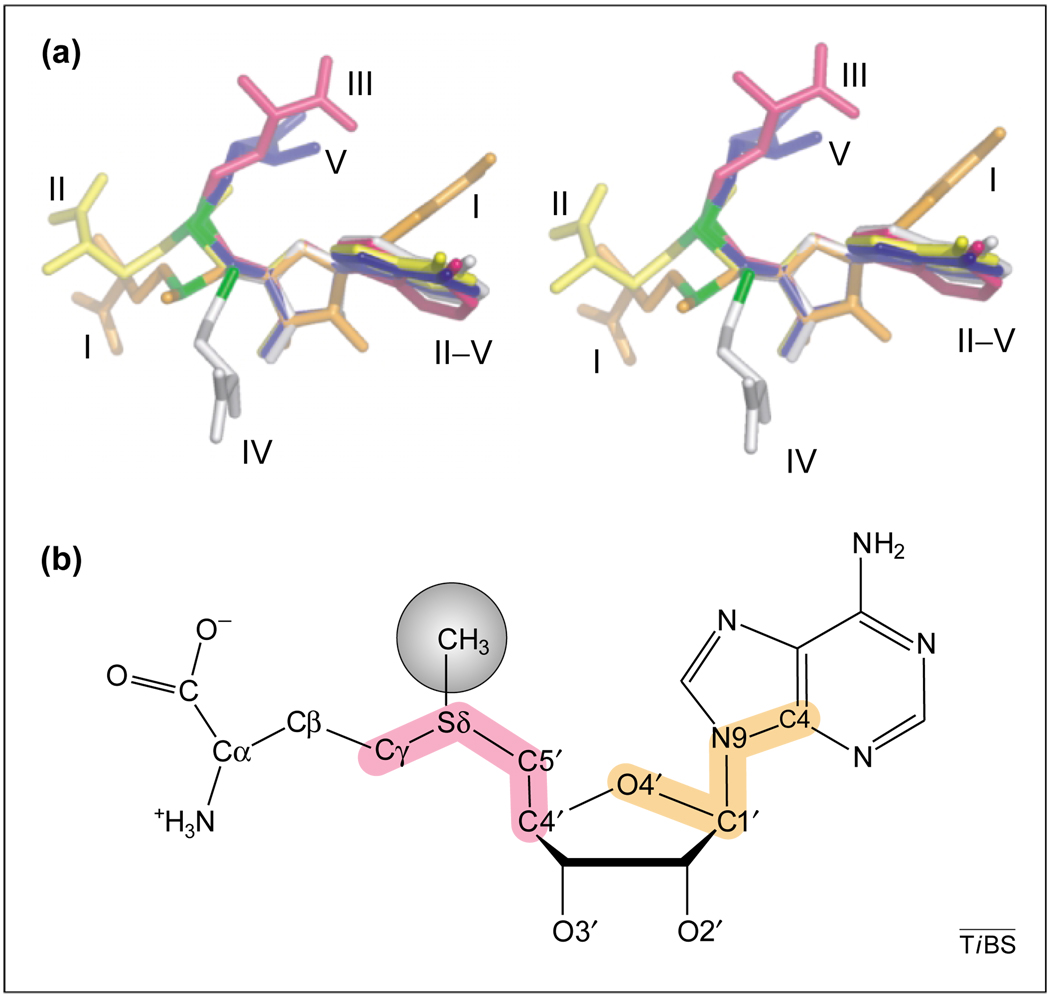
The bound AdoMet or AdoHcy ligand exhibits significantly different conformations in the five structural classes, which emphasizes its flexibility. Figure 3a compares the prototypical AdoMet and AdoHcy conformations of each structural class by aligning the molecules via their ribose moieties. The ribose ring of AdoMet in Class I adopts an envelope 2′-endo conformation, with its base in the anti position at ~135° (defined by the O4′–C1′–N9–C4 dihedral; Fig. 3b). The ribose rings of the other AdoMet classes adopt an envelope 2′-exo conformation with the base in the anti position at ~180°. These differences, although significant, are small compared with the differences in the O4′–C4′–C5′–Sδ dihedral angle, which begins to define whether the nucleotide is extended or folded. An extended trans conformation (~180°) is adopted by the Class I MTases, an angle of approximately −90° is adopted by Classes II, III and V, whereas the AdoHcy twists in the opposite direction with an angle of 80° in Class IV. The next dihedral angle (C4′–C5′–Sδ–Cγ; Fig. 3b) further separates the classes such that, overall, the Class I and Class II ligands are relatively extended, but Class III–V MTases bind the methyl donor in tightly folded conformations (Fig. 3a). Because AdoMet is an exchanged substrate, its solvent accessibility might be directly related to the rate of catalytic turnover, or could at least provide an indication of structural flexibility of the catalytic core. The classes of MTase surround their methyl donor to differing degrees, such that in catechol-O-MTase (Class I) <1% of the AdoMet surface area is exposed to solvent, whereas this exposure is 8–21% in the other classes.
Fig. 3.
AdoMet. (a) Overlay of the S-adenosyl-l-methionine (AdoMet) or S-adenosyl-l-homocysteine (AdoHcy) conformations in different methyltransferase (MTase) classes. The molecules are aligned via the ribose rings, and show varying degrees of bend at the intersection between the adenosine and methionine moieties. Described in order of structural class, indicating enzyme PDB code:Class I, M.HhaI:6MHT (orange); Class II, MetH:1MSK (yellow); Class III, CbiF:1CBF (pink); Class IV, YbiK:1K2X (white), Class V, Rubisco:1MVL (blue). (b) AdoMet atomic nomenclature. The O4′-C1′-N9-C4 and C4′-C5′-Sδ-Cγ dihedrals are highlighted yellow and pink, respectively.
A diverse set of mechanisms for a conserved class of MTase
Substrate-bound complexes have been determined mainly for Class I structures, although recently a Class V (SET) MTase in complex with a lysine-containing peptide has also been determined [36]. All MTases are thought to proceed with direct transfer of the methyl group to substrate with inversion of symmetry in an SN2-like mechanism [37,38]. This reaction also requires that a proton be removed before, concurrent with, or after methyl transfer. Even within the structurally conserved family of Class I MTases, a wide variety of mechanisms have evolved to activate the catalytic nucleophile, dependent on the polarizability of the target atom.
N-methylation
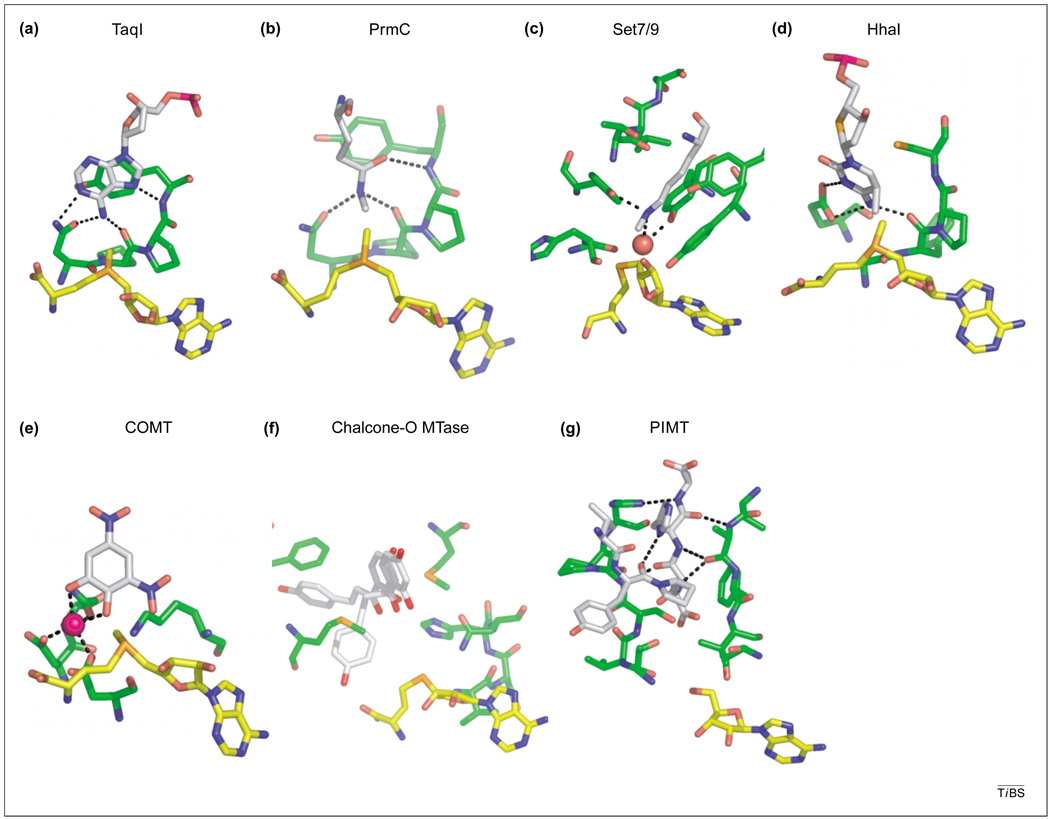
A common mode of substrate binding is the use of an [D/N/S]PP[Y/F] motif (DPPY, for brevity), employed by DNA N6-adenine [39] and DNA N4-cytosine MTases [16] and by the newly described protein N5-glutamine MTase, PrmC/HemK [40]. This set of diverse substrates indicates that the DPPY motif is not nucleotide specific, but is selective for nitrogens conjugated to a planar system such as an amide moiety or a nucleotide base. DPPY motifs extend from the C terminus of β-strand four in these Class I structures (Fig. 1a), in which the di-proline bends the polypeptide towards the surface of the active site. Two hydrogen bonds are formed between the nucleophilic nitrogen and both the oxygen atom of the [D/N/S] side-chain and the carbonyl oxygen of the first proline (Fig. 4a,b). These hydrogen bonds position the substrate such that the lone-pair electrons on the nitrogen nucleophile point towards the incoming methyl group [41]. A charged methylamine intermediate results after methyl transfer, but resolves into the neutral sp2-hybridized amide upon proton loss to solvent during product release [38,40,42].Other Class I N-MTases – including protein arginine MTase [15], small molecule glycine-N MTase [12] and phenylethanolamine-N MTase [43] – do not contain the DPPY motif, but instead use acidic residue(s) to neutralize the positive charge on the substrate amino group.
Fig. 4.
Substrate-bound Class I methyltransferase (MTase) structures [protein, green; S-adenosyl-l-methionine (AdoMet) or S-adenosyl-l-homocysteine (AdoHcy), yellow; substrate, white]. (a) DNA adenine N6-MTase M.TaqI bound to adenosine (a combination of two PDB files: 2ADM and 1G38). (b) Protein glutamine N5-MTase PrmC/HemK bound to an AdoMet–AdoHcy mixture and partially methylated glutamine (1NV8). (c) Class V SET7/9 N-MTase bound to the lysine residue of a histone-tail peptide (1O9S). (d) DNA C5-cytosine MTase M.HhaI bound to a mixture of AdoMet and AdoHcy and partially methylated cytosine (6MHT). (e) Catechol O-MTase bound to magnesium (magenta) and 3,5-dinitrocatechol (1VID). (f) Chalcone O-MTase bound to two alternate conformations of 2′,4,4′-trihydroxychalcone (1FP1). (g) Protein isoaspartate O-MTase facilitates protein repair by binding the residues surrounding the mutant isoaspartate as if it were a β strand within the MTase structure (1JG3).
Using entirely different structural scaffolding, the Class V SET-domain MTases bind to a kinked AdoMet molecule on the opposite side of a small channel from the N5-nitrogen of a peptide lysine substrate [36] (Fig. 4c). The C-terminal tail of the domain forms a pseudo knot and provides an integral part of the hydrophobic active-site pocket, including tyrosine residues implicated in the catalytic mechanism.
C-methylation
Many substrate-bound structures are known for DNA C5-cytosine MTases, including M.HhaI [19,44]. A ProCys dipeptide is universally conserved within the active site of C5-cytosine MTases, and structurally resembles the PY portion of the DPPY motif, whereas an aspartic acid residue from a neighboring portion of the enzyme functionally replaces the [D/N/S] residue of the DPPY motif. In M.HhaI, the N4 nitrogen of the cytosine residue is positioned further away from the AdoMet by hydrogen bonds, such that the C5 atom is presented as a methylation target (Fig. 4d). Because methylation on carbon atoms is more difficult than on polarizable nitrogens, the nucleotide must first be activated by covalent-bond formation between the conserved Cys thiol and carbon C6 [45]. This generates a negative charge on C5 that facilitates methyl transfer.
The design flexibility of the Class I structure is further illustrated by a large family of RNA C5-cytosine MTases [46]. Our threading analysis (not shown) suggests that these enzymes adopt a Class I fold; but although a cysteine nucleophile [47] is predicted to be in the active site similar to M.HhaI, it would be contributed by the end of β-strand five.
O-methylation
Several O-MTase structures have been determined, including catechol O-MTase (COMT) [13], the dimeric structures of two plant MTases [48] and the glutamate O-MTase CheR [49]. A Mg2+ ion is required to bind and orient the two catechol hydroxyls in COMT (Fig. 4e). Molecular simulation and pKa studies suggest that Mg2+ acts primarily to organize the substrate-binding site and not as a general base [50,51]. Instead, a nearby lysine residue appears to deprotonate the substrate hydroxyl before attack on the AdoMet methyl group. The chalcone (Fig. 4f) and isoflavone O-MTases do not require a metal ion, but do require a histidine residue to deprotonate the hydroxyls of plant metabolites [48].
Glutamic acid O-methylation by CheR differs in that the methyl group is transferred to a carboxylic acid rather than to a hydroxyl moiety. The active site of CheR contains the side chains of arginine and tyrosine residues [49], which might position the glutamic acid substrate and facilitate the methyl-transfer reaction. The negatively charged carboxyl group might attack the methyl group unassisted. The isoaspartate O-MTase recognizes damaged asparagine residues as part of an essential repair process. This enzyme binds a VYP(L-isoAsp)HA peptide by forming a series of hydrogen bonds to the surrounding peptide main-chain, as if the substrate were a β strand within the enzyme itself [52] (Fig. 4g). The extended polypeptide chains of other protein MTases (e.g. PRMT, CheR, PrmC and the Class V SET-domain lysine N-MTases) might form similar interactions.
Concluding remarks
Evolution has independently achieved AdoMet-dependent MTase activity at least five times, producing five unique structural MTase classes. Most of the other examples of analogous enzyme families also use substrates, such as ATP or NAD, that include a nucleotide ‘handle’ for binding. The Class I and Class IV MTases are plausibly derived from Rossmann-fold proteins, and even the Class III CbiF structure contains a GxGxG nucleotide-binding motif, but uses it unconventionally. The Class II and Class V MTases do not appear to have structural analogs, so their evolutionary history is not yet clear. If the Class I MTases are actually derived from the ubiquitous Rossmann-fold proteins, then multiple independent evolutionary sublineages might explain the predominance of the Class I enzyme family. The limited sequence similarity between Class I proteins could even be consistent with independent evolution from a generic GxGxG-containing nucleotide-binding domain.
Catalysis of AdoMet-dependent methyltransfer does not appear to be rigidly restricted by tertiary structure or local spatial requirements. Even within the structural constraints of the Class I family, many different methods of binding substrate and activating a nucleophile have been described. One important question that remains is whether chance or functional constraints define which reactions are carried out by which classes of MTase. It could be argued that precorrin MTases have adopted a novel conformation because of their rigid, planar substrates, however, it appears that not all tetrapyrrole biosynthetic MTases are in Class III. Precorrin-C6 MTase CbiT has [53] and protoporphyrin IX O-MTase is predicted to have a Class I structure [54]. Similarly, not all histone-lysine N-MTases contain the Class V SET domain; the Dot1 histone H3-Lys79 N-MTase belongs to Class I [55].
New methyltransferase activities are still being described, and although the genome projects are providing large lists of enzymes orthologous to the five known structures, it would not be a great shock to find the members of MTase Classes VI and up among uncharacterized open reading frames. In fact, given what we now know about enzyme evolution, these new MTase classes might already have been (mis)annotated based on their similarity to characterized enzymes [9]. The challenge will be for biochemists to test annotation claims, or to discover the real identity of these new genes, whereas the structural biologists can begin to address substrate recognition and catalytic mechanisms, particularly in the newer structural classes.
Acknowledgments
We thank Osnat Herzberg and Steve Gamblin for early release of coordinates. H.L.S. was supported by grants from NIH (GM56775 and DK02794), R.M.B. was supported by a grant from the U.S. National Science Foundation (MCB-9904523), and X.C. was supported by NIH (GM49245 and GM61355) and the Georgia Research Alliance.
References
- 1.Cantoni GL. Biological methylation: selected aspects. Annu. Rev. Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 2.Ohsawa N, et al. Purification and characterization of a monohalomethane-producing enzyme S-adenosyl-l-methionine: halide ion methyltransferase from a marine microalga, Pavlova pinguis. Biosci. Biotechnol. Biochem. 2001;65:2397–2404. doi: 10.1271/bbb.65.2397. [DOI] [PubMed] [Google Scholar]
- 3.Attieh JM, et al. Purification and characterization of a novel methyltransferase responsible for biosynthesis of halomethanes and methanethiol in Brassica oleracea. J. Biol. Chem. 1995;270:9250–9257. doi: 10.1074/jbc.270.16.9250. [DOI] [PubMed] [Google Scholar]
- 4.Cheng X, et al. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 5.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 6.Galperin MY, et al. Analogous enzymes: independent inventions in enzyme evolution. Genome Res. 1998;8:779–790. doi: 10.1101/gr.8.8.779. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 8.Anantharaman V, et al. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr. Opin. Chem. Biol. 2003;7:12–20. doi: 10.1016/s1367-5931(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 9.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 10.Cheek S, et al. Sequence and structure classification of kinases. J. Mol. Biol. 2002;320:855–881. doi: 10.1016/s0022-2836(02)00538-7. [DOI] [PubMed] [Google Scholar]
- 11.Fauman EB, et al. Structure and evolution of AdoMet-dependent methyltransferases. In: Cheng X, Blumenthal RM, editors. S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific Publishing; 1999. pp. 1–38. [Google Scholar]
- 12.Huang Y, et al. Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes. J. Mol. Biol. 2000;298:149–162. doi: 10.1006/jmbi.2000.3637. [DOI] [PubMed] [Google Scholar]
- 13.Vidgren J, et al. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–357. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 14.Skinner MM, et al. Crystal structure of protein isoaspartyl methyltransferase: a catalyst for protein repair. Structure. 2000;8:1189–1201. doi: 10.1016/s0969-2126(00)00522-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong W, et al. Structure of Pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scavetta RD, et al. Structure of RsrI methyltransferase, a member of the N6-adenine β class of DNA methyltransferases. Nucleic Acids Res. 2000;28:3950–3961. doi: 10.1093/nar/28.20.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bujnicki JM, et al. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J. Mol. Evol. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- 19.Klimasauskas S, et al. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 20.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon MM, et al. The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure. 1996;4:1263–1275. doi: 10.1016/s0969-2126(96)00135-9. [DOI] [PubMed] [Google Scholar]
- 22.Jarrett JT, et al. Methionine synthase exists in two distinct conformations that differ in reactivity toward methyltetrahydrofolate, adenosylmethionine, and flavodoxin. Biochemistry. 1998;37:5372–5382. doi: 10.1021/bi9730893. [DOI] [PubMed] [Google Scholar]
- 23.Schubert HL, et al. The X-ray structure of a cobalamin biosynthetic enzyme, cobalt- precorrin-4 methyltransferase. Nat. Struct. Biol. 1998;5:585–592. doi: 10.1038/846. [DOI] [PubMed] [Google Scholar]
- 24.Anantharaman V, et al. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- 25.Michel G, et al. The structure of the RlmB 23S rRNA methyltransferase reveals a new methyltransferase fold with a unique knot. Structure. 2002;10:1303–1315. doi: 10.1016/s0969-2126(02)00852-3. [DOI] [PubMed] [Google Scholar]
- 26.Nureki O, et al. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1129–1137. doi: 10.1107/s0907444902006601. [DOI] [PubMed] [Google Scholar]
- 27.Lim K, et al. Structure of YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot. Proteins. 2003;51:56–67. doi: 10.1002/prot.10323. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Structure of the neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trievel R, et al. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 30.Wilson J, et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 31.Min J, et al. Structure of the SET domain histone lysine methyltransferase Clr4. Nat. Struct. Biol. 2002;9:828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs SA, et al. The active site of the SET domain is constructed on a knot. Nat. Struct. Biol. 2002;9:833–838. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- 33.Kwon T, et al. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doerks T, et al. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeates T. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002;111:5–7. doi: 10.1016/s0092-8674(02)01010-3. [DOI] [PubMed] [Google Scholar]
- 36.Xiao B, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 37.Woodard RW, et al. Stereochemical course of the transmethylation catalyzed by catechol O-methyltransferase. J. Biol. Chem. 1980;255:9124–9127. [PubMed] [Google Scholar]
- 38.Coward JK. Chemical mechanisms of methyl transfer reactions: comparison of methylases with nonenzymic ‘model reactions’. In: Salvatore F, editor. The Biochemistry of Adenosylmethionine. Columbia University Press; 1977. pp. 127–144. [Google Scholar]
- 39.Goedecke K, et al. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 40.Schubert HL. Structures along the catalytic pathway of PrmC/HemK, an N5-glutamine AdoMet-dependent methyltransferase. Biochemistry. doi: 10.1021/bi034026p. (in press) [DOI] [PubMed] [Google Scholar]
- 41.Malone T, et al. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 42.Newby ZE, et al. A theoretical examination of the factors controlling the catalytic efficiency of the DNA-(adenine-N6)-methyltransferase from Thermus aquaticus. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7922–7927. doi: 10.1073/pnas.122231499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin JL, et al. Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT. Structure. 2001;9:977–985. doi: 10.1016/s0969-2126(01)00662-1. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, et al. DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 46.Reid R, et al. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Santi DV. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8263–8265. doi: 10.1073/pnas.97.15.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zubieta C, et al. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 2001;8:271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- 49.Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y-J, Bruice TC. A theoretical examination of the factors controlling the catalytic efficiency of a transmethylation enzyme: catechol O-methyltransferase. J. Am. Chem. Soc. 1997;119:8137–8145. [Google Scholar]
- 51.Thakker DR, et al. Regioselectivity of catechol O-methyltransferase. The effect of pH on the site of O-methylation of fluorinated norepinephrines. J. Biol. Chem. 1986;261:178–184. [PubMed] [Google Scholar]
- 52.Griffith SC, et al. Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its l-isoaspartyl peptide substrate. J. Mol. Biol. 2001;313:1103–1116. doi: 10.1006/jmbi.2001.5095. [DOI] [PubMed] [Google Scholar]
- 53.Keller JP, et al. The crystal structure of MT0146/CbiT suggests that the putative precorrin-8w decarboxylase is a methyltransferase. Structure. 2002;10:1475–1487. doi: 10.1016/s0969-2126(02)00876-6. [DOI] [PubMed] [Google Scholar]
- 54.Shepherd M, et al. Purification and kinetic characterisation of the magnesium protoporphyrin IX methyltransferase from Synechocystis PCC6803. Biochem. J. 371:351–360. doi: 10.1042/BJ20021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Min J, et al. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]